Abstract
CRISPR-Cas-mediated genome editing relies on guide RNAs that direct site-specific DNA cleavage facilitated by the Cas endonuclease. Here we report that chemical alterations to synthesized single guide RNAs (sgRNAs) enhance genome editing efficiency in human primary T cells and CD34+ hematopoietic stem and progenitor cells. Co-delivering chemically modified sgRNAs with Cas9 mRNA or protein is an efficient RNA- or ribonucleoprotein (RNP)-based delivery method for the CRISPR-Cas system, without the toxicity associated with DNA delivery. This approach is a simple and effective way to streamline the development of genome editing with the potential to accelerate a wide array of biotechnological and therapeutic applications of the CRISPR-Cas technology.
The type II bacterial clustered, regularly interspaced, short palindromic repeats (CRISPR) and the CRISPR-associated protein 9 (Cas9), known as CRISPR-Cas9 (refs. 1–4), consisting of an RNA-guided nuclease (Cas9) and a short guide RNA (gRNA), generates site-specific DNA breaks, which are repaired by endogenous cellular mechanisms. Possible outcomes of the approach include mutating a specific site through mutagenic non-homologous end-joining (NHEJ), creating insertions or deletions (indels) at the site of the break, and precise change of a genomic sequence through homologous recombination (HR) using an exogenously introduced donor template5. The guide RNA is composed of two RNAs termed CRISPR RNA (crRNA) and trans-activating crRNA, which can be combined in a chimeric single guide RNA (sgRNA). The sgRNAs are typically about 100 nucleotides (nt) long. Twenty nt at the 5′ end hybridize to a target DNA sequence by Watson-Crick base pairing and guide the Cas endonuclease to cleave the target genomic DNA, and the remaining double-stranded structure at the 3′ side is critical for Cas9 recognition (Fig. 1a). The sgRNA has been delivered into cells as RNA (e.g., prepared by in vitro transcription), or by using a DNA vector expressing the sgRNA.
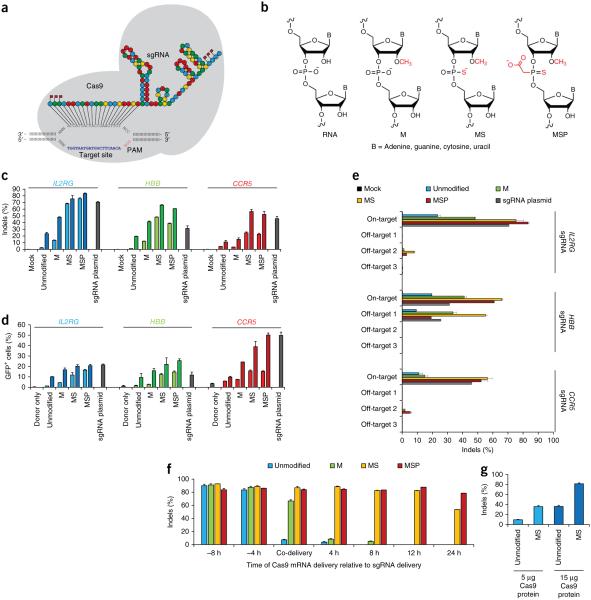
Figure 1.
Synthesized and chemically modified sgRNAs facilitate high frequencies of indels and HR in the human cell line K562. (a) Sequence and schematic secondary structure of the IL2RG sgRNA loaded into Cas9 and bound to its genomic target site. Nucleotides with chemical modifications are marked with red flags. (b) Structures of chemical modifications incorporated during chemical synthesis of sgRNAs (sequences can be found in Supplementary Table 1). (c,d amplicons (c) or gene addition by HR at the three loci IL2RG, HBB and CCR5 with synthetic sgRNAs (d). The synthetic sgRNAs were delivered at 1 μg (light shade) or 20 μg (dark shade) per 1 million cells. Cas9 was expressed from a plasmid (2 μg) and for HR experiments 5 μg of GFP-encoding donor plasmid was included. As a positive control, 2 μg of sgRNA plasmid encoding both the sgRNA and the Cas9 protein was used (gray bars). Bars represent average values + s.e.m., n = 3. (e) Specificity of targeted cleavage mediated by synthetic sgRNAs as performed in c for 20 μg of sgRNA. Indel frequencies were measured by deep sequencing of PCR amplicons of the targeted genomic loci and three bioinformatically predicted off-target loci for each gene. Bars represent average values + s.e.m., n = 3. (f) Staggered delivery of 15 μg Cas9 mRNA and 10 μg IL2RG synthetic sgRNAs into 1 million K562 cells using electroporation. Bars represent average indel frequencies + s.e.m., n = 3, as measured by tracking of indels by decomposition (TIDE) analysis of PCR amplicons spanning the sgRNA target sites, using a mock-treated sample as reference control. (g) Cas9 protein was complexed with a 2.5 molar excess of the indicated synthetic IL2RG sgRNAs and nucleofected into 1 million K562 cells at the indicated amounts. Indel frequencies were measured by TIDE analysis as above and bars represent average indel frequencies + s.e.m., n = 3.
Although genome editing using the CRISPR-Cas system is highly efficient in human cell lines, CRISPR-Cas genome editing in primary human cells is generally more challenging. The reasons for this remain elusive, but differences in transfection rates, promoter activity, exonuclease activity, interferon production when delivering nucleic acids, and DNA repair fidelity may contribute. Here we demonstrate that chemically synthesized sgRNAs can induce high levels of genome editing and that chemical alterations of the sgRNAs can enhance genome editing in both human primary T cells and CD34+ hematopoietic stem and progenitor cells (HSPCs). The increase in genome editing is further improved by delivering Cas9 as mRNA or protein rather than through a DNA expression plasmid, thus generating a simple and complete RNA- or RNP-based delivery method for the CRISPR-Cas system.
