Abstract
Complementarity determining region 3 of the immunoglobulin (Ig) H chain (CDR-H3) lies at the center of the antigen binding site where it often plays a decisive role in antigen recognition and binding. Amino acids encoded by the diversity (DH) gene segment are the main component of CDR-H3. Each DH has the potential to rearrange into one of six DH reading frames (RFs), each of which exhibits a characteristic amino acid hydrophobicity signature that has been conserved among jawed vertebrates by natural selection. A preference for use of RF1 promotes the incorporation of tyrosine into CDR-H3 while suppressing the inclusion of hydrophobic or charged amino acids. To test the hypothesis that these evolutionary constraints on DH sequence influence epitope recognition, we used mice with a single DH that has been altered to preferentially use RF2 or inverted RF1. B cells in these mice produce a CDR-H3 repertoire that is enriched for valine or arginine in place of tyrosine. We serially immunized this panel of mice with gp140 from HIV-1 JR-FL isolate and then used ELISA or peptide microarray to assess antibody binding to key or overlapping HIV-1 envelope epitopes. By ELISA, serum reactivity to key epitopes varied by DH sequence. By microarray, sera with Ig CDR-H3s enriched for arginine bound to linear peptides with a greater range of hydrophobicity, but had a lower intensity of binding than sera containing Ig CDR-H3s enriched for tyrosine or valine. We conclude that patterns of epitope recognition and binding can be heavily influenced by DH germline sequence. This may help explain why antibodies in HIV infected patients must undergo extensive somatic mutation in order to bind to specific viral epitopes and achieve neutralization.
Keywords: epitope, immunoglobulin DH gene, complementarity determining region (CDR-H3), HIV-1 envelope protein
Introduction
Although the mechanisms of combinatorial VDJ rearrangement, junctional exonucleolytic loss, and N nucleotide addition would appear to permit a repertoire of randomly created immunoglobulin antigen binding sites, specific biases in amino acid usage are observed in the mature CDR-H3 repertoire (Fugmann et al., 2000; Ivanov et al., 2002; West et al., 2014). CDR-H3 lies at the center of the antigen binding site and often dictates the specificity of the antibody. The non-random use of amino acids in CDR-H3 reflects natural selection of the amino acids encoded by germline DH gene segments, somatic regulation of reading frame preference, and somatic selection of CDR-H3 content as B cells pass through sequential quality control and tolerance checkpoints in the primary and secondary lymphoid organs (Meffre et al., 2001; Schroeder et al., 2010a; Verkoczy et al., 2013). We have previously shown that violation of germline constraints on DH sequence, and thus CDR-H3, can alter susceptibility to infectious diseases (Nguyen et al., 2007). In this work, we sought to address whether the changes in the B cell receptor antigen binding site repertoire created by altering DH sequence could influence patterns of antigen, or ligand, recognition.
The immune system appears to recognize individual antigens as a collection of epitopes (Alt et al., 1987; Vinuesa and Chang, 2013). For non-repetitive antigens, production of antigen-specific antibody by B cells typically requires recognition of a minimum of two epitopes on that antigen. One epitope is recognized by the BCR on B cells (Swanson et al., 2010). The second epitope is typically recognized either by innate receptors [such as the toll-like receptors (Vinuesa and Chang, 2013)] in the case of T independent (TI) responses, or by the TCR on T cells as an MHC:peptide complex in the case of T dependent (TD) responses. Functionally, however, not all epitopes are equal in biologic importance. Antigen neutralization, for example, typically depends on the production of antibodies with antigen binding sites that recognize and bind to a subset of potential epitopes on the antigen. Antibodies that bind elsewhere can innocuously decorate the antigen, interfere with binding to critical epitopes due to steric hindrance, or even enhance pathogenicity (Wells et al., 2014). Thus patterns of epitope recognition can have significant consequences to the health of the individual and the likelihood of successfully bearing children and living long enough to raise them to maturity.
HIV-1 is an RNA virus that accumulates a host of mutant viral proteins by means of an error prone reverse transcriptase. The broadly neutralizing antibodies (bNAbs) that are needed to provide protection against such a variable virus are often characterized by uncommon CDR-H3s. For example, the average length of CDR-H3 in the normal adult pre-immune repertoire is 16 amino acids (Tiller et al., 2007), yet in order to penetrate the viral glycan shield (McLellan et al., 2011) some envelope protein-binding bNAb contain CDR-H3s with lengths of up to 33 amino acids (West et al., 2014). Other bNAbs, such as 2F5, contain CDR-H3s that extend a hydrophobic loop that tethers the antibody onto the viral membrane in the membrane proximal region (MPER) of gp41 (Ofek et al., 2010). To test whether the restrictions in CDR-H3 content placed by natural selection of DH gene sequence might specifically influence patterns of epitope recognition and perhaps help explain why it can be difficult to generate adequately protective antibodies in HIV as well as other infections, we analyzed serum antibody reactivity to the envelope protein of HIV-1 in mice expressing altered gradients of CDR-H3 amino acid content.
We had previously generated a panel of D-altered mice (Schroeder et al., 2010a) containing IgH loci limited to one wild type or one sequence altered DH (Fig 1A). The antibody repertoires produced by these D-altered mice contain alternative CDR-H3 sequence gradients of charge or hydrophobicity that include an amplified representation of categories of sequences that are present normally but only at very low prevalence.
