SUMMARY
Environmental heterogeneity is thought to be an important process maintaining genetic variation in populations [1–4]: if alternative alleles are favored in different environments, a stable polymorphism can be maintained [1, 5, 6]. This situation has been hypothesized to occur in genes encoding multi-substrate enzymes [7], in which changes that increase activity with one substrate typically decrease activity with others [8–10], but examples of polymorphisms maintained by this mechanism are rare. Here we show that a polymorphism in an enzyme gene in Drosophila melanogaster is maintained by such a trade-off. The mitochondrially-localized aldehyde dehydrogenase in D. melanogaster has two important functions: detoxifying acetaldehyde derived from dietary ethanol [11], and detoxifying larger aldehydes produced as byproducts of oxidative phosphorylation [12]. A derived variant of the enzyme, Leu479Phe, is present in moderate frequencies in most temperate populations, but is rare in more ethanol-averse tropical populations. Using purified recombinant protein, we show that the Leu-Phe substitution increases turnover rate of acetaldehyde but decreases turnover rate of larger aldehydes. Further, using transgenic fly lines, we show that the substitution increases lifetime fitness on medium supplemented with an ecologically relevant ethanol concentration, but decreases fitness on medium lacking ethanol. The strong, opposing selection pressures, coupled with documented highly variable ethanol concentrations in breeding sites of temperate populations, implicate an essential role for environmental heterogeneity in maintaining the polymorphism.
RESULTS AND DISCUSSION
Aldehyde dehydrogenases are a family of enzymes that convert aldehydes to their corresponding acids. The product of Drosophila Aldh, DmALDH, like its human orthologue ALDH2 (with which it shares 70% sequence identity), is a mitochondrially-localized enzyme essential for detoxifying acetaldehyde derived from the oxidation of dietary ethanol [11] (the ethanol to acetaldehyde conversion is carried out by a different enzyme, the well-studied alcohol dehydrogenase). Because D. melanogaster breeds in fermenting fruits in which ethanol concentrations often exceed 4% [13–15], this function of DmALDH is likely to be important in natural populations. It is clear, however, that aldehyde dehydrogenases did not evolve to detoxify ethanol, because their origin long predates the appearance of flowering plants and fermentative yeasts [16, 17]. The major ancestral, and still important, role of DmALDH and its orthologues in vertebrates appears to be detoxification of harmful endogenous aldehydes generated by lipid peroxidation, a side-effect of aerobic respiration within mitochondria [12, 18, 19]. The most damaging of these aldehydes are larger than the two-carbon acetaldehyde, typically containing six or more carbons [20]. Because size is known to be a major determinant of ALDH substrate specificity [16, 21], the optimal ALDH for detoxifying acetaldehyde is not likely to be optimal for detoxifying lipid peroxidation products.
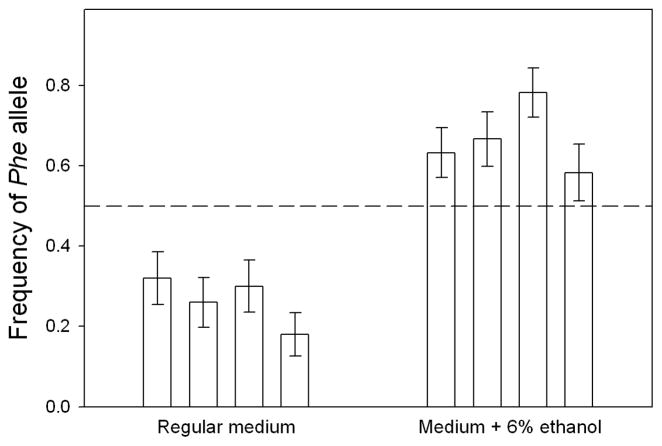
