Abstract
Simultaneous detection of multiple biomarkers, such as extracellular signaling molecules, is a critical aspect in disease profiling and diagnostics. Precise positioning of antibodies on surfaces, especially at the micro- and nano- scale, is important for the improvement of assays, biosensors, and diagnostics on the molecular level, and therefore, the pursuit of device miniaturization for parallel, fast, low-volume assays is a continuing challenge. Here, we describe a multiplexed cytokine immunoassay utilizing electron beam lithography and a trehalose glycopolymer as a resist for the direct writing of antibodies on silicon substrates allowing for micro- and nano-scale precision of protein immobilization. Specifically, anti-interleukin 6 (IL-6) and anti-tumor necrosis factor alpha (TNFα) antibodies were directly patterned. Retention of the specific binding properties of the patterned antibodies was shown by the capture of secreted cytokines from stimulated RAW 264.7 macrophages. A sandwich immunoassay was employed using gold nanoparticles and enhancement with silver for the detection and visualization of bound cytokines to the patterns by localized surface plasmon resonance detected with dark field microscopy. Multiplexing with both IL-6 and TNFα on a single chip was also successfully demonstrated with high specificity and in relevant cell culture conditions and at different times after cell stimulation. The direct fabrication of capture antibody patterns for cytokine detection described here could be useful for biosensing applications.
Keywords: electron beam lithography, antibody patterning, trehalose glycopolymer, biosensor, localized surface plasmon resonance, dark field microscopy
Introduction
Extracellular signaling molecules are secreted by cells in response to a variety of factors, including temperature, addition of drugs, chemicals, and any other changes in the surrounding environment. Therefore, knowing what types of molecules and the amounts secreted would provide important insight and information on the functional state of the cell. For example, interleukin-6 (IL-6) and tumor necrosis factor alpha (TNFα) are common cell signaling proteins (cytokines) that influence many aspects of the immune and inflammatory response. These molecules are also important biomarkers; elevated IL-6 or TNFα serum levels are associated with various diseases, such as rheumatoid arthritis and prostate cancer.1, 2 Thus, detection of these molecules secreted from cells is an important undertaking.
Currently available and commonly used methods for cytokine detection include intracellular cytokine cytometry (ICC) and enzyme-linked immunospot (ELISPOT) assays.3 ICC involves inhibiting stimulated cells from secreting cytokines, and then allowing labeled anti-cytokine antibodies to permeate into the cells. This is coupled by analysis by flow cytometry. In the ELISPOT assay, cells are plated directly into wells coated with anti-cytokine antibodies, and the released cytokines are visualized by an enzyme-labeled detection antibody. An important contribution towards improving biosensor technology aims to miniaturize sample volumes and for inline detection from live cells, where micro- and nano-fabrication approaches become highly desired. The resulting benefits for miniaturization are better sensitivities and high-throughput analyses. Examples of developing technologies for the detection of secreted cytokines from living cells include a microfluidics platform for immobilizing antibodies,4, 5 photolithography to fabricate gold electrodes for modification with aptamers,6 and nanoimprint lithography to generate structures for cytokine biosensing.7
There are a variety of patterning techniques that have been used for biomolecule immobilization with micro- and nano- scale features.8–10 Some commonly used techniques include ink-jet printing,11 stamping techniques such as microcontact printing12, 13 and nanoimprint lithography,14, 15 scanning probe microscopy techniques such as dip-pen nanolithography (DPN),16, 17 polymer pen lithography (PPL),18 and electron beam lithography (EBL).19–21 Expectedly, each technique has advantages and limitations. Stamping techniques allow for the rapid generation of patterns, but require template fabrication and preparing dense patterns with different biomolecules can be difficult. DPN and PPL enable direct write methods with nanoscale spacing between features, but are limited in that most often they are serial processes. EBL is also a serial process and suffers from significantly longer exposure times; however it allows for the generation of user-designed patterns with high resolution and nanoscale alignment capability, allowing for multiplexing and high density patterns.22
Direct writing of proteins by EBL presents a challenge due to the nature of the harsh processing conditions. EBL requires high vacuum and involves high energy irradiation with electrons for patterning, which are not ideal for maintaining protein bioactivity. Previously, we reported that polymers with trehalose side chains were able to stabilize various proteins against environmental stresses such as heat and lyophilization.23–25 Recently, we have reported the use of a trehalose glycopolymer (PolyProtek) as a novel aqueous-based resist material that enables the direct writing of proteins by EBL.26 PolyProtek was shown to stabilize proteins under high vacuum and direct exposure to high energy irradiation. Another material, silk, was also used as a positive or negative resist for protein patterning with EBL as shown by Kim et al.27 Herein, we demonstrate the ability to use PolyProtek for direct write EBL of IL-6 or TNFα antibodies for capture of these important biomolecules secreted from stimulated macrophages. The patterns were visualized via dark field microscopy exploiting the surface plasmon resonance of silver enhanced gold nanoparticle secondary antibodies.