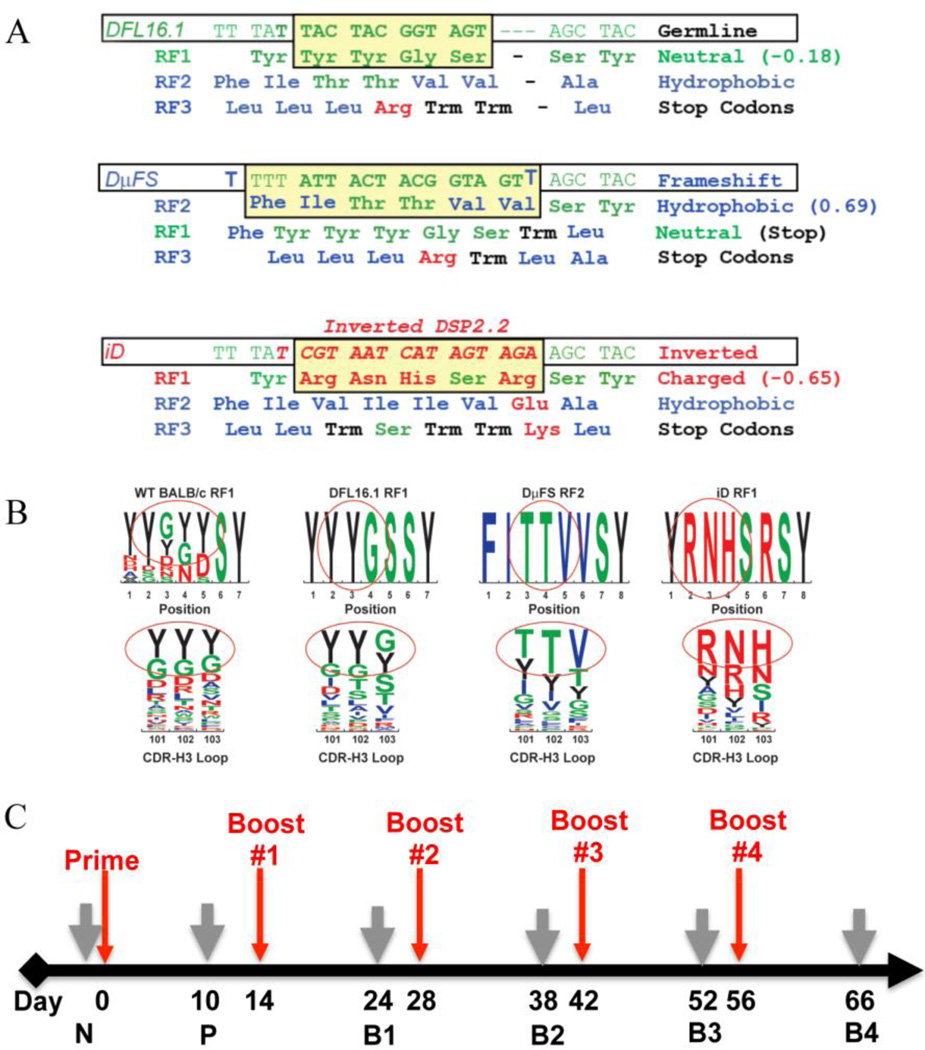
Fig. 1. Sequences of the altered DH alleles and the immunization strategy used.
(A) Sequences of the single DH gene segments present in the D-altered strains. Germline DFL16.1 in the ΔD-DFL allele is shown at the top, frameshifted DFL16.1 in the ΔD-DμFS allele is shown in the middle, and inverted DSP2.2 replacing the middle sequences of DFL16.1 in the ΔD-iD allele is shown at the bottom. (B) Impact of DH germline sequence on the amino acid content of the CDR-H3 loop. The derivation of the most common amino acids at positions 101–103 is circled and compared to the sequence of the progenitor DH. Color code, charged amino acids are marked in red, hydrophobic amino acids are marked in blue, neutral amino acids are marked in green, and tyrosine is marked in black. (C) Immunization scheme for all studies described in this manuscript. The mice were primed at day 0 and boosted at the time points indicated by red arrows. Grey arrows indicate blood collection time points. N, non-immunized. P, prime. B1, B2, B3, and B4; boost #1, #2, #3 and #4, respectively. Details of the experimental protocol and immunogen formulations are provided in Materials and Methods.
We had sorted developing B cells from these mice, characterized the CDR-H3 repertoire, and obtained evidence that DH sequence biases CDR-H3 amino acid content at positions that can interact directly with antigen (Schroeder et al., 2010b) (Fig 1B). We had found that divergence from wild-type DH sequence was often associated with increased susceptibility to a number of infectious agents (Nguyen et al., 2007; Vale et al., 2013)
Serial immunization with HIV-1-gp140 envelope protein (containing the soluble extracellular portion of HIV-1 gp160) allowed us to test the relationship and consequences of patterns of CDR-H3 content on responses to specific epitopes. After immunization we used ELISA and peptide microarrays to examine serum antibody reactivity to key and overlapping linear epitopes of the HIV-1 envelope protein. We found that DH alteration modified the pattern of antibody production against individual HIV-1 envelope epitopes. The diverse patterns of epitope recognition that we observed in the D-altered mice may help explain why some antigen epitopes elicit a greater antibody response than others, and why HIV-1-infected individuals may need access to rare CDR-H3s in order to bind to specific HIV epitopes and achieve broad neutralization.
Results
Ig DH germline sequence influences B-cell primary epitope recognition
We immunized cohorts of mice homozygous for wild-type (WT) or altered DH alleles (Fig. 1) with vaccinia-derived, purified recombinant HIV-1 JR-FL gp140 (Verkoczy et al., 2013). Gp140, the external portion of the HIV-1 envelope protein, is a 140 kDa soluble oligomeric Env protein that contains gp120 and approximately 20 kDa of gp41 (Liao et al., 2006b), all of which are normally displayed on the surface of the viral membrane (Stamatatos et al., 2000; Stamatos et al., 1998). Gp140 is often used as an antigen to induce anti-HIV-1 humoral immune responses (Harris et al., 2011). Homozygous mice carrying ΔD-DFL, ΔD-DμFS or ΔD-iD IgH alleles are limited to use of a single normal DFL16.1 gene segment, a single frame-shifted DFL16.1 gene segment, or a single DH containing inverted DSP2.2 gene sequence, respectively (Fig. 1). The CDR-H3 repertoire of each of the three mouse strains is quite different: ΔD-DFL mice produce a DFL16.1-specific repertoire enriched for DH reading frame 1 (RF1)-encoded neutral tyrosine, as well as serine and glycine. ΔD-DμFS mice produce a DFL16.1-RF2-specific repertoire enriched for hydrophobic valine, as well as isoleucine and hydrophilic threonine (Ippolito et al., 2006). ΔD-iD mice produce a repertoire that includes inverted RF1 DSP2.2 arginine, as well as histidine and asparagine, charged amino acids that can form salt bridges with antigen.
