Abstract
Recurrent copy number variations (CNVs) are genetic alterations commonly observed in human tumors. One of the most frequent CNVs in human tumors involves copy number gains (CNGs) at chromosome 3q26, which is estimated to occur in >20% of human tumors. The high prevalence and frequent occurrence of 3q26 CNG suggest that it drives the biology of tumors harboring this genetic alteration. The chromosomal region subject to CNG (the 3q26 amplicon) spans from chromosome 3q26 to q29, a region containing ~200 protein-encoding genes. The large number of genes within the amplicon makes it difficult to identify relevant oncogenic target(s). Whereas a number of genes in this region have been linked to the transformed phenotype, recent studies indicate a high level of cooperativity among a subset of frequently amplified 3q26 genes. Here we use a novel bioinformatics approach to identify potential driver genes within the recurrent 3q26 amplicon in lung squamous cell carcinoma (LSCC). Our analysis reveals a set of 35 3q26 amplicon genes that are coordinately amplified and overexpressed in human LSCC tumors, and that also map to a major LSCC susceptibility locus identified on mouse chromosome 3 that is syntenic with human chromosome 3q26. Pathway analysis reveals that 21 of these genes exist within a single predicted network module. Four 3q26 genes, SOX2, ECT2, PRKCI and PI3KCA occupy the hub of this network module and serve as nodal genes around which the network is organized. Integration of available genetic, genomic, biochemical and functional data demonstrates that SOX2, ECT2, PRKCI and PIK3CA are cooperating oncogenes that function within an integrated cell signaling network that drives a highly aggressive, stem-like phenotype in LSCC tumors harboring 3q26 amplification. Based on the high level of genomic, genetic, biochemical and functional integration amongst these 4 3q26 nodal genes, we propose that they are the key oncogenic targets of the 3q26 amplicon and together define a “3q26 OncCassette” that mediates 3q26 CNG-driven tumorigenesis. Genomic analysis indicates that the 3q26 OncCassette also operates in other major tumor types that exhibit frequent 3q26 CNGs, including head and neck squamous cell carcinoma (HNSCC), ovarian serous cancer and cervical cancer. Finally, we discuss how the 3q26 OncCassette represents a tractable target for development of novel therapeutic intervention strategies that hold promise for improving treatment of 3q26-driven cancers.
Keywords: cancer, copy number gain (CNG), amplification, OncCassette, 3q26 amplicon, oncogenic drivers
Introduction
Cancer is a disease of progressive genetic alterations that conspire to drive a cellular phenotype characterized by uncontrolled growth, aggressive invasive behavior, enhanced survival, evasion of immune surveillance, and resistance to therapeutic interventions. Recent analysis of the genetic landscape of human cancers reveals that tumors can be classified into two main groups based upon the nature of the genetic alterations they harbor (Ciriello et al., 2013). In M-type tumors, somatic mutations that either activate oncogenes or inactivate tumor suppressor genes are the predominant genetic alterations. In contrast, in C-type tumors recurrent gene copy number variations (CNVs) are the predominant genetic alterations (Ciriello, Miller, 2013). Many of the key oncogenic driver mutations occurring in M-type tumors have been identified, molecularly characterized, functionally validated, and in some cases therapeutically targeted; prominent examples include EGFR, BRAF and PI3KCA mutations that are key drivers of oncogenic phenotypes (Samuels and Velculescu, 2004, Serra et al., 2008, Thomas et al., 2005, Tie and Desai, 2015). Likewise, prevalent recurring inactivating mutations in tumor suppressor genes, including those in TP53 (Hrstka et al., 2009), CDKN2A (p16) (Li et al., 2011), PTEN (Hollander et al., 2011) and APC (Polakis, 1997), have been well-documented and molecularly characterized. However, much less is known about whether and how tumor-specific CNVs drive C-type cancers. Comparative Genomic Hybridization (CGH) studies and more recent global genomic initiatives, such as The Cancer Genome Atlas (TCGA) project, have revealed that C-type tumors exhibit frequent and recurrent CNVs, both CNGs, or deletions, often encompassing large chromosomal regions (including, in some cases, entire chromosomal arms). As a consequence, these CNV events involve genetic loss or gain of large numbers of genes within the affected region. Recurrent CNVs involving loss of specific chromosomal regions are in some cases associated with loss of specific tumor suppressor genes; the most prevalent CNV in human tumors involves loss of chromosome 9p21 containing the prominent tumor suppressor target CDKN2A (reviewed in (Liggett and Sidransky, 1998)). However, for many recurrent CNVs involving chromosome loss, the relevant tumor suppressor(s) remain to be conclusively identified and functionally validated.
Recurrent CNGs suggest the presence of oncogenic driver(s) within these amplified regions. It is generally assumed that CNGs result in increased expression of one or more genes in the amplified region, thereby promoting or enhancing their oncogenic potential. Examples of CNGs driving oncogene activation through increased expression include amplification of CMYC at chromosome 8q24 (Little et al., 1983), cyclin D1 (CCND1) at chromosome 11q23 (Nishida et al., 1994), and EGFR at chromosome 7p11 (Wong et al., 1987). However, unlike the situation involving specific oncogenic somatic mutations, in which a distinct mutation specifically defines the molecular target of the genetic alteration, the relevant target(s) of many recurrent cancer-associated CNGs are not readily apparent in the context of tumor biology. Herein, we describe the genomic, genetic and functional characterization of the most common CNG event in human tumors, 3q26 amplification.
Materials and Methods
The prevalence of chromosome 3q26 CNGs across human tumor types was estimated in TCGA datasets using the cBioPortal for Cancer Genomics (http://www.cbioportal.org/). PRKCI and SOX2 were used as core 3q26 amplicon genes for this analysis. The cBioPortal analysis tool was used to interrogate GISTIC scores for these two genes in TCGA datasets for individual tumor types. A GISTIC score of +1 or +2 was considered to indicate the presence of significant CNG in that gene. Tumors exhibiting a GISTIC score of +1 or +2 at both loci were considered positive for CNG at 3q26. Prevalence was calculated as the number of tumors exhibiting CNG at these loci divided by all tumors in the dataset.
All RNA-encoding genes mapping within human chromosome 3q26-q29 (194 genes) were obtained and annotated using the Ensembl human gene database (GRCh38.p3; http://useast.ensembl.org/). Mouse homologs for these genes (164 direct mouse homolog genes) were identified and assigned chromosomal locations using the vertebrate homology tool of the Mouse Genome Informatics database (http://www.informatics.jax.org/). The subset of these genes mapping to the identified lung squamous cell carcinoma susceptibility locus on mouse chromosome 3 (Wang et al., 2004), which is syntenic with human chromosome 3q26, were identified (50 genes). The cBioPortal analysis tool was used to interrogate the TCGA lung squamous cell carcinoma dataset containing 178 primary tumors for expression and CNG (using GISTIC) of each of the 50 genes identified above that have direct mouse homologs mapping to mouse chromosome 3. Tumors were segregated into two groups; those exhibiting CNG at the locus for the gene being interrogated and those exhibiting no CNG at that locus. RNA expression for each gene was analyzed in these two groups and assessed for differential expression based on CNG of the gene using a two-sided t-test to assess significance. P-values of <0.05 were considered as statistically significant. 35 of the 50 genes were found to exhibit expression that was significantly higher in tumors harboring CNG at that gene than in those not exhibiting CNG. These 35 genes, whose expression correlate positively with CNG in LSCC tumors, were interrogated using the gene set analysis tool at the ConsensusPathDB maintained by the Max Planck Institute for Molecular Genetics (http://consensuspathdb.org/). Analysis revealed that this set of genes form a single induced network module containing 21 of the 35 genes. No other induced network modules were identified.
Results and Discussion
Copy number gain at 3q26: the most common CNG in human tumors
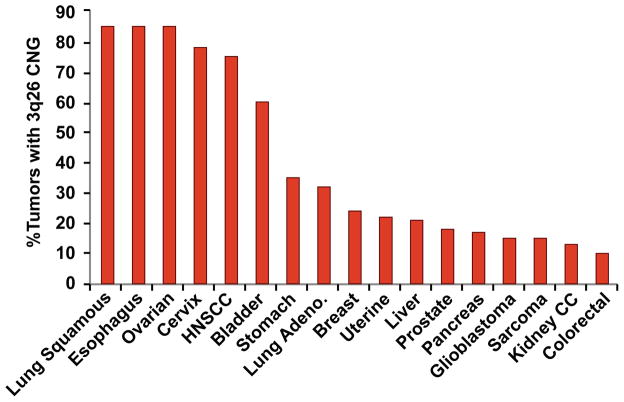
CNGs involving the long arm of chromosome 3 (3q) were first reported in human cancers nearly 20 years ago in HNSCC (Speicher et al., 1995). Subsequent CGH and deep sequencing analyses identified 3q as one of the most common CNG events in human tumors, being observed in >16% of solid tumors arising from 27 different tissue types (Ciriello, Miller, 2013, Rooney et al., 1999). Independent CGH analyses revealed prevalent 3q26 CNVs in cervical (77–90%) (Heselmeyer et al., 1997, Heselmeyer et al., 1996), esophageal (50–69%) (Bandla et al., 2012, Pack et al., 1999, Sakai et al., 2010), HNSCC (50–91%) (Bockmuhl et al., 1998, Brzoska et al., 1995, Hashimoto et al., 2001, Speicher, Howe, 1995), LSCC (77–85%) (Balsara et al., 1997, Bjorkqvist et al., 1998, Brass et al., 1996, Massion et al., 2002, Petersen et al., 1997), ovarian serous (36–51%) (Arnold et al., 1996, Iwabuchi et al., 1995, Sonoda et al., 1997) and endometrial serous cancers (50%)(Pere et al., 1998). 3q26 CNGs were also observed in 76% of small cell lung cancers (SCLC), though the estimate of prevalence in SCLC must be considered tentative given the extremely small sample size of these studies (Ried et al., 1994, Sugita et al., 2000). We therefore assessed the prevalence of 3q26 CNVs across major human tumor types using TCGA datasets available through the cBioPortal (http://www.cbioportal.org/index.do) (Figure 1). To estimate 3q26 CNGs, we interrogated the TCGA datasets for CNGs in two genes within the core of the consensus 3q26 amplicon (PRKCI and SOX2) using the GISTIC algorithm. These genes were chosen to represent 3q26 CNGs since they: 1) exhibit virtually identical GISTIC scores across the examined tumor types indicating near perfect co-occurrence of CNGs at these genes; and 2) routinely exhibit the highest CNG frequency of any 3q26 genes in the affected tumor types, indicating that these genes are essentially invariably part of the recurrent 3q26 amplicon. Analysis revealed extremely prevalent chromosome 3q26 CNGs in LSCC (85%), esophageal (85%), ovarian serous (85%), cervical (78%), HNSCC (75%) and bladder (60%) cancers. 3q26 CNGs were of intermediate prevalence (20–35%) in stomach, lung adenocarcinoma, breast, uterine and liver cancers, and of lower, but significant, prevalence (10–18%) in prostate, pancreatic, sarcoma, glioblastoma, kidney and colorectal cancers (Figure 1). Our analysis confirms earlier CGH studies demonstrating very high frequency 3q26 CNGs in many tumors types (i.e. LSCC, esophageal, HNSCC, ovarian and cervical cancers) (Chujo et al., 2002, Sakai, Kajiyama, 2010, Sonoda, Palazzo, 1997, Speicher, Howe, 1995, Sugita, Tanaka, 2000) and also revealed somewhat less prevalent, but significant CNGs in tumor types not previously identified as harboring this alteration. These data provide a comprehensive snapshot of the sweeping prevalence and distribution of 3q26 CNGs across major human cancer types.
Figure 1. Prevalence and distribution of 3q26 CNGs across human tumor types.
The prevalence of 3q26 CNGs were assessed by interrogation of TCGA datasets for major tumor types using the cBioPortal tools as described in Materials and Methods.
3q26 amplification is an oncogenic driver of tumorigenesis
A major question with regard to the involvement of CNGs in human cancer is whether these genetic alterations represent oncogenic “drivers” of tumorigenesis. Several lines of experimental evidence indicate that 3q26 CNG is an oncogenic driver in affected tumors. First, 3q26 CNGs are observed in precancerous lesions, demonstrating that this is an early event in tumorigenesis. Using fluorescence bronchoscopy, sequential biopsies of a carcinoma in situ (CIS) lesion, and a LSCC identified at the site of the CIS lesion approximately 1.5 years after initial identification were analyzed (Foster et al., 2005). Both the CIS and the subsequent LSCC lesion harbored specific 3q26 CNG, indicating occurrence of this event prior to progression to malignant LSCC (Foster, Banerjee, 2005). Similarly, 3q26 CNGs were found to be present in both early stage pre-invasive lung squamous cell lesions and in invasive LSCCs, albeit at a higher frequency in the more advanced lesions (Pelosi et al., 2007). These results demonstrate that 3q26 CNGs occur early during transformation, in some cases prior to the acquisition of a frank malignant phenotype, and that CNGs are enriched in more malignant lesions. Since 3q26 CNGs are detected in pre-invasive lesions that progress to LSCC, 3q26 CNG, and/or increased expression of 3q26 target genes, has been proposed as a biomarker of early pre-cancerous lesions with a high potential for progression to invasive LSCC. Similar findings of 3q26 CNGs in early malignant or preneoplastic lesions have been observed in cervical cancer (Heselmeyer, Schrock, 1996) and HNSCC (Singh et al., 2002). CGH analysis of normal cervical epithelium, dysplasias (subcategorized as mild, moderate and severe), and stage I invasive carcinomas, revealed that 3q26 CNGs are the most consistent chromosomal aberration occurring in dysplastic cells that progress to invasive cervical carcinoma (Heselmeyer, Schrock, 1996). Likewise, a study of HNSCC found 3q26 CNGs in 3% of normal mucosa, 25% of premalignant tissue, and 56% of invasive HNSCCs (Singh, Stoffel, 2002), while a second report found frequent 3q CNGs in low grade HNSCCs (Bockmuhl et al., 1996). The finding that 3q26 CNGs occur in pre-neoplastic lesions, and at higher frequency in subsequent malignant lesions, suggests that 3q26 CNGs play an important functional role in the transition from the pre-malignant to malignant state, at least in LSCC, HNSCC and cervical cancer, three tumor types harboring frequent 3q26 CNGs.