In previous work, we identified a replacement SNP in Aldh, Leu479Phe, in which the derived phenylalanine (henceforth, “Phe”) allele is present at modest, albeit variable, frequencies (5–30%) in most temperate populations, but absent or rare (<5%) in tropical populations [22] (see also Figure S1). Patterns of geographic variation in ethanol resistance give evidence that temperate populations experience stronger selection for ethanol resistance than tropical populations [23–27], possibly because they have higher preference for ethanol in feeding and oviposition [28–30]. Thus, the higher frequency of the Phe allele in temperate populations suggests that the allele may be beneficial in flies on high-ethanol diets, but disadvantageous in the absence of ethanol. To test this hypothesis, we created replicate transgenic lines (n = 3 per genotype) containing an insert of either a natural leucine (“Leu”) allele, or the same allele in which the leucine codon had been mutated to phenylalanine, at the same genomic location in an Aldh-null background (the inserts were on the third chromosome, while Aldh is on the second chromosome). Leu lines were crossed to Phe lines to establish populations with equal frequencies of the two alleles, which were maintained for nine generations on either normal medium or medium supplemented with 6% ethanol, a concentration within the range encountered by natural populations [13–15]. The frequency of the Phe allele decreased in each replicate population on normal medium, whereas it increased in each population on ethanol-supplemented medium (Figure 1). Assuming intermediate dominance, the allele frequency changes indicate that Leu homozygotes have 26% (95% confidence interval: 13–43%) higher fitness than Phe homozygotes in the absence of ethanol. In contrast, Phe homozygotes have 16% (2.8–36%) higher fitness than Leu homozygotes in the presence of ethanol. Although the point estimates of selection coefficients change somewhat if the favored allele is assumed to be partly dominant or partly recessive, the lower bounds of the confidence intervals are little affected (Table 1).
Figure 1. Change in frequency of the Phe allele in experimental populations reared in the absence or presence of ethanol.
Populations started with equal frequencies of Leu and Phe transgenic alleles (dashed line), and were genotyped after nine generations. Bars show ±1 binomial standard error. See also Figure S1.
Table 1. Selection coefficients on Aldh replacement polymorphism estimated from allele frequency changes in the experimental populations.
Representing the favored allele by A2, genotypes A1A1, A1A2, and A2A2 are assumed to have relative fitness values of 1, ehs (≈1+ hs), and es (≈ 1+ s), respectively. 95% confidence limits are given in parentheses.
| h = 0.1 | h = 0.5 | h = 0.9 | |
|---|---|---|---|
| s favoring Leu on regular medium | 0.19 (0.11, 0.28) | 0.23 (0.12, 0.36) | 0.30 (0.14, 0.59) |
| s favoring Phe on ethanol-supplemented medium | 0.13 (0.028, 0.25) | 0.15 (0.028, 0.31) | 0.18 (0.029, 0.47) |
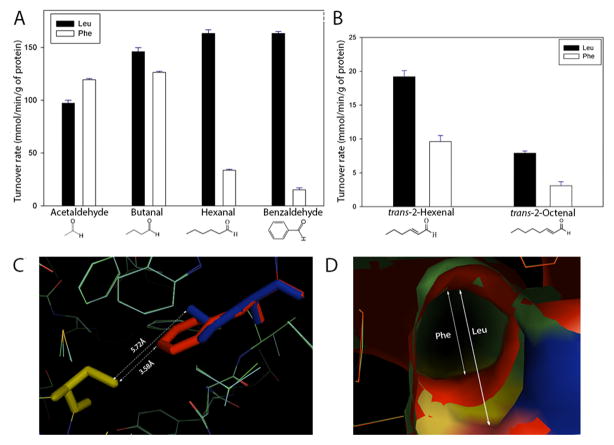
The Leu and Phe lines showed no difference in Aldh expression (Figure S2), suggesting that the fitness effects of the polymorphism result from differences in substrate specificity. To test this hypothesis, we overexpressed the variants in vitro, purified the recombinant enzymes, and measured turnover rates with saturating levels of a range of aldehydes (Figure 2A). The Phe variant detoxifies acetaldehyde significantly faster than the Leu variant, by about 20%, consistent with its advantage on ethanol-supplemented medium (similarly, extracts of Phe line flies showed 20% higher ALDH activity with acetaldehyde as a substrate than extracts of Leu line flies; Figure S3). With larger aldehydes, this difference is strikingly reversed (Figure 2A, 2B). Particularly notable is the 2–3 fold faster detoxification by the Leu variant than the Phe variant of trans-2-hexenal and trans-2-octenal (Figure 2B), two known toxic products of lipid peroxidation [31].