Results and Discussion
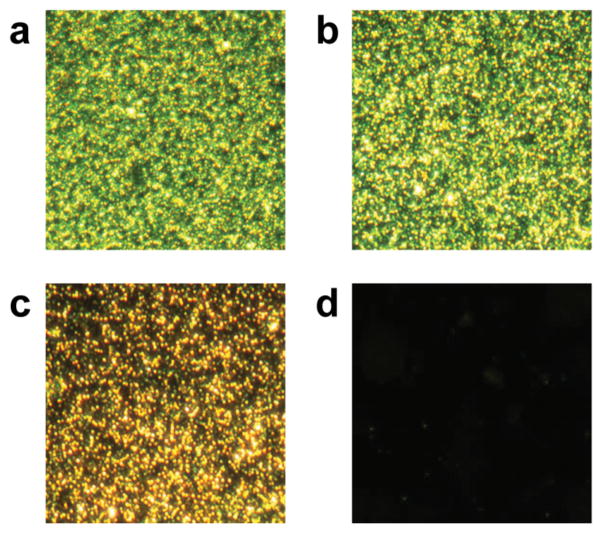
The amount of non-specific adsorption is an important consideration for any biosensor application because background adsorption increases noise and decreases signal thus reducing sensitivity.28 Therefore, surfaces were first passivated to prevent non-specific adsorption and ensure specific recognition. This was achieved by the single-step surface modification of grafting poly(ethylene glycol) (PEG)-silane onto silicon surfaces. This passivation strategy allows for ultrathin and stable protein-repellent PEG monolayers.29 The surface immunoassay described in this report, employed gold nanoparticle-labeled analytes for detection. Gold nanoparticles generate a localized surface plasmon resonance signal that can be simply visualized using a dark field optical microscope. Reducing the non-specific adsorption of gold nanoparticles and primary antibodies helps eliminate background noise and thus improve signal and sensitivity. The amount of non-specifically adsorbed neutravidin-conjugated gold nanoparticles was evaluated for plain silicon substrates or silicon coated with PEG-silane of different molecular weights (Figure 1). All surfaces were incubated with 30 nm neutravidin-conjugated gold nanoparticle solutions for 18 hours. Compared to the piranha-cleaned silicon substrates (Figure 1a), the PEG 400 coating also had considerable fouling (Figure 1b) while PEG 2000 showed a slight reduction in protein adsorption (Figure 1c). A PEG-silane of molecular weight 5 kDa was required to significantly reduce non-specific adsorption to barely detectable levels (Figure 1d). Specifically, a 95% reduction in protein adsorption was observed compared to the bare silicon substrate. Therefore, the 5 kDa PEG silane coating was utilized in all subsequent experiments.
Figure 1.
Non-specific adsorption of neutravidin-conjugated 30 nm gold nanoparticles to treated silicon substrates: a) bare silicon, b) PEG 400-silane, c) PEG 2000-silane, and d) PEG 5000-silane.
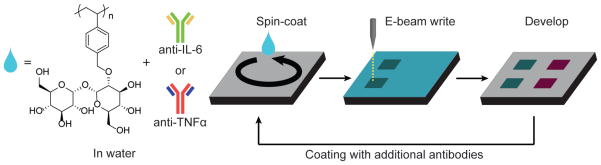
The process of fabricating antibody patterns by direct electron beam lithography is depicted in Figure 2. PEG-silane coated silicon substrates were spin-coated with an aqueous solution containing the antibody, 0.5% wt/vol PolyProtek, and 1 mM ascorbic acid. The resulting thickness for PEG-silane was 26.8 ± 1.1 Å, and the spin-coated antibody layer was 205.3 ± 1.9 Å, as measured by ellipsometry. When irradiated by electrons, the trehalose polymer cross-links to the PEG-coated silicon substrate by a mechanism similarly observed for other polymers.30, 31 The cross-linking results in a trehalose-based hydrogel, which helps to stabilize and immobilize the protein. Ascorbic acid is a known radical scavenger,32 and therefore can help protect proteins from e-beam radiation damage.26 We verified this in our system by patterning without or with 1 and 2 mM ascorbic acid, finding that 1 mM ascorbic acid provided the highest signal by dark field microscopy (Figure S1). Thus, 1 mM ascorbic acid was added to the resist/protein solutions hereafter. For multiplexed antibody patterns, the chips were fabricated by re-subjecting the substrate to another round of e-beam processing of spin-coating with a different antibody, aligning, and writing steps.
Figure 2.
Direct electron beam patterning process for generating antibody patterns. An aqueous solution containing PolyProtek, ascorbic acid, and target antibody is spin-coated on a PEG-silane coated substrate and subsequently patterned by electron beam lithography. Uncross-linked areas are washed away. Multicomponent antibody patterns can be generated by spin-coating a new layer with a different antibody, alignment and subsequent e-beam patterning and rinsing.
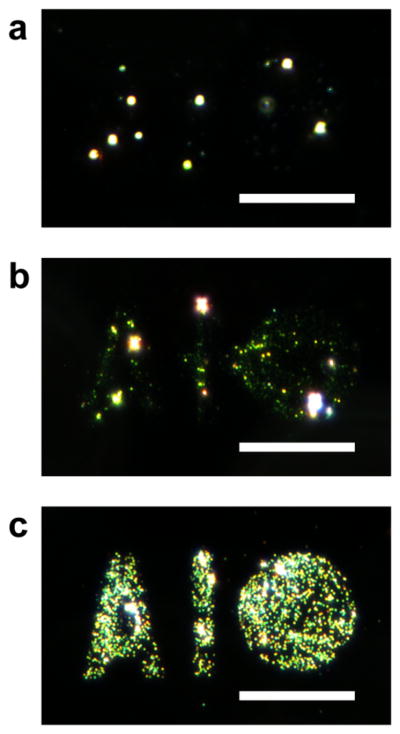
The direct e-beam writing and assay of antibodies was first evaluated using human immunoglobulin G (IgG) to optimize various parameters and components of the immunoassay system. Silicon wafers were spin-coated with a solution containing 0.5% wt/vol PolyProtek, 1 mM ascorbic acid, and 1 mg/mL human IgG. The human IgG coated surfaces was patterned in an “A1/Circle” shape (Figure 3c) with an area dose of 25 μC/cm2 and after processing, the chips were sequentially incubated with biotinylated anti-human IgG and neutravidin-conjugated 30 nm gold nanoparticles, followed by silver enhancement. Dark field microscopy images were taken from a reference pattern without neutravidin-conjugated gold nanoparticles (Figure 3a), with bound gold nanoparticles (Figure 3b) and the silver enhanced gold nanoparticles pattern (Figure 3c). The localized surface plasmon resonance (LSPR) signal was observed for the gold nanoparticle sample; however silver enhancement greatly increased signal and thus sensitivity, and thus this process was utilized for all further studies.