To test the utility of chemically synthesized sgRNAs for genome editing, we synthesized full-length sgRNAs of 100 nt using 2′-O-thionocarbamate-protected nucleoside phosphoramidites6. We also synthesized sgRNAs with three different chemical modifications at both termini to evaluate their effects on efficacy (Fig. 1a,b and Supplementary Table 1). Chemical modifications comprising 2′-O-methyl (M), 2′-O-methyl 3′phosphorothioate (MS), or 2′-O-methyl 3′thioPACE (MSP) were incorporated at three terminal nucleotides at both the 5′ and 3′ ends. We selected these modifications for evaluation owing to their previously reported stability to serum and snake venom phosphodiesterases and for their reported effects on the immunostimulatory properties of nucleic acids7–9.
We selected three human genes that have been targeted with sgRNAs with high gene editing frequencies in cell lines10,11: (i) IL2RG, which can harbor mutations responsible for severe combined immunodeficiency (SCID)-X1, (ii) HBB, which can harbor mutations responsible for sickle cell anemia and thalassemia, and (iii) CCR5, which encodes a co-receptor of HIV and is currently being investigated as a target for therapeutic gene editing in anti-HIV clinical trials12.
We first tested each of the synthetic sgRNAs in the presence of recombinant Cas9 protein in a biochemical in vitro DNA cleavage assay and found that all sgRNAs mediated targeted DNA cleavage efficiently (Supplementary Figs. 1 and 2). We next examined whether the synthesized sgRNAs could induce targeted indels indicative of mutagenic NHEJ and gene disruption in human cell lines. We delivered each sgRNA together with a DNA plasmid encoding Cas9 into K562 cells by nucleofection and analyzed indel frequencies. Delivery of 1 μg of synthetic unmodified sgRNA targeting the IL2RG locus generated targeted indel frequencies of 2.4% (Fig. 1c). For the M-modified sgRNA, we observed a small increase in indel frequencies to 13.5%, suggesting a modest improvement in stability over the unmodified sgRNA. Notably, the same amount of MS- and MSP-modified sgRNAs increased the indel frequency to 68.0% and 75.7%, respectively. Increasing the amount of modified sgRNAs by 20-fold further increased the indel frequency in all cases, bringing the efficiency of MS- and MSP-modified sgRNAs to 75.3% and 83.3%, respectively (Fig. 1c), which was comparable to frequencies obtained by expressing the CRISPR-Cas system from a plasmid. Similar results were obtained for the HBB and CCR5 targets (Fig. 1c).
We next determined whether the synthetic sgRNAs could stimulate gene targeting by HR. We designed targeting vectors for each of the three loci with ~0.8 kb arms of homology 5′ and 3′ of the CRISPR target site. Between the homology arms, we included a GFP expression cassette that can be stably integrated upon successful HR at the targeted locus13,14. At all three targeted loci, the MS- and MSP-modified sgRNAs stimulated higher levels of HR than the unmodified and M-modified sgRNAs (Fig. 1d). At higher sgRNA levels (20 μg), the HR frequencies at IL2RG, HBB and CCR5 were 20.6%, 25.5% and 50.0%, respectively, for the MSP-modified sgRNAs. These frequencies are comparable to or higher than those obtained by expressing the CRISPR-Cas system entirely from a plasmid.
To investigate whether the sgRNA chemical modifications affected off-target activity, we used deep sequencing to measure off-target mutation frequencies at three different loci for each sgRNA. Eight of these off-target sites were predicted by in silico prediction tools15,16, and one for HBB was included because it has previously been shown to have high levels of off-target activity11. For four of the eight predicted sites, we found near-background off-target activity for all chemically synthesized sgRNAs, despite detecting high levels of on-target activity for the modified sgRNAs (Fig. 1e, Supplementary Fig. 3 and Supplementary Table 2). Chemical modification of the sgRNAs tended to result in higher off-target activity at the other four predicted sites but the levels were variable. In some cases, the ratio of on-target to off-target indel frequencies was improved with the modified sgRNAs, but this impact varied among targets (Fig. 1e and Supplementary Table 2). We detected off-target activity for the unmodified sgRNA at only two sites (IL2RG ‘off-target 2’ and HBB ‘off-target 1’, which had been previously reported as an off-target site for the HBB sgRNA). At the IL2RG site, the on-target:off-target ratio was 5.8-fold better for the unmodified sgRNA compared to the MSP-modified sgRNA (Fig. 1e and Supplementary Table 2), whereas for the HBB site this ratio was 2.6-fold and 1.5-fold better (for 1 μg and 20 μg, respectively) for the MSP-modified sgRNA compared to the unmodified sgRNA (Fig. 1e, Supplementary Fig. 3 and Supplementary Table 2). Comparing the on-target:off-target ratios of the modified sgRNAs to on-target:off-target ratios of the sgRNA plasmid, the sgRNA plasmid had a better ratio at CCR5 ‘off-target 2’ and the IL2RG ‘off-target 2’, whereas for the HBB ‘off-target 1’ the MSP-modified sgRNA had the better on-target:off-target ratio (Fig. 1e and Supplementary Table 2).
Taken together, these results suggest that typically the chemically modified sgRNAs retain high specificity. The differences observed in on-target:off-target ratios suggest the possibility that chemical alterations to the sgRNA may have the potential to modulate on-target: off-target activity; however, the impact of a given chemical alteration appears to depend on the sequence and perhaps also on other factors such as cell type and delivery conditions. Whether these observations are generalizable to other sgRNAs targeting different loci in different species will require further studies.
To further explore the performance of chemically modified sgRNAs in human cell lines, we turned to an all-RNA delivery platform co-delivering the sgRNAs with mRNA encoding Cas9. We measured indel frequencies at the IL2RG locus using varying amounts of Cas9 mRNA (1–15 μg) together with varying amounts of the MSP-modified sgRNA (1–20 μg) (Supplementary Fig. 4). We observed indel frequencies of 81–90% for all concentrations tested except for a modest decrease to 70% indels when using 1 μg each of the Cas9 mRNA and MSP-modified sgRNA, demonstrating the high efficiency of this all-RNA approach.
