Abstract
Previous studies from our laboratories have shown that the tissue specificity of α1-antitrypsin (AT) expression differs among closely related mouse species. In laboratory mice (Mus domesticus), AT mRNA is found almost exclusively in the liver. In the wild-derived species Mus caroli, the mRNA is expressed not only in the liver but also in the kidney, where it is regulated by androgens during post-natal development. We presently show that the tissue specificity, the species specificity, and the developmental regulation of AT mRNA levels correlate with the transcription rate of the AT gene, as measured by nuclear run-on assays. During the course of these experiments, we found that some AT-specific probes are complementary to constitutively synthesized RNAs that do not accumulate and that are unrelated to functional AT mRNA expression. These RNAs, which result from both sense and anti-sense transcription, may derive from aberrant initiation events within certain regions of the AT gene.
α1-Antitrypsin (AT)1 is a serine protease inhibitor that is found in the bloodstream of all mammals and that functions in controlling the activity of polymorphonuclear leucocyte elastase within the circulation. AT synthesis occurs predominantly in hepatocytes; however, recent evolutionary modifications in the tissue specificity of AT expression have occurred, as exemplified by the murine species Mus caroli, which contains high concentrations of AT mRNA in kidney as well as in liver (1). The unique ability of M. caroli to accumulate AT mRNA in the kidney is a consequence of variation in a cis-acting genetic element (1).
A striking feature of kidney AT expression is its regulation during post-natal development. A large increase in the AT mRNA concentration occurs during puberty, between 25 and 40 days of age (2). This developmental pattern, which appears to be mediated by testosterone (2), is in sharp contrast to the situation in liver, where AT expression reaches maximal levels during gestation and is maintained through adulthood (2, 3).
Our earlier studies showed that the AT gene is hypomethylated in the liver of Mus domesticus and in the liver and kidney of M. caroli (1), suggesting that in both species the tissue-specific accumulation of AT mRNA is generated at the level of transcription. In the present communication, we have used nuclear run-on assays (4, 5) to test this suggestion directly. Our results show that AT mRNA levels do, in general, reflect gene transcription rates in various tissues within both M. domesticus and M. caroli. During the course of these studies, we observed constitutive transcription within some regions of the gene; this may be a consequence of aberrant RNA synthesis that does not reflect functional AT mRNA expression.
MATERIALS AND METHODS
Animals
Mice of M. domesticus strain DBA/2J were obtained from the Jackson Laboratory, Bar Harbor, ME. M. caroli mice were from the University of South Carolina’s colony, and were established with animals provided by Dr. V. Chapman of Roswell Park Memorial Institute, Buffalo, NY. Males were used in all experiments.
Plasmids
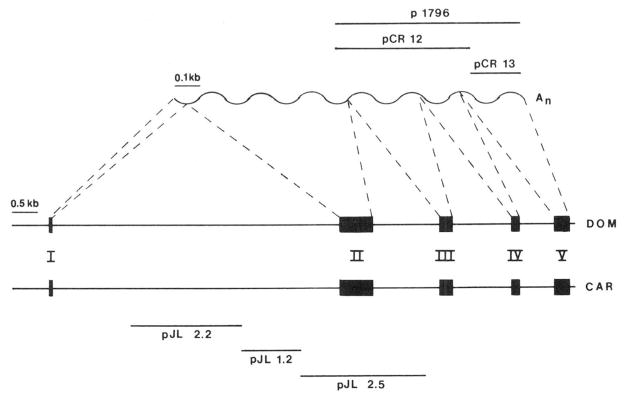
pJL2.2, pJL1.2, and pJL2.5 are subclones of three consecutive fragments of the M. caroli AT gene in the vector pUC13 (2). Northern blot analyses, as well as hybridization studies with cDNA probes, indicate that the inserts of pJL2.2 and pJL1.2 comprise 2.2-and 1.2-kilobase pair regions, respectively, of the first intron, while the insert of pJL2.5 corresponds to a 2.5-kilobase pair fragment containing the second exon (Fig. 1). Plasmid pCR24 contains the insert of pJL2.5 in the vector pT3T7-18. Plasmids pCR12 and pCR13 contain the 503- and 215-base pair fragments, respectively, of M. domesticus AT cDNA plasmid p1796 (3) in the vector pT3T7-18 (Fig. 1). Plasmid pCR1 contains the 1.1-kilobase pair insert of the RP2 cDNA plasmid pMK908 (7) in the vector pT3T7-18. To generate sense and antisense probes, the appropriate inserts were subcloned into M13mp8 DNA in both orientations.