Figure 3.
Dark field micrographs for the sandwich assay of human IgG patterns. IgG patterns were incubated with biotin anti-human IgG, neutravidin-conjugated gold nanoparticles, and silver enhanced. Patterns are shown a) before gold nanoparticle incubation, b) after gold nanoparticle incubation, and c) after silver enhancement. Scale bars = 35 μm.
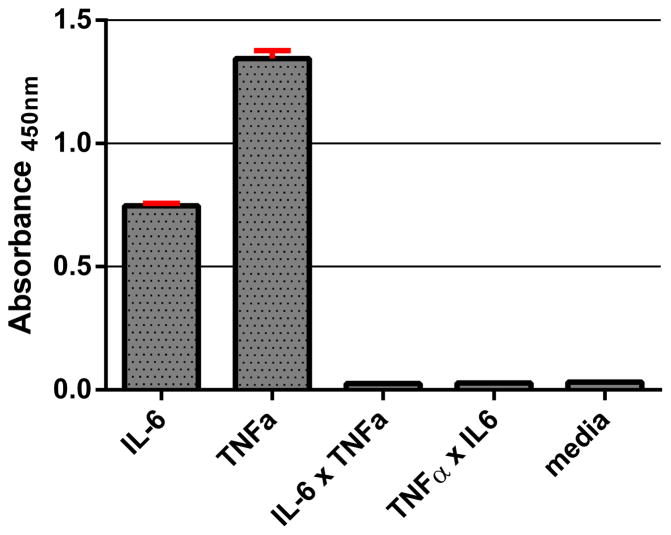
Cytokines IL-6 and TNFα were chosen for multiplexed detection by the surface immunoassay because these secreted cell signaling molecules are commonly associated with macrophage activation. Cultured macrophages readily secrete the signaling proteins to regulate the immune response upon stimulation. RAW 264.7 mouse macrophages were stimulated with lipopolysaccharide (LPS), a component of the outer membrane in Gram-negative bacteria, to secrete IL-6, TNFα, and many other cytokines into the cell culture media. An enzyme-linked immunosorbent assay (ELISA) was performed to detect the presence of IL-6 and TNFα in media (Figure 4). Importantly, the results show that there are undetectable levels of cytokines from unstimulated macrophages and that the secondary antibodies have high specificity for the target antibody and no cross-reactivity. These properties are necessary for translating the components towards a multiplex surface immunoassay system.
Figure 4.
ELISA detection of IL-6 and TNFα from LPS-stimulated RAW 264.7 macrophages (5 × 105 cells). IL-6 × TNFα represents anti-IL-6 capture antibody and biotinylated anti-TNFα antibody, and TNFα × IL-6 represents anti-TNFα capture antibody and biotinylated anti-IL-6 antibody. Media represents the amount of detected cytokines from unstimulated macrophages.
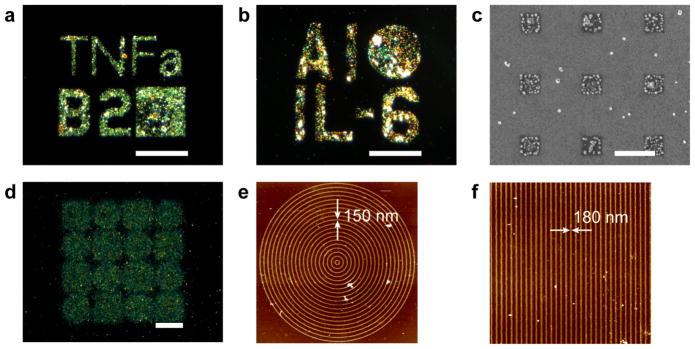
Electron beam lithography was first used for direct writing of anti-IL-6 and anti-TNFα antibody patterns on separately patterned chips. The antibody patterns were sequentially incubated with undiluted cell culture media from RAW 264.7 macrophages that were stimulated with LPS for 24 hours. Then, either biotinylated anti-IL-6 or biotinylated anti-TNFα was added, followed by 30 nm neutravidin-conjugated gold nanoparticles and the silver enhancement step. The signal depended on the concentration of antibody used during spin-coating. The concentration of antibody tested ranged from 1 to 5 μM, with 5 μM resulting in the highest observed signal (Figure S2). Not surprisingly, with increased antibody concentrations, there was an increase in signal, which is likely related to a higher density of bioactive antibodies following electron beam processing. Both TNFα and IL-6 capture was observed by dark field microscopy with silver-enhanced gold nanoparticles bound to micro sized patterns of defined texts and shapes (Figure 5a, b). The gold nanoparticles were visualized on submicron patterns with scanning electron microscopy, where the gold antibodies were observed as dots on 500×500 nm-sized squares (Figure 5c). Furthermore, nanopatterns were fabricated. Anti-TNFα nanopatterns with line widths of 150–180 nm were observed by atomic force microscopy (AFM, Fig 5e, f). The line patterns were fabricated in concentric shapes that were visualized by dark field microscopy as squares and circles (Figure 5d) demonstrating that that nanopatterns of active anti-TNFα were achieved. These results confirm that the immunoassay following the direct write of antibodies by EBL can be used for the detection of cytokines in complex cell culture media. Further, these patterns of shapes, letters, and numbers demonstrate the capability to generate arbitrary patterns with high resolution.