To explore whether the chemical modifications had an effect on the half-life of the sgRNA activity, we co-delivered or sequentially delivered Cas9 mRNA and the various synthetic sgRNAs targeting IL2RG by nucleofection (Supplementary Fig. 5). Co-delivery of Cas9 mRNA and either the MS- or MSP-modified sgRNA resulted in high editing frequencies of 87% and 84% indels, respectively (Fig. 1f). Whereas the M-modified sgRNA gave rise to 66% indels, the unmodified sgRNA gave rise to a modest 7.0%, demonstrating the importance of the sgRNA modification when it is co-delivered with Cas9 mRNA. Delivery of Cas9 mRNA first and the sgRNAs 4 or 8 h later produced high and similar levels of indels (83.1–92.4%) across all four sgRNAs with no difference in efficiencies between the unmodified and chemically modified sgRNAs (Fig. 1f). In contrast, when we delivered the sgRNA first, followed by delivery of Cas9 mRNA 4, 8, 12 or 24 h later, indel frequencies obtained with the unmodified sgRNA had already dropped to near-background levels by the 4-h time point. For the M-modified sgRNA we also observed a decrease in indel frequencies to near-background levels, but this drop was slightly delayed, suggesting a modest improvement in stability over the unmodified sgRNA. For the MS-modified sgRNA, we did not observe a decrease in indel frequencies until the 24-h time point, at which frequencies dropped to 53%. Notably, for the MSP-modified sgRNA we did not detect a significant decline in activity even after a 24-h delay between sgRNA and Cas9 delivery. These results are consistent with a model in which sgRNA end-modifications enhance intracellular stability, thus enabling increased efficacy of genome editing when Cas9 mRNA and sgRNAs are co-delivered into human cells. The fact that indel frequencies directed by the unmodified sgRNA were not reduced when Cas9 was delivered first, suggests that Cas9 protein protects sgRNAs from degradation.
To investigate this hypothesis we complexed the unmodified or the MS-modified IL2RG sgRNA with recombinant Cas9 protein before electroporating this active RNP into K562 cells at two different amounts (Fig. 1g and Supplementary Fig. 6). Comparing MS-modified sgRNA to the unmodified sgRNA, we observed 3.8-fold higher indel frequencies (35.7% vs. 9.5%) at the lower dose and 2.3-fold higher (81.0% vs. 35.9%) for the higher dose. This ratio between MS-modified and unmodified sgRNA was, however, significantly lower than that observed when co-delivering the sgRNAs with Cas9 mRNA (Fig. 1f), indicating that the Cas9 protein does partially protect the unmodified sgRNA from degradation. Nonetheless, modifications to the sgRNA improve gene editing efficiencies when delivering the sgRNA complexed with Cas9 protein.
Next, we compared the off-target activity of the unmodified and MS-modified IL2RG sgRNAs at the three previously investigated off-target sites when delivered with either Cas9 plasmid or Cas9 mRNA, or complexed with Cas9 protein. With the unmodified sgRNA we detected low levels of indels (<0.37%) across all three Cas9 sources and all three off-target sites (Supplementary Fig. 7). For the MS-modified sgRNA, we observed improved on-target:off-target ratios at all three off-target sites when delivering Cas9 plasmid compared to Cas9 mRNA (2.6–3.0-fold). Notably, for Cas9 RNP delivery with MS-modified sgRNA we detected significantly better on-target: off-target ratios compared to Cas9 plasmid and mRNA for all three sites, with near-background off-target activity at two of the sites.
In summary, the chemically modified sgRNAs demonstrate an advantage over the unmodified sgRNA for gene editing in human cell lines when co-delivered with Cas9 mRNA or delivered as RNP.
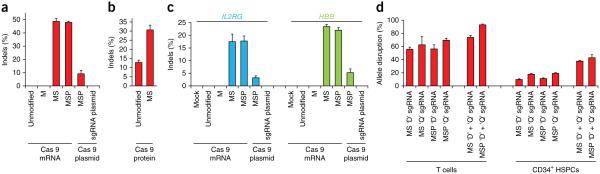
We next tested the chemically modified sgRNAs in primary cells. We tested the chemically modified CCR5 sgRNAs in stimulated human primary T cells co-delivered with Cas9-encoding mRNA. Notably, using GFP mRNA we consistently observed more than 98% nucleofection efficiency in T cells as measured by flow cytometry (Supplementary Fig. 8), obviating the need to enrich for transfected cells. Nucleofection of the plasmid encoding both the sgRNA and Cas9 did not give rise to allele modification frequencies above background (Fig. 2a). However, co-transfection of the MSP-modified sgRNA with DNA expression plasmid for Cas9 was able to bring activity up to 9.3% indel frequency. Delivery of Cas9 mRNA with the unmodified or the M-modified sgRNA did not give rise to allele modification frequencies above background. In contrast, Cas9 mRNA with either the MS- or MSP-modified sgRNA generated 48.7% and 47.9% indel frequencies, respectively. Increasing the amounts of sgRNA or Cas9 mRNA did not yield higher indel frequencies (Supplementary Fig. 9). For cells treated with Cas9 mRNA and MSP-modified sgRNA, we found similar allele modification frequencies in CD4+, CD8+ and total T-cell populations (Supplementary Fig. 10a). The observed high modification frequencies were stable when measured for the MSP-modified sgRNA over 21 d (Supplementary Fig. 10b), and we only observed a minor impact on cell viability and proliferation after delivery of the modified sgRNAs, in contrast to plasmid nucleofection, which caused considerable cell death and decreased proliferative potential (Supplementary Figs. 11 and 12).
Figure 2.