Figure 2. Kinetic and structural properties of purified ALDH variants.
(A) Turnover rates (± S.E.M.) with saturating levels of aldehydes of a range of molecular weights (1 mM substrate). The difference between Leu and Phe variants is statistically significant for each substrate (acetaldehyde, P < 0.01; butanal, P < 0.05; others, P < 0.001). (B) Turnover rates with two damaging aldehydes generated in mitochondria by lipid peroxidation (1 uM substrate). The difference between Leu and Phe variants is statistically significant for each (P < 0.002). (C) Superposition of Phe479 (red) on Leu479 (blue) in model structure of D. melanogaster ALDH. The catalytic residue, Cys322, is colored in yellow. Distances between Cys322 and Phe479 or Leu479 sidechains are shown to demonstrate the relative occupancy of the two sidechains within the substrate entry channel (SEC). (D) The Leu479Phe substitution constricts the SEC proximal to the substrate binding site, reducing the volume of the channel from 535Å3 to 456Å3. The inner ring (red) represents the SEC diameter in ALDHPhe479 and the outer ring (blue and green) represents the SEC diameter in ALDHLeu479. See also Figures S2 and S3.
To gain insight into the molecular basis of the differences in substrate specificity, we used protein structure modeling software [32] to investigate the effect of the Leu-Phe substitution on the diameter of the substrate entry channel (SEC), the intramolecular tunnel that guides the substrate to the active site. A smaller SEC could facilitate binding of acetaldehyde to the active site, while hindering access of larger aldehydes [16]. Indeed, compared to the ancestral leucine at position 479, the bulky phenylalanine group protrudes into the SEC (Figure 2C, D), reducing its volume by an estimated 15%. Although for technical reasons [33] we did not attempt to estimate Km, constriction of the SEC would be expected to have a larger effect on Km than on our measure of enzyme activity [34], which is proportional to kcat. Hence, differences between the Leu and Phe variants in kcat/Km, which is arguably a more relevant measure of catalytic efficiency than kcat alone, could be larger than reflected in our estimates.
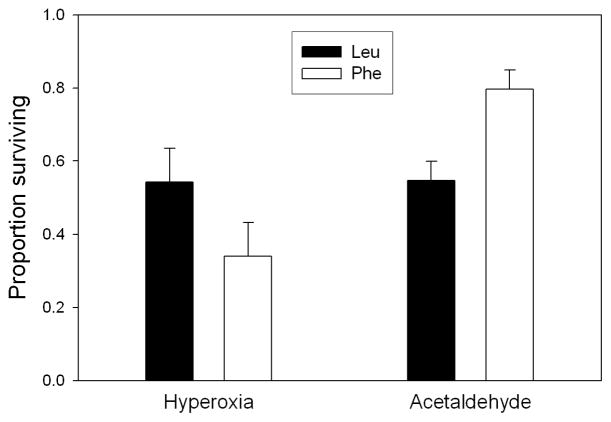
To test whether the Leu variant is more effective than the Phe variant at detoxifying lipid peroxidation products in vivo, we compared the transgenic lines in resistance to hyperoxia, a manipulation that increases ROS production, lipid peroxidation, and resulting damage to mitochondrial proteins due to formation of adducts with aldehydes [12, 20, 35]. The Leu lines survived significantly better than the Phe lines under this challenge (Figure 3; P = 0.048 one-tailed). Because lipid peroxidation products are continuously produced even under normal conditions, this result gives evidence that the fitness disadvantage of the Phe variant on normal medium is at least partly the result of its lower ability to detoxify these aldehydes. Conversely, when challenged with acetaldehyde vapor, the Phe lines had significantly higher survival than the Leu lines (Figure 3; P = 0.017 one-tailed). Apparently, on ethanol-supplemented medium, this advantage of the Phe variant more than compensates for its disadvantage in detoxifying lipid peroxidation products.