Figure 5.
Cytokine detection with a bound silver enhanced gold nanoparticles immunoassay on surface immobilized micro- and nano- patterns. Two cytokines, a) TNFα and b) IL-6 were detected from cell media of RAW 264.7 macrophages, scale bars = 35 μm. c) electron micrograph of anti-TNFα submicron patterns, scale bar = 1 μm. Anti-TNFα nanopatterns showing d) dark field micrograph cytokine detection immunoassay of circles and squares with nanometer line widths, scale bar = 20 μm, e) AFM of one of the circle patterns with 150 nm line widths, and f) AFM of one of the square patterns with 180 nm line widths indicated by the arrows.
Sensitivity to cytokine concentrations and detection limit are both important considerations for immunoassays. The levels of TNFα and IL-6 in LPS-stimulated macrophage culture media were quantified by ELISA and used as a benchmark. From 5 × 105 cultured macrophages, the amount of TNFα and IL-6 detected was 43 ng/mL and 53 ng/mL, respectively. Patterns of anti-TNFα and anti-IL-6 were incubated in varying dilutions ranging from 200 ng/mL to 0 of TNFα or IL-6 from cell media of stimulated macrophages and further developed. Figure 6 and Figure S3 shows the sensitivity of the immunoassay to TNFα and IL-6, respectively. Signals were strongly observed at TNFα and IL-6 of 200 ng/mL and 40 ng/mL concentrations, while detection was visible above 5 pg/mL for TNFα and 50 pg/mL for IL-6. This is comparable to the detection limit of ELISA, which from the manufacturer’s kit (eBioscience) has a standard curve range of 4 – 500 pg/mL.
Figure 6.
Detection sensitivity of anti-TNFα patterns to varying concentrations of TNFα in media: a) 200 ng/mL, b) 40 ng/mL, c) 500 pg/mL, d) 50 pg/mL, e) 5 pg/mL, and f) 0. Scale bars = 20 μm.
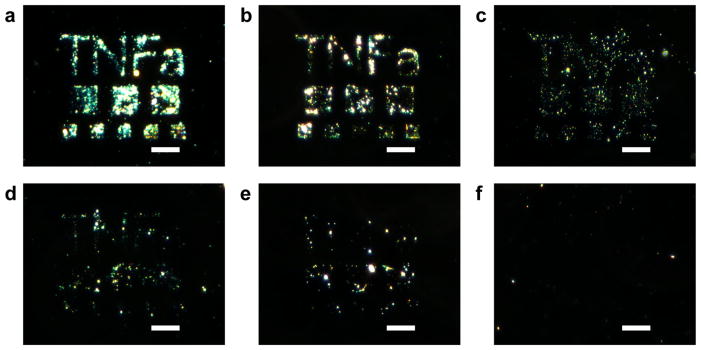
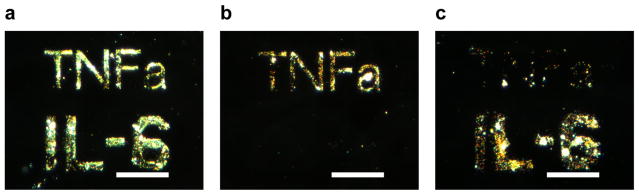
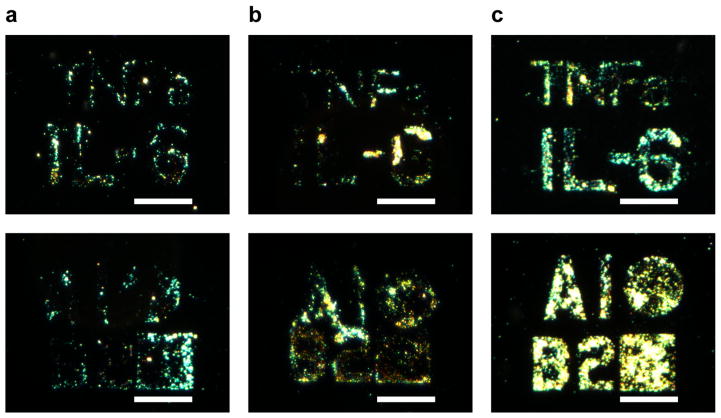
Multiplexed antibody patterns to detect both IL-6 and TNFα were then investigated. First, anti- TNFα patterns were generated and the process of spin-coating with anti-TNFα, writing, and rinsing on the same substrate was repeated four times. The signal-to-noise of the patterns was comparable between the four processing and exposure cycles (Figure S4) suggesting that the process is amenable to multiplex patterning. Taking advantage of the alignment capabilities of electron beam lithography, next anti-IL-6 and anti-TNFα patterns of “TNFa” and “IL-6” were generated in close proximity. Following the direct writing of anti-TNFα, another round of patterning was performed with anti-IL-6 to achieve multicomponent antibody patterns. The detection of both IL-6 and TNFα was observed by visualization of their associated patterns (Figure 7a). The surface immunoassay also demonstrates high specificity towards both cytokines, with slight cross-reactivity against IL-6. The corresponding patterns were visible when stained for only TNFα (Figure 7b) or for only IL-6 (Figure 7c).
Figure 7.