Chemically modified sgRNAs facilitate high frequencies of gene disruption in stimulated primary human T cells and CD34+ hematopoietic stem and progenitor cells (HSPCs). (a) 1 million primary human T cells were nucleofected with 10 μg of the indicated synthetic CCR5 sgRNAs and either 15 μg Cas9 mRNA or 1 μg Cas9-encoding plasmid. 1 μg sgRNA plasmid encoding both the sgRNA and Cas9 protein was included for comparison. Bars represent average indel frequencies for three different donors + s.e.m., n = 6, as measured by TIDE analysis of PCR amplicons spanning the sgRNA target site, and using a mock-treated sample as control reference. (b) Stimulated T cells were nucleofected as above, but with 15 μg Cas9 protein complexed with a 2.5 molar excess of the indicated synthetic CCR5 sgRNAs. Indel frequencies were measured by TIDE analysis as above. Bars represent average indel frequencies for three different donors + s.e.m., n = 6. (c) 500,000 mobilized human peripheral blood CD34+ HSPCs were nucleofected with 10 μg of the indicated synthetic sgRNAs targeting IL2RG or HBB and either 15 μg Cas9 mRNA or 1 μg Cas9 plasmid. 1 μg of sgRNA plasmid encoding both the sgRNA and Cas9 protein was included for comparison. Bars represent average indel frequencies + s.e.m., n = 3, as measured by T7 endonuclease cleavage assay. (d) 1 million stimulated T cells or mobilized human peripheral blood CD34+ HSPCs were nucleofected with 15 μg Cas9 mRNA and 10 μg of the indicated synthetic CCR5 sgRNAs. When used in combination the amount of each sgRNA was 5 μg. Indel frequencies for samples with single sgRNAs were measured by TIDE analysis as above and allele disruption frequencies for samples with two sgRNAs were measured by sequencing of cloned PCR products (Supplementary Fig. 15). Bars represent average indel frequencies + s.e.m., n = 3.
We also tested the MS-modified sgRNA in unstimulated T cells, which have been shown to be more difficult to edit than stimulated T cells17. In contrast to stimulated T cells, indel frequencies in unstimulated T cells ranged from 6.6% to 22.2%, showing higher variability across donors (Supplementary Fig. 13). These editing frequencies may still have utility, particularly in engineering T cells in which activation and prolonged culture may affect the subsequent biological functionality of the cells. We next tested RNP delivery using unmodified and MS-modified sgRNAs in stimulated primary T cells. Similarly to the results obtained in K562 cells, we observed a 2.4-fold improvement in indel frequencies of the MS-modified sgRNA over the unmodified sgRNA (30.7% vs. 12.8%, Fig. 2b), further supporting the use of chemically modified sgRNAs when delivered complexed with Cas9 protein.
Gene therapy in HSPCs has been explored extensively for treating genetic or acquired disorders of the hematopoietic system. We tested whether the chemically modified sgRNAs targeting IL2RG and HBB are functional in CD34+ HSPCs isolated from mobilized peripheral blood. Again, the sgRNA plasmid expressing both sgRNA and Cas9 did not give rise to detectable indel frequencies, but co-transfection of the MSP-modified sgRNA with the Cas9 DNA expression plasmid brought indel frequencies to 3.2% and 5.2% for IL2RG and HBB, respectively (Fig. 2c and Supplementary Fig. 14). As in T cells, we did not detect indels at either locus using the unmodified or M-modified sgRNAs when co-transfected with Cas9 mRNA. However, the IL2RG MS- and MSP-modified sgRNAs showed 17.5% and 17.7% indel frequencies, respectively, and 23.4% and 22.0%, respectively, for the HBB MS- and MSP-modified sgRNAs. Further studies could elucidate whether genetic modifications of HSPCs by chemically modified sgRNAs affect their multipotent capacity, or if editing efficiencies differ between long-term repopulating stem cells and lineage-committed progenitor cells.
A recent study showed that the simultaneous use of two sgRNAs could improve gene disruption in human primary T cells and in CD34+ HSPCs18. We chemically synthesized MS- and MSP-modified CCR5 sgRNAs with the sequences reported in that study (termed ‘D’ and ‘Q’), which cut 205 bp apart. We tested them in T cells and in CD34+ HSPCs co-delivered with Cas9 mRNA. When the two sgRNAs were used individually, we quantified allele modification frequencies using TIDE (tracking of indels by decomposition) analysis. When both were used, we quantified allele modification frequencies by sequencing of cloned PCR products, which also allowed us to quantify the full spectrum of editing events, including the previously reported high incidence of deletions of sequence between the two sgRNA target sites. In T cells, the ‘D’ sgRNA alone gave rise to 56.0% and 56.3% indels for the MS- and MSP-modified sgRNA, respectively, and the ‘Q’ sgRNA gave rise to 62.6% and 69.6%, respectively (Fig. 2d and Supplementary Fig. 15). When used in combination, the frequencies of allele modification increased, as we observed 73.9% and 93.1% for the MS- and MSP-modified sgRNAs, respectively, of which the majority of the modification events were deletions between the two sgRNA target sites (Supplementary Fig. 15). In CD34+ HSPCs, our observations were similar though the overall frequencies were lower. For the ‘D’ sgRNA we observed 9.8% and 11.2% allele modification frequencies for the MS- and MSP-modified sgRNA, respectively, and 17.8% and 19.2% for the ‘Q’ sgRNA (Fig. 2d and Supplementary Fig. 15). When used in combination the frequencies increased to 37.8% and 43.0% for the MS- and MSP-modified sgRNAs, respectively. We conclude that the use of two chemically modified sgRNAs is a highly effective way to facilitate gene disruption in primary human T cells and CD34+ HSPCs.