Cohorts of ten mice, each homozygous for either wild-type (WT) or altered DH alleles, were immunized at two week intervals for a total of one primary immunization and four boosters (Fig. 1). Ten days after each immunization, sera were collected and tested by endpoint ELISA for IgM or IgG binding to HIV-1 JR-FL gp140, to peptides derived from V1V2 loop (or V1V2-NQ156Q) of gp120, to the gp41 immunodominant epitope (Sp400), and to the epitopes recognized by the MPER-specific broadly neutralizing antibodies 2F5 and 4E10. These latter peptides differ in their charged amino acid (q-aa) composition and degree of hydrophobicity, as determined by the Kyte-Doolittle hydrophobicity scale as modified by Eisenberg (Eisenberg, 1984; Kyte and Doolittle, 1982). The MPER-peptide that contains the 2F5 and 4E10 epitopes is the most charged (KD = −1.42, q-aa = 60%), and the immunodominant Sp400 epitope is the least charged (KD = −0.041, q-aa = 32%) peptides (Liao et al., 2006a; Verkoczy et al., 2011; Verkoczy et al., 2013).
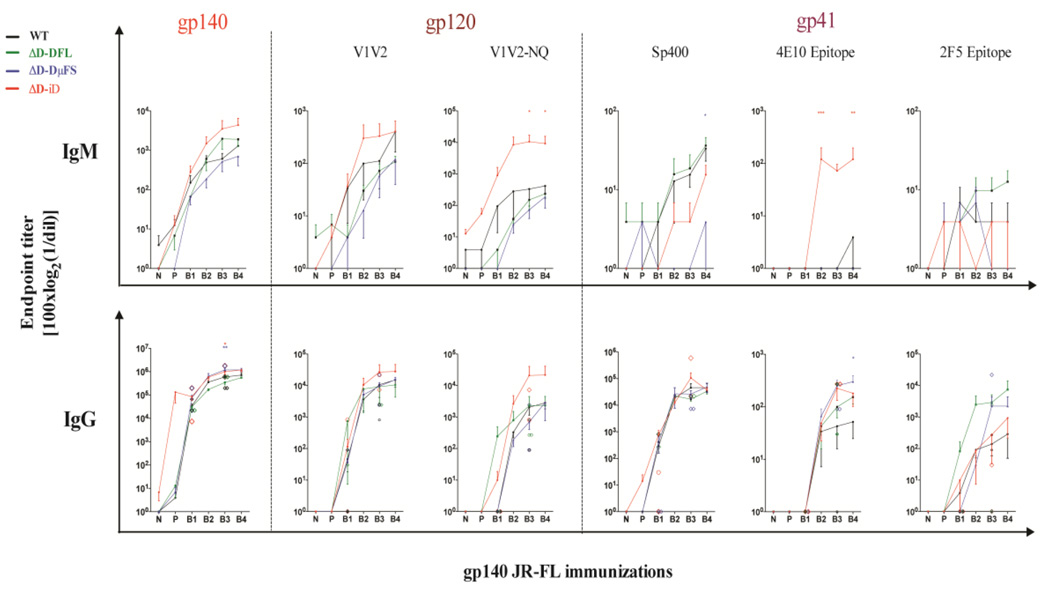
Endpoint titers to the full-length gp140 protein demonstrated different trends by DH allele, but these trends did not achieve statistical significance (Fig. 2). Thus, the total amount of antibody that was produced in response to immunization was similar irrespective of global CDR-H3 content. However, statistically significant differences in endpoint titer were observed when we evaluated serum reactivity to specific epitopes (Fig. 2). Within gp120, variable regions 1 and 2 (V1/V2) contain critical epitopes that are targets for both antibody dependent cellular cytotoxicity (ADCC) and broadly neutralizing antibodies. These variable regions are protected both by extraordinary sequence diversity and N-linked glycosylation (McLellan et al., 2011). Two peptides, one encoding the V1V2 epitope and the other the V1V2-NQ156Q, N160 variant, were examined. For both peptides and for both IgM and IgG, ΔD-iD mice demonstrated the highest endpoint titers. For the IgM response to the V1V2-N156Q, N160 peptide, the differences between ΔD-iD and the other three strains was significant (p<0.01 in pairwise comparisons, and p<0.05 when adjusted for multiple comparisons).
Fig. 2. Alteration of DH sequence biases anti-HIV-1 gp140 epitope preference.
Specific-IgG and IgM binding to HIV-1-gp140 and neutralization-associated epitopes were assessed in sera from WT (black) and the three strains of D-altered mice [ΔD-DFL (green), ΔD-DμFS (blue), and ΔD-iD (red)]. Individual mean endpoint titers obtained after 1–5 JR-FL gp140 i.p. immunizations are shown. The diamond symbols indicate the individual endpoint titers in the two mice/genotype selected for positive linear epitope identification by PEPperPRINT® chip (see Fig. 4). The mean and standard error of the mean are shown for responder mice. Statistical significance was determined by a p value <0.01 (**), <0.05(*), respectively, ANOVA followed by Tukey HSD. Time points are as presented in Fig. 1.
Within gp41, amino acids 570 to 613 comprise the immunodominant epitope, which is represented by peptide Sp400 (Banerjee et al., 2010). The endpoint titer against Sp400 also differed significantly by DH content. For IgM, the anti-Sp400 response was highest among WT and ΔD-DFL mice, followed by ΔD-iD and then ΔD-DμFS. The differences between ΔD-DμFS and WT & ΔD-DFL achieved statistical significance (p<0.001 in pairwise comparisons and p<0.005 when adjusted for multiple comparisons). After the 3rd boost, the IgG endpoint titer between ΔD-iD mice and each of the other three strains reached significance at p<0.05 in pairwise comparisons, but failed to achieve significance when adjusted for multiple comparisons. This difference disappeared after the 4th boost.
The broadly neutralizing antibody 2F5 recognizes the DKW (single letter amino acid code) core epitope within amino acids 643 to 662 of the MPER of gp41, which is represented by the SP62 peptide (Alam et al., 2008). ΔD-DFL mice exhibited the highest endpoint titer for both IgM and IgG, achieving significance (p<0.05) when compared to the IgG titer of WT and ΔD-iD.