Dark field micrographs of multiple antibody patterning and cytokine detection of IL-6 and TNFα. The generated patterns were stained to detect for a) both IL-6 and TNFα, or selectively for: b) TNFα, and c) IL-6. Scale bars = 35 μm.
Finally, the multiplexed patterns were used to monitor IL-6 and TNFα secretion over time. Multicomponent IL-6 and TNFα patterns were directly incubated with RAW 264.7 macrophages. Upon addition of LPS to the cultured macrophages, the patterned chips (with “TNFa”/B2square shape for TNFα and “IL-6”/A1circle shape for IL-6) were then removed from cell cultures at various time points over the course of 24 hours. Subsequent development of the immunoassay showed an expected increase in LSPR signals and thus developing visualization of the patterns incubated from 2 to 24 hours (Figure 8). This steady increase in IL-6 and TNFα secretion over 24 hours is in agreement with levels reported in literature.33
Figure 8.
Multiplexed detection of IL-6 and TNFα from stimulated RAW 264.7 macrophages over time. Chips were directly incubated in LPS-stimulated macrophages at time points: a) 2 hr, b) 12 hr, and c) 24 hr. Scale bars = 35 μm.
The detection of both cytokines, as well as the capability to selectively detect one specific signaling protein from cellular milieu, is essential for a multiplexed immunoassay system. Using a trehalose glycopolymer, the antibodies retained their binding activity even after subjection to repeated electron beam processing conditions, such as vacuum and rinsing cycles, exemplifying the use of PolyProtek as a stabilizing resist. Furthermore, low amounts of non-specific adsorption were observed on the e-beam generated patterns. Specifically, the amount of gold nanoparticles bound to the opposing cytokine pattern was greatly reduced (Figure 7b,c). The direct write assay was capable of detection as low as cytokine concentrations of 5 pg/mL and could detect increasing concentrations of the secreted proteins over time from live cells. The combination of all these properties demonstrates that multiplexed patterns for the detection of two different cytokines can be successfully patterned with this technique, and detection in relevant cell culture conditions on these patterns is possible. Electron beam lithography is a serial process, but there are several new techniques in development. For example, stencil masks or character project technology allow for generation of complex shapes with one exposure, and multiple electron beams for parallel processes are being investigated.34, 35 The process we describe herein should also be applicable to these technologies.
Conclusions
We have successfully demonstrated a cytokine detection method by directly patterning antibodies onto surfaces utilizing PolyProtek and EBL. The patterned IL-6 and TNFα antibodies retained their recognition properties, allowing for binding to the respective cytokines released from LPS-stimulated macrophages. Capture of the cytokines was successfully detected via a sandwich immunoassay visualized from LSPR of silver-enhanced gold nanoparticles bound to the patterns. Multiplexed cytokine detection was also shown to specifically visualize IL-6 and TNFα concentrations in relevant cell culture conditions. These results demonstrate the potential of this patterning technique for developing biosensors and diagnostic assays.
Materials and Methods
Materials
5 kDa mPEG-silane was purchased from Creative PEGWorks. Silicon wafers were purchased from Cemat Silicon S.A. Human IgG and Alexa Fluor 488 donkey anti-human IgG were purchased from Jackson ImmunoResearch. Rat anti-mouse IL-6 and biotinylated rat anti-mouse IL-6 IgGs were purchased from eBioscience. Rabbit anti-mouse TNFα IgG was purchased from Thermo Fisher Scientific, and biotinylated rabbit anti-mouse TNFα IgG was from Invitrogen. Antibodies for patterning were concentrated with Centriprep MWCO 100 kDa prior to use. Mouse IL-6 and mouse TNF-alpha ELISA Ready-SET-Go!® reagent sets were purchased from eBioscience. Neutravidin-conjugated 30 nm gold nanoparticles was purchased from Nanopartz. LI Silver Enhancement Kit was purchased from Molecular Probes. Lipopolysaccharides from Escherichia coli (055:B5) were from Sigma-Aldrich. RAW 264.7 murine macrophage cells were kindly provided by Professor Tian Xia from UCLA.
Synthesis of Trehalose Glycopolymer (PolyProtek)
The synthesis of a styrenyl ether-based trehalose glycopolymer was modified from a previously reported procedure.24 Styrenyl ether trehalose monomer (375.2 mg, 8.18 × 10−1 mmol) and AIBN (3.13 mg, 1.91 × 10−2 mmol) were dissolved in H2O (2.73 mL) and DMF (1.36 mL), respectively. Both solutions were added to a reaction flask and subjected to five cycles of freeze-pump-thawing. The polymerization was started by immersing the flask in a 75 °C oil bath. After 8.33 hr, the polymerization was stopped exposing the solution to oxygen and cooling with liquid nitrogen. Residual monomer was removed by dialysis against H2O (MWCO 3,500 g/mol) for 3 days and lyophilized to obtain a white powder, with number average molecular weight Mn (GPC) = 15.7 kDa, and molecular weight dispersity Ð = 3.25.
PEG-silane Coating of Si Chips
Silicon substrates were cleaned by immersing into freshly prepared piranha solution (3:1 H2SO4 to 30% H2O2) and heated at 70 °C for 15 min. The chips were extensively rinsed with Milli-Q water and dried under a stream of filtered air. The cleaned chips were immediately incubated in a 1% wt/vol mPEG-silane solution in anhydrous toluene for at least 24 hours. The PEG-silane coated chips were then rinsed with methanol, followed by a large excess of Milli-Q water, and then dried under a stream of air. The substrates were immediately used for subsequent patterning experiments.