Figure 3. Survival under hyperoxia and acetaldehyde stress.
Leu lines have significantly higher survival than Phe lines (P < 0.05) under hyperoxia, but significantly lower survival (P < 0.02) in the presence of acetaldehyde vapor. P-values are one-tailed.
Studies of D. melanogaster in temperate regions show that ethanol is an important, but highly variable, constituent of the species’ breeding sites [13–15]. In naturally fermenting grapes and pears containing D. melanogaster larvae, ethanol concentrations averaged 2%, but ranged up to 8%, more than high enough to result in selection for the Phe allele. (For comparison, ethanol concentrations in the vials in our experimental populations, although initially 6%, were likely to have declined to ~5% by the time eggs hatched, with further declines throughout larval development [36]). In contrast, in other fruits used as breeding sites by D. melanogaster, such as melons and tomatoes, mean ethanol concentrations were only 0.1%, likely resulting in the Leu allele being favored.
In theoretical models, whether environmental heterogeneity can maintain a polymorphism depends on the values of several parameters, including the carrying capacity of the different habitats, the relative fitness values of the genotypes in each habitat, and the rate of migration between habitats [6]. Because these parameters would be difficult or impossible to estimate in the wild for D. melanogaster, we cannot formally show that the conditions for the maintenance of polymorphism by environmental heterogeneity are met. (As proof of principle, however, we can show that it is possible for the fitness estimates in Table 1 with h = 0.9 to result in a stable polymorphism; see Supplemental Experimental Procedures). Nonetheless, the following considerations suggest that the Phe-Leu polymorphism is maintained by selection, and that variation in ethanol levels among breeding sites plays an important role in its maintenance. First, although the polymorphism is not ancient, neither is it extremely young: using the length of ancestral haplotypes on chromosomes carrying the Phe allele [37], we estimate its age as 19,390 (95% C.I.: 9,530–40,100) generations. Given this amount of time and the strong selection pressures acting on the polymorphism (Table 1), it is likely that the Phe allele would either have been fixed or eliminated unless a mechanism existed to maintain the polymorphism. Second, the strong disadvantage of the Phe allele in the absence of ethanol indicates that high ethanol levels in a subset of breeding sites is critical for maintaining the allele in populations. Giving additional evidence that the unusually “ethanolphilic” lifestyle of temperate D. melanogaster populations [15, 28, 38] is critical to the maintenance of the Phe allele, among 21 Dipteran species for which genome sequences are available, ranging from other Drosophila species to distantly related mosquitoes, all have leucine at the homologous position. (Moreover, although only a single reference genome is available for most of these species, 270 D. simulans sampled from the wild were all homozygous for leucine; see Supplemental Experimental Procedures).
It is also possible that heterozygous advantage plays a role, in addition to environmental heterogeneity, in the maintenance of the Phe-Leu polymorphism. In particular, because lipid peroxidation products are generated irrespective of diet, having both the Leu and Phe alleles might result in the highest fitness in flies feeding on ethanol. More work will be needed to test this hypothesis. On the other hand, we see little reason to expect heterozygous advantage in flies not feeding on ethanol, in which a copy of the Phe allele would compromise ability to detoxify lipid peroxidation products, without providing a compensating benefit.
The Phe-Leu polymorphism illustrates an adaptive constraint: the optimal protein structure for one function is likely to differ from that for a different function [9, 39–41]. One way such a constraint can be overcome is by gene duplication [39, 41–43]. Indeed, the presence of a polymorphism maintained by balancing selection has been hypothesized to favor fixation of gene duplicates [44–46]. To determine whether Aldh duplicates might be segregating in natural populations, we examined genome sequences of 171 temperate and 110 African strains; none showed evidence for Aldh copy number variation. Given the age of the Phe allele, it is unclear why gene duplications have not occurred and risen to high frequencies. For comparison, in a variety of pest insect species, duplicates of genes involved in insecticide resistance have occurred and reached high frequencies in a matter of decades [47, 48].