The second line of evidence that 3q26 CNG is an oncogenic driver is the strong association of this alteration with poor clinical outcome. In primary HNSCC tumors, 3q26 CNG is associated with significantly higher rates of tumor recurrence and cancer-related death (Singh, Stoffel, 2002), and with shorter disease-specific survival (Bockmuhl et al., 2000) when compared to tumors not harboring 3q26 CNG. Consistent with this observation, 3q26 CNG is more frequently observed in high grade ovarian serous tumors when compared to low grade tumors, suggesting an association with disease progression and acquisition of an aggressive phenotype in ovarian cancer (Balsara, Sonoda, 1997). Similarly, 3q26 CNGs were found at significantly higher frequencies in metastatic esophageal squamous cell carcinomas when compared to those tumors that did not metastasize (Sakai, Kajiyama, 2010). These findings support a role for 3q26 CNG in tumor aggressiveness, progression and/or clinical outcome in essentially all of the major tumor types that frequently harbor this alteration.
The third line of evidence that 3q26 CNG is an oncogenic driver comes from a genetic analysis of strain-specific differences in susceptibility to tobacco smoke carcinogen-induced LSCC in the mouse (Wang, Zhang, 2004). In this study, an extensive genomic analysis was conducted to identify strain-specific genomic regions associated with increased susceptibility to the tobacco smoke carcinogen N-nitroso-tris-chloroethylurea (NNTC)-induced LSCC. Interestingly, 3 strain-specific LSCC susceptibility loci were identified that are significantly associated with LSCC tumor development (Wang, Zhang, 2004). One of these loci localized to a region of mouse chromosome 3 that is syntenic with human chromosome 3q26; in contrast, the other two susceptibility loci do not map to regions of frequent CNG in human tumors. These data provide strong genetic evidence that 3q26-specific CNGs are involved in LSCC initiation in both mice and humans.
A majority of 3q26 target genes map to a single predicted network module
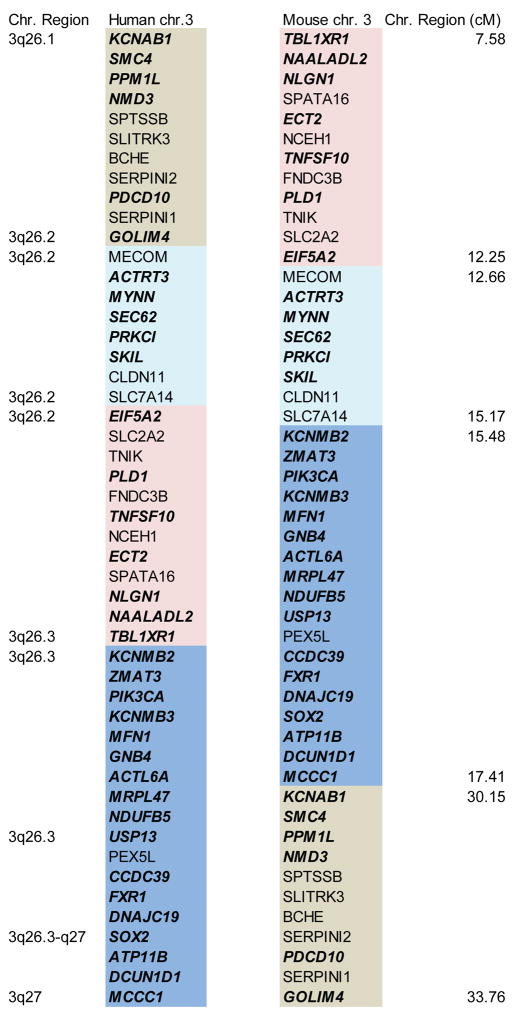
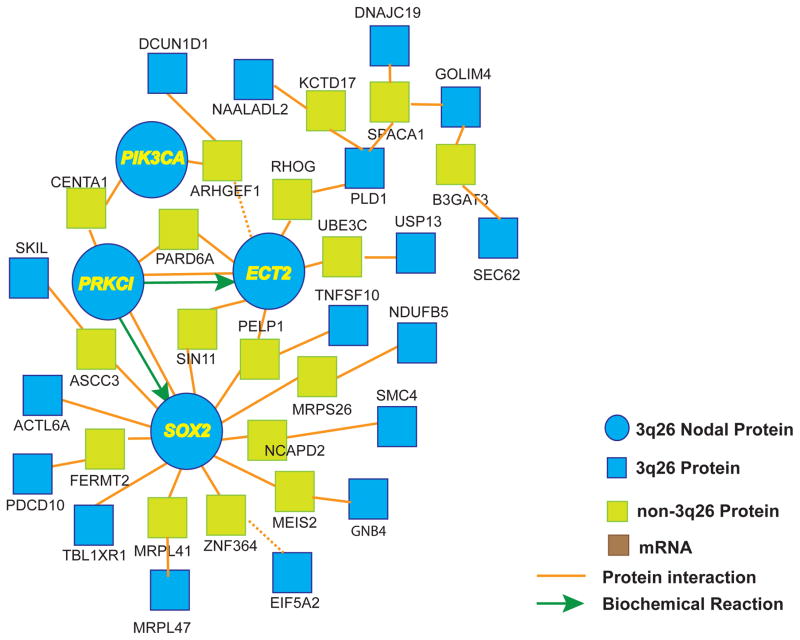
CGH analyses have demonstrated that tumors with 3q CNGs arising at different anatomic sites share an overlapping area of chromosomal gain centered around 3q26 (Brass, Ukena, 1996, Heselmeyer, Macville, 1997, Singh, Stoffel, 2002, Speicher, Howe, 1995). Depending on the study, the amplified region was found to extend from 3q26 to as far as the telomere at 3q29 (Wang et al., 2013a), a region encompassing 194 protein-encoding genes according to the Ensembl human gene database (Ensembl Genes-Build 37; http://www.ensembl.org) (Flicek et al., 2014). In order to identify potential driver genes within this large affected region, we first identified genes within this region that also map to the syntenic mouse chromosome 3 region previously identified as a major susceptibility locus for carcinogen-induced LSCC in the mouse (Wang, Zhang, 2004). Interrogation of the Mouse Genome Informatics database revealed that 146 of the 194 human genes mapping to human chromosome 3q26-29 have direct mouse homologs. Of these genes, 50 map to mouse chromosome 3. Interestingly, these genes are restricted to human chromosome 3q26.1-3q27, and span remarkably similar chromosomal expanses in human and mouse chromosome 3 (54.1 Mb in human; 55.9 Mb in mouse) (Figure 2). Interestingly, this region appears to have undergone several rearrangement/inversion events through evolution but the gene cluster remains intact (Figure 2). These data are consistent with several reports mapping the core of the 3q26 amplicon to chromosome 3q26.1-3q27 (Hagerstrand et al., 2013, Hussenet et al., 2010, Sugita, Tanaka, 2000). We reasoned that since these 50 genes are within the recurrent human 3q26 amplicon and map to a major LSCC susceptibility locus in the mouse, they are likely to be enriched in relevant targets of the 3q26 amplicon that drive LSCC development and maintenance. We further reasoned that the most critical target genes within this group would exhibit expression that is driven by 3q26 CNG in human tumors. Thus, to further focus our analysis, we interrogated the TCGA gene expression dataset consisting of 178 human LSCC tumor samples for expression of these 50 candidate 3q26 driver genes. For each gene, the LSCC tumor samples were divided into two sets; those harboring CNG for that gene as determined using the GISTIC algorithm (CNG defined as a GISTIC score of +1 or +2), and those not harboring CNG (GISTIC score of 0 or −1). We then determined whether the expression of that gene was significantly different between the groups (p≤0.05 by T-test). This analysis yielded 35 genes that satisfied the following criteria: 1) resides within the recurrent 3q26 amplicon; 2) maps within the centenic LSCC susceptibility locus on mouse chromosome 3; and 3) exhibits expression that is significantly associated with CNGs of the gene in human LSCC tumors (Figure 2; genes indicated in bold italics). Remarkably, geneset analysis using the ConsensuPathDB-human (CPDB; http://cpdb.molgen.mpg.de/CPDB) tool revealed a single induced network module containing 21 of these 35 genes (Figure 3). The network module has at its core major hubs centered on SOX2, PRKCI, ECT2 and PIK3CA. These data suggest that the 3q26 amplicon serves to drive expression of a set of genes whose function is integrated around these nodal genes.
Figure 2. Identification of core 3q26 amplicon genes.
50 genes corresponding to the core 3q26 amplicon in human tumors that also map to mouse chromosome 3 LSCC susceptibility locus are shown (see text for details). The color coded regions indicate sub-sections of the syntenic human and mouse chromosomes indicating evolutionary rearrangements and inversions in this region. Genes indicated in bold, italics correspond to genes within this syntenic region whose expression correlates with the presence of CNGs at that locus in human LSCC tumors. These genes were subjected to pathway analysis as described in the text.
Figure 3. A proposed 3q26 network module.
The core 3q26 amplicon genes identified in Figure 2 were subjected to gene set analysis using the ConsensuPathDB-human (CPDB; http://cpdb.molgen.mpg.de/CPDB) tool. 21 of the 35 core 3q26 genes reside within a single induced network module consisting of four nodal hub genes, SOX2, ECT2, PRKCI and PIK3CA, around which the module is organized.
The core nodal 3q26 genes SOX2, PRKCI, ECT2 and PIK3CA are functionally involved in 3q26-driven cancers
The genomic analysis above indicates that 3q26 CNG drives tumorigenesis by coordinately overexpressing a set of core genes that functionally interact within a signaling network. A survey of the primary literature regarding the involvement of the 21 network genes identified above indicate that, for at least for some of these genes, there is good evidence for involvement in 3q26-driven cancers. These data are summarized in Table 1. Perhaps not surprisingly, of the genes within this group, the most convincing evidence for a functional role in 3q26-driven cancers applies to the four nodal genes SOX2, PRKCI, ECT2 and PIK3CA occupying the core of the identified network. Below, we will focus our discussion on the published literature regarding the role of each of these nodal genes in 3q26-driven cancers.
Table 1.