In this study, we show that chemically synthesized sgRNAs can be used effectively for targeted genome editing, and we demonstrate that chemically modified sgRNAs significantly enhance genome editing efficiencies in human primary T cells and CD34+ HSPCs. Chemically synthesized and modified sgRNAs offer advantages over expressed or in vitro transcribed sgRNAs, including (i) increased efficacy, (ii) robust and scalable production of highly pure sgRNAs for biotechnological and therapeutic applications, (iii) greater flexibility in the sgRNA design in contrast to constraints on the first transcribed nucleotides imposed by the U6 or T7 promoters typically used for plasmid expression or in vitro transcription of sgRNAs, respectively, and (iv) enabling of a highly active RNA-only or RNP CRISPR platform with lower cytotoxicity in primary cells than DNA plasmid-based systems.
The simplification of the CRISPR-Cas system to a purely RNA or RNP CRISPR system lends itself to formulation in different nanoparticle vectors for delivery in vivo such that the nuclease will not be expressed continuously, as it would be when delivered as part of a viral vector. Furthermore, we anticipate that chemically modified sgRNAs will enhance multiplexed genome editing as well as high-throughput, multiwell experiments. We also anticipate that modified sgRNAs will refine a wide range of CRISPR-associated technologies such as the CRISPRi/CRISPRa systems for inhibition and activation of gene expression19, the CRISPR imaging tool for dynamic visualization of genomic loci20, and CRISPR-mediated RNA recognition and cleavage21. The technology may be further developed to improve intracellular delivery to target cells or tissues, and enable conjugation to various molecules for imaging and biochemical studies. Future studies could investigate a larger variety of chemical modifications, explore different locations of the modifications for rational design of optimized sgRNAs, as well as the mechanism for the enhanced activity of modified sgRNAs. In conclusion, this study suggests that chemically modified sgRNAs such as those presented here have the potential to substantially improve a wide array of CRISPR/Cas bio-technological and therapeutic applications.
ONLINE METHODS
sgRNA synthesis
All RNA oligomers were synthesized on an ABI 394 Synthesizer (Life Technologies, Carlsbad, CA, USA) using 2′-O-thionocarbamate-protected nucleoside phosphoramidites (Sigma-Aldrich, St. Louis, MO, USA or Thermo Fisher, Waltham, MA, USA) according to previously described procedures6. 2′-O-methyl phosphoramidites were purchased from Thermo Scientific, Grand Island, NY, and incorporated into RNA oligomers under the same conditions as the 2′-O-thionocarbamate protected phosphoramidites. The 2′-O-methyl-3′-O-(diisopropylamino)phosphinoacetic acid-1,1-dimethylcyanoethyl ester-5′-O-dimethoxytrityl nucleosides used for synthesis of thiophosphonoacetate (thioPACE)-modified RNAs were synthesized essentially according to published methods9,22. For phosphorothioate containing oligomers, the iodine oxidation step after the coupling reaction was replaced by a sulfurization step using a 0.05 M solution of 3-((N,N-dimethylamino methylidene)amino)-3H-1,2,4-dithiazole-5-thione in a pyridine-acetonitrile (3:2) mixture for 6 min. Unless noted otherwise, reagents for solid phase RNA synthesis were purchased from Glen Research (Sterling, VA, USA).
All oligonucleotides were purified using reversed phase high-performance liquid chromatography (HPLC) and analyzed by liquid chromatography– mass spectrometry (LC-MS) using an Agilent 1290 Infinity series LC system coupled to an Agilent 6520 Q-TOF (time-of-flight) mass spectrometer (Agilent Technologies, Santa Clara, CA, USA). The yields for the synthesis and purification of the sgRNAs were estimated using deconvolution of mass spectra obtained from LC-MS-derived total ion chromatograms. The chemical synthesis of the 100-mer sgRNAs typically yielded 25–35% full-length product from a nominal 1 micromole scale synthesis. Reversed-phase HPLC purification using ion pairing buffer conditions typically gave 20% yield from the crude product with an estimated purity of the final sgRNA in the range of 90% to 95%. Supplementary Table 1 shows the sequences of all sgRNAs used and the masses obtained from deconvolution of the multiple charge state series of peaks found from analysis of the purified sgRNAs. The deconvolution was done using Mass Hunter Qualitative Analysis (version B.06.00) software (Agilent).
Biochemical in vitro DNA cleavage assays
4 kb PAM-addressable targets were prepared by preparative PCR amplification of plasmid-borne human sequences. In a 20 μl reaction volume, 50 fmoles of linearized DNA target in the presence of 50 nM sgRNA, 39 nM recombinant purified Cas9 protein (Agilent) and 10 mM MgCl2 at pH 7.6 was incubated at 37 °C for 30 min. Upon completion, 0.5 μl of RNace-It (Agilent) was added, and incubation was continued at 37 °C for 5 min and then at 70 °C for 15 min. Subsequently 0.5 μl of Proteinase K (Mol. Bio. grade, New England Biolabs) was added and incubated at 37 °C for 15 min. Aliquots were loaded into a DNA 7500 LabChip and were analyzed on a Bioanalyzer 2200. Cleavage frequencies were calculated by the formula: (a/b) × 100 where ‘a’ is the sum of the band intensities of the two cleavage products and b is the sum of band intensities of cleaved and uncleaved DNA.
Plasmids
sgRNA expression vectors were constructed by cloning of 20 bp oligonucleotide target sequences into px330 (Addgene plasmid #42230) containing a human codon-optimized SpCas9 expression cassette and a human U6 promoter driving the expression of the chimeric sgRNA (Supplementary Table 1 for sgRNA sequences).
All three plasmid targeting vectors carry ~2 × 800 bp arms of homology, which were generated by PCR amplification of the corresponding loci using genomic DNA isolated from K562 cells. The homology arms were then cloned into a ~2,900 base pair vector based on pBluescript SK+ using standard cloning methods. Between the homology arms, both the HBB and CCR5 donors contain the EF1α promoter driving expression of GFP. The IL2RG donor lacks a promoter and relies on endogenous activity of the IL2RG gene to drive GFP expression. Complete plasmid sequences can be found in Supplementary Note 1.