The broadly neutralizing antibody 4E10 recognizes the HIV-1 gp41 core epitope sequence NWFDIT (Brunel et al., 2006; Cardoso et al., 2005; Kunert et al., 2004). For IgM, ΔD-iD mice exhibited the highest endpoint titer of the four mouse strains for the 4E10 peptide, achieving statistical significance (p<0.01) when compared to WT, ΔD-DFL, and ΔD-DμFS at the third and fifth immunizations and adjusted for multiple comparisons. For IgG, the ΔD-DμFS mice continued to express higher endpoint titers than WT after five immunizations (p<0.05). However, the ΔD-iD titers had dropped to ΔD-DFL levels, which were intermediate between ΔD-DμFS and WT. For both 4E10 and Sp400 described above, while recognition was clearly affected by the 3rd boost, the limited duration of antibody production and the lack of persistence of antibody with boosting could be affected by mechanisms totally different from epitope recognition, e.g., the inability of the ΔD-iD mice to be further boosted or to mount a long lived plasma cell response (Trad et al., 2014).
Each of the studies above evaluated individual epitopes using end-point titers, which are discrete values derived from the OD values at a series of dilutions from the ELISAs. The OD values are continuous and correlate with antibody concentration. In order to evaluate the role of the DH on antibody production more globally, we summed all the OD values at all dilutions for each sample and modeled the effect of genotype (G), antibody type (IgM or IgG) (A), and epitope (E) after the prime immunization (P) and after each of the four booster immunizations (B1–B4) (Table 1).
Table 1.
Use of OD sums to globally evaluate the role of DH on antibody production.
| Tested effect |
P | B1 | B2 | B3 | B4 | Null model | Alternative model |
|---|---|---|---|---|---|---|---|
| G | 0.00056 | 0.349 | 0.0472 | 0.0433 | 0.187 | Y=A+E+ε | Y=A+E+G+ε |
| G*E | 2.00E-08 | 3.26E-08 | 7.71E-06 | 0.00132 | 0.00733 | Y=G+A+E+G*A+A*E+ε | Y=G+A+E+G*A+A*E+G*E+ε |
| G*A | 0.0199 | 0.269 | 0.0395 | 0.00618 | 0.00472 | Y=G+A+E+A*E+G*E+ε | Y=G+A+E+A*E+G*E+G*A+ε |
| G*A*E | 1.78E-15 | 3.21E-07 | 2.58E-05 | 0.00334 | 0.00014 | Y=G+A+E+A*E+G*E+G*A+ε | Y=G+A+E+A*E+G*E+G*A+G*A*E+ε |
OD (optical density) values in ELISAs are continuous and correlate with antibody concentration. To evaluate the global role of DH sequence on antibody production, the OD values at all dilutions for each sample were summed. Mixed effect models were used for comparisons. Variables used in the comparison included the effect of genotype (G), antibody type (IgM or IgG) (A), and epitope (E) after the prime immunization (P) and after each of the four booster immunizations (B1–B4). As per common practice in statistical analyses of this type, these studies considered both a null model and an alternative model, which contains the additional variable to be tested. E.g., when testing for genotype, the null model lacked this variable and the alternative model included it In addition to the fixed effects provided in the models, both included mouse as a random effect. The residual, ε, is the difference between the actual and predicted ODsums for each model.