Although the hypothesis that polymorphisms causing shifts in substrate specificity can be maintained by environmental heterogeneity was first proposed more than 40 years ago [7], there appear to be few documented examples of such polymorphisms. The clearest case of which we are aware occurs in the sheep blowfly, Lucilia cuprina (reviewed in [48]), in which a mutant carboxylesterase confers metabolic resistance to organophosphate insecticides, at the expense of both the native carboxylesterase activity and (for unknown reasons) overall fitness. In contrast to the Phe allele, however, the resistance allele has risen to high frequency, aided in part by duplication events that have combined resistance and susceptibility alleles on the same chromosome [47, 48]. Our work also differs in that the selective agent favoring the novel allele, ethanol, is more natural and less novel than insecticides.
Our results give direct experimental evidence that selection on a widespread single nucleotide polymorphism is strong, and switches direction depending on the environment. These results add to recent evidence from genome-wide surveys that loci under balancing selection, broadly defined to include spatially- and temporally-varying selection, may be more common than often supposed [49, 50]. For example, by analogy to our results, we might expect variation in plant secondary chemistry to be able to maintain polymorphisms in populations of phytophagous insects [51]. We suggest that experimental approaches such as ours, which allow the phenotypic and fitness effects of a polymorphism to be measured unconfounded by differences in genetic background, should be used to explore the possible role of balancing selection in maintaining other polymorphisms.
Supplementary Material
Acknowledgments
Supported by NIH grant R01AA016178 and NSF grant DEB-0623268 to J.D.F. We thank J. Zhu for technical assistance, and A. Long, M. Rausher, and three anonymous reviewers for helpful comments on the manuscript.
Footnotes
AUTHOR CONTRIBUTIONS
Conceptualization, J.D.F.; Methodology, M.C. and J.D.F.; Formal analysis, J.D.F. and M.C.; Investigation: M.C.; Writing – Original Draft, J.D.F., M.C.; Writing – Review and Editing, J.D.F.; Funding Acquisition, J.D.F.; Resources, J.D.F.; Supervision, J.D.F.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Levene H. Genetic equilibrium when more than one ecological niche is available. Am Nat. 1953;87:331–333. [Google Scholar]
- 2.Gillespie JH, Turelli M. Genotype-environment interactions and the maintenance of polygenic variation. Genetics. 1989;121:129–138. doi: 10.1093/genetics/121.1.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hedrick PW. Genetic polymorphism in heterogeneous environments: The age of genomics. Ann Rev Ecol Evol Syst. 2006;37:67–93. [Google Scholar]
- 4.Delph LF, Kelly JK. On the importance of balancing selection in plants. New Phytol. 2014;201:45–56. doi: 10.1111/nph.12441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dempster ER. Maintenance of genetic heterogeneity. Cold Spring Harbor Symp Quant Biol. 1955;20:25–32. doi: 10.1101/sqb.1955.020.01.005. [DOI] [PubMed] [Google Scholar]
- 6.Felsenstein J. The theoretical population genetics of variable selection and migration. Ann Rev Genet. 1976;10:253–280. doi: 10.1146/annurev.ge.10.120176.001345. [DOI] [PubMed] [Google Scholar]