Film Thickness Measurements
Film thicknesses from the PEG-silane layer and spin-coated solutions of antibody layers were measured using a Gaertner LSE ellipsometer equipped with a 633 nm HeNe laser fired at a 70° incidence angle. The silicon oxide on the piranha-cleaned silicon wafer was measured and fitted using the refractive index of Palik (n1 = 0.54264, k1 = 0.00) and silicon as substrate (n1 = 3.589, k1 = 0.016). The measurement was repeated on the same sample after PEG-silane coating and spin-coating the protein and PolyProtek solution. The subsequent protein and polymer layer was fitted using values for the previously obtained silicon oxide thickness and an additional Cauchy layer model (n1 = 1.45, k1 = 0.01). A minimum of 15 measurements were performed at three different locations and the values were then averaged.
Electron Beam Lithography
Silicon substrates were spin-coated using Spin Coater Model ACE-200 (Dong-Ah). Aqueous solutions were spin-coated at 500 rpm for 5 sec, ramped to 1000 rpm for 5 sec, then ramped to 2000 rpm for 20 sec, and finally to 4000 rpm for 10 sec. PEG-silane coated silicon substrates were first spin-coated with Milli-Q H2O. Then, the substrates were spin-coated with a solution comprised of anti-IL-6 or anti-TNFα antibody, 0.5% wt/vol styrenyl ether-based trehalose glycopolymer, and 1 mM L-ascorbic acid in H2O. Patterns for electron beam lithography were designed in DesignCAD Express 16 software, and were generated using JC Nabity Lithography System (Nanometer Pattern Generation System, Ver. 9.0) modified from a JEOL JSM-6610 scanning electron microscope. An accelerating voltage of 30 kV, a spot size of 34 nm, and a beam current of 15 pA were used (dosage 25 μC/cm2). Following electron beam irradiation, any non-crosslinked polymer was rinsed away with wash buffer (0.05% Tween 20 in D-PBS). Alignment silicon wafers were fabricated via standard photolithography, metal evaporation and lift-off techniques as previously described.19 To generate multicomponent antibody patterns, the second antibody was spin-coated onto the same substrate. The chips were aligned by the prefabricated gold features, and patterned in close proximity to the first antibody. Non-crosslinked polymer was removed by rinsing with wash buffer.
Atomic Force Microscopy
AFM characterization of patterns was performed on a Bruker Dimension Icon AFM using Peak Force tapping mode with ScanAsyst Air probes. AFM imaging was performed on a scan size of 25 μm with a scan rate of 0.7 Hz and 512 samples per line.
Stimulation of RAW 264.7 Macrophages
RAW 264.7 murine macrophages were cultured in RPMI 1640 medium supplemented with 10% fetal bovine serum (FBS), and 1% penicillin-streptomycin. The cells were seeded in 6-well plates at a density of 5 × 105 cells per well and incubated at 37 °C and 5% CO2. After 24 hours, the culture media was replaced with 10 μg/mL lipopolysaccharide in the supplemented RPMI 1640 media. After incubation for 24 hours, the cell culture media was collected and centrifuged at 1,500 rpm for 5 min at 4 °C. Aliquots of the supernatant were stored at −80 °C until use.
Surface Immunoassay
Following electron beam lithography, the antibody-patterned surfaces were incubated in cytokine-containing media for 2 hours. The chips were then rinsed with wash buffer for 5 min, followed by incubation with biotinylated anti-IL-6 or anti-TNFα (5 μg/ml in D-PBS), or both, for 2 hours. After rinsing with wash buffer, neutravidin-conjugated 30 nm gold nanoparticles (1:100 in D-PBS) were allowed to bind to the patterns. The chips were rinsed again, and the gold nanoparticles were silver-enhanced, following the procedure as described by the manufacturer, for 5 min prior to a final rinsing step. The developed substrates and patterns were then imaged under aqueous conditions (D-PBS) using an Olympus BX51 microscope equipped with QImaging Retiga-2000R camera under dark field.
Measurement of Cytokine Detection by ELISA
Anti-IL-6 or anti-TNFα antibodies (2 μg/mL in D-PBS) were incubated in 96-well plates for 16 h at 23 °C. The plate was washed between each step with wash buffer. The plate was then blocked with 1% BSA in D-PBS for 2 hours at 23 °C. Supernatant from RAW 264.7 cell culture media were added to the wells and incubated for 2 hours. Biotin anti-IL6 or biotin anti-TNFα conjugate antibodies (0.25 μg/mL in 1% BSA D-PBS) were added and incubated for 2 hours, followed by incubation with streptavidin-HRP (1:200 in 1% BSA D-PBS) for 20 min. The plate was developed with 1-Step™ Ultra TMB solution (Pierce Biotechnology, Rockford) for 7 min prior to adding 1 M H2SO4 as the stop solution. The absorbance signals were measured at 450 nm and the background at 630 nm was subtracted.
Cytokine Detection Sensitivity and Time Studies
Concentrated solutions of IL-6 and TNFα from LPS-stimulated macrophage cell culture media were quantified using ELISA. Serial dilutions of the cell culture media in D-PBS were prepared with the cytokine concentrations at 200 ng/ml, 50 ng/ml, 500 pg/ml, 50 pg/ml and 5 pg/ml. The dilutions were used to incubate onto anti-IL-6 and anti-TNFα patterned surfaces. Subsequent processing of the chips with biotinylated antibodies, neutravidin-conjugated gold conjugates and the silver enhancement steps were performed and imaged under dark field as described above.