3q26 Gene Network
| Gene | Product and function | Role in cancer | Impact of overexpression/amplification | References |
|---|---|---|---|---|
| ACTL6A | Actin-Like 6A, (aka: BAF53, ARP4) is an actin binding protein that plays a role in ATP-dependent chromatin remodeling. | ACTL6A binds the SWI/SNF complex in progenitor cells to prevent expression of differentiation genes. ACTL6A has a potential role in cancer stem cell function. |
None described. | (Bao et al., 2013) |
| DCUN1D1 | Defective in cullin neddylation 1, domain-containing 1 (aka: Squamous cell carcinoma-related oncogene, SCRO) regulates neddylation of E3 complex and regulates complex assembly and activation. | DCUN1D1 is required for survival of 3q amplified cancer cell lines. DCUN1D1 binds and activates GLI1 promoter in SCC cells. |
DCUN1D1 is overexpressed in tumors with 3q copy number gains. DCUN1D1 amplification and expression correlates with poor clinical outcome in SCC. DCUN1D1 is predicted to be a driver mutation in 3q26-29 amplified LSCC. |
(Kim et al., 2008) (Sarkaria et al., 2006) (Wang, Qian, 2013a) |
| DNAJC19 | DNAJ (Hsp40) Homolog, Subfamily C, Member 19-(aka: Mitochondrial Import Inner Membrane Translocase Subunit TIM14) is a member of the co-chaperone system that imports proteins to the inner mitochondrial membrane. Mutations in this gene are associated with cardiomyopathy. | DNAJC19 mRNA expression is reduced in breast cancer ALDH1+ cancer stem cells. DNAJC19 mRNA expression is elevated in ovarian tumor compared to normal ovarian epithelium. |
RNAi-mediated knockdown (KD) of DNAJC19 in 3q26 amplified ESCC and LSCC cell lines has no effect on anchorage-independent growth. | (Charafe-Jauffret et al., 2009) (Chen et al., 2012b) (Bass, Watanabe, 2009) |
| ECT2 | Epithelial cell transforming 2 is a guanine nucleotide exchange factor for the Rho family of GTPases. | ECT2 is highly expressed in many tumor types, and required for lung cancer and glioma cell growth. | ECT2 expression is associated with poor prognosis in HNSCC, glioblastoma and ESCC. | (Hirata, Yamabuki, 2009) (Salhia, Tran, 2008) (Sano, Genkai, 2006) (Justilien and Fields, 2009) (Fields and Justilien, 2010) |
| EIF5A2 | Eukaryotic translation initiation factor 5A2 binds mRNA and functions in the initiation and elongation processes of protein synthesis. | EIF5A2 KD in ESCC cell lines increases sensitivity to chemotherapy. EIF5A2 KD in gastric cancer cells suppresses growth, migration and invasion. |
EIF5A2 expression predicts poor prognosis in NSCLC, ovarian, gastric, liver and esophageal cancers. | (Mathews and Hershey, 2015) (Yang et al., 2015) (Meng et al., 2015) |
| GNB4 | Guanine Nucleotide Binding Protein (G Protein), Beta Polypeptide 4 encodes a heterotrimeric G-protein beta subunit. Functions downstream of G-protein coupled receptors (GPCR) and mutation in this gene impacts GPCR signaling in some inherited neuropathies. |
High expression of GNB4 predicts survival in tamoxifen-treated recurrent breast cancer patients. Intronic haplotypes of GNB4 predict survival of colorectal and bladder cancer patients. |
None described. | (Soong et al., 2013) (Umar et al., 2009) (Riemann et al., 2009) (Riemann et al., 2008) |
| GOLIM4 | Golgi integral membrane protein 4, (aka: Golgi phosphoprotein of 130 kDa, GPP130) is a member of the Golgi complex that sorts and modifies proteins exported from the endosome to the Golgi. GOLIM4 cycles between endosome and Golgi in a pH-dependent manner. |
None described. | None described. | |
| MRPL47 | Mitochondrial Ribosomal Protein L47 is a mitochondrial ribosomal protein member of the large 39S subunit. | None described. | None described. | |
| NDUFB5 | NADH Dehydrogenase (Ubiquinone) 1 Beta sub-complex 5 is an oxidative phosphorylation member of mitochondrial complex 1. NDUFB5 transfers electrons from NADH to ubiquinone. | NDUFB5 expression is decreased in ErbB2/Neu-initiated mammary tumor cells. | None described. | Klimcakova E, Cancer Res 2012 |
| NAALADL2 | N-acetylated alpha-linked acidic dipeptidase-like 2 is a member of the glutamate carboxypeptidase II family. | NAALADL2 was identified as a novel loci associated with prostate cancer aggressiveness. NAALADL2 expression predicts poor survival after prostatectomy. NAALADL2 positively regulates migration, invasion and anchorage-independent growth in a prostate cancer cell line. |
None described. | (Berndt et al., 2015) (Whitaker et al., 2014) |
| PDCD10 | Programmed cell death 10 (aka: cerebral cavernous malformation 3; CCM3) is associated with apoptosis; functions in vascular stability. | PDCD10 is upregulated in prostate cancer. PDCD10 is proposed to function in the PI3K pathway via interaction with PtdIns(3,4,5)P3. |
None described. | (Hilder et al., 2007) (You et al., 2013) (Zhang et al., 2014) |
| PIK3CA | Phosphatidy-linositol-4,5-Bisphosphate 3-Kinase, Catalytic Subunit Alpha encodes the catalytic subunit of class 1 phosphatidyl-inositol 3-kinase (PI3K); phosphorylates PtdIns4P and PtdIns (4,5)P2. | PI3K signaling is elevated in cancers with mutant and amplified PIK3CA. PI3K is activated by oncogenic K-ras or receptor tyrosine kinases in many cancers to promote proliferative signaling, cell motility, invasion and protein synthesis. |
PIK3CA copy number gains correlate with increased expression in 3q-amplified tumors (endometrial, gastric, LSCC, ovarian). PIK3CA mutation or amplification predicts cervical cancer response to cisplatin. PIK3CA copy number predicts poor survival in HNSCC and gastric cancer. |
(Salvesen, Carter, 2009) (Byun, Cho, 2003) (Yamamoto, Shigematsu, 2008) (Wang et al., 2015a) (Suda et al., 2012) (Shi et al., 2012) (Shayesteh, Lu, 1999) |
| PLD1 | Phospholipase D1 is a phosphatidylcholine (PC)-specific phospholipase that catalyzes hydrolysis of phosphatidylcholine to form phosphatidic acid and choline. | PLD1 functions in a positive feedback loop to promote Wnt/beta-catenin signaling in cancer cells. PLD1 inhibition promotes autophagic flux and sensitizes cancer to inhibition of autophagy. PLD1 in microenvironment promotes tumor metastasis and tumor vascularization. |
None described. | (Kang et al., 2010) (Dall’Armi et al., 2010) (Chen et al., 2012a) |
| PRKCI | Protein kinase C, iota (PKCι) is a serine/threonine protein kinase. | PKCι regulates cancer cell proliferation and transformed phenotype in lung, pancreatic, ovarian cancers and glioma. PKCι is required for cancer stem cell phenotype in ovarian and LSCC. |
PRKCI amplification is associated with elevated expression and poor prognosis in LSCC, HNSCC and ESCC, as well as other 3q amplified tumors. | (Regala et al., 2005a) (Regala, Weems, 2005b) (Scotti, Bamlet, 2010) (Justilien, Walsh, 2014) (Desai et al., 2011) (Wang, Hill, 2013b) (Murray, Kalari, 2011) |
| SEC62 | SEC62 Homolog (aka: Translocation protein 1, TLOC1) regulates protein translation and processing in the endoplasmic reticulum (ER) and transport across the ER membrane. | SEC62 is a prognostic marker in NSCLC. SEC62 mediates resistance to ER stress in prostate cancer and NSCLC cell lines. |
SEC62 KD reduces transformed growth of 3q26 amplified cancer cell lines. High SEC62 mRNA and protein expression correlates with 3q26 copy number gains. Elevated expression of SEC62 correlates with lymph node metastasis and poor differentiation in LSCC. |
(Hagerstrand, Tong, 2013) (Linxweiler et al., 2012) (Linxweiler et al., 2013) (Greiner et al., 2011) (Jung et al., 2006) |
| SKIL | SKI-like proto-oncogene (aka: SNO) is a negative regulator of TGFβ-Smad signaling that also stabilizes p53 and induces senescence. | Overexpression of SKIL may contribute to tumor cell resistance to the growth-inhibitory effects of TGFβ. SKIL promotes invasion via up-regulation of SLUG, and induces xenograft tumor growth. |
SKIL expression correlates with copy number in ESSC cell lines. SKIL expression correlates with poor prognosis in ESCC. |
(Imoto, Pimkhaokham, 2001) (Hagerstrand, Tong, 2013) (Akagi et al., 2008) |
| SMC4 | Structural maintenance of chromosomes 4 is an ATPase involved in chromosome condensation in early mitosis. SMC4 regulates chromosomal segregation and assembly. | SMC4 regulates JAK/STAT signaling. SMC4 KD in HCC reduces in vitro and in vivo growth. |
Elevated expression of SMC4 in HCC is associated with progression. | (Zhou et al., 2012) (Zhou et al., 2014) |
| SOX2 | SRY (Sex Determining Region Y)-Box 2 is a transcription factor that regulates gene expression in development and stem cell maintenance. | SOX2 is required for the cancer stem cell phenotype in LSCC and HNSCC. | SOX2 expression in HNSCC predicts poor survival. | (Justilien, Walsh, 2014) (Lee, Oh, 2014) (Dong, Liu, 2014) |
| TBL1XR1 | Transducin-beta-like 1 X-linked receptor 1 is a member of nuclear receptor corepressor that represses or activates gene expression, depending on the regulatory complexes involved. | TBL1XR1 regulates β-catenin-mediated gene expression and breast cancer cell proliferation. TBL1XR1 activates NF-κB and cisplatin resistance in nasopharyngeal cancer. TBL1XR1 promotes EMT in cervical cancer and promotes VEGF-C expression and lymph-angiogenesis and lymph node metastasis. |
TBL1XR1 is highly expressed and prognostic for poor survival in breast, cervical, nasopharyngeal and ESCC cancers. | (Li et al., 2014) (Li and Wang, 2008) (Chen et al., 2014b) (Liu et al., 2015) |
| TNFSF10 | Tumor necrosis factor ligand superfamily member 10 (aka: TNF-related apoptosis-inducing ligand: TRAIL) is a cytokine belonging to the TNF ligand family that binds pro-apoptotic death receptors to transduce an apoptotic signal. | Loss of TNFSF10 expression in cervical cancer is associated with worse differentiation. | TNFSF10 expression in cervical cancer cell lines is down-regulated despite gene amplification. | (Yao et al., 2015) (Vazquez-Mena et al., 2012) |
| USP13 | Ubiquitin specific peptidase 13 (aka: isopeptidase T-3) is a deubiquitinase enzyme implicated in tumor promotion and suppression by regulating stability of tumor suppressors and autophagy mediators. | USP13 de-ubiquitinates and stabilizes MITF to promote melanoma growth and tumorigenesis. USP13 stabilizes PTEN in breast cancer to suppress tumorigenesis and stabilizes autophagy regulators beclin1 and USP10. |
None described. | (Zhao et al., 2011) (Zhang et al., 2013) (Liu et al., 2011a) |
SOX2 is a major transcriptional regulator of gene expression and is critical for both embryonic development and maintenance of the “stem” phenotype of pluripotent cells in adult tissues (Liu et al., 2013). SOX2 also plays a key role in the stem-like TIC phenotype, particularly in SCC (Bass et al., 2009, Liu, Lin, 2013). SOX2 is the most highly upregulated transcription factor in skin SCC TICs where it regulates genes associated with stemness (Boumahdi et al., 2014). Conditional ablation of SOX2 in existing skin SCC tumors leads to tumor regression and decreased tumor transplantation, supporting a critical role for SOX2 in TIC cell maintenance (Boumahdi, Driessens, 2014). Likewise, ectopic expression of SOX2 in HNSCC cells promotes “stemness” while SOX2 knockdown (KD) in HNSCC TICs reduces self-renewal, chemoresistance and tumorigenicity, demonstrating a functional role for SOX2 in 3q26 amplified cancers (Lee et al., 2014).
SOX2 has been identified as a lineage-restricted oncogene in LSCC and HNSCC (Bass, Watanabe, 2009) that functions to maintain a stem-like phenotype in LSCC and HNSCC cells (Boumahdi, Driessens, 2014, Hussenet, Dali, 2010, Hussenet and du Manoir, 2010, Justilien et al., 2014, Lee, Oh, 2014, Liu, Lin, 2013, Lu et al., 2010, Nakatsugawa et al., 2011, Yuan et al., 2010). Genetic silencing of SOX2 expression leads to apoptosis, loss of tumorgienicity and decreased stemness in lung cancer cells (Chen et al., 2014a, Lee, Oh, 2014, Xiang et al., 2011). Gene amplification drives SOX2 expression in lung, esophageal and other squamous cell cancers (Bass, Watanabe, 2009, Justilien, Walsh, 2014, Maier et al., 2011). ShRNA-mediated knockdown of SOX2 expression and candidate neighboring genes in 3q26 amplified and non-amplified cancer cell lines revealed that inhibition of SOX2 expression had the largest differential anti-proliferative effect on 3q26 amplified SCC cell lines (Bass, Watanabe, 2009). Meta-analysis of SOX2 expression and clinical/prognostic features of HNSCC patients revealed that elevated SOX2 predicts poor overall and disease-free survival, and SOX2 expression significantly correlates with higher tumor grade, and presence of lymph node and distant metastases (Dong et al., 2014). SOX2 expression is similarly prognostic of poor outcome in lung adenocarcinoma (LADC) (Sholl et al., 2010) and small cell lung cancer (Xiang, Liao, 2011). However, somewhat paradoxically, SOX2 expression has been associated with better clinical outcome for NSCLC patients, suggesting that multigenic factors drive aggressive behavior and poor outcome in 3q26-driven tumors (Chen et al., 2013, Iijima et al., 2015, Toschi et al., 2014, Velcheti et al., 2013, Wang et al., 2015b). These studies strongly support a driver role for SOX2 in 3q26 amplified tumors and perhaps other tumor types.
ECT2 is a guanine nucleotide exchange factor for the Rho family of GTPases (Rho, Cdc42 and Rac) that functions primarily in the control of cytokinesis in non-transformed cells (Tatsumoto et al., 1999) Ect2 was originally identified as a gene that can transform mouse fibroblasts (Miki et al., 1993); reviewed in (Fields and Justilien, 2009). Subsequent studies have demonstrated a role for ECT2 in the transformed phenotype of multiple tumor types including glioma (Salhia et al., 2008), LSCC (Justilien and Fields, 2009, Justilien et al., 2011) and ovarian cancers (Huff et al., 2013). Though ECT2 has been demonstrated to play a functional role in transformed behavior in a number of tumor types, the mechanism by which ECT2 participates in transformation is still incompletely understood. Recent experimental evidence strongly suggests that the oncogenic function of ECT2 is distinct from its role in regulating cytokinesis (Justilien, Jameison, 2011) and appears to involve as yet unidentified nuclear function(s) for this protein (Huff, Decristo, 2013, Justilien, Jameison, 2011).
Ect2 is highly expressed in a variety of human tumors including brain (Salhia, Tran, 2008, Sano et al., 2006) lung (Fields and Justilien, 2010, Hirata et al., 2009), esophageal (Hirata, Yamabuki, 2009), pancreatic (Zhang et al., 2008) and ovarian tumors (Haverty et al., 2009). Ect2 overexpression is associated with poor prognosis in patients with NSCLC and ESCC (Hirata, Yamabuki, 2009). Specifically, NSCLC and ESCC patients whose tumors exhibit strong Ect2 staining had a poorer prognosis than patients whose tumors showed weak or no Ect2 staining. In ESCC patients, high Ect2 expression also positively correlated with tumor size and tumor metastasis to lymph nodes (Hirata, Yamabuki, 2009), suggesting that Ect2 expression is important for these clinicopathologic features of tumor progression. Ect2 expression may also serve as a prognostic indicator in glioblastoma patients, where high Ect2 expression was associated with a shorter survival time (Salhia, Tran, 2008, Sano, Genkai, 2006). Further investigation is required to determine if Ect2 expression is useful as a prognotic marker in other human cancer types.
ECT2 CNGs frequently occur in LSCC (>70%), but rarely in lung adenocarcinoma (LADC) (Justilien and Fields, 2009). These findings are consistent with the prevalence of chromosome 3q26 CNGs in LSCCs and the relatively rare occurrence of 3q26 CNGs in LADCs (Balsara, Sonoda, 1997, Brass, Ukena, 1996). Ect2 mRNA expression in LSCCs correlates with ECT2 CNGs, demonstrating that tumor specific ECT2 CNG is an important mechanism driving Ect2 overexpression in LSCCs (Justilien and Fields, 2009). In addition to LSCC, ovarian tumors also show tumor-specific amplification at 3q26 (Eder et al., 2005, Zhang et al., 2006). Ect2 is overexpressed in ovarian tumors harboring ECT2 amplification compared to whole normal ovary (Haverty, Hon, 2009) indicating that tumor-specific ECT2 amplification also drives Ect2 expression in ovarian tumors. Thus, a major mechanism driving Ect2 expression is tumor-specific amplification of ECT2 as part of the 3q26 amplicon.