When the mice were tested by genotype after controlling for isotype and epitope, the antibody response as measured by the sum of ODs was found to be significant after the prime immunization (p=0.00056) and the second (B2, p=0.047) and third (B3, p=0.043) boosts. When tested by genotype and epitope, the differences between groups was significant after all five of the immunizations with p values ranging from p=0.0073 to p=3.3E-08. This is in agreement with the results of the endpoint titer analysis (Fig. 2) in that the difference in the pattern of response by genotype varies from epitope to epitope. When tested by genotype and isotype, the differences between groups was significant after the priming immunization (p<0.020) and after the 2nd (B2), 3rd (B3) and 4th (B4) boosts (p=0.040, =0.0062, and =0.0047, respectively). This is also in agreement with the results of the endpoint titer analysis (Fig. 2) in that the difference in the pattern of response by genotype varies between IgM and IgG. Finally, when the genotype, isotype, and epitope were all taken together, the difference between groups was highly statistically significant (p=1.8E-15, 3.2E-07, 2.6E-05, 3.3E-03, and 1.4E-04, respectively). This emphasizes that the differences in antibody production among the DH genotypes is a function of both the epitope, which is fixed, and the isotype for which IgG is associated with somatic hypermutation and then clonal selection.
D-altered mice display altered recognition of gp140 linear peptides
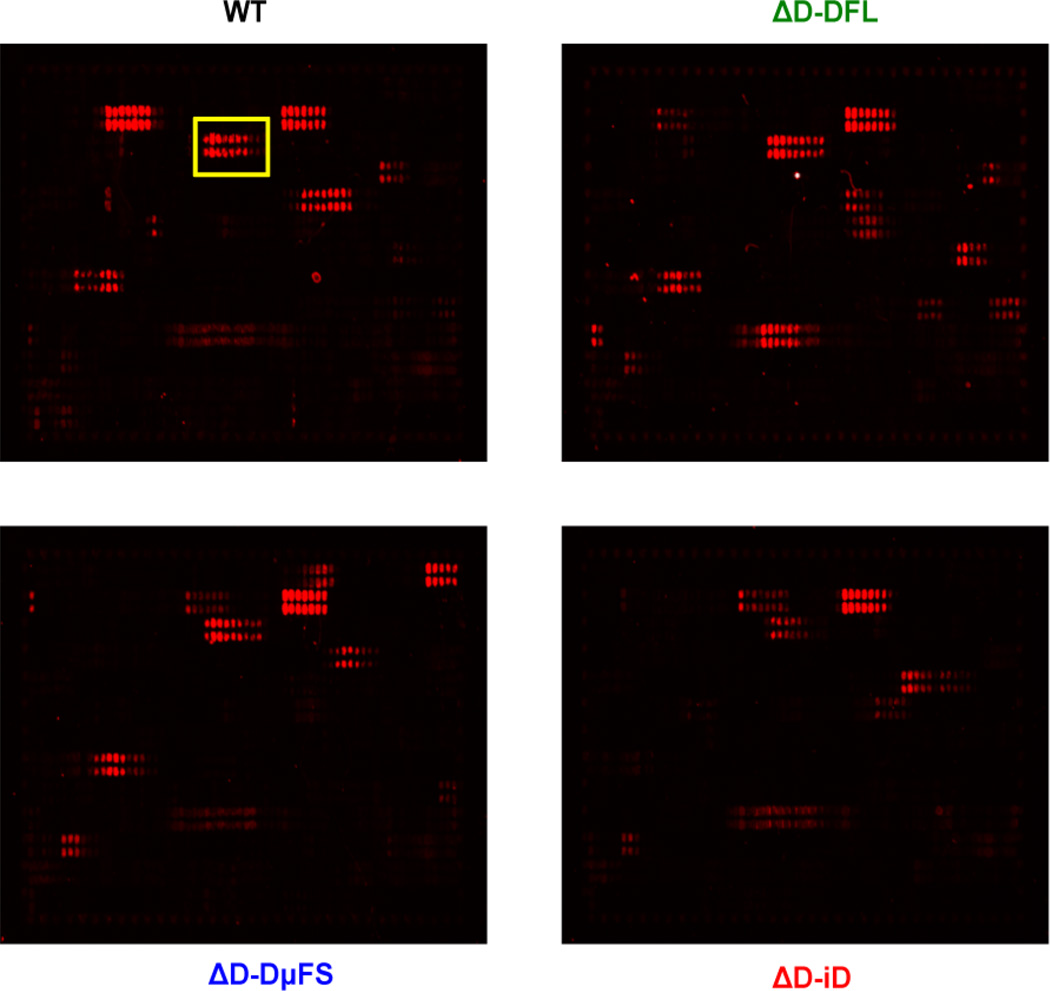
To further assess patterns of epitope recognition in the D-altered mice versus WT, we turned to high throughput technology. The PEPperPRINT® chip allows an assessment of binding to 849 possible overlapping 13 amino acid linear epitopes within the protein sequence of gp160 (Heilkenbrinker et al., 2013; Shukla et al., 2012). Based on averaged median foreground intensities, intensity maps were generated and antibody binding to the peptide map was highlighted by an intensity color code with red for high spot intensities (Fig. 3). Unlike the previously examined peptides, which are of sufficient length that they could display in solution the conformational structures found on the intact molecule, these smaller peptides most likely remain linear. We selected two mice at random from each 10 mice cohort and tested peptide reactivity from pre-immune sera and after the first (B1) and third (B3) booster immunizations. Our subsequent analyses were based on the peptides that reacted with both first boost (B1) and third boost (B3) immunized serum.
Fig. 3. Representative peptide microarray images.
One of three chips is shown as an example. The chips contained thirteen amino acid peptides with a twelve amino acid overlap spanning the entire gp160 sequence. Peptides were spotted in duplicate. The chips were incubated with WT, ΔD-DFL, ΔD-DμFS and ΔD-iD immune sera (1:1000 dilution) followed by staining with the secondary antibody F(ab´)2 goat anti-mouse IgG(H+L) conjugate at a dilution of 1:5000 and read-out at a scanning intensity of 5 (red). Read-out was performed with a LI-COR Odyssey Imaging System. The red dots represent the peptides with bound antibody, e.g. the boxed area shows the V1V2 epitope. Quantification of spot intensities and peptide annotation were done with PepSlide® Analyzer.
We plotted the averaged spot intensities from each pre-immune/immunized assay against the linear peptide sequence of gp160 from the N-terminus to the C-terminus to visualize overall spot intensities. Intensity was also overlaid onto the structure of gp160 (Fig. 4A, B). In those cases where it was not clear if a certain amino acid contributed to a consensus motif, the corresponding letters have been outlined in gray. The reactivities of the mice assayed in the microarray (Fig. 3 and 4) were largely representative of the average reactivity of the cohorts against the specified antigens (Fig. 2). (The only exception was the ΔD-DμFS IgG response to the 2F5 epitope),
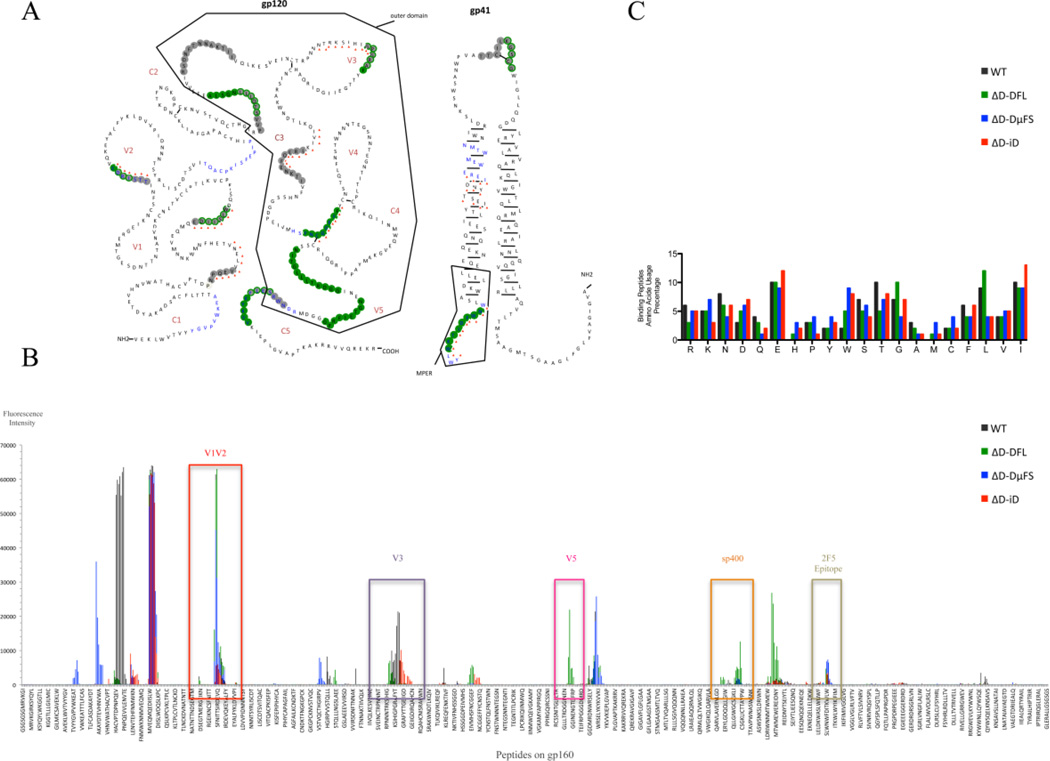
Fig. 4. Antibody binding to HIV-1 JR-FL gp140 linear epitopes.