Fig. 1. Structure of the AT gene and mRNA.
The exon/intron structure at the AT gene from M. domesticus (DOM) is taken from Krauter et al. (20); the corresponding mRNA, and the cDNA subclones derived from it, are indicated. The structure of the M. caroli (CAR) gene was partially characterized by hybridization studies with appropriate genomic and cDNA clones, which indicate that the M. caroli exons are located in homologous positions to the M. domesticus exons (J. Latimer, unpublished experiments); only exon I of the M. caroli gene has been sequenced. The genomic subclones derived from the M. caroli AT gene are indicated, kb, kilo-base.
Northern Blot Analysis
Preparations of total RNA (15 μg) were denatured in formamide, fractionated by agarose gel electrophoresis in the presence of 2.2 M formaldehyde, transferred to nitrocellulose, and hybridized to the p1796 DNA (3). Hybridizing RNA was observed by autoradiography.
Nuclear Run-on Assays
Nuclei were isolated by the method of Lamers et al. (6). Yields were usually 5–10 × 107 nuclei/g tissue. In vitro transcription was done according to Lamers et al. (6), using 20–30 × 106 nuclei and 200 μCi of [α-32P]UTP (800 Ci/mmol; Du Pont-New England Nuclear) per assay. Reactions were allowed to proceed at 25 °C for 60 min. (In some experiments, reactions ran for 15 min, which did not significantly change the results.) Following the incubation, the 32P-labeled RNA was purified according to methods described by Mahajan and Thompson (8), as communicated by H. Lieberman.2 In brief, the transcription reaction was treated with DNase I and proteinase K, extracted with phenol, and the RNA was precipitated and washed in trichloroacetic acid followed by rinsing in ethanol. Generally, a total of 4–10 × 106 cpm of RNA/assay were obtained. Plasmid or single-stranded DNAs (2 μg) were denatured in 1 ml of 0.3 N NaOH for 1 h at 65 °C, neutralized by the addition of 1 ml of 2 M ammonium acetate, pH 7.0, and applied to a nylon membrane (HYBOND-N; Amersham Corp.) in 1 M ammonium acetate using a Mini-fold apparatus (Schleicher & Schuell). The membranes were irradiated with UV light for 3.5 min, air-dried, baked for 2 h at 80 °C, and washed for 1 h in 10 mM Tris-Cl, pH 7.4, 2 mM Na2EDTA, 0.3 M NaCl, and 0.1% sodium dodecyl sulfate. Prehybridization of the filters was for 1 h at 65 °C in 10 mM HEPES, pH 7.5, 0.5 M NaCl, 10 mM Na2EDTA, 0.2% sodium dodecyl sulfate, 0.2% Ficoll, and 0.02% polyvinylpyrollidone. The solution was changed, and 1–2 × 106 cpm of [32P]RNA was added. Hybridization was for 48 h at 65 °C, after which the membranes were washed at 65 °C in 5 x SSC (1 x SSC is 15 mM sodium citrate, pH 7.0, 0.15 M NaCl), 0.5 mM Na2EDTA, and 0.5% sodium dodecyl sulfate, and then in 2 x SSC containing 5 mM Na2EDTA. Radioactivity was detected by autoradiography.
RESULTS
AT Gene Transcription in Mouse Tissues
Nuclear run-on assays (4, 5) were utilized to measure gene transcription rates in several tissues from both M. domesticus (strain DBA/2J) and M. caroli. Isolated nuclei were incubated in vitro in the presence of [α-32P]UTP, and the nascent RNA was purified and hybridized to various plasmid DNAs on nylon filters. AT transcription was measured using plasmid pJL2.5, which contains a subcloned fragment of the M. caroli AT gene, and includes the 3′-end of the first intron, the second exon, and the 5′-end of the second intron (Fig. 1). As a control, transcription of the RP2 gene (7) was measured using plasmid pCR1; RP2 is expressed in all mouse tissues (9, 10) and is constitutively transcribed.3 To monitor nonspecific hybridization, various vector DNAs were included on the filters.
Fig. 2 shows results of analysis of AT transcription in nuclei from liver and kidney. In M. domesticus, where AT mRNA expression is liver-specific, transcription was observed in liver nuclei, while none was detected in kidney. In contrast, for M. caroli, where AT mRNA is produced in liver and kidney, transcription was observed in both liver and kidney nuclei. No transcription was detected in brain nuclei from either species (data not shown). Kidney and liver nuclei from both species exhibited RP2 transcription (Fig. 2). Thus, the tissue specificity of AT mRNA accumulation in both M. domesticus and M. caroli is paralleled by the tissue specificity of AT gene transcription.