PIK3CA encodes p110α, the catalytic subunit of phosphatidylinositol 3-kinase (PI3K). PI3-kinases mediate production of 3-phosphorylated phosphoinositides (PtdIns3P, PtdIns(3,5)P2 and PtdIns(3,4,5)P3) that serve as required second messengers for many physiological processes, including cell growth and proliferation, cell motility, survival, differentiation, cell adhesion, invasion, intracellular vesicular traffic, and protein synthesis. PIK3CA is frequently targeted for oncogenic activation either by somatic mutation or CNG as part of the 3q26 amplicon. PIK3CA is frequently mutated in breast, endometrial, colorectal, urinary tract and ovarian cancers (reviewed in (Yuan and Cantley, 2008)). These mutations cluster in two conserved regions encoding the kinase and helical domains of the protein. These “hot spot” mutations, H1047R, E545K and E542K, are non-synonymous missense mutations that confer constitutive kinase activity (Samuels et al., 2004). Amplification of PIK3CA has been found in primary ovarian tumors and ovarian cancer cell lines (Shayesteh et al., 1999), primary cervical tumors and cervical cancer cell lines (Ma et al., 2000), HNSCC primary tumors (Woenckhaus et al., 2002) and LSCC (Brass, Ukena, 1996). PIK3CA gene amplification was initially observed in ovarian cancer where an increase in PIK3CA copy number was found to associate with an increased in transcription, p110α protein expression and PI3K activity (Shayesteh, Lu, 1999). Similarly, in cervical tumors PIK3CA gene copy drives its overexpression (Ma, Wei, 2000). CNGs in PIK3CA have been shown to correlate with its mRNA expression in HNSCC cell primary tumors (Redon et al., 2001, Woenckhaus, Steger, 2002). CNGs in PIK3CA are prevalent in thyroid cancer, particularly follicular thyroid carcinoma (FTC) and anaplastic thyroid carcinoma (ATC) sub-types which exhibit 24% and 42% CNG, respectively (Wu et al., 2005). Several studies have shown that PIK3CA amplification is associated with overexpression of p110α in thyroid tumors (Hou et al., 2007, Wu, Mambo, 2005). Endometrial tumors with 3q amplification overexpress PIK3CA when compared with unamplified tumors (Salvesen et al., 2009). In a separate study, 9 of 15 (60.0%) gastric cancer cell lines and 17 of 55 (30.9%) primary gastric carcinomas harbored PIK3CA amplification, whereas no normal and benign tumor tissues showed abnormal amplification (Byun et al., 2003). In addition, amplification of PIK3CA in gastric tumor cell lines was strongly associated with increased transcript level (Byun, Cho, 2003). LSCC frequently harbor CNGs in PIK3CA (Massion, Kuo, 2002) and NSCLC cell lines harboring PIK3CA CNGs express higher PIK3CA mRNA levels when compared to cell lines without an increase in copy number (Yamamoto et al., 2008).
PRKCI encodes a member of the atypical sub-class of the protein kinase C gene family, protein kinase C iota (PKCι). PKCι is a component of the cell polarity complex that functions to establish epithelial cell polarity, cell fate and tissue integrity of non-transformed cells, through regulation and subcellular localization of Rac1 and cdc42 (reviewed in (Vorhagen and Niessen, 2014). PRKCI is overexpressed in many tumor types including breast, brain, cervix, chronic myelogenous leukemia (CML), colorectal, esophageal, gastric, head and neck, hepatocellular, kidney, lung, melanoma, ovarian, pancreatic, prostate, rhabdomyosarcoma and uterine (reviewed in (Murray et al., 2011)). The overexpression of PKCι in tumors is associated with poor prognosis and reduced patient survival. PKCι expression is predictive of poor outcome in patients with bile duct (Li et al., 2008), lung (Regala et al., 2005b), ovarian (Eder, Sui, 2005, Weichert et al., 2003), pancreatic (Scotti et al., 2010) and prostate cancers (Ishiguro et al., 2011). In some tumor types such as bile duct (Li, Wang, 2008), breast (Kojima et al., 2008), ESCC (Yang et al., 2008), liver (Du et al., 2009) and ovarian (Zhang, Huang, 2006), PKCι expression is higher in late stage cancers, suggesting that PKCι may promote higher grade, and a more aggressive phenotype in these tumors. In contrast, PKCι expression levels do not correlate with stage in NSCLC in which PKCι overexpression was frequently observed in early stage (stage I and II) tumors (Regala, Weems, 2005b). Interestingly, patients with gastric tumors that have higher PKCι expression are more likely to relapse (Takagawa et al., 2010). Thus PKCι expression may serve as an important prognostic marker to identify patients who may be at elevated risk for tumor progression and relapse, and these patients may benefit from early and more aggressive adjuvant anti-tumor therapies.
PRKCI is frequently amplified in NSCLC, and amplification drives PKCι expression in NSCLC primary tumors and established cell lines (Regala, Weems, 2005b). Disruption of PKCι signaling blocks transformed growth of NSCLC cell lines with PRKCI amplification, demonstrating that PKCι is a critical target of 3q26 amplification in NSCLC. PRKCI amplification is associated with lymph node metastases in ESCC, and PKCι promotes metastasis of ESCC cell lines (Liu et al., 2011b, Yang, Chu, 2008). Likewise, PKCι has been characterized as an oncogene in ovarian cancer. PRKCI is frequently amplified and overexpressed in ovarian serous cancer and PRKCI amplification predicts poor patient survival (Eder, Sui, 2005). In summary, PKCι is highly expressed in 3q26 amplified tumors, suggesting an oncogenic role for PKCι in these tumors. Interestingly however, PKCι is overexpressed in a vast majority of tumor types, including many that do not harbor frequent 3q26 CNGs, indicating that multiple mechanisms are at play to ensure high oncogenic PKCι expression in the majority of human cancer types.
The core 3q26 nodal genes act as a highly coordinated “OncCassette” that drives tumorigenesis
As described above, each of the four 3q26 nodal genes, SOX2, ECT2, PIK3CA and PRKCI, have individually been shown to function in the establishment and maintenance of the transformed phenotype of 3q26 driven tumors. It is also quite evident that these genes operate in a tumor environment in which they are coordinately amplified and overexpressed in tumors harboring 3q26 CNGs, demonstrating that these genes are genetically integrated with the oncogenic behavior of these tumors. Interestingly however, our genomic analysis suggests an even higher level of integration among these genes; specifically that these genes reside within a single network module in the context of 3q26 amplified tumors (Figure 3). Indeed, accumulating evidence from a number of biochemical, cellular signaling and cell biological studies provide compelling evidence that these four genes are functionally and biochemically linked in these tumors. Below we highlight major studies demonstrating this functional integration among 3q26 nodal genes.
The 3q26 genes EVI1 and PIK3CA regulate expression of a third 3q26 gene, SKIL/SNO
SKIL (alias SNO) (ski-related novel gene) is a member of the Ski family of nuclear oncoproteins that repress transforming growth factor-beta (TGF-beta) signaling through inhibition of transcriptional activity of Smad proteins (Sarker et al., 2005). SKIL/SNO, which resides at 3q26, was found to be consistently over-expressed in ESCC cell lines and primary tumors that exhibit 3q26 amplification, suggesting that this gene is a potential target of 3q26 amplification (Imoto et al., 2001). A study to identify candidate target genes for yet another 3q26 gene EVI1, that may play a role in EVI1-dependent transformation revealed that EVI1 can occupy the SKIL/SNO promoter and regulate SKIL expression (Yatsula et al., 2005). In ovarian cancer cells, the PI3K/AKT signaling pathway transcriptionally up-regulates SKIL expression in response to As2O3 (a drug exhibiting anti-tumor activity), and expression of siRNA targeting PIK3CA abrogated this response in ovarian cancer cells (Kodigepalli et al., 2013).
PKC;ι and Ect2 cooperate in Rac1 activation and lung transformation
In NSCLC cells, the protein product of the 3q26 gene PRKCI, PKCι resides in an oncogenic complex with the polarity associated protein Par6, an interaction that functions to drive anchorage-independent growth and invasion through activation of the Rho family GTPase Rac1 (Frederick et al., 2008). In an effort to biochemically characterize the mechanism by which the PKCι-Par6 complex regulates Rac1 activity in LSCC cells, we performed a proteomic screen to identify proteins that associate with the PKCι-Par6 complex and participate in PKCι signaling in NSCLC (Justilien and Fields, 2009). We identified Ect2, a gene co-localized on the frequently amplified 3q26 chromosomal region (Justilien and Fields, 2009), as a component of the oncogenic PKCι-Par6 complex in LSCC cells. Like PKCι and Par6, RNAi-mediated knockdown of Ect2 in NSCLC cells inhibits Rac1 activation and transformation. We further demonstrated that the oncogenic potential of Ect2 requires its interaction with PKCι-Par6 (Justilien and Fields, 2009). Thus, two 3q26 nodal genes, PRKCI and ECT2 are coordinately amplified and overexpressed in LSCC tumors, and the protein products of these genes biochemically interaction in an oncogenic complex in these cells.
The transformed behavior of LSCC cells requires both formation of the PKCι-Par6 complex and PKCι kinase activity (Frederick, Matthews, 2008). We therefore reasoned that substrates important for PKCι-mediated transformation might be identified within the PKCι-Par6 complex. Indeed, in a companion study, we demonstrated that PKCι can directly phosphorylate Ect2 (Justilien, Jameison, 2011). Mass spectrometry (MS) analysis of recombinant Ect2 incubated with PKCι in vitro revealed that Ect2 was phosphorylated on T328, a consensus PKCι phosphorylation site predicted by the kinase phosphorylation site prediction program Net-PhosK (Justilien and Fields, 2009). Furthermore, we showed that this phosphorylation event regulates Ect2 activation of Rac1 and transforming potential to maintain the transformed phenotype of NSCLC cells (Justilien, Jameison, 2011). Therefore, Ect2 and PKCι drive tumor cell proliferation through formation of an oncogenic PKCι-Par6-Ect2 complex.
Studies in primary LSCC tumors demonstrated that PRKCI and ECT2 are coordinately amplified and overexpressed in LSCC tumors (Justilien and Fields, 2009). Thus, Ect2 and PKCι are genetically linked through coordinate gene amplification in LSCC, and biochemically and functionally linked in LSCC through formation of an oncogenic PKCι-Par6-Ect2 complex that drives NSCLC cell transformation by activating Rac1 (Justilien and Fields, 2009, Justilien, Jameison, 2011). Finally, PKCι utilizes Ect2 as a direct substrate and through direct, site-specific phosphorylation, regulates the oncogenic activity of Ect2.
PKCι and SOX2 cooperate to regulate cancer stem cells
Human tumors contain a small population of highly tumorigenic cells (alternatively termed cancer stem cells (CSCs) or tumor initiating cells (TICs)) that are thought to be responsible for tumor initiation, maintenance, metastasis and relapse. We have demonstrated that PKCι is required for lung tumor formation in a mouse model of oncogenic KRAS-mediated lung adenocarcinoma (LADC) (Regala et al., 2009). In this model, PKCι is required for the transformation and growth of TICs that give rise to LADC (Regala, Davis, 2009). This study led us to hypothesize that the oncogenic function of PKCι is to establish and maintain a TIC phenotype in LSCC tumors harboring 3q26 CNG as well. In a subsequent study, we demonstrated that indeed PKCι functions to maintain a TIC phenotype in human LSCC (Justilien, Walsh, 2014). Functional analysis demonstrated that PRKCI cooperates with SOX2, a second 3q26 nodal gene, to drive a TIC phenotype in LSCC cells and primary LSCC tumors (Justilien, Walsh, 2014). PKCι maintains the TIC phenotype in LSCC through SOX2-mediated regulation of Hh signaling (Justilien, Walsh, 2014). Mechanistically, PKCι phosphorylates SOX2 at T118, a phosphorylation event that drives SOX2-mediated transcriptional activation of Hedgehog acetyl transferase (HHAT), an enzyme that catalyzes a critical step in the maturation of Hh ligand. Thus PRKCI and SOX2 cooperate to regulate a cell autonomous Hh signaling axis that maintains the TIC phenotype in LSCC.
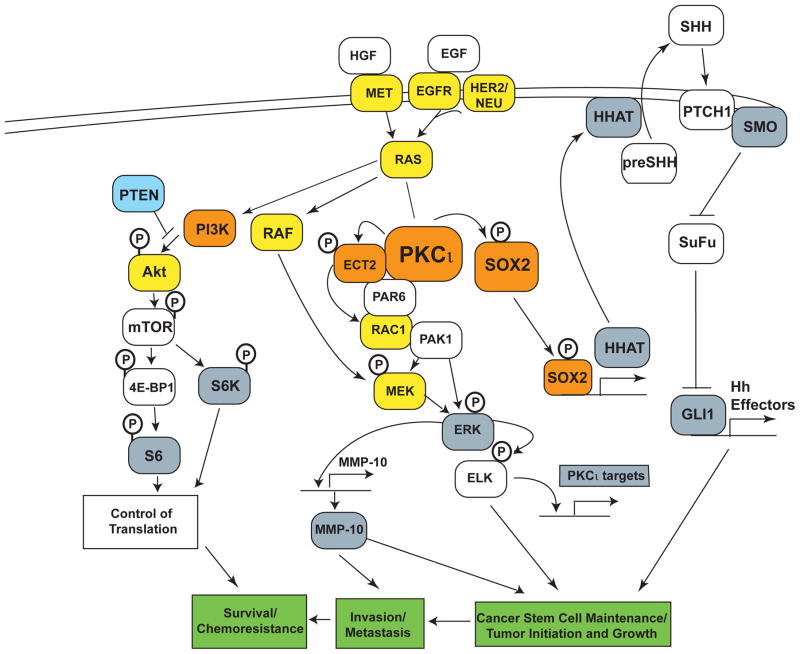
Taken together, these biochemical and functional studies provide a compelling picture of the 3q26 amplicon as a highly coordinated, functionally integrated 3q26 OncCassette consisting of 4 core 3q26 nodal genes, SOX2, ECT2, PRKCI and PIK3CA. The 3q26 OncCassette drives tumorigenesis through coordinated overexpression of these 4 oncogenic genes that reside in a single integrated signaling network that functions to establish and maintain a population of highly aggressive and tumorigenic stem-like cells that drive tumor formation, survival, proliferation, and progression to a highly metastatic phenotype (Figure 4). In addition, many the non-3q26 components of the 3q26 OncCassette induced signaling pathways outlined in Figure 4 have been implicated in the establishment and maintenance of a cancer stem-like phenotype including EGFR, KRAS and PTEN (Ciuffreda et al., 2014, Fitzgerald et al., 2015, McCubrey et al., 2015).
Figure 4. The 3q26 OncCassette.
A schematic diagram is shown illustrating the integrated functional and biochemical relationships between the four 3q26 nodal genes SOX2, ECT2, PRKCI and PIK3CA. Functional studies demonstrate that these four nodal genes constitute a 3q26 OncCassette that establishes and maintains a highly tumorigenic tumor-initiating cell (TIC) phenotype that drives LSCC tumorigenesis. Genes in yellow indicate non 3q26 oncogenes; Genes in orange indicate the four 3q26 nodal genes; genes in blue indicate tumor suppressor genes; genes in gray indicate potential surrogate biomarkers of 3q26 OncCassette activity.