Peptide binding data from sera obtained after the second and fourth immunizations were combined for these analyses. (A) Illustration of the peptides bound by the various sera superimposed on the JR-FL gp140 secondary structure. The residues bound by sera from the different mouse strains are color-coded: WT (grey), DFL16.1 (green), ΔD-DμFS (blue), ΔD-iD (red triangle). The symbolic spatial structures of HIV-1 JR-FL gp120 and gp41 are plotted based on Binley’s published research (Binley et al., 2000). (B) Response intensity chart of the bound peptides for each mouse strain. The bound peptides, which are located in the positions of V1V2, V3, V5, Sp400 and 2F5 epitopes, are indicated in the figure to illustrate the overlapping binding of the positive natural epitopes in Fig. 2. (C) Amino acid usage percentage of the bound peptides. The average frequency of the different amino acids present in the bound peptides was calculated by Prism GraphPad® 5. The measurement methods are in a supplementary Microsoft Excel table.
A comparison of binding profiles revealed that both mice with a full complement of wild type DH (WT) and mice limited to a single, wild type DH (ΔD-DFL) demonstrated a broad and intense pattern of epitope recognition. Of these, the WT mice generated the greatest intensity (3 peaks >60,000 fold over baseline and 2 peaks >20,000 fold over baseline) to epitopes in the gp120 region, particularly in region C1, V1V2, V1V2, V3, V5, and C5. There was also a modest reaction with gp41 region peptides (WNNMTWMEWEREIDNYT). ΔD-DFL showed the strongest intensity against gp41 (2 peaks >60,000 fold over baseline and 3 peaks >20,000 fold over baseline. Included within these response peptides is the gp41 immunodominant region Sp400.
By linear peptide array, antibodies in ΔD-DFL serum were found to strongly bind to peptides N terminal to V1V2, to V1V2, and to V5 (Fig. 4A, B). ΔD-DμFS serum antibodies also reacted with peptides N terminal to V1V2 (3 peaks with >30,000 fold intensity over baseline), V1V2 and the region C terminal to V1V2 and N terminal to V3. ΔD-iD serum antibodies responded strongly to only one region, V1V2. Thus, varying the sequence of the DH substantially altered the focus of the antibody response.
To test whether there was a correlation between the enrichment for specific ranges of hydrophobicity in the CDR-H3 repertoire in the D-altered mice and the amino acid content and average hydrophobicity of the peptides assayed in the microarray, we examined the amino composition of the peptides (Fig. 4C) (Ippolito et al., 2006; Mahmoud et al., 2011; Zemlin et al., 2008). The peptides contained a mixture of charged, neutral and hydrophobic amino acids. No clear associations between the amino acid content of the altered DH and the overall amino acid content of the peptides recognized were observed, so the bias in epitope recognition cannot entirely be explained simply by alterations in the physico-chemical structure of the antigen binding site.
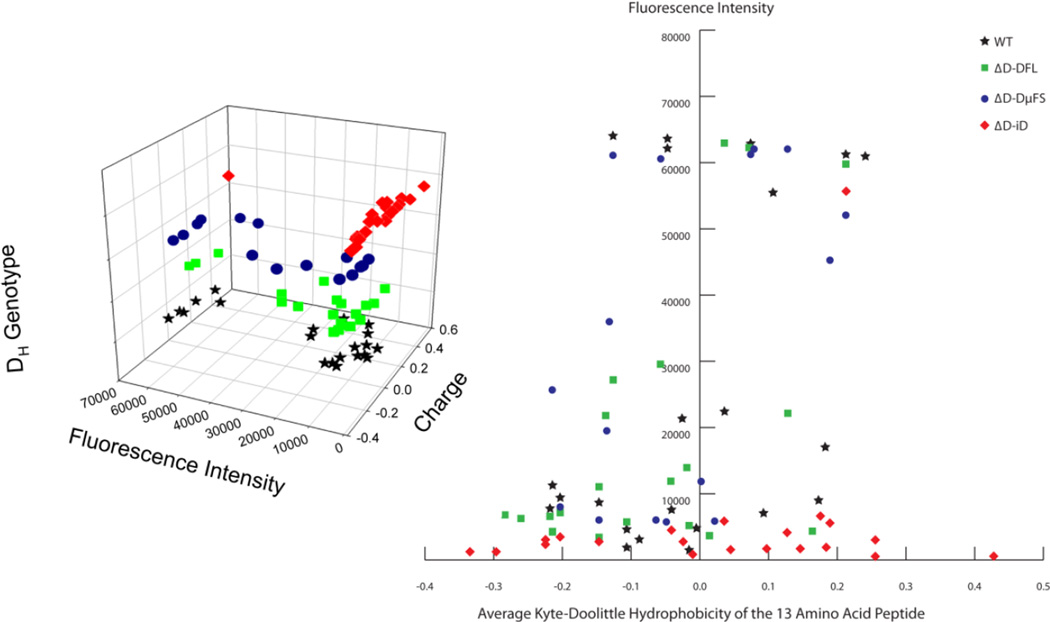
We then examined the relationship of the charge of the linear peptide to the intensity of the microarray response. These data were visualized in both 3D (Fig. 5A) and 2D (Fig. 5B) representations. Each peptide was assigned an average hydrophobicity based on the Kyte-Doolittle scale. A clear distinction could be drawn between the ΔD-iD mice and the other three strains. Using a cut-off of 10,000 for intensity, ΔD-iD sera showed intense binding to only one of 21 peptides, whereas intense binding was observed in 11 of 21 peptides for WT mice, ten of 20 peptides for ΔD-FL mice, and eleven of 17 peptides for ΔD-DμFS mice (p=0.0014, p=0.0014, and p<0.0001; respectively). Thus, while the ΔD-iD mice produced serum antibodies that were able to bind to peptides with a wider range of average hydrophobicities then the other three strains, all but one of the reactivities were of a low response intensity.
Fig. 5. Responses of mice of different genotypes versus intensity and charge of the bound peptides.
(Left) 3-D visualization of the response intensity to HIV envelope peptides with their charge and corresponding DH genotype. (Right) 2-D visualization of the mouse strain preference, charge and reaction intensity.
Discussion
We found that changing the sequence of DH altered patterns of gp140 epitope recognition after both primary and secondary immunization. Differences in epitope recognition were apparent both before and after class switching.