- 7.Gillespie JH, Langley CH. A general model to account for enzyme variation in natural populations. Genetics. 1974;76:837–884. doi: 10.1093/genetics/76.4.837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tokuriki N, Jackson CJ, Afriat-Jurnou L, Wyganowski KT, Tang R, Tawfik DS. Diminishing returns and tradeoffs constrain the laboratory optimization of an enzyme. Nat Commun. 2012;3:1257. doi: 10.1038/ncomms2246. [DOI] [PubMed] [Google Scholar]
- 9.Miller SP, Lunzer M, Dean AM. Direct demonstration of an adaptive constraint. Science. 2006;314:458–461. doi: 10.1126/science.1133479. [DOI] [PubMed] [Google Scholar]
- 10.McLoughlin SY, Copley SD. A compromise required by gene sharing enables survival: Implications for evolution of new enzyme activities. Proc Natl Acad Sci USA. 2008;105:13497–13502. doi: 10.1073/pnas.0804804105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fry JD, Saweikis M. Aldehyde dehydrogenase is essential for both adult and larval ethanol resistance in Drosophila melanogaster. Genet Res. 2006;87:87–92. doi: 10.1017/S0016672306008032. [DOI] [PubMed] [Google Scholar]
- 12.Chakraborty M, Fry JD. Drosophila lacking a homologue of mammalian ALDH2 have multiple fitness defects. Chem-Biol Interact. 2011;191:296–302. doi: 10.1016/j.cbi.2011.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gibson JB, May TW, Wilks AV. Genetic variation at the alcohol dehydrogenase locus in Drosophila melanogaster in relation to environmental variation: ethanol levels in breeding sites and allozyme frequencies. Oecologia. 1981;51:191–198. doi: 10.1007/BF00540600. [DOI] [PubMed] [Google Scholar]
- 14.Oakeshott JG, May TW, Gibson JB, Willcocks DA. Resource partitioning in five domestic Drosophila species and its relationship to ethanol metabolism. Austral J Zool. 1982;30:547–556. [Google Scholar]
- 15.McKenzie JA, McKechnie SW. A comparative study of resource utilization in natural populations of Drosophila melanogaster and D. simulans. Oecologia. 1979;40:299–309. doi: 10.1007/BF00345326. [DOI] [PubMed] [Google Scholar]
- 16.Sobreira TJP, Marletaz F, Simoes-Costa M, Schechtman D, Pereira AC, Brunet F, Sweeney S, Pani A, Aronowicz J, Lowe CJ, et al. Structural shifts of aldehyde dehydrogenase enzymes were instrumental for the early evolution of retinoid-dependent axial patterning in metazoans. Proc Natl Acad Sci USA. 2011;108:226–231. doi: 10.1073/pnas.1011223108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ashburner M. Speculations on the subject of alcohol dehydrogenase and its properties in Drosophila and other flies. BioEssays. 1998;20:949–954. doi: 10.1002/(SICI)1521-1878(199811)20:11<949::AID-BIES10>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- 18.Ohta S, Ohsawa I, Kamino K, Ando F, Shimokata H. Mitochondrial ALDH2 deficiency as an oxidative stress. Ann New York Acad Sci. 2004;1011:36–44. doi: 10.1007/978-3-662-41088-2_4. [DOI] [PubMed] [Google Scholar]
- 19.Ebert AD, Kodo K, Liang P, Wu H, Huber BC, Riegler J, Churko J, Lee J, de Almeida P, Lan F, et al. Characterization of the molecular mechanisms underlying increased ischemic damage in the aldehyde dehydrogenase 2 genetic polymorphism using a human induced pluripotent stem cell model system. Sci Trans Med. 2014;6:255ra130. doi: 10.1126/scitranslmed.3009027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Esterbauer H, Schaur RJ, Zollner H. Chemistry and biochemistry of 4-hydroxynonenal, malonaldehyde and related aldehydes. Free Radical Biol Med. 1991;11:81–128. doi: 10.1016/0891-5849(91)90192-6. [DOI] [PubMed] [Google Scholar]
- 21.Sophos NA, Vasiliou V. Aldehyde dehydrogenase gene superfamily: the 2002 update. Chem-Biol Interact. 2003;143–144:5–22. doi: 10.1016/s0009-2797(02)00163-1. [DOI] [PubMed] [Google Scholar]