Therapeutic Implications: Targeting the 3q26 OncCassette
The realization that 3q26 CNG drives oncogenesis through coordinated overexpression of biochemically and functionally linked genes presents both challenges and opportunities for therapeutic intervention. Inspection of the nodal and integrated 3q26 genes (Figure 3) and their experimentally validated signaling interactions (Figure 4) reveals multiple avenues for drug development. Several 3q26 nodal genes are direct druggable targets (i.e. PIK3KCA and PRKCI). Other 3q26 genes belonging to the predicted network module are also potentially druggable enzymes including PLD1 (Gomez-Cambronero, 2014) and the PTEN signaling axis (Ciuffreda, Falcone, 2014). Furthermore, the four nodal 3q26 OncCassette genes function within the context of signaling pathways that represent even more druggable targets for therapeutic exploitation. The most developmentally advanced inhibitors targeting a 3q26 gene are the PIK3CA inhibitors (reviewed in (Garcia-Echeverria and Sellers, 2008, Rodon et al., 2013)). Though PI3K inhibitors showed great promise in pre-clinical studies, their clinical use has been limited by unacceptable toxicity profiles and detrimental off-target effects. Second generation inhibitors targeting various points in the PI3K signaling axis, including inhibitors of AKT, specific PI3K isoforms which serve distinct roles in tumorigenesis (Baer et al., 2015), and dual inhibitors of PI3K and mTOR, and PI3K and PLK1, have shown promise as anti-tumor agents (Bowles et al., 2014, Roychowdhury et al., 2010, Serra, Markman, 2008).
More recently, PKCι inhibitors have been identified and validated in pre-clinical studies (Butler et al., 2015, Erdogan et al., 2006, Fields et al., 2007, Fields and Murray, 2008, Fields and Regala, 2007, Kjaer et al., 2013, Parker et al., 2014, Stallings-Mann et al., 2006). We identified the anti-rheumatoid gold salts aurothiomalate (ATM) and auranofin (ANF) as potent and selective PKCι inhibitors that function through disruption of the interaction between oncogenic PKCι and its binding partner Par6 (Erdogan, Lamark, 2006, Stallings-Mann, Jamieson, 2006). A major advantage of these compounds is that they are FDA-approved and have favorable toxicity profiles that make them suitable for use in the oncology setting. Both ATM and ANF exhibit potent anti-tumor activity in vitro and in vivo in lung, ovarian and pancreatic tumor models (Butler, Scotti Buzhardt, 2015, Erdogan, Lamark, 2006, Stallings-Mann, Jamieson, 2006, Wang et al., 2013b). Interestingly, response to ATM and ANF correlates directly with PRKCI CNG and elevated PKCι expression, indicating that 3q26 CNG-driven tumors may be particularly sensitive to PKCι inhibitor therapy (Regala et al., 2008). Our initial clinical experience with these agents has demonstrated proof of principle for PKCι inhibition using ATM or ANF as a viable therapeutic approach in both LSCC and ovarian serous cancer, tumor types exhibiting frequent 3q26 CNGs (Jatoi et al., 2014, Mansfield et al., 2013). These agents were well tolerated in the oncology setting at concentrations that exhibit effective anti-tumor activity, and these agents have shown promising initial indications of clinical activity in heavily pre-treated patients with advanced disease. More recently, we have demonstrated synergistic anti-tumor activity when combining ANF with mTOR inhibition, a combination that effectively targets both the PKCι and PI3K-AKT-mTOR pathways shown in Figure 4. This combination has shown good synergistic anti-tumor activity in pre-clinical models of 3q26 CNG-driven LSCC in vitro and in vivo (manuscript in preparation). An ongoing phase 1B/II clinical trial using ANF combined with the mTOR inhibitor sirolimus is currently accruing patients with advanced LSCC. Similarly, ANF exhibits synergistic anti-tumor activity in combination with the Smoothened inhibitor erismodegib (Justilien and Fields, 2015), a drug combination that effectively targets the recently identified PKCι-SOX2-Hh signaling axis that maintains a stem-like phenotype in 3q26 CNG LSCC (Justilien, Walsh, 2014). These findings provide a compelling rationale for therapeutic approaches involving PKCι inhibition in combination with other targeted agents against signaling pathways that are activated and driven by the 3q26 OncCassette. This functionally rationalized approach to combined therapy for 3q26 driven tumors may be paradigm shifting with regard to the clinical treatment of the large number of human tumors harboring this common genetic alteration.
In conclusion, characterization of a functionally integrated, multigenic 3q26 OncCassette reveals a novel network module comprised of a genetically induced and functionally integrated set of oncogenic drivers. Combination therapies targeting nodal genes within this network show promise for enhancing clinical outcome of 3q26 amplified cancers. Further functional characterization of the 3q26 OncCassette holds considerable promise of revealing further opportunities for development of novel combined therapeutic intervention strategies that target unique vulnerabilities of 3q26 CNG driven tumors.
Acknowledgments
The authors wish to thank all of the members of the Fields laboratory who have contributed to many of the studies described herein. This work was supported by grants from the National Institutes of Health (R01 CA081436-16 and R21 CA151250-02 to APF; R01 CA14090-05 to NRM); the James and Esther King Biomedical Research Program (1KG-05-33971) and the Mayo Clinic Center for Individualized Medicine (CIM) to APF; and a National Institutes of Health Research Supplement to Promote Diversity in Health-related Research Award from the National Cancer Institute to VJ. APF is the Monica Flynn Jacoby Endowed Professor of Cancer Research.
Footnotes
Conflicts of Interest: None to declare
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Akagi I, Miyashita M, Makino H, Nomura T, Hagiwara N, Takahashi K, et al. SnoN overexpression is predictive of poor survival in patients with esophageal squamous cell carcinoma. Ann Surg Oncol. 2008;15:2965–75. doi: 10.1245/s10434-008-9986-y. [DOI] [PubMed] [Google Scholar]
- Arnold N, Hagele L, Walz L, Schempp W, Pfisterer J, Bauknecht T, et al. Overrepresentation of 3q and 8q material and loss of 18q material are recurrent findings in advanced human ovarian cancer. Genes Chromosomes Cancer. 1996;16:46–54. doi: 10.1002/(SICI)1098-2264(199605)16:1<46::AID-GCC7>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- Baer R, Cintas C, Therville N, Guillermet-Guibert J. Implication of PI3K/Akt pathway in pancreatic cancer: When PI3K isoforms matter? Adv Biol Regul. 2015;59:19–35. doi: 10.1016/j.jbior.2015.05.001. [DOI] [PubMed] [Google Scholar]
- Balsara BR, Sonoda G, du Manoir S, Siegfried JM, Gabrielson E, Testa JR. Comparative genomic hybridization analysis detects frequent, often high-level, overrepresentation of DNA sequences at 3q, 5p, 7p, and 8q in human non-small cell lung carcinomas. Cancer Res. 1997;57:2116–20. [PubMed] [Google Scholar]
- Bandla S, Pennathur A, Luketich JD, Beer DG, Lin L, Bass AJ, et al. Comparative genomics of esophageal adenocarcinoma and squamous cell carcinoma. Ann Thorac Surg. 2012;93:1101–6. doi: 10.1016/j.athoracsur.2012.01.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao X, Tang J, Lopez-Pajares V, Tao S, Qu K, Crabtree GR, et al. ACTL6a enforces the epidermal progenitor state by suppressing SWI/SNF-dependent induction of KLF4. Cell Stem Cell. 2013;12:193–203. doi: 10.1016/j.stem.2012.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bass AJ, Watanabe H, Mermel CH, Yu S, Perner S, Verhaak RG, et al. SOX2 is an amplified lineage-survival oncogene in lung and esophageal squamous cell carcinomas. Nat Genet. 2009;41:1238–42. doi: 10.1038/ng.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berndt SI, Wang Z, Yeager M, Alavanja MC, Albanes D, Amundadottir L, et al. Two susceptibility loci identified for prostate cancer aggressiveness. Nat Commun. 2015;6:6889. doi: 10.1038/ncomms7889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjorkqvist AM, Husgafvel-Pursiainen K, Anttila S, Karjalainen A, Tammilehto L, Mattson K, et al. DNA gains in 3q occur frequently in squamous cell carcinoma of the lung, but not in adenocarcinoma. Genes Chromosomes Cancer. 1998;22:79–82. [PubMed] [Google Scholar]
- Bockmuhl U, Schluns K, Kuchler I, Petersen S, Petersen I. Genetic imbalances with impact on survival in head and neck cancer patients. Am J Pathol. 2000;157:369–75. doi: 10.1016/S0002-9440(10)64549-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bockmuhl U, Schwendel A, Dietel M, Petersen I. Distinct patterns of chromosomal alterations in high- and low-grade head and neck squamous cell carcinomas. Cancer Res. 1996;56:5325–9. [PubMed] [Google Scholar]
- Bockmuhl U, Wolf G, Schmidt S, Schwendel A, Jahnke V, Dietel M, et al. Genomic alterations associated with malignancy in head and neck cancer. Head Neck. 1998;20:145–51. doi: 10.1002/(sici)1097-0347(199803)20:2<145::aid-hed8>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- Boumahdi S, Driessens G, Lapouge G, Rorive S, Nassar D, Le Mercier M, et al. SOX2 controls tumour initiation and cancer stem-cell functions in squamous-cell carcinoma. Nature. 2014;511:246–50. doi: 10.1038/nature13305. [DOI] [PubMed] [Google Scholar]
- Bowles DW, Diamond JR, Lam ET, Weekes CD, Astling DP, Anderson RT, et al. Phase I study of oral rigosertib (ON 01910.Na), a dual inhibitor of the PI3K and Plk1 pathways, in adult patients with advanced solid malignancies. Clin Cancer Res. 2014;20:1656–65. doi: 10.1158/1078-0432.CCR-13-2506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brass N, Ukena I, Remberger K, Mack U, Sybrecht GW, Meese EU. DNA amplification on chromosome 3q26.1-q26.3 in squamous cell carcinoma of the lung detected by reverse chromosome painting. Eur J Cancer. 1996;32A:1205–8. doi: 10.1016/0959-8049(96)00016-0. [DOI] [PubMed] [Google Scholar]
- Brzoska PM, Levin NA, Fu KK, Kaplan MJ, Singer MI, Gray JW, et al. Frequent novel DNA copy number increase in squamous cell head and neck tumors. Cancer Res. 1995;55:3055–9. [PubMed] [Google Scholar]
- Butler AM, Scotti Buzhardt ML, Erdogan E, Li S, Inman KS, Fields AP, et al. A small molecule inhibitor of atypical protein kinase C signaling inhibits pancreatic cancer cell transformed growth and invasion. Oncotarget. 2015;6:15297–310. doi: 10.18632/oncotarget.3812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byun DS, Cho K, Ryu BK, Lee MG, Park JI, Chae KS, et al. Frequent monoallelic deletion of PTEN and its reciprocal associatioin with PIK3CA amplification in gastric carcinoma. Int J Cancer. 2003;104:318–27. doi: 10.1002/ijc.10962. [DOI] [PubMed] [Google Scholar]
- Charafe-Jauffret E, Ginestier C, Iovino F, Wicinski J, Cervera N, Finetti P, et al. Breast cancer cell lines contain functional cancer stem cells with metastatic capacity and a distinct molecular signature. Cancer Res. 2009;69:1302–13. doi: 10.1158/0008-5472.CAN-08-2741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q, Hongu T, Sato T, Zhang Y, Ali W, Cavallo JA, et al. Key roles for the lipid signaling enzyme phospholipase d1 in the tumor microenvironment during tumor angiogenesis and metastasis. Sci Signal. 2012a;5:ra79. doi: 10.1126/scisignal.2003257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Li X, Lu D, Xu Y, Mou W, Wang L, et al. SOX2 regulates apoptosis through MAP4K4-survivin signaling pathway in human lung cancer cells. Carcinogenesis. 2014a;35:613–23. doi: 10.1093/carcin/bgt371. [DOI] [PubMed] [Google Scholar]
- Chen SP, Yang Q, Wang CJ, Zhang LJ, Fang Y, Lei FY, et al. Transducin beta-like 1 X-linked receptor 1 suppresses cisplatin sensitivity in Nasopharyngeal Carcinoma via activation of NF-kappaB pathway. Mol Cancer. 2014b;13:195. doi: 10.1186/1476-4598-13-195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Thiaville MM, Chen L, Stoeck A, Xuan J, Gao M, et al. Defining NOTCH3 target genes in ovarian cancer. Cancer Res. 2012b;72:2294–303. doi: 10.1158/0008-5472.CAN-11-2181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Huang Y, Chen J, Wang S, Zhou J. The prognostic value of SOX2 expression in non-small cell lung cancer: a meta-analysis. PLoS One. 2013;8:e71140. doi: 10.1371/journal.pone.0071140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chujo M, Noguchi T, Miura T, Arinaga M, Uchida Y, Tagawa Y. Comparative genomic hybridization analysis detected frequent overrepresentation of chromosome 3q in squamous cell carcinoma of the lung. Lung Cancer. 2002;38:23–9. doi: 10.1016/s0169-5002(02)00151-4. [DOI] [PubMed] [Google Scholar]