In BALB/c mice, the in developing B cells for RF1 and the amino acids it encodes is a product of a predilection for rearrangement by deletion, the frequent occurrence of stop codons in RF3, a bias towards rearrangement at sites of sequence microhomology between the 3’ end of the DH and the 5’ end of the JH that favors rearrangement into RF1, and an ATG start site upstream of RF2 that permits production of a truncated Dμ protein and by allelic exclusion impairs subsequent V→DRF2J rearrangement (Gu et al., 1990; Nadel and Feeney, 1995). To study the role that these biases may play in antibody production, we previously generated a panel of mice with DH loci that promote alternative reading frame usage. In all three strains, twelve of the thirteen D gene segments have been deleted (Schroeder et al., 2010a). The ΔD-DFL strain retains the most frequently expressed DH gene, DFL16.1, with its normal preference for tyrosine-enriched RF1. The ΔD-DμFS strain uses a modified DFL16.1 gene segment with two flanking nucleotide insertions. The first shifts the upstream Dμ ATG to be in frame with RF1, and the second shifts the terminal microhomology between 3’DH and 5’JH to be in frame with RF2. These frameshifts enhance use of hydrophobic RF2 in place of hydrophilic RF1. The ΔD-iD strain contains a modified DFL16.1 gene segment wherein the central core of DFL16.1 has been replaced by inverted DSP DSP2.2. The iRF1 DSP2.2 sequence is in frame with and retains terminal DFL16.1 RF1 DH and JH microhomology, and the upstream ATG start site is in frame with the iRF2 sequence of DSP2.2. iRF1 enriches for arginine in place of tyrosine.
B cells in D-altered mice undergo the normal processes of VDJ recombination and N addition to create large, diverse, and polyclonal H chain repertoires. The representation of V gene segment utilization appears minimally affected by these changes in CDR-H3 content (Schroeder et al., 2010a). B cells using D-altered alleles pass through all the normal checkpoints of B cell development and they respond to antigenic challenge by undergoing class switching, somatic hypermutation and affinity maturation. Deletion of the 3’ terminal DQ52 gene segment leads to doubling of the use of JH1 and halving of the use of JH4 when compared to wild type (Nitschke et al., 2001). The effect of this deletion on JH, which was common to all three D altered strains, may have contributed to some of the differences between the D-altered strains and wild-type. However, the similarity in JH usage among the three D-altered strains allowed us to attribute the differences in antibody responses to the change in the pattern of amino acids contributed by the DH to CDR-H3, which lies at the center of the antigen binding site, and is the starting point for affinity maturation.
In previous studies we have shown that both the total amount of antibody produced in response to immunization and the ability of the antibodies that are produced to neutralize pathogens can be affected by changes in the sequence of the DH (Nguyen et al., 2007). For example, in the response to influenza virus A the total amount of antibody produced by ΔD-DFL and ΔD-iD mice was similar to WT. However, whereas heterosubtypic immunity, the cross-protection against a serotype different from the one used for immunization, was maintained in the ΔD-DFL mice, ΔD-iD mice exhibited increased morbidity and mortality. This suggested to us that either the epitopes being bound by the ΔD-iD mice were different from those bound by WT or ΔD-DFL, or the quality of binding differed between the strains.
To test the hypothesis that global epitope recognition might be influenced by the sequence of the DH, for this work we serially immunized our panel of mice with JR-FL gp140, an HIV envelope antigen. We then assessed end-point titers to well-recognized epitopes on that antigen. In accordance with our hypothesis, serum reactivity to the various epitopes differed between the strains. Although with a total of ten mice tested a number of the differences that we observed did not reach statistical significance; statistical significance was achieved for reactivity against the V1V2 NQ epitopes and, to a lesser extent, the 4E10 MPER epitope. For these epitopes, the response of the mice biased towards a rarely used inverted RF1 that encodes charged residues exceeded that of wild type. We then re-analyzed the data using OD as a continuous variable and testing for the global effect of DH genotype, antibody type (IgM and IgG), and epitope. In each case, highly significant differences were found as a result of the change in DH gene sequence and thus global CDR-H3 content.
In two instances, reactivity with the entire gp140 and with the 4E10 epitope, the serum IgM response of the ΔD-DμFS mice was very low, but the IgG response became very high (Fig. 2). It is possible that RF2, whose usage is enhanced in these mice, provided an easier path for somatic mutation to generate more highly reactive antibodies with T cell help. Testing of this hypothesis will require cloning and sequencing of the IgM and IgG antibodies in these mice.
To gain a greater insight into these patterns of epitope recognition, we turned to peptide printing technology. This enabled us to not only see overlapping or discordant epitopes on the envelope antigen, but also to assess the intensity of the binding. The epitopes bound by WT and ΔD-DFL serum distributed in high and low intensity areas, with modest variation in hydrophobicity. The epitopes bound by ΔD-DμFS sera showed a sparse distribution pattern. Markedly, the epitopes bound by ΔD-iD sera had a broader reactivity by charge/hydrophobicity than the other three strains, but fewer reactivities of high response intensity (Fig. 5).
The three strains that we have created all use DH sequence that is normally present in the germline; we simply altered the frequency at which the sequences belonging to the various reading frames were used. We have not yet created mice with totally novel DH sequence, thus the three strains underestimate the range of diversity of CDR-H3 that could be generated. It seems likely that the use of synthetic DH segments containing sequences not seen in the normal germline would yield an even greater divergence in epitope recognition than what we observed in our extant panel of mice.
We selected HIV gp140 as our test antigen because it has been extensively studied and because it has been very difficult to elicit effective antibodies against this pathogen (Lutje Hulsik et al., 2013; West et al., 2014). Our studies here were not designed to test for the functional effects of antibody production by the different D-altered mice on the ability of the mice to neutralize HIV. However, we would suggest that our findings do offer one potential explanation for why broadly neutralizing antibodies against HIV often contain extensive somatic hypermutation. We propose that the wild-type germline antibody repertoire has been selected by evolution to respond preferentially to a relatively restricted range of epitopes with specific physico-chemical properties. The epitopes that drive this phenomenon are yet to be defined, but could include self-antigens, common components of the microbiota, or common environmental antigens. It would appear that HIV has responded to this evolutionary pressure by ensuring that its neutralizing epitopes contain sequences or structures that are difficult for the normal antibody repertoire to respond to. We speculate that these pressures may also explain why it has been difficult to create protective vaccines against certain other pathogens as well.
In summary, our findings suggest that natural selection of DH gene sequence can influence the likelihood and intensity of the production of antibody against individual epitopes. These findings may help in the interpretation of immune responses to various vaccines and could help develop tools for predicting the likelihood of responses to vaccines under development.