Fig. 2. Transcription rates in liver and kidney nuclei.

AT and RP2 transcription rates were measured in M. caroli (CAR) and M. domesticus (DOM) kidney (KD) and liver (LV) nuclei by the run-on assay, as described under “Materials and Methods.” The AT probe was pJL2.5, while the RP2 probe was pCR1; specificity was assured by using pBR322 and pT3T7-18 (pT3T7) DNAs.
In M. caroli kidney, AT mRNA concentrations undergo an androgen-mediated developmental induction between 25 and 40 days of age; in the liver, AT mRNA levels are adult-like at birth (2). This difference between liver and kidney is depicted in Fig. 3. To determine if the developmental patterns are generated at the transcriptional level, assays were performed with liver and kidney nuclei from 13-day-old, 30-day-old, and adult (>60-day-old) animals. The results (Fig. 4) show that in kidney nuclei, AT gene transcription is barely detectable at 13 days, is increased at 30 days, and is maximal in adults. In liver nuclei, transcription is high throughout post-natal development. As expected, transcription of the RP2 gene is about equal in both tissues throughout development. Thus, the developmental increase in AT mRNA concentrations in M. caroli kidney is correlated with a specific induction of transcription. The inability to measure accurately the low level of transcription in 13-day nuclei precludes determining whether or not an increased rate of transcription totally accounts for the mRNA induction.
Fig. 3. Developmental regulation of AT mRNA expression in M. caroli.

Total RNA from 13-day-old and adult M. caroli liver and kidney were fractionated on agarose gels, transferred to and fixed onto a nylon membrane, hybridized to the AT cDNA plasmid p1796, and observed by autoradiography. Lane A, 13-day liver; lane B, adult liver; lane C, 13-day kidney; lane D, adult kidney.
Fig. 4. Developmental regulation of transcription rates.

Nuclei from 13-day-old (lanes a and d), 30-day-old (lanes b and e), and adult (lanes c and f) M. caroli kidney (KD) and liver (LV) were analyzed for AT and RP2 transcription, as described under “Materials and Methods.” The AT probe was pCR24, while the RP2 probe was pCR1; pT3T7-18 (pT3T7) and pBR322 (pBR) served as controls.
Constitutive Transcription within the AT Gene
The results presented above suggest that the tissue specificity, the species specificity, and the developmental regulation of AT mRNA levels are determined, at least in part, by the transcription rate of the AT gene. To substantiate this conclusion, run-on assays were performed using a variety of other AT-specific probes, including genomic (pJL2.2, pJL1.2) as well as cDNA (pCR12, pCR13) clones (Fig. 1). Surprisingly, the results showed striking discrepancies, especially with regard to M. domesticus kidney (Fig. 5). While pJL2.2 hybridized to sequences whose transcription rates paralleled tissue- and species-specific AT mRNA levels, pJL1.2 and pCR12 hybridized to abundantly transcribed RNAs that were synthesized constitutively. Very little transcription was measurable with pCR13, probably due to the small size of its insert. In other experiments,3 pCR12 hybridized to transcription products in nuclei from brain, which accumulates no AT mRNA in either murine species. Thus, several regions of the AT gene are complementary to sequences that are constitutively transcribed in isolated nuclei in vitro and that do not reflect AT mRNA levels in the corresponding tissues.
Fig. 5. Analysis of AT gene transcription using several cDNA and genomic probes.

Nuclei from M. domesticus (DOM) and M. caroli (CAR) liver (LV) and kidney (KD) were analyzed for AT gene transcription by the run-on assay. The AT-specific probes were genomic clones pJL1.2 and pJL2.2 and cDNA clones pCR12 and pCR13 (see Fig. 1).
It is generally assumed that nuclear run-on assays measure elongation of preexisting transcripts and that no re-initiation occurs (4, 5). However, it is formally possible that constitutive transcription within the AT gene results from initiation events in vitro. To test this, run-on transcription was measured in kidney nuclei in the presence of 0.015% Sarkosyl, which inhibits transcription initiation but has no effect upon elongation (11). As shown in Fig. 6, in kidney nuclei, preincubation with Sarkosyl had no effects upon transcription from any region of the AT gene. Importantly, the high levels of transcription observed in M. domesticus kidney nuclei with pCR12 and pJL1.2 are resistant to Sarkosyl, indicating that they are derived from pre-initiated transcription complexes.