- Ciriello G, Miller ML, Aksoy BA, Senbabaoglu Y, Schultz N, Sander C. Emerging landscape of oncogenic signatures across human cancers. Nat Genet. 2013;45:1127–33. doi: 10.1038/ng.2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciuffreda L, Falcone I, Incani UC, Del Curatolo A, Conciatori F, Matteoni S, et al. PTEN expression and function in adult cancer stem cells and prospects for therapeutic targeting. Adv Biol Regul. 2014;56:66–80. doi: 10.1016/j.jbior.2014.07.002. [DOI] [PubMed] [Google Scholar]
- Dall’Armi C, Hurtado-Lorenzo A, Tian H, Morel E, Nezu A, Chan RB, et al. The phospholipase D1 pathway modulates macroautophagy. Nat Commun. 2010;1:142. doi: 10.1038/ncomms1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai S, Pillai P, Win-Piazza H, Acevedo-Duncan M. PKC-iota promotes glioblastoma cell survival by phosphorylating and inhibiting BAD through a phosphatidylinositol 3-kinase pathway. Biochim Biophys Acta. 2011;1813:1190–7. doi: 10.1016/j.bbamcr.2011.03.007. [DOI] [PubMed] [Google Scholar]
- Dong Z, Liu G, Huang B, Sun J, Wu D. Prognostic significance of SOX2 in head and neck cancer: a meta-analysis. Int J Clin Exp Med. 2014;7:5010–20. [PMC free article] [PubMed] [Google Scholar]
- Du GS, Wang JM, Lu JX, Li Q, Ma CQ, Du JT, et al. Expression of P-aPKC-iota, E-cadherin, and beta-catenin related to invasion and metastasis in hepatocellular carcinoma. Ann Surg Oncol. 2009;16:1578–86. doi: 10.1245/s10434-009-0423-7. [DOI] [PubMed] [Google Scholar]
- Eder AM, Sui X, Rosen DG, Nolden LK, Cheng KW, Lahad JP, et al. Atypical PKCiota contributes to poor prognosis through loss of apical-basal polarity and cyclin E overexpression in ovarian cancer. Proc Natl Acad Sci U S A. 2005;102:12519–24. doi: 10.1073/pnas.0505641102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erdogan E, Lamark T, Stallings-Mann M, Lee J, Pellecchia M, Thompson EA, et al. Aurothiomalate inhibits transformed growth by targeting the PB1 domain of protein kinase Ciota. J Biol Chem. 2006;281:28450–9. doi: 10.1074/jbc.M606054200. [DOI] [PubMed] [Google Scholar]
- Fields AP, Frederick LA, Regala RP. Targeting the oncogenic protein kinase Ciota signalling pathway for the treatment of cancer. Biochem Soc Trans. 2007;35:996–1000. doi: 10.1042/BST0350996. [DOI] [PubMed] [Google Scholar]
- Fields AP, Justilien V. The guanine nucleotide exchange factor (GEF) Ect2 is an oncogene in human cancer. Adv Enzyme Regul. 2009 doi: 10.1016/j.advenzreg.2009.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fields AP, Justilien V. The guanine nucleotide exchange factor (GEF) Ect2 is an oncogene in human cancer. Adv Enzyme Regul. 2010;50:190–200. doi: 10.1016/j.advenzreg.2009.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fields AP, Murray NR. Protein kinase C isozymes as therapeutic targets for treatment of human cancers. Adv Enzyme Regul. 2008;48:166–78. doi: 10.1016/j.advenzreg.2007.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fields AP, Regala RP. Protein kinase C iota: human oncogene, prognostic marker and therapeutic target. Pharmacol Res. 2007;55:487–97. doi: 10.1016/j.phrs.2007.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzgerald TL, Lertpiriyapong K, Cocco L, Martelli AM, Libra M, Candido S, et al. Roles of EGFR and KRAS and their downstream signaling pathways in pancreatic cancer and pancreatic cancer stem cells. Adv Biol Regul. 2015;59:65–81. doi: 10.1016/j.jbior.2015.06.003. [DOI] [PubMed] [Google Scholar]
- Flicek P, Amode MR, Barrell D, Beal K, Billis K, Brent S, et al. Ensembl 2014. Nucleic Acids Res. 2014;42:D749–55. doi: 10.1093/nar/gkt1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster NA, Banerjee AK, Xian J, Roberts I, Pezzella F, Coleman N, et al. Somatic genetic changes accompanying lung tumor development. Genes Chromosomes Cancer. 2005;44:65–75. doi: 10.1002/gcc.20223. [DOI] [PubMed] [Google Scholar]
- Frederick LA, Matthews JA, Jamieson L, Justilien V, Thompson EA, Radisky DC, et al. Matrix metalloproteinase-10 is a critical effector of protein kinase Ciota-Par6alpha-mediated lung cancer. Oncogene. 2008;27:4841–53. doi: 10.1038/onc.2008.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Echeverria C, Sellers WR. Drug discovery approaches targeting the PI3K/Akt pathway in cancer. Oncogene. 2008;27:5511–26. doi: 10.1038/onc.2008.246. [DOI] [PubMed] [Google Scholar]
- Gomez-Cambronero J. Phosphatidic acid, phospholipase D and tumorigenesis. Adv Biol Regul. 2014;54:197–206. doi: 10.1016/j.jbior.2013.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greiner M, Kreutzer B, Lang S, Jung V, Cavalie A, Unteregger G, et al. Sec62 protein level is crucial for the ER stress tolerance of prostate cancer. Prostate. 2011;71:1074–83. doi: 10.1002/pros.21324. [DOI] [PubMed] [Google Scholar]
- Hagerstrand D, Tong A, Schumacher SE, Ilic N, Shen RR, Cheung HW, et al. Systematic interrogation of 3q26 identifies TLOC1 and SKIL as cancer drivers. Cancer Discov. 2013;3:1044–57. doi: 10.1158/2159-8290.CD-12-0592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto Y, Oga A, Kawauchi S, Furuya T, Shimizu N, Nakano T, et al. Amplification of 3q26 approximately qter correlates with tumor progression in head and neck squamous cell carcinomas. Cancer Genet Cytogenet. 2001;129:52–6. doi: 10.1016/s0165-4608(01)00425-3. [DOI] [PubMed] [Google Scholar]
- Haverty PM, Hon LS, Kaminker JS, Chant J, Zhang Z. High-resolution analysis of copy number alterations and associated expression changes in ovarian tumors. BMC Med Genomics. 2009;2:21. doi: 10.1186/1755-8794-2-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heselmeyer K, Macville M, Schrock E, Blegen H, Hellstrom AC, Shah K, et al. Advanced-stage cervical carcinomas are defined by a recurrent pattern of chromosomal aberrations revealing high genetic instability and a consistent gain of chromosome arm 3q. Genes Chromosomes Cancer. 1997;19:233–40. [PubMed] [Google Scholar]
- Heselmeyer K, Schrock E, du Manoir S, Blegen H, Shah K, Steinbeck R, et al. Gain of chromosome 3q defines the transition from severe dysplasia to invasive carcinoma of the uterine cervix. Proc Natl Acad Sci U S A. 1996;93:479–84. doi: 10.1073/pnas.93.1.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilder TL, Malone MH, Bencharit S, Colicelli J, Haystead TA, Johnson GL, et al. Proteomic identification of the cerebral cavernous malformation signaling complex. J Proteome Res. 2007;6:4343–55. doi: 10.1021/pr0704276. [DOI] [PubMed] [Google Scholar]
- Hirata D, Yamabuki T, Miki D, Ito T, Tsuchiya E, Fujita M, et al. Involvement of epithelial cell transforming sequence-2 oncoantigen in lung and esophageal cancer progression. Clin Cancer Res. 2009;15:256–66. doi: 10.1158/1078-0432.CCR-08-1672. [DOI] [PubMed] [Google Scholar]
- Hollander MC, Blumenthal GM, Dennis PA. PTEN loss in the continuum of common cancers, rare syndromes and mouse models. Nat Rev Cancer. 2011;11:289–301. doi: 10.1038/nrc3037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou P, Liu D, Shan Y, Hu S, Studeman K, Condouris S, et al. Genetic alterations and their relationship in the phosphatidylinositol 3-kinase/Akt pathway in thyroid cancer. Clin Cancer Res. 2007;13:1161–70. doi: 10.1158/1078-0432.CCR-06-1125. [DOI] [PubMed] [Google Scholar]
- Hrstka R, Coates PJ, Vojtesek B. Polymorphisms in p53 and the p53 pathway: roles in cancer susceptibility and response to treatment. J Cell Mol Med. 2009;13:440–53. doi: 10.1111/j.1582-4934.2008.00634.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huff LP, Decristo MJ, Trembath D, Kuan PF, Yim M, Liu J, et al. The Role of Ect2 Nuclear RhoGEF Activity in Ovarian Cancer Cell Transformation. Genes Cancer. 2013;4:460–75. doi: 10.1177/1947601913514851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussenet T, Dali S, Exinger J, Monga B, Jost B, Dembele D, et al. SOX2 is an oncogene activated by recurrent 3q26.3 amplifications in human lung squamous cell carcinomas. PLoS One. 2010;5:e8960. doi: 10.1371/journal.pone.0008960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussenet T, du Manoir S. SOX2 in squamous cell carcinoma: amplifying a pleiotropic oncogene along carcinogenesis. Cell Cycle. 2010;9:1480–6. doi: 10.4161/cc.9.8.11203. [DOI] [PubMed] [Google Scholar]
- Iijima Y, Seike M, Noro R, Ibi T, Takeuchi S, Mikami I, et al. Prognostic significance of PIK3CA and SOX2 in Asian patients with lung squamous cell carcinoma. Int J Oncol. 2015;46:505–12. doi: 10.3892/ijo.2014.2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imoto I, Pimkhaokham A, Fukuda Y, Yang ZQ, Shimada Y, Nomura N, et al. SNO is a probable target for gene amplification at 3q26 in squamous-cell carcinomas of the esophagus. Biochem Biophys Res Commun. 2001;286:559–65. doi: 10.1006/bbrc.2001.5428. [DOI] [PubMed] [Google Scholar]
- Ishiguro H, Akimoto K, Nagashima Y, Kagawa E, Sasaki T, Sano JY, et al. Coexpression of aPKClambda/iota and IL-6 in prostate cancer tissue correlates with biochemical recurrence. Cancer Sci. 2011;102:1576–81. doi: 10.1111/j.1349-7006.2011.01972.x. [DOI] [PubMed] [Google Scholar]
- Iwabuchi H, Sakamoto M, Sakunaga H, Ma YY, Carcangiu ML, Pinkel D, et al. Genetic analysis of benign, low-grade, and high-grade ovarian tumors. Cancer Res. 1995;55:6172–80. [PubMed] [Google Scholar]
- Jatoi A, Radecki Breitkopf C, Foster NR, Block MS, Grudem M, Wahner Hendrickson A, et al. A Mixed-Methods Feasibility Trial of Protein Kinase C Iota Inhibition with Auranofin in Asymptomatic Ovarian Cancer Patients. Oncology. 2014;88:208–13. doi: 10.1159/000369257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung V, Kindich R, Kamradt J, Jung M, Muller M, Schulz WA, et al. Genomic and expression analysis of the 3q25-q26 amplification unit reveals TLOC1/SEC62 as a probable target gene in prostate cancer. Mol Cancer Res. 2006;4:169–76. doi: 10.1158/1541-7786.MCR-05-0165. [DOI] [PubMed] [Google Scholar]
- Justilien V, Fields AP. Ect2 links the PKCiota-Par6alpha complex to Rac1 activation and cellular transformation. Oncogene. 2009;28:3597–607. doi: 10.1038/onc.2009.217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Justilien V, Fields AP. Molecular pathways: novel approaches for improved therapeutic targeting of hedgehog signaling in cancer stem cells. Clin Cancer Res. 2015;21:505–13. doi: 10.1158/1078-0432.CCR-14-0507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Justilien V, Jameison L, Der CJ, Rossman KL, Fields AP. Oncogenic activity of Ect2 is regulated through protein kinase C iota-mediated phosphorylation. J Biol Chem. 2011;286:8149–57. doi: 10.1074/jbc.M110.196113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Justilien V, Walsh MP, Ali SA, Thompson EA, Murray NR, Fields AP. The PRKCI and SOX2 oncogenes are coamplified and cooperate to activate Hedgehog signaling in lung squamous cell carcinoma. Cancer Cell. 2014;25:139–51. doi: 10.1016/j.ccr.2014.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang DW, Lee SH, Yoon JW, Park WS, Choi KY, Min do S. Phospholipase D1 drives a positive feedback loop to reinforce the Wnt/beta-catenin/TCF signaling axis. Cancer Res. 2010;70:4233–42. doi: 10.1158/0008-5472.CAN-09-3470. [DOI] [PubMed] [Google Scholar]
- Kim AY, Bommelje CC, Lee BE, Yonekawa Y, Choi L, Morris LG, et al. SCCRO (DCUN1D1) is an essential component of the E3 complex for neddylation. J Biol Chem. 2008;283:33211–20. doi: 10.1074/jbc.M804440200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kjaer S, Linch M, Purkiss A, Kostelecky B, Knowles PP, Rosse C, et al. Adenosine-binding motif mimicry and cellular effects of a thieno[2,3-d]pyrimidine-based chemical inhibitor of atypical protein kinase C isoenzymes. Biochem J. 2013;451:329–42. doi: 10.1042/BJ20121871. [DOI] [PubMed] [Google Scholar]
- Kodigepalli KM, Dutta PS, Bauckman KA, Nanjundan M. SnoN/SkiL expression is modulated via arsenic trioxide-induced activation of the PI3K/AKT pathway in ovarian cancer cells. FEBS Lett. 2013;587:5–16. doi: 10.1016/j.febslet.2012.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kojima Y, Akimoto K, Nagashima Y, Ishiguro H, Shirai S, Chishima T, et al. The overexpression and altered localization of the atypical protein kinase C lambda/iota in breast cancer correlates with the pathologic type of these tumors. Hum Pathol. 2008;39:824–31. doi: 10.1016/j.humpath.2007.11.001. [DOI] [PubMed] [Google Scholar]