For detection of IL-6 and TNFα secretion over time, multi-antibody patterns were first fabricated and stored in D-PBS until use. RAW 264.7 macrophages were seeded in 6-well plates at a density of 5 × 105 cells per well and incubated for 24 hours. Then, the culture media was replaced with 10 μg/mL LPS in the working media. Prepared chips were incubated in the wells following LPS addition at the time points: 2, 12, 18, and 24 hr at 37 °C for 30 min. After rinsing, the immunoassay was continued as described above.
Supplementary Material
Acknowledgments
This work was supported by funds from the Center for Scalable and Integrated NanoManufacturing (SINAM, CMMI-0751621). The authors thank Professor Yu Huang (UCLA) for the use of her optical microscope. U.Y.L. thanks the NIH Chemistry-Biology Interface Training Program for support (T32 GM 008496). J.L. thanks the NIH Biotechnology Training Grant (T32 GM067555) for funding. S.S. thanks the Swiss National Science Foundation for the fellowship (PBEZP2-133211). E.B. thanks the Netherlands Organization for Scientific Research and Marie Curie Cofund Action for the financial support (Rubicon Grant 680-50-1101).
Footnotes
Conflict of Interest: The authors declare that a patent application has been filed on the use of PolyProtek as a resist for EBL.
Supporting Information Available: Optimization of anti-TNFα concentration for the immunoassay; effects of ascorbic acid concentration; detection sensitivity of patterned anti-IL-6; multiple processing and exposure cycles of patterned anti-TNFα.
References
- 1.Feldmann M, Brennan FM, Maini RN. Role of cytokines in rheumatoid arthritis. Annu Rev Immunol. 1996;14:397–440. doi: 10.1146/annurev.immunol.14.1.397. [DOI] [PubMed] [Google Scholar]
- 2.Michalaki V, Syrigos K, Charles P, Waxman J. Serum levels of IL-6 and TNF-alpha correlate with clinicopathological features and patient survival in patients with prostate cancer. Br J Cancer. 2004;90:2312–2316. doi: 10.1038/sj.bjc.6601814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Letsch A, Scheibenbogen C. Quantification and characterization of specific T-cells by antigen-specific cytokine production using ELISPOT assay or intracellular cytokine staining. Methods. 2003;31:143–149. doi: 10.1016/s1046-2023(03)00124-5. [DOI] [PubMed] [Google Scholar]
- 4.Ma C, Fan R, Ahmad H, Shi Q, Comin-Anduix B, Chodon T, Koya RC, Liu CC, Kwong GA, Radu CG, Ribas A, Heath JR. A clinical microchip for evaluation of single immune cells reveals high functional heterogeneity in phenotypically similar T cells. Nat Med. 2011;17:738–743. doi: 10.1038/nm.2375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen PY, Chung MT, McHugh W, Nidetz R, Li YW, Fu JP, Cornell TT, Shanley TP, Kurabayashi K. Multiplex Serum Cytokine Immunoassay Using Nanoplasmonic Biosensor Microarrays. ACS Nano. 2015;9:4173–4181. doi: 10.1021/acsnano.5b00396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu Y, Kwa T, Revzin A. Simultaneous detection of cell-secreted TNF-alpha and IFN-gamma using micropatterned aptamer-modified electrodes. Biomaterials. 2012;33:7347–7355. doi: 10.1016/j.biomaterials.2012.06.089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang S, Ota S, Guo B, Ryu J, Rhodes C, Xiong Y, Kalim S, Zeng L, Chen Y, Teitell MA, Zhang X. Subcellular Resolution Mapping of Endogenous Cytokine Secretion by Nano-Plasmonic-Resonator Sensor Array. Nano Lett. 2011;11:3431–3434. doi: 10.1021/nl2018838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barbulovic-Nad I, Lucente M, Sun Y, Zhang MJ, Wheeler AR, Bussmann M. Bio-microarray fabrication techniques - A review. Crit Rev Biotechnol. 2006;26:237–259. doi: 10.1080/07388550600978358. [DOI] [PubMed] [Google Scholar]
- 9.Christman KL, Enriquez-Rios VD, Maynard HD. Nanopatterning proteins and peptides. Soft Matter. 2006;2:928–939. doi: 10.1039/b611000b. [DOI] [PubMed] [Google Scholar]
- 10.Blawas AS, Reichert WM. Protein patterning. Biomaterials. 1998;19:595–609. doi: 10.1016/s0142-9612(97)00218-4. [DOI] [PubMed] [Google Scholar]
- 11.Derby B. Bioprinting: inkjet printing proteins and hybrid cell-containing materials and structures. J Mater Chem. 2008;18:5717–5721. [Google Scholar]
- 12.Bernard A, Delamarche E, Schmid H, Michel B, Bosshard HR, Biebuyck H. Printing patterns of proteins. Langmuir. 1998;14:2225–2229. [Google Scholar]
- 13.Volcke C, Gandhiraman RP, Basabe-Desmonts L, Iacono M, Gubala V, Cecchet F, Cafolla AA, Williams DE. Protein pattern transfer for biosensor applications. Biosens Bioelectron. 2010;25:1295–1300. doi: 10.1016/j.bios.2009.10.016. [DOI] [PubMed] [Google Scholar]