Cell culture and nucleofections
K562 (ATCC) and T cells were cultured at 37 °C, 5% CO2, and ambient oxygen levels. CD34+ hematopoietic stem/ progenitor cells (HSPCs) were cultured at 37 °C, 5% CO2 and 5% O2. K562 cells were maintained in RPMI 1640 (HyClone) supplemented with 10% bovine growth serum, 100 mg/ml streptomycin, 100 units/ml penicillin, and 2 mM l-glutamine. K562 cells were nucleofected using the Lonza Nucleofector 2b (program T-016) and a nucleofection buffer containing 100 mM KH2PO4, 15 mM NaHCO3, 12 mM MgCl2 × 6H2O, 8 mM ATP, 2 mM glucose (pH 7.4) Nucleofection conditions: 100 μl nucleofection solution, 106 cells, 1 to 20 μg chemically modified sgRNA, 1 to 15 μg Cas9 mRNA (Cas9 mRNA, 5meC, Ψ, Product Code: L-6125, TriLink BioTechnologies, San Diego, CA, USA), 2 μg sgRNA/Cas9-encoding plasmid, or 5 μg HR donor plasmid. CD3+ T cells were isolated from buffy coats obtained from the Stanford School of Medicine Blood Center using a human Pan T Cell Isolation Kit (Miltenyi Biotec, San Diego, CA, USA). CD3+ cells were maintained in X-VIVO 15 (Lonza, Walkersville, MD, USA) supplemented with 5% human serum (Sigma-Aldrich, St. Louis, MO, USA), 100 IU/ml human recombinant IL-2 (Peprotech, Rocky Hill, NJ, USA), and 10 ng/ml human recombinant IL-7 (BD Biosciences, San Jose, CA, USA). Before nucleofection, T cells were activated for 3 d with immobilized anti-CD3 antibodies (clone: OKT3, eBioscience, San Diego, CA, USA) and soluble anti-CD28 antibodies (clone: CD28.2, eBioscience). For non-activated CD3+ T cells, cells were nucleofected immediately after isolation. T cells were nucleofected using the Lonza Nucleofector 2b (program U-014) and the Human T Cell Nucleofector Kit (VPA-1002, Lonza). Nucleofection conditions: 100 μl nucleofection solution, 106 cells, 10 to 20 μg chemically modified sgRNA, 15 to 30 μg Cas9 (or 15 μg eGFP mRNA, TriLink BioTechnologies, San Diego, CA, USA), 1 μg sgRNA/Cas9-encoding plasmid. Mobilized human peripheral blood CD34+ HSPCs were purchased from AllCells and thawed according to manufacturer’s instructions. CD34+ HSPCs were maintained in X-VIVO 15 (Lonza) supplemented with SCF (100 ng/ml), TPO (100 ng/ml), Flt3-Ligand (100 ng/ml), IL-6 (100 ng/ml) and StemRegenin1 (0.75 mM). CD34+ HSPCs were nucleofected using the Lonza 4D-Nucleofector (program EO-100) and the P3 Primary Cell 4D-Nucleofector Kit (V4XP-3024). Nucleofection conditions: 100 μl nucleofection solution, 5 × 105 cells, 10 μg chemically modified sgRNA, 15 μg Cas9 mRNA, 1 μg plasmid. For Cas9 RNP experiments, Cas9 protein was purchased from PNA Bio (Thousand Oaks, CA, USA) or Life Technologies (Carlsbad, CA, USA). For all RNP experiments except for Supplementary Figure 6, Cas9 protein from PNA Bio was used. Cas9 protein was complexed with sgRNAs in a Cas9:sgRNA molar ratio of 1:2.5 for 10 min at 25 °C. RNPs were nucleofected into K562 cells or T cells as described above with 106 cells in 100 μl of the respective nucleofection solutions. For the dual sgRNA experiments, the total sgRNA amount was 10 μg (10 μg when used individually and 2 × 5 μg when used together). For both T cells and CD34+ HSPCs, sgRNAs were nucleofected with 15 μg Cas9 mRNA into 106 cells. T-cell nucleofections were performed as above whereas nucleofection of CD34+ HSPCs were similar to the T-cell nucleofections using the Lonza Nucleofector 2b (program U-014) and the Human T Cell Nucleofector Kit (VPA-1002, Lonza). Directly after nucleofection CD34+ HSPCs were incubated at 30 °C for 24 h after which they were transferred to 37 °C until harvest of genomic DNA.
Flow cytometry and fluorescence-activated cell sorting (FACS)
For HR experiments, cells were analyzed 2–3 weeks after nucleofection depending on the donor plasmid when there was no remaining eGFP expression from episomal plasmid. eGFP expression was measured on an Accuri C6 flow cytometer (BD Biosciences, San Jose, CA, USA). Cell death was measured with the LIVE/DEAD Fixable Red Dead Cell Stain Kit (Life Technologies, Carlsbad, CA, USA) according to manufacturer’s instructions and cells were analyzed on the Accuri C6 flow cytometer. For sorting of CD3+ T cells into CD4+ and CD8+ populations cells were stained with a mix of PE-Cy7 anti-human CD4 (clone: RPA-T4, Tonbo Biosciences, San Diego, CA, USA) and an APC anti-human CD8a (clone: RPA-T8, Tonbo Biosciences) and the two populations were sorted on a FACS Aria II SORP.