- Lee SH, Oh SY, Do SI, Lee HJ, Kang HJ, Rho YS, et al. SOX2 regulates self-renewal and tumorigenicity of stem-like cells of head and neck squamous cell carcinoma. Br J Cancer. 2014;111:2122–30. doi: 10.1038/bjc.2014.528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Poi MJ, Tsai MD. Regulatory mechanisms of tumor suppressor P16(INK4A) and their relevance to cancer. Biochemistry. 2011;50:5566–82. doi: 10.1021/bi200642e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Wang CY. TBL1-TBLR1 and beta-catenin recruit each other to Wnt target-gene promoter for transcription activation and oncogenesis. Nat Cell Biol. 2008;10:160–9. doi: 10.1038/ncb1684. [DOI] [PubMed] [Google Scholar]
- Li Q, Wang JM, Liu C, Xiao BL, Lu JX, Zou SQ. Correlation of aPKC-iota and E-cadherin expression with invasion and prognosis of cholangiocarcinoma. Hepatobiliary Pancreat Dis Int. 2008;7:70–5. [PubMed] [Google Scholar]
- Li X, Liang W, Liu J, Lin C, Wu S, Song L, et al. Transducin (beta)-like 1 X-linked receptor 1 promotes proliferation and tumorigenicity in human breast cancer via activation of beta-catenin signaling. Breast Cancer Res. 2014;16:465. doi: 10.1186/s13058-014-0465-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liggett WH, Jr, Sidransky D. Role of the p16 tumor suppressor gene in cancer. J Clin Oncol. 1998;16:1197–206. doi: 10.1200/JCO.1998.16.3.1197. [DOI] [PubMed] [Google Scholar]
- Linxweiler M, Linxweiler J, Barth M, Benedix J, Jung V, Kim YJ, et al. Sec62 bridges the gap from 3q amplification to molecular cell biology in non-small cell lung cancer. Am J Pathol. 2012;180:473–83. doi: 10.1016/j.ajpath.2011.10.039. [DOI] [PubMed] [Google Scholar]
- Linxweiler M, Schorr S, Schauble N, Jung M, Linxweiler J, Langer F, et al. Targeting cell migration and the endoplasmic reticulum stress response with calmodulin antagonists: a clinically tested small molecule phenocopy of SEC62 gene silencing in human tumor cells. BMC Cancer. 2013;13:574. doi: 10.1186/1471-2407-13-574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Little CD, Nau MM, Carney DN, Gazdar AF, Minna JD. Amplification and expression of the c-myc oncogene in human lung cancer cell lines. Nature. 1983;306:194–6. doi: 10.1038/306194a0. [DOI] [PubMed] [Google Scholar]
- Liu J, Xia H, Kim M, Xu L, Li Y, Zhang L, et al. Beclin1 controls the levels of p53 by regulating the deubiquitination activity of USP10 and USP13. Cell. 2011a;147:223–34. doi: 10.1016/j.cell.2011.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu K, Lin B, Zhao M, Yang X, Chen M, Gao A, et al. The multiple roles for Sox2 in stem cell maintenance and tumorigenesis. Cell Signal. 2013;25:1264–71. doi: 10.1016/j.cellsig.2013.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Lin C, Liang W, Wu S, Liu A, Wu J, et al. TBL1XR1 promotes lymphangiogenesis and lymphatic metastasis in esophageal squamous cell carcinoma. Gut. 2015;64:26–36. doi: 10.1136/gutjnl-2013-306388. [DOI] [PubMed] [Google Scholar]
- Liu SG, Wang BS, Jiang YY, Zhang TT, Shi ZZ, Yang Y, et al. Atypical protein kinase Ciota (PKCiota) promotes metastasis of esophageal squamous cell carcinoma by enhancing resistance to Anoikis via PKCiota-SKP2-AKT pathway. Mol Cancer Res. 2011b;9:390–402. doi: 10.1158/1541-7786.MCR-10-0359. [DOI] [PubMed] [Google Scholar]
- Lu Y, Futtner C, Rock JR, Xu X, Whitworth W, Hogan BL, et al. Evidence that SOX2 overexpression is oncogenic in the lung. PLoS One. 2010;5:e11022. doi: 10.1371/journal.pone.0011022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma YY, Wei SJ, Lin YC, Lung JC, Chang TC, Whang-Peng J, et al. PIK3CA as an oncogene in cervical cancer. Oncogene. 2000;19:2739–44. doi: 10.1038/sj.onc.1203597. [DOI] [PubMed] [Google Scholar]
- Maier S, Wilbertz T, Braun M, Scheble V, Reischl M, Mikut R, et al. SOX2 amplification is a common event in squamous cell carcinomas of different organ sites. Hum Pathol. 2011;42:1078–88. doi: 10.1016/j.humpath.2010.11.010. [DOI] [PubMed] [Google Scholar]
- Mansfield AS, Fields AP, Jatoi A, Qi Y, Adjei AA, Erlichman C, et al. Phase I dose escalation study of the PKCiota inhibitor aurothiomalate for advanced non-small-cell lung cancer, ovarian cancer, and pancreatic cancer. Anticancer Drugs. 2013;24:1079–83. doi: 10.1097/CAD.0000000000000009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massion PP, Kuo WL, Stokoe D, Olshen AB, Treseler PA, Chin K, et al. Genomic copy number analysis of non-small cell lung cancer using array comparative genomic hybridization: implications of the phosphatidylinositol 3-kinase pathway. Cancer Res. 2002;62:3636–40. [PubMed] [Google Scholar]
- Mathews MB, Hershey JW. The translation factor eIF5A and human cancer. Biochim Biophys Acta. 2015;1849:836–44. doi: 10.1016/j.bbagrm.2015.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCubrey JA, Abrams SL, Fitzgerald TL, Cocco L, Martelli AM, Montalto G, et al. Roles of signaling pathways in drug resistance, cancer initiating cells and cancer progression and metastasis. Adv Biol Regul. 2015;57:75–101. doi: 10.1016/j.jbior.2014.09.016. [DOI] [PubMed] [Google Scholar]
- Meng QB, Kang WM, Yu JC, Liu YQ, Ma ZQ, Zhou L, et al. Overexpression of eukaryotic translation initiation factor 5A2 (EIF5A2) correlates with cell aggressiveness and poor survival in gastric cancer. PLoS One. 2015;10:e0119229. doi: 10.1371/journal.pone.0119229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miki T, Smith CL, Long JE, Eva A, Fleming TP. Oncogene ect2 is related to regulators of small GTP-binding proteins. Nature. 1993;362:462–5. doi: 10.1038/362462a0. [DOI] [PubMed] [Google Scholar]
- Murray NR, Kalari KR, Fields AP. Protein kinase Ciota expression and oncogenic signaling mechanisms in cancer. J Cell Physiol. 2011;226:879–87. doi: 10.1002/jcp.22463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakatsugawa M, Takahashi A, Hirohashi Y, Torigoe T, Inoda S, Murase M, et al. SOX2 is overexpressed in stem-like cells of human lung adenocarcinoma and augments the tumorigenicity. Lab Invest. 2011;91:1796–804. doi: 10.1038/labinvest.2011.140. [DOI] [PubMed] [Google Scholar]
- Nishida N, Fukuda Y, Komeda T, Kita R, Sando T, Furukawa M, et al. Amplification and overexpression of the cyclin D1 gene in aggressive human hepatocellular carcinoma. Cancer Res. 1994;54:3107–10. [PubMed] [Google Scholar]
- Pack SD, Karkera JD, Zhuang Z, Pak ED, Balan KV, Hwu P, et al. Molecular cytogenetic fingerprinting of esophageal squamous cell carcinoma by comparative genomic hybridization reveals a consistent pattern of chromosomal alterations. Genes Chromosomes Cancer. 1999;25:160–8. doi: 10.1002/(sici)1098-2264(199906)25:2<160::aid-gcc12>3.0.co;2-u. [DOI] [PubMed] [Google Scholar]
- Parker PJ, Justilien V, Riou P, Linch M, Fields AP. Atypical Protein Kinase Ciota as a human oncogene and therapeutic target. Biochem Pharmacol. 2014;88:1–11. doi: 10.1016/j.bcp.2013.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelosi G, Del Curto B, Trubia M, Nicholson AG, Manzotti M, Veronesi G, et al. 3q26 Amplification and polysomy of chromosome 3 in squamous cell lesions of the lung: a fluorescence in situ hybridization study. Clin Cancer Res. 2007;13:1995–2004. doi: 10.1158/1078-0432.CCR-06-2483. [DOI] [PubMed] [Google Scholar]
- Pere H, Tapper J, Wahlstrom T, Knuutila S, Butzow R. Distinct chromosomal imbalances in uterine serous and endometrioid carcinomas. Cancer Res. 1998;58:892–5. [PubMed] [Google Scholar]
- Petersen I, Bujard M, Petersen S, Wolf G, Goeze A, Schwendel A, et al. Patterns of chromosomal imbalances in adenocarcinoma and squamous cell carcinoma of the lung. Cancer Res. 1997;57:2331–5. [PubMed] [Google Scholar]
- Polakis P. The adenomatous polyposis coli (APC) tumor suppressor. Biochim Biophys Acta. 1997;1332:F127–47. doi: 10.1016/s0304-419x(97)00008-5. [DOI] [PubMed] [Google Scholar]
- Redon R, Muller D, Caulee K, Wanherdrick K, Abecassis J, du Manoir S. A simple specific pattern of chromosomal aberrations at early stages of head and neck squamous cell carcinomas: PIK3CA but not p63 gene as a likely target of 3q26-qter gains. Cancer Res. 2001;61:4122–9. [PubMed] [Google Scholar]
- Regala RP, Davis RK, Kunz A, Khoor A, Leitges M, Fields AP. Atypical protein kinase C{iota} is required for bronchioalveolar stem cell expansion and lung tumorigenesis. Cancer Res. 2009;69:7603–11. doi: 10.1158/0008-5472.CAN-09-2066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regala RP, Thompson EA, Fields AP. Atypical protein kinase C iota expression and aurothiomalate sensitivity in human lung cancer cells. Cancer Res. 2008;68:5888–95. doi: 10.1158/0008-5472.CAN-08-0438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regala RP, Weems C, Jamieson L, Copland JA, Thompson EA, Fields AP. Atypical protein kinase Ciota plays a critical role in human lung cancer cell growth and tumorigenicity. J Biol Chem. 2005a;280:31109–15. doi: 10.1074/jbc.M505402200. [DOI] [PubMed] [Google Scholar]
- Regala RP, Weems C, Jamieson L, Khoor A, Edell ES, Lohse CM, et al. Atypical protein kinase C iota is an oncogene in human non-small cell lung cancer. Cancer Res. 2005b;65:8905–11. doi: 10.1158/0008-5472.CAN-05-2372. [DOI] [PubMed] [Google Scholar]
- Ried T, Petersen I, Holtgreve-Grez H, Speicher MR, Schrock E, du Manoir S, et al. Mapping of multiple DNA gains and losses in primary small cell lung carcinomas by comparative genomic hybridization. Cancer Res. 1994;54:1801–6. [PubMed] [Google Scholar]
- Riemann K, Struwe H, Alakus H, Obermaier B, Schmitz KJ, Schmid KW, et al. Association of GNB4 intron-1 haplotypes with survival in patients with UICC stage III and IV colorectal carcinoma. Anticancer Res. 2009;29:1271–4. [PubMed] [Google Scholar]
- Riemann K, Struwe H, Eisenhardt A, Obermaier B, Schmid KW, Siffert W. Characterization of intron-1 haplotypes of the G protein beta 4 subunit gene--association with survival and progression in patients with urothelial bladder carcinoma. Pharmacogenet Genomics. 2008;18:999–1008. doi: 10.1097/FPC.0b013e3283117d79. [DOI] [PubMed] [Google Scholar]
- Rodon J, Dienstmann R, Serra V, Tabernero J. Development of PI3K inhibitors: lessons learned from early clinical trials. Nature reviews Clinical oncology. 2013;10:143–53. doi: 10.1038/nrclinonc.2013.10. [DOI] [PubMed] [Google Scholar]
- Rooney PH, Murray GI, Stevenson DA, Haites NE, Cassidy J, McLeod HL. Comparative genomic hybridization and chromosomal instability in solid tumours. Br J Cancer. 1999;80:862–73. doi: 10.1038/sj.bjc.6690433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roychowdhury A, Sharma R, Kumar S. Recent advances in the discovery of small molecule mTOR inhibitors. Future medicinal chemistry. 2010;2:1577–89. doi: 10.4155/fmc.10.233. [DOI] [PubMed] [Google Scholar]
- Sakai N, Kajiyama Y, Iwanuma Y, Tomita N, Amano T, Isayama F, et al. Study of abnormal chromosome regions in esophageal squamous cell carcinoma by comparative genomic hybridization: relationship of lymph node metastasis and distant metastasis to selected abnormal regions. Dis Esophagus. 2010;23:415–21. doi: 10.1111/j.1442-2050.2009.01026.x. [DOI] [PubMed] [Google Scholar]
- Salhia B, Tran NL, Chan A, Wolf A, Nakada M, Rutka F, et al. The guanine nucleotide exchange factors trio, Ect2, and Vav3 mediate the invasive behavior of glioblastoma. Am J Pathol. 2008;173:1828–38. doi: 10.2353/ajpath.2008.080043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salvesen HB, Carter SL, Mannelqvist M, Dutt A, Getz G, Stefansson IM, et al. Integrated genomic profiling of endometrial carcinoma associates aggressive tumors with indicators of PI3 kinase activation. Proc Natl Acad Sci U S A. 2009;106:4834–9. doi: 10.1073/pnas.0806514106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuels Y, Velculescu VE. Oncogenic mutations of PIK3CA in human cancers. Cell Cycle. 2004;3:1221–4. doi: 10.4161/cc.3.10.1164. [DOI] [PubMed] [Google Scholar]
- Samuels Y, Wang Z, Bardelli A, Silliman N, Ptak J, Szabo S, et al. High frequency of mutations of the PIK3CA gene in human cancers. Science. 2004;304:554. doi: 10.1126/science.1096502. [DOI] [PubMed] [Google Scholar]
- Sano M, Genkai N, Yajima N, Tsuchiya N, Homma J, Tanaka R, et al. Expression level of ECT2 proto-oncogene correlates with prognosis in glioma patients. Oncol Rep. 2006;16:1093–8. [PubMed] [Google Scholar]
- Sarkaria I, Oc P, Talbot SG, Reddy PG, Ngai I, Maghami E, et al. Squamous cell carcinoma related oncogene/DCUN1D1 is highly conserved and activated by amplification in squamous cell carcinomas. Cancer Res. 2006;66:9437–44. doi: 10.1158/0008-5472.CAN-06-2074. [DOI] [PubMed] [Google Scholar]
- Sarker KP, Wilson SM, Bonni S. SnoN is a cell type-specific mediator of transforming growth factor-beta responses. J Biol Chem. 2005;280:13037–46. doi: 10.1074/jbc.M409367200. [DOI] [PubMed] [Google Scholar]