Fig. 6. Transcription of the AT gene in the presence of Sarkosyl.

Run-on assay mixtures, using nuclei from M. domesticus (DOM) and M. caroli (CAR) kidney, were preincubated for 5 min in 0.015% Sarkosyl (SARK) in the absence of nucleotides; nucleotides were added, and the transcription rates were measured in the usual manner, using the four AT-specific probes described in Fig. 5. Untreated controls, indicated by minus (−), were run in parallel with the treated samples, indicated by plus (+).
Strand Specificity of Transcription
To distinguish sense from antisense transcription, the nuclear run-on assays were performed with single-stranded probes corresponding to each of the four plasmids used in Fig. 5. It can be seen in Fig. 7 that sense transcription predominated within all regions of the AT gene. As observed with the double-stranded probes, RNA synthesis corresponding to the sense strand of pJL2.2 paralleled mRNA levels, while that corresponding to the sense strand of pCR12 or pJL1.2 was constitutive. Antisense transcription, although low, was detectable with all probes (Fig. 7). Thus, although significant antisense transcription occurs within the AT gene, it does not fully account for the constitutive expression observed with some probes. Sense transcription is resistant to Sarkosyl (Fig. 8), suggesting that it arises from pre-initiated RNA chains.
Fig. 7. Sense and antisense transcription of the AT gene.

Run-on assays were conducted with liver (LV) and kidney (KD) nuclei from both M. domesticus (DOM) and M. caroli (CAR). Single-stranded M13-derived probes were used to detect RNA made in the sense or antisense direction from AT gene regions corresponding to each of the four plasmids described in the legend to Fig. 5.
Fig. 8. Transcription of sense RNA in kidney nuclei in the presence of Sarkosyl.

Run-on transcription was measured in kidney nuclei from M. domesticus (DOM) and M. caroli (CAR), using sense-specific probes. Nuclei were pretreated for 5 min with Sarkosyl (SARK) (see Fig. 6). Untreated controls (−) and treated samples (+) were run in parallel.
DISCUSSION
The results presented in this report indicate that the tissue and species specificity of AT mRNA accumulation is controlled at the transcriptional level. Liver-specific expression of both the human and the mouse AT genes has been shown to be determined by cis-acting elements located immediately upstream from each gene (12–14); this is consistent with the high rate of AT transcription in M. domesticus liver nuclei, as compared to kidney or brain nuclei (Fig. 2). Of particular interest is the transcription of the AT gene in M. caroli kidney (Fig. 1), which parallels the abundant, species-specific expression of AT mRNA found in this tissue (1, 2). Thus, during evolution of the Mus genus, transcriptional activation of the kidney AT gene occurred and was fixed in M. caroli. The element responsible for this activation is cis-acting (1), which suggests that it lies within or near the gene. Most likely, the AT promoter harbors kidney-specific, as well as liver-specific, control elements; these elements are regulated differently during development (2) and probably interact with different tissue-specific trans-acting factors. It will be of interest to determine the structural and functional relationships between the liver-specific and kidney-specific elements and to understand the nature of the changes that are responsible for the interspecies variation in kidney AT expression.
An unexpected finding was that some AT-specific probes hybridize to constitutively transcribed RNAs that do not reflect mRNA levels. These apparently aberrant RNAs are not cross-hybridizing products of other genes, since none of the probes in the present study represents repetitive sequences, as judged by Southern blot analyses (data not shown). The constitutive RNAs are derived from pre-initiated transcripts, as indicated by their synthesis in the presence of Sarkosyl (Figs. 6 and 8). Thus, they do not result from artifactural initiation events occurring in vitro, but must be a consequence of initiation in vivo. Most of these RNAs are transcribed in the sense direction, although significant anti-sense transcription was observed (Fig. 7).
The biological significance, if any, of this phenomenon is not known. Similar findings have been reported for the c-myc gene, where it appears that functional mRNA synthesis occurs in a “background” of sense and antisense transcription from specific regions within the gene (15–17). Sense transcription of c-myc is regulated by an attenuation-like mechanism, which explains constitutive transcription of its 5′-proximal region (15, 16); the extent of antisense RNA synthesis varies throughout the gene and plays an unknown role in regulation. For the AT gene, there is no correlation between the location of a particular probe within the gene and its correspondence to either constitutive or tissue-specific RNAs. Constitutive AT gene transcription generates RNAs that must be rapidly turned over and therefore do not serve as precursors to cytoplasmic mRNA. Interestingly, detailed kinetic studies of nuclear RNA metabolism have led others to postulate that a substantial fraction of the nuclear RNA pool may not give rise to mRNA (18, 19).