- 22.Fry JD, Donlon K, Saweikis M. A world-wide polymorphism in Aldehyde dehydrogenase in Drosophila melanogaster: evidence for selection mediated by dietary ethanol. Evolution. 2008;62:66–75. doi: 10.1111/j.1558-5646.2007.00288.x. [DOI] [PubMed] [Google Scholar]
- 23.David JR, Bocquet C. Similarities and differences in latitudinal adaptation of two Drosophila sibling species. Nature. 1975;257:588–590. doi: 10.1038/257588a0. [DOI] [PubMed] [Google Scholar]
- 24.Anderson D. Alcohol dehydrogenase activity and ethanol tolerance along the Adh cline in Australia. In: Lakovaara S, editor. Advances in Genetics, Development, and Evolution of Drosophila. New York: Plenum; 1982. pp. 263–272. [Google Scholar]
- 25.David JR, Merçot H, Capy P, McEvey SF, van Herrewege J. Alcohol tolerance and Adh gene frequencies in European and African populations of Drosophila melanogaster. Genet Sel Evol. 1986;18:405–416. doi: 10.1186/1297-9686-18-4-405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Parkash R, Karan D, Munjal AK. Geographical variation in AdhF and alcoholic resource utilization in Indian populations of Drosophila melanogaster. Biol J Linn Soc. 1999;66:205–214. [Google Scholar]
- 27.Montooth KL, Siebenthall KT, Clark AG. Membrane lipid physiology and toxin catabolism underlie ethanol and acetic acid tolerance in Drosophila melanogaster. J Exp Biol. 2006;209:3837–3850. doi: 10.1242/jeb.02448. [DOI] [PubMed] [Google Scholar]
- 28.Zhu J, Fry JD. Preference for ethanol in feeding and oviposition in temperate and tropical populations of Drosophila melanogaster. Ent Exp Appl. 2015;155:64–70. doi: 10.1111/eea.12285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Parsons PA. Adaptive strategies in natural populations of Drosophila. Theor Appl Genet. 1980;57:257–266. doi: 10.1007/BF00264952. [DOI] [PubMed] [Google Scholar]
- 30.Parsons PA. Larval reaction to alcohol as an indicator of resource utilization differences between Drosophila melanogaster and D. simulans. Oecologia. 1977;30:141–146. doi: 10.1007/BF00345417. [DOI] [PubMed] [Google Scholar]
- 31.Novotny MV, Yancey MF, Stuart R, Wiesler D, Peterson RG. Inhibition of glycolytic enzymes by endogenous aldehydes -- a possible relation to diabetic neuropathies. Biochim Biophys Acta. 1994;1226:145–150. doi: 10.1016/0925-4439(94)90022-1. [DOI] [PubMed] [Google Scholar]
- 32.Offman MN, Tournier AL, Bates PA. Alternating evolutionary pressure in a genetic algorithm facilitates protein model selection. BMC Struct Biol. 2008;8:34. doi: 10.1186/1472-6807-8-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Klyosov AA. Kinetics and specificity of human liver aldehyde dehydrogenases toward aliphatic, aromatic, and fused polycyclic aldehydes. Biochemistry. 1996;35:4457–4467. doi: 10.1021/bi9521102. [DOI] [PubMed] [Google Scholar]
- 34.Moore SA, Baker HM, Blythe TJ, Kitson KE, Kitson TM, Baker EN. Sheep liver cytosolic aldehyde dehydrogenase: the structure reveals the basis for the retinal specificity of class 1 aldehyde dehydrogenases. Structure. 1998;6:1541–1551. doi: 10.1016/s0969-2126(98)00152-x. [DOI] [PubMed] [Google Scholar]
- 35.Berlett BS, Stadtman ER. Protein oxidation in aging, disease, and oxidative stress. J Biol Chem. 1997;272:20313–20316. doi: 10.1074/jbc.272.33.20313. [DOI] [PubMed] [Google Scholar]
- 36.Hageman J, Eisses KT, Jacobs PJM, Scharloo W. Ethanol in Drosophila cultures as a selective factor. Evolution. 1990;44:447–454. doi: 10.1111/j.1558-5646.1990.tb05213.x. [DOI] [PubMed] [Google Scholar]