- Scotti ML, Bamlet WR, Smyrk TC, Fields AP, Murray NR. Protein kinase Ciota is required for pancreatic cancer cell transformed growth and tumorigenesis. Cancer Res. 2010;70:2064–74. doi: 10.1158/0008-5472.CAN-09-2684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serra V, Markman B, Scaltriti M, Eichhorn PJ, Valero V, Guzman M, et al. NVP-BEZ235, a dual PI3K/mTOR inhibitor, prevents PI3K signaling and inhibits the growth of cancer cells with activating PI3K mutations. Cancer Res. 2008;68:8022–30. doi: 10.1158/0008-5472.CAN-08-1385. [DOI] [PubMed] [Google Scholar]
- Shayesteh L, Lu Y, Kuo WL, Baldocchi R, Godfrey T, Collins C, et al. PIK3CA is implicated as an oncogene in ovarian cancer. Nat Genet. 1999;21:99–102. doi: 10.1038/5042. [DOI] [PubMed] [Google Scholar]
- Shi J, Yao D, Liu W, Wang N, Lv H, Zhang G, et al. Highly frequent PIK3CA amplification is associated with poor prognosis in gastric cancer. BMC Cancer. 2012;12:50. doi: 10.1186/1471-2407-12-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sholl LM, Barletta JA, Yeap BY, Chirieac LR, Hornick JL. Sox2 protein expression is an independent poor prognostic indicator in stage I lung adenocarcinoma. Am J Surg Pathol. 2010;34:1193–8. doi: 10.1097/PAS.0b013e3181e5e024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh B, Stoffel A, Gogineni S, Poluri A, Pfister DG, Shaha AR, et al. Amplification of the 3q26.3 locus is associated with progression to invasive cancer and is a negative prognostic factor in head and neck squamous cell carcinomas. Am J Pathol. 2002;161:365–71. doi: 10.1016/S0002-9440(10)64191-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonoda G, Palazzo J, du Manoir S, Godwin AK, Feder M, Yakushiji M, et al. Comparative genomic hybridization detects frequent overrepresentation of chromosomal material from 3q26, 8q24, and 20q13 in human ovarian carcinomas. Genes Chromosomes Cancer. 1997;20:320–8. [PubMed] [Google Scholar]
- Soong BW, Huang YH, Tsai PC, Huang CC, Pan HC, Lu YC, et al. Exome sequencing identifies GNB4 mutations as a cause of dominant intermediate Charcot-Marie-Tooth disease. Am J Hum Genet. 2013;92:422–30. doi: 10.1016/j.ajhg.2013.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speicher MR, Howe C, Crotty P, du Manoir S, Costa J, Ward DC. Comparative genomic hybridization detects novel deletions and amplifications in head and neck squamous cell carcinomas. Cancer Res. 1995;55:1010–3. [PubMed] [Google Scholar]
- Stallings-Mann M, Jamieson L, Regala RP, Weems C, Murray NR, Fields AP. A novel small-molecule inhibitor of protein kinase Ciota blocks transformed growth of non-small-cell lung cancer cells. Cancer Res. 2006;66:1767–74. doi: 10.1158/0008-5472.CAN-05-3405. [DOI] [PubMed] [Google Scholar]
- Suda T, Hama T, Kondo S, Yuza Y, Yoshikawa M, Urashima M, et al. Copy number amplification of the PIK3CA gene is associated with poor prognosis in non-lymph node metastatic head and neck squamous cell carcinoma. BMC Cancer. 2012;12:416. doi: 10.1186/1471-2407-12-416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugita M, Tanaka N, Davidson S, Sekiya S, Varella-Garcia M, West J, et al. Molecular definition of a small amplification domain within 3q26 in tumors of cervix, ovary, and lung. Cancer Genet Cytogenet. 2000;117:9–18. doi: 10.1016/s0165-4608(99)00135-1. [DOI] [PubMed] [Google Scholar]
- Takagawa R, Akimoto K, Ichikawa Y, Akiyama H, Kojima Y, Ishiguro H, et al. High expression of atypical protein kinase C lambda/iota in gastric cancer as a prognostic factor for recurrence. Ann Surg Oncol. 2010;17:81–8. doi: 10.1245/s10434-009-0708-x. [DOI] [PubMed] [Google Scholar]
- Tatsumoto T, Xie X, Blumenthal R, Okamoto I, Miki T. Human ECT2 is an exchange factor for Rho GTPases, phosphorylated in G2/M phases, and involved in cytokinesis. J Cell Biol. 1999;147:921–8. doi: 10.1083/jcb.147.5.921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas RK, Greulich H, Yuza Y, Lee JC, Tengs T, Feng W, et al. Detection of oncogenic mutations in the EGFR gene in lung adenocarcinoma with differential sensitivity to EGFR tyrosine kinase inhibitors. Cold Spring Harb Symp Quant Biol. 2005;70:73–81. doi: 10.1101/sqb.2005.70.056. [DOI] [PubMed] [Google Scholar]
- Tie J, Desai J. Targeting BRAF mutant metastatic colorectal cancer: clinical implications and emerging therapeutic strategies. Target Oncol. 2015;10:179–88. doi: 10.1007/s11523-014-0330-0. [DOI] [PubMed] [Google Scholar]
- Toschi L, Finocchiaro G, Nguyen TT, Skokan MC, Giordano L, Gianoncelli L, et al. Increased SOX2 gene copy number is associated with FGFR1 and PIK3CA gene gain in non-small cell lung cancer and predicts improved survival in early stage disease. PLoS One. 2014;9:e95303. doi: 10.1371/journal.pone.0095303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umar A, Kang H, Timmermans AM, Look MP, Meijer-van Gelder ME, den Bakker MA, et al. Identification of a putative protein profile associated with tamoxifen therapy resistance in breast cancer. Mol Cell Proteomics. 2009;8:1278–94. doi: 10.1074/mcp.M800493-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazquez-Mena O, Medina-Martinez I, Juarez-Torres E, Barron V, Espinosa A, Villegas-Sepulveda N, et al. Amplified genes may be overexpressed, unchanged, or downregulated in cervical cancer cell lines. PLoS One. 2012;7:e32667. doi: 10.1371/journal.pone.0032667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velcheti V, Schalper K, Yao X, Cheng H, Kocoglu M, Dhodapkar K, et al. High SOX2 levels predict better outcome in non-small cell lung carcinomas. PLoS One. 2013;8:e61427. doi: 10.1371/journal.pone.0061427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vorhagen S, Niessen CM. Mammalian aPKC/Par polarity complex mediated regulation of epithelial division orientation and cell fate. Exp Cell Res. 2014;328:296–302. doi: 10.1016/j.yexcr.2014.08.008. [DOI] [PubMed] [Google Scholar]
- Wang J, Chai YL, Wang T, Liu JH, Dai PG, Liu Z. Genetic alterations of PIK3CA and tumor response in patients with locally advanced cervical squamous cell carcinoma treated with cisplatin-based concurrent chemoradiotherapy. Exp Mol Pathol. 2015a;98:407–10. doi: 10.1016/j.yexmp.2015.03.014. [DOI] [PubMed] [Google Scholar]
- Wang J, Qian J, Hoeksema MD, Zou Y, Espinosa AV, Rahman SM, et al. Integrative genomics analysis identifies candidate drivers at 3q26-29 amplicon in squamous cell carcinoma of the lung. Clin Cancer Res. 2013a;19:5580–90. doi: 10.1158/1078-0432.CCR-13-0594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Tie J, Wang R, Hu F, Gao L, Wang W, et al. SOX2, a predictor of survival in gastric cancer, inhibits cell proliferation and metastasis by regulating PTEN. Cancer Lett. 2015b;358:210–9. doi: 10.1016/j.canlet.2014.12.045. [DOI] [PubMed] [Google Scholar]
- Wang Y, Hill KS, Fields AP. PKCiota maintains a tumor-initiating cell phenotype that is required for ovarian tumorigenesis. Mol Cancer Res. 2013b;11:1624–35. doi: 10.1158/1541-7786.MCR-13-0371-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Zhang Z, Yan Y, Lemon WJ, LaRegina M, Morrison C, et al. A Chemically Induced Model for Squamous Cell Carcinoma of the Lung in Mice: Histopathology and Strain Susceptibility. Cancer Res. 2004;64:1647–54. doi: 10.1158/0008-5472.can-03-3273. [DOI] [PubMed] [Google Scholar]
- Weichert W, Gekeler V, Denkert C, Dietel M, Hauptmann S. Protein kinase C isoform expression in ovarian carcinoma correlates with indicators of poor prognosis. Int J Oncol. 2003;23:633–9. [PubMed] [Google Scholar]
- Whitaker HC, Shiong LL, Kay JD, Gronberg H, Warren AY, Seipel A, et al. N-acetyl-L-aspartyl-L-glutamate peptidase-like 2 is overexpressed in cancer and promotes a pro-migratory and pro-metastatic phenotype. Oncogene. 2014;33:5274–87. doi: 10.1038/onc.2013.464. [DOI] [PubMed] [Google Scholar]
- Woenckhaus J, Steger K, Werner E, Fenic I, Gamerdinger U, Dreyer T, et al. Genomic gain of PIK3CA and increased expression of p110alpha are associated with progression of dysplasia into invasive squamous cell carcinoma. J Pathol. 2002;198:335–42. doi: 10.1002/path.1207. [DOI] [PubMed] [Google Scholar]
- Wong AJ, Bigner SH, Bigner DD, Kinzler KW, Hamilton SR, Vogelstein B. Increased expression of the epidermal growth factor receptor gene in malignant gliomas is invariably associated with gene amplification. Proc Natl Acad Sci U S A. 1987;84:6899–903. doi: 10.1073/pnas.84.19.6899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G, Mambo E, Guo Z, Hu S, Huang X, Gollin SM, et al. Uncommon mutation, but common amplifications, of the PIK3CA gene in thyroid tumors. J Clin Endocrinol Metab. 2005;90:4688–93. doi: 10.1210/jc.2004-2281. [DOI] [PubMed] [Google Scholar]
- Xiang R, Liao D, Cheng T, Zhou H, Shi Q, Chuang TS, et al. Downregulation of transcription factor SOX2 in cancer stem cells suppresses growth and metastasis of lung cancer. Br J Cancer. 2011;104:1410–7. doi: 10.1038/bjc.2011.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto H, Shigematsu H, Nomura M, Lockwood WW, Sato M, Okumura N, et al. PIK3CA mutations and copy number gains in human lung cancers. Cancer Res. 2008;68:6913–21. doi: 10.1158/0008-5472.CAN-07-5084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Li XD, Zhou Y, Ban X, Zeng TT, Li L, et al. Stemness and chemotherapeutic drug resistance induced by EIF5A2 overexpression in esophageal squamous cell carcinoma. Oncotarget. 2015 doi: 10.18632/oncotarget.4581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang YL, Chu JY, Luo ML, Wu YP, Zhang Y, Feng YB, et al. Amplification of PRKCI, located in 3q26, is associated with lymph node metastasis in esophageal squamous cell carcinoma. Genes Chromosomes Cancer. 2008;47:127–36. doi: 10.1002/gcc.20514. [DOI] [PubMed] [Google Scholar]
- Yao Q, Du J, Lin J, Luo Y, Wang Y, Liu Y, et al. Prognostic significance of TRAIL signalling molecules in cervical squamous cell carcinoma. J Clin Pathol. 2015 doi: 10.1136/jclinpath-2014-202811. [DOI] [PubMed] [Google Scholar]
- Yatsula B, Lin S, Read AJ, Poholek A, Yates K, Yue D, et al. Identification of binding sites of EVI1 in mammalian cells. J Biol Chem. 2005;280:30712–22. doi: 10.1074/jbc.M504293200. [DOI] [PubMed] [Google Scholar]
- You C, Sandalcioglu IE, Dammann P, Felbor U, Sure U, Zhu Y. Loss of CCM3 impairs DLL4-Notch signalling: implication in endothelial angiogenesis and in inherited cerebral cavernous malformations. J Cell Mol Med. 2013;17:407–18. doi: 10.1111/jcmm.12022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan P, Kadara H, Behrens C, Tang X, Woods D, Solis LM, et al. Sex determining region Y-Box 2 (SOX2) is a potential cell-lineage gene highly expressed in the pathogenesis of squamous cell carcinomas of the lung. PLoS One. 2010;5:e9112. doi: 10.1371/journal.pone.0009112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan TL, Cantley LC. PI3K pathway alterations in cancer: variations on a theme. Oncogene. 2008;27:5497–510. doi: 10.1038/onc.2008.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Ma X, Peng S, Nan X, Zhao H. Differential expression of MST4, STK25 and PDCD10 between benign prostatic hyperplasia and prostate cancer. Int J Clin Exp Pathol. 2014;7:8105–11. [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Zhang P, Wei Y, Piao HL, Wang W, Maddika S, et al. Deubiquitylation and stabilization of PTEN by USP13. Nat Cell Biol. 2013;15:1486–94. doi: 10.1038/ncb2874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Huang J, Yang N, Liang S, Barchetti A, Giannakakis A, et al. Integrative genomic analysis of protein kinase C (PKC) family identifies PKCiota as a biomarker and potential oncogene in ovarian carcinoma. Cancer Res. 2006;66:4627–35. doi: 10.1158/0008-5472.CAN-05-4527. [DOI] [PubMed] [Google Scholar]
- Zhang ML, Lu S, Zhou L, Zheng SS. Correlation between ECT2 gene expression and methylation change of ECT2 promoter region in pancreatic cancer. Hepatobiliary Pancreat Dis Int. 2008;7:533–8. [PubMed] [Google Scholar]
- Zhao X, Fiske B, Kawakami A, Li J, Fisher DE. Regulation of MITF stability by the USP13 deubiquitinase. Nat Commun. 2011;2:414. doi: 10.1038/ncomms1421. [DOI] [PubMed] [Google Scholar]
- Zhou B, Chen H, Wei D, Kuang Y, Zhao X, Li G, et al. A novel miR-219-SMC4-JAK2/Stat3 regulatory pathway in human hepatocellular carcinoma. J Exp Clin Cancer Res. 2014;33:55. doi: 10.1186/1756-9966-33-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou B, Yuan T, Liu M, Liu H, Xie J, Shen Y, et al. Overexpression of the structural maintenance of chromosome 4 protein is associated with tumor de-differentiation, advanced stage and vascular invasion of primary liver cancer. Oncol Rep. 2012;28:1263–8. doi: 10.3892/or.2012.1929. [DOI] [PubMed] [Google Scholar]