- 14.Falconnet D, Pasqui D, Park S, Eckert R, Schift H, Gobrecht J, Barbucci R, Textor M. A novel approach to produce protein nanopatterns by combining nanoimprint lithography and molecular self-assembly. Nano Lett. 2004;4:1909–1914. [Google Scholar]
- 15.Hoff JD, Cheng LJ, Meyhofer E, Guo LJ, Hunt AJ. Nanoscale protein patterning by imprint lithography. Nano Lett. 2004;4:853–857. [Google Scholar]
- 16.Hyun J, Ahn SJ, Lee WK, Chilkoti A, Zauscher S. Molecular recognition-mediated fabrication of protein nanostructures by dip-pen lithography. Nano Lett. 2002;2:1203–1207. [Google Scholar]
- 17.Lee KB, Lim JH, Mirkin CA. Protein nanostructures formed via direct-write dip-pen nanolithography. J Am Chem Soc. 2003;125:5588–5589. doi: 10.1021/ja034236p. [DOI] [PubMed] [Google Scholar]
- 18.Huo F, Zheng Z, Zheng G, Giam LR, Zhang H, Mirkin CA. Polymer pen lithography. Science. 2008;321:1658–1660. doi: 10.1126/science.1162193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Christman KL, Schopf E, Broyer RM, Li RC, Chen Y, Maynard HD. Positioning multiple proteins at the nanoscale with electron beam cross-linked functional polymers. J Am Chem Soc. 2009;131:521–527. doi: 10.1021/ja804767j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hong Y, Krsko P, Libera M. Protein surface patterning using nanoscale PEG hydrogels. Langmuir. 2004;20:11123–11126. doi: 10.1021/la048651m. [DOI] [PubMed] [Google Scholar]
- 21.Kolodziej CM, Kim SH, Broyer RM, Saxer SS, Decker CG, Maynard HD. Combination of integrin-binding peptide and growth factor promotes cell adhesion on electron-beam-fabricated patterns. J Am Chem Soc. 2012;134:247–255. doi: 10.1021/ja205524x. [DOI] [PubMed] [Google Scholar]
- 22.Kolodziej CM, Maynard HD. Electron-Beam Lithography for Patterning Biomolecules at the Micron and Nanometer Scale. Chem Mater. 2012;24:774–780. [Google Scholar]
- 23.Mancini RJ, Lee J, Maynard HD. Trehalose Glycopolymers for Stabilization of Protein Conjugates to Environmental Stressors. J Am Chem Soc. 2012;134:8474–8479. doi: 10.1021/ja2120234. [DOI] [PubMed] [Google Scholar]
- 24.Lee J, Lin EW, Lau UY, Hedrick JL, Bat E, Maynard HD. Trehalose glycopolymers as excipients for protein stabilization. Biomacromolecules. 2013;14:2561–2569. doi: 10.1021/bm4003046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee J, Ko JH, Lin EW, Wallace P, Ruch F, Maynard HD. Trehalose hydrogels for stabilization of enzymes to heat. Polym Chem. 2015;6:3443–3448. doi: 10.1039/C5PY00121H. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bat E, Lee J, Lau UY, Maynard HD. Trehalose glycopolymer resists allow direct writing of protein patterns by electron-beam lithography. Nat Commun. 2015;6:1–8. doi: 10.1038/ncomms7654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim S, Marelli B, Brenckle MA, Mitropoulos AN, Gil ES, Tsioris K, Tao H, Kaplan DL, Omenetto FG. All-water-based electron-beam lithography using silk as a resist. Nat Nanotechnol. 2014;9:306–310. doi: 10.1038/nnano.2014.47. [DOI] [PubMed] [Google Scholar]
- 28.Zhang M, Desai T, Ferrari M. Proteins and cells on PEG immobilized silicon surfaces. Biomaterials. 1998;19:953–960. doi: 10.1016/s0142-9612(98)00026-x. [DOI] [PubMed] [Google Scholar]
- 29.Papra A, Gadegaard N, Larsen NB. Characterization of ultrathin poly(ethylene glycol) monolayers on silicon substrates. Langmuir. 2001;17:1457–1460. [Google Scholar]
- 30.Zhang LH, Zhang WD, Zhang ZC, Yu L, Zhang HF, Qi YC, Chen DG. Radiation Effects on Crystalline Polymers .1. Gamma-Radiation-Induced Cross-Linking and Structural Characterization of Polyethylene Oxide. Radiat Phys Chem. 1992;40:501–505. [Google Scholar]
- 31.Dey RK, Cui B. Effect of molecular weight distribution on e-beam exposure properties of polystyrene. Nanotechnology. 2013;24:245302. doi: 10.1088/0957-4484/24/24/245302. [DOI] [PubMed] [Google Scholar]
- 32.Niki E. Action of Ascorbic-Acid as a Scavenger of Active and Stable Oxygen Radicals. Am J Clin Nutr. 1991;54:S1119–S1124. doi: 10.1093/ajcn/54.6.1119s. [DOI] [PubMed] [Google Scholar]
- 33.Thorley AJ, Ford PA, Giembycz MA, Goldstraw P, Young A, Tetley TD. Differential regulation of cytokine release and leukocyte migration by lipopolysaccharide-stimulated primary human lung alveolar type II epithelial cells and macrophages. J Immunol. 2007;178:463–473. doi: 10.4049/jimmunol.178.1.463. [DOI] [PubMed] [Google Scholar]
- 34.Yuan K, Yu B, Pan DZ. E-Beam Lithography Stencil Planning and Optimization with Overlapped Characters. IEEE T Comput Aid D. 2012;31:167–179. [Google Scholar]
- 35.Wieland MJ, de Boer G, ten Berge GF, van Kervinck M, Jager R, Peijster JJM, Slot E, Steenbrink SWHK, Teepen TF, Kampherbeek BJ. MAPPER: High throughput maskless lithography. Alternative Lithographic Technologies Ii. 2010;7637 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.