Measuring allele modification frequencies using TIDE, T7 assay and sequencing of TOPO-cloned PCR fragments
For TIDE and T7 assays, genomic DNA was extracted from cells 3 d after nucleofection (if not otherwise indicated) using QuickExtract DNA Extraction Solution (Epicentre, Madison, WI, USA) following manufacturer’s instructions. PCR amplicons spanning the sgRNA genomic target sites were generated using the iProof High-Fidelity Master Mix (Bio-Rad, Hercules, CA, USA) with the following primer pairs: IL2RG_fw: 5′-TCACACAGCACATATTTGCCACACCCTCTG-3′; IL2RG_RV: 5′-TGCCCACATGATTGTAATGGCCAGTGG-3′; HBB_fw: 5′-CCAACTCCT AAGCCAGTGCCAGAAGAG-3′; HBB_rv: 5′-AGTCAGTGCCTATCAGAAAC CCAAGAG-3′; CCR5_fw: 5′-GCACAGGGTGGAACAAGATGG-3′; CCR5_rv: 5′-CACCACCCCAAAGGTGACCGT-3′. For T7 assays, PCR amplicons were purified and 200 ng was denatured and re-annealed in a thermocycler and digested with T7 Endonuclease I (New England Biolabs, Waltham, MA, USA) according to manufacturer’s protocol. Digested DNA was run on a 4–20% TBE polyacrylamide gel, stained with Diamond Nucleic Acid Dye (Promega, Madison, WI, USA), and visualized on a ChemiDoc XRS+ (Bio-Rad). Band intensities were analyzed using the Image Lab Software (Bio-Rad) and allele modification frequencies were calculated with the formula: 100 × (1 – (1 – fraction cleaved)^0.5). For analyzing allele modification frequencies using TIDE (Tracking of In/dels by Decomposition)23, the purified PCR products were Sanger-sequenced using both PCR primers and each sequence chromatogram was analyzed with the online TIDE software available at http:// tide.nki.nl. Analyses were performed using a reference sequence from a mock-transfected sample. Parameters were set to the default maximum indel size of 10 nucleotides and the decomposition window to cover the largest possible window with high quality traces. All TIDE analyses below the detection sensitivity of 3.5% were set to 0%. For sequencing of TOPO-cloned PCR fragments a 2.1 kb amplicon (WT size) spanning the cleavage site(s) was generated using iProof High-Fidelity Master Mix and primers 5′-ggctgttgtcatctatgaccttccc-3′ and 5′-tgtaaactgagcttgctcgctcg-3′ with 25 cycles including 72 °C annealing temperature, and 2 min elongation time. The PCR reaction products were subcloned into a plasmid directly using the Zero Blunt TOPO PCR Cloning Kit (Life Technologies) according to the manufacturer’s protocol. TOPO reactions were transformed into XL-1 Blue competent cells, plated on agar plates with kanamycin, and single colonies were sequenced directly from the plates by McLab (South San Francisco, CA, USA) by rolling circle amplification followed by sequencing using primer 5′-gcacagggtggaacaagatgg-3′.
Proliferation assay
For measuring proliferation of T cells following nucleofection, the CellTiter-Glo 2.0 Assay (Promega, Madison, WI, USA) was used. Directly after nucleofection, T cells were transferred to multiple 96-well U-bottom 96-well plates at 3 × 104 cells/well. Directly after nucleofection and at 24-h intervals, cells were transferred to white 96-well plates in 100 μl medium and adding 100 μL CellTiter-Glo 2.0 per manufacturer’s guidelines. Luminescence was read on a Tecan Infinite 200 PRO (Tecan, Männedorf, Switzerland) using a 1 s integration time.
Deep sequencing to quantify efficiency and specificity of genome modifications
For each gene targeting experiment, genomic DNA was extracted from the various CRISPR-treated and control K562 cells 72 h post-transfection. Genomic regions flanking the CRISPR target and three off-targets (Supplementary Table 3) were amplified by two rounds of PCR to attach (treatment-specific) barcodes and Illumina sequencing adaptors (Supplementary Tables 4 and 5). Barcoded PCR amplicons were pooled equimolarly, purified by a spin-column and sequenced on the Illumina MiSeq DNA sequencer platform. See Supplementary Note 2 for details.
For analysis of sequencing data, reads from different treatments were binned by their corresponding treatment barcodes and were mapped to the genome using BWA-MEM24 (bwa-0.7.10) with default parameters. Inconsistent paired end mappings (>1 kbp apart) were filtered from the analysis along with low quality mapped and secondary aligned reads as defined by the default parameters in BWA-MEM. For each of the on- and off-target regions we calculated the % of reads with indels surrounding the predicted cut-site, where the predicted cut-site is estimated to be between the 3rd and the 4th position upstream of the PAM site. The indel % for each position in the genome was calculated by using indel[i] = (I[i] + D[i])/C[i], where D[i] and I[i] indicates the number of reads with a deletion or an insertion of any size at position i, respectively, and C[i] indicates the number of reads mapped to any genomic interval containing position i. The indel % for each target was then calculated by indel[c], where c is the expected cut-site which is the 4th position upstream the PAM site. In cases with homo-nucleotide sequence at position 4, ‘AA’ in IL2RG and ‘CCC’ in CCR5, and an insertion event of the same nucleotide composition or a deletion event shorter than homo-nucleotide sequence, the BWA_MEM aligner cannot resolve the position of the inserted/deleted nucleotide(s). This was corrected by taking the relevant position in the alignment report instead of position 4, specifically, taking positions 3 and 6 for IL2RG and CCR5 off-targets, respectively.
Supplementary Material
ACKNOWLEDGMENTS
A.H. was supported by the Myotonic Dystrophy Foundation. R.O.B. was supported through an Individual Postdoctoral grant (DFF–1333-00106B) and a Sapere Aude, Research Talent grant (DFF–1331-00735B), both from the Danish Council for Independent Research, Medical Sciences. M.H.P. gratefully acknowledges the support of the Amon Carter Foundation, the Laurie Krauss Lacob Faculty Scholar Award in Pediatric Translational Research and US National Institutes of Health grant support PN2EY018244 and R01-AI097320. We thank R. Perriman, and Porteus laboratory members and C. Carstens, S. Laderman and Agilent laboratories members for helpful input, comments and discussion.