- 37.Gandolfo LC, Bahlo M, Speed TP. Dating rare mutations from small samples with dense marker data. Genetics. 2014;197:1315–1327. doi: 10.1534/genetics.114.164616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Merçot H, Defaye D, Capy P, Pla E, David JR. Alcohol tolerance, ADH activity, and ecological niche of Drosophila species. Evolution. 1994;48:746–757. doi: 10.1111/j.1558-5646.1994.tb01358.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.DePristo MA. The subtle benefits of being promiscuous: adaptive evolution potentiated by enzyme promiscuity. HFSP J. 2007;1:94–98. doi: 10.2976/1.2754665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.DePristo MA, Weinreich DM, Hartl DL. Missense meanderings in sequence space: a biophysical view of protein evolution. Nat Rev Genet. 2005;6:678–687. doi: 10.1038/nrg1672. [DOI] [PubMed] [Google Scholar]
- 41.Huang R, Hippauf F, Rohrbeck D, Haustein M, Wenke K, Feike J, Sorrelle N, Piechulla B, Barkman TJ. Enzyme functional evolution through improved catalysis of ancestrally nonpreferred substrates. Proc Natl Acad Sci USA. 2012;109:2966–2971. doi: 10.1073/pnas.1019605109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Des Marais DL, Rausher MD. Escape from adaptive conflict after duplication in an anthocyanin pathway gene. Nature. 2008;454:762–U785. doi: 10.1038/nature07092. [DOI] [PubMed] [Google Scholar]
- 43.Sikosek T, Chan HS, Bornberg-Bauer E. Escape from adaptive conflict follows from weak functional trade-offs and mutational robustness. Proc Natl Acad Sci USA. 2012;109:14888–14893. doi: 10.1073/pnas.1115620109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Spofford JB. Heterosis and the evolution of duplications. Am Nat. 1969;103:407–432. [Google Scholar]
- 45.Proulx SR, Phillips PC. Allelic divergence precedes and promotes gene duplication. Evolution. 2006;60:881–892. [PubMed] [Google Scholar]
- 46.Innan H, Kondrashov F. The evolution of gene duplications: classifying and distinguishing between models. Nat Rev Genet. 2010;11:97–108. doi: 10.1038/nrg2689. [DOI] [PubMed] [Google Scholar]
- 47.Newcomb RD, Gleeson DM, Yong CG, Russell RJ, Oakeshott JG. Multiple mutations and gene duplications conferring organophosphorus insecticide resistance have been selected at the Rop-1 locus of the sheep blowfly, Lucilia cuprina. J Mol Evol. 2005;60:207–220. doi: 10.1007/s00239-004-0104-x. [DOI] [PubMed] [Google Scholar]
- 48.Russell RJ, Scott C, Jackson CJ, Pandey R, Pandey G, Taylor MC, Coppin CW, Liu JW, Oakeshott JG. The evolution of new enzyme function: lessons from xenobiotic metabolizing bacteria versus insecticide-resistant insects. Evol Appl. 2011;4:225–248. doi: 10.1111/j.1752-4571.2010.00175.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bergland AO, Behrman EL, O’Brien KR, Schmidt PS, Petrov DA. Genomic evidence of rapid and stable adaptive oscillations over seasonal time scales in Drosophila. PLoS Genet. 2014;10:e1004775. doi: 10.1371/journal.pgen.1004775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Leffler EM, Gao ZY, Pfeifer S, Segurel L, Auton A, Venn O, Bowden R, Bontrop R, Wall JD, Sella G, et al. Multiple instances of ancient balancing selection shared between humans and chimpanzees. Science. 2013;339:1578–1582. doi: 10.1126/science.1234070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gloss AD, Dittrich ACN, Goldman-Huertas B, Whiteman NK. Maintenance of genetic diversity through plant-herbivore interactions. Curr Opin Plant Biol. 2013;16:443–450. doi: 10.1016/j.pbi.2013.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.