In the least, our results indicate that care should be exercised when using isolated nuclei in the in vitro analysis of transcription rates. The potential masking of functional RNA synthesis by aberrant transcription may be considerable and could lead to erroneous conclusions.
Acknowledgments
We thank Drs. Aubrey Thompson and Michael Felder for advice and discussion during the course of this work. We also thank Debra Williams for secretarial assistance.
Footnotes
This work was supported by Grant DK33886 from the National Institutes of Diabetes, Digestive, and Kidney Diseases.
The abbreviations used are: AT, α1-antitrypsin; HEPES, 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid.
H. Lieberman, personal communication (to E. A. Thompson (8)).
C. Rheaume, unpublished experiments.
References
- 1.Berger FG, Baumann H. J Biol Chem. 1985;260:1160–1165. [PubMed] [Google Scholar]
- 2.Latimer JJ, Berger FG, Baumann H. J Biol Chem. 1987;262:12641–12646. [PMC free article] [PubMed] [Google Scholar]
- 3.Barth RK, Gross KW, Gremke LC, Hastie ND. Proc Natl Acad Sci U S A. 1982;79:500–504. doi: 10.1073/pnas.79.2.500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Evans RM, Fraser N, Ziff E, Weber J, Wilson M, Darnell JE. Cell. 1977;12:733–739. doi: 10.1016/0092-8674(77)90273-2. [DOI] [PubMed] [Google Scholar]
- 5.Derman E, Krauter K, Walling L, Weinberger C, Ray M, Darnell JE. Cell. 1981;23:731–739. doi: 10.1016/0092-8674(81)90436-0. [DOI] [PubMed] [Google Scholar]
- 6.Lamers WH, Hanson RW, Meisner HM. Proc Natl Acad Sci U S A. 1982;79:5137–5141. doi: 10.1073/pnas.79.17.5137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Berger FG, Gross KW, Watson G. J Biol Chem. 1981;256:7006–7013. [PubMed] [Google Scholar]
- 8.Mahajan PB, Thompson EA., Jr J Biol Chem. 1987;262:16150–16156. [PubMed] [Google Scholar]
- 9.Snider LD, King D, Lingrel JB. J Biol Chem. 1985;260:9884–9893. [PubMed] [Google Scholar]
- 10.Tseng-Crank J, Berger FG. Genetics. 1987;113:593–599. doi: 10.1093/genetics/116.4.593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hawley DK, Roeder RG. J Biol Chem. 1985;260:8163–8172. [PubMed] [Google Scholar]
- 12.Ciliberto G, Dente L, Cortese R. Cell. 1985;41:531–540. doi: 10.1016/s0092-8674(85)80026-x. [DOI] [PubMed] [Google Scholar]
- 13.DeSimone V, Ciliberto G, Hardon E, Paonessa G, Palla F, Lundberg L, Cortese R. EMBO J. 1987;6:2759–2766. doi: 10.1002/j.1460-2075.1987.tb02570.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grayson DR, Costa RH, Xanthopoulos KG, Darnell JE. Mol Cell Biol. 1988;8:1055–1066. doi: 10.1128/mcb.8.3.1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bentley DL, Groudine M. Nature. 1986;321:702–706. doi: 10.1038/321702a0. [DOI] [PubMed] [Google Scholar]
- 16.Nepveu A, Marcu KB. EMBO J. 1986;5:2859–2865. doi: 10.1002/j.1460-2075.1986.tb04580.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kindy MS, McCormack JE, Buckler AJ, Levine RA, Sonenshein GE. Mol Cell Biol. 1987;7:2857–2862. doi: 10.1128/mcb.7.8.2857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Harpold M, Evans RM, Saldit-Georgieff M, Darnell JE. Cell. 1979;17:1025–1035. doi: 10.1016/0092-8674(79)90341-6. [DOI] [PubMed] [Google Scholar]
- 19.Salditt-Georgieff M, Harpold MM, Wilson MC, Darnell JE. Mol Cell Biol. 1981;1:179–187. doi: 10.1128/mcb.1.2.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Krauter KS, Citron BA, Hsu MT, Powell O, Darnell JE. DNA (N Y) 1986;5:29–36. doi: 10.1089/dna.1986.5.29. [DOI] [PubMed] [Google Scholar]