Footnotes
Note: Any Supplementary Information and Source Data files are available in the online version of the paper.
AUTHOR CONTRIBUTIONS
A.H., R.O.B., J.T.C., A.B.W., R.B., A.B.K. and D.E.R. performed and designed experiments. D.D., D.E.R. and R.J.K. chose the specific modification types and the positions for their incorporation into the sgRNAs. S.R., B.D.L. and R.J.K. performed synthesis and purification of all the sgRNAs. I.S. and A.T. developed and applied the sequencing data analysis pipeline used to analyze the deep sequencing data. L.B. and M.H.P. directed the research. A.H. and R.O.B. wrote the manuscript with help from all authors.
COMPETING FINANCIAL INTERESTS
The authors declare competing financial interests: details are available in the online version of the paper.
References
- 1.Jinek M, et al. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mali P, et al. RNA-guided human genome engineering via Cas9. Science. 2013;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cong L, et al. Multiplex genome engineering using CRISPR-Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hsu PD, Lander ES, Zhang F. Development and applications of CRISPR-Cas9 for genome engineering. Cell. 2014;157:1262–1278. doi: 10.1016/j.cell.2014.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hendel A, Fine EJ, Bao G, Porteus MH. Quantifying on- and off-target genome editing. Trends Biotechnol. 2015;33:132–140. doi: 10.1016/j.tibtech.2014.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dellinger DJ, et al. Streamlined process for the chemical synthesis of RNA using 2′-O-thionocarbamate-protected nucleoside phosphoramidites in the solid phase. J. Am. Chem. Soc. 2011;133:11540–11556. doi: 10.1021/ja201561z. [DOI] [PubMed] [Google Scholar]
- 7.Deleavey GF, Damha MJ. Designing chemically modified oligonucleotides for targeted gene silencing. Chem. Biol. 2012;19:937–954. doi: 10.1016/j.chembiol.2012.07.011. [DOI] [PubMed] [Google Scholar]
- 8.Eckstein F. Phosphorothioates, essential components of therapeutic oligonucleotides. Nucleic Acid Ther. 2014;24:374–387. doi: 10.1089/nat.2014.0506. [DOI] [PubMed] [Google Scholar]
- 9.Dellinger DJ, Sheehan DM, Christensen NK, Lindberg JG, Caruthers MH. Solid-phase chemical synthesis of phosphonoacetate and thiophosphonoacetate oligodeoxynucleotides. J. Am. Chem. Soc. 2003;125:940–950. doi: 10.1021/ja027983f. [DOI] [PubMed] [Google Scholar]
- 10.Hendel A, et al. Quantifying genome-editing outcomes at endogenous loci with SMRT sequencing. Cell Reports. 2014;7:293–305. doi: 10.1016/j.celrep.2014.02.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cradick TJ, Fine EJ, Antico CJ, Bao G. CRISPR-Cas9 systems targeting beta-globin and CCR5 genes have substantial off-target activity. Nucleic Acids Res. 2013;41:9584–9592. doi: 10.1093/nar/gkt714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tebas P, et al. Gene editing of CCR5 in autologous CD4 T cells of persons infected with HIV. N. Engl. J. Med. 2014;370:901–910. doi: 10.1056/NEJMoa1300662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lombardo A, et al. Site-specific integration and tailoring of cassette design for sustainable gene transfer. Nat. Methods. 2011;8:861–869. doi: 10.1038/nmeth.1674. [DOI] [PubMed] [Google Scholar]
- 14.Voit RA, Hendel A, Pruett-Miller SM, Porteus MH. Nuclease-mediated gene editing by homologous recombination of the human globin locus. Nucleic Acids Res. 2014;42:1365–1378. doi: 10.1093/nar/gkt947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hsu PD, et al. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat. Biotechnol. 2013;31:827–832. doi: 10.1038/nbt.2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cradick TJ, Qiu P, Lee CM, Fine EJ, Bao G. COSMID: a web-based tool for identifying and validating CRISPR-Cas off-target sites. Mol. Ther. Nucleic Acids. 2014;3:e214. doi: 10.1038/mtna.2014.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yi G, et al. CCR5 gene editing of resting CD4(+) T cells by transient ZFN expression from HIV envelope pseudotyped nonintegrating lentivirus confers HIV-1 resistance in humanized mice. Mol. Ther. Nucleic Acids. 2014;3:e198. doi: 10.1038/mtna.2014.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mandal PK, et al. Efficient ablation of genes in human hematopoietic stem and effector cells using CRISPR-Cas9. Cell Stem Cell. 2014;15:643–652. doi: 10.1016/j.stem.2014.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gilbert LA, et al. Genome-scale CRISPR-mediated control of gene repression and activation. Cell. 2014;159:647–661. doi: 10.1016/j.cell.2014.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen B, et al. Dynamic imaging of genomic loci in living human cells by an optimized CRISPR-Cas system. Cell. 2013;155:1479–1491. doi: 10.1016/j.cell.2013.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.O’Connell MR, et al. Programmable RNA recognition and cleavage by CRISPR-Cas9. Nature. 2014;516:263–266. doi: 10.1038/nature13769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Threlfall RN, Torres AG, Krivenko A, Gait MJ, Caruthers MH. Synthesis and biological activity of phosphonoacetate- and thiophosphonoacetate-modified 2′-O-methyl oligoribonucleotides. Org. Biomol. Chem. 2012;10:746–754. doi: 10.1039/c1ob06614e. [DOI] [PubMed] [Google Scholar]
- 23.Brinkman EK, Chen T, Amendola M, van Steensel B. Easy quantitative assessment of genome editing by sequence trace decomposition. Nucleic Acids Res. 2014;42:e168. doi: 10.1093/nar/gku936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li H, Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics. 2010;26:589–595. doi: 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.