Methods
Mice
We previously altered the DH locus in BALB/c mice by deleting all but one DH and forcing use of an alternative reading frame in that DH (Ippolito et al., 2006; Ivanov et al., 2005; Schelonka et al., 2005; Schelonka et al., 2008; Zemlin et al., 2008) (Fig. 1A).
Immunization
All experiments with mice were approved by and performed in compliance with Institutional Animal Care Use Committee (IACUC) regulation. Cohorts of ten each of homozygous WT, ΔD-DFL, ΔD-DμFS, and ΔD-iD BALB/c mice were serially immunized with vaccinia-derived HIV-1 JR-FL gp140. Each mouse was injected intraperitoneally with antigenic protein (8ul JRFL peptide with 162ul saline) and 10µl of 20-mer oCpG (5’ TCCATGACGTTCCTGACGTT 3’) (Midland Certified Reagent Company, Midland, TX) mixed with 20µl Emulsigen (MVP laboratories, Omaha, NE) as the adjuvant (200µl total volume/mouse). The primary injection was followed by four booster injections at two weeks intervals. The mice were bled 10 days after each immunization to collect serum for the antibody-binding assays (Liao et al., 2006a; Verkoczy et al., 2011; Verkoczy et al., 2013) (Fig. 1C).
Serum Ab binding assays
Binding to the original recombinant JR-FL gp140 and to selected peptide epitopes was examined by ELISA, as previously described (Liao et al., 2006a; Verkoczy et al., 2011; Verkoczy et al., 2013). Recombinant gp140 (group M CON-S gp140CFI) was produced in 293T cells by recombinant vaccinia viruses. The two tested peptides contained within gp120 are the V1V2 peptide and a variant V1V2-N156Q, N160Q produced as described (Liao et al., 2013; Yates et al., 2014). Within gp41, the three tested peptides are Sp400-BAL (RVLAVERYLRDQQLLGIWGCSGKLICTTAVPWNASWSNKSLNKI) from the immunodominant region; and from the MPER region the 2F5 epitope sp62 (QQEKNEQELLELDKWASLWN) and the 4E10 epitope (SLWNWFNITNWLWYIK). The data was collected and measured by endpoint titer 1000 × Log2 (l/dil). The Y-axis was transformed into log10 scale.
Linear peptide microarray analysis
Each mouse of each strain was numbered 1–10. We selected mice 4 and 5 in each strain to test. Sera obtained prior to immunization and after the 2nd and 4th immunizations were used for the PEPperPRINT® Chip assay to assess binding to small linear epitopes from HIV-1 JR-FL gp160. A total of 849 peptides of 13aa length with 12aa length overlap were used, covering the complete gp160. Incubation of the peptide arrays with pre-immune and immune sera at dilutions of 1:1,000 was followed by staining with secondary goat anti-mouse IgG (H+L) IRDye680 antibody and read-out at a scanning intensity of 5. A software algorithm breaks down fluorescence intensities of each spot into foreground and background signal, and calculates the standard deviation of median foreground intensities. Based on averaged median foreground intensities, intensity maps were generated and antibody binding to the peptide map was highlighted by an intensity color code with red for high for low spot intensities (Fig. 3). The peptides bound by each mouse strain B1 and B3 sera were analyzed, and the collective binding peptides bound by both No.4 and No.5 mouse sera at both B1 and B3 were chosen for analysis (Supplementary Excel Table). Fluorescence intensity plots were made using OriginLab Origin V8.0. The normalized Kyte-Doolittle hydrophobicity scale (Eisenberg, 1984) was used to calculate average hydrophobicity of each 13 amino acid peptide (Supplementary Excel Table). Although this scale ranges from −1.3 (arginine) to + 1.7 (isoleucine), only the range from −0.4 (charged) to +0.5 (hydrophobic) is shown (Fig. 5).
Statistical analyses
One way ANOVA tests were performed in Prism Graphpad 6.0 in order to compare serum IgG and IgM binding to the HIV-1 JR-FL natural antigen. In addition, Tukey’s HSD tests were applied to all the analysis to adjust the multiple variables comparisons. Statistical significance was determined by a p value <0.001 (***), <0.01 (**), <0.05 (*). In addition to analyzing the titer values, which are discrete, we summed up all the OD values at all dilutions, which are continuous and correlated to antibody concentrations. We used the “lme4” R package (version 3.1.0) for fitting a mixed effect ANOVA model with DH genotype, antibody isotype, and epitope as predictors. Mouse was included as a random effect to capture the correlation among measurements from the same mouse. All three two-way interactions and the three-way interaction were included in the full model. Significance level was set as 0.05. For differences in the distribution of intensity, we used Fisher’s exact test.
Supplementary Material
Acknowledgments
The work was supported by the National Institute of Allergy and Infectious Diseases, National Institutes of Health Grants AI07051, AI48115, and AI090742.
References
- Alam SM, Scearce RM, Parks RJ, Plonk K, Plonk SG, Sutherland LL, Gorny MK, Zolla-Pazner S, Vanleeuwen S, Moody MA, et al. Human immunodeficiency virus type 1 gp41 antibodies that mask membrane proximal region epitopes: antibody binding kinetics, induction, and potential for regulation in acute infection. Journal of virology. 2008;82:115–125. doi: 10.1128/JVI.00927-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alt FW, Blackwell TK, Yancopoulos GD. Development of the primary antibody repertoire. Science. 1987;238:1079–1087. doi: 10.1126/science.3317825. [DOI] [PubMed] [Google Scholar]
- Banerjee K, Klasse PJ, Sanders RW, Pereyra F, Michael E, Lu M, Walker BD, Moore JP. IgG subclass profiles in infected HIV type 1 controllers and chronic progressors and in uninfected recipients of Env vaccines. AIDS Res Hum Retroviruses. 2010;26:445–458. doi: 10.1089/aid.2009.0223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binley JM, Sanders RW, Clas B, Schuelke N, Master A, Guo Y, Kajumo F, Anselma DJ, Maddon PJ, Olson WC, et al. A recombinant human immunodeficiency virus type 1 envelope glycoprotein complex stabilized by an intermolecular disulfide bond between the gp120 and gp41 subunits is an antigenic mimic of the trimeric virion-associated structure. Journal of virology. 2000;74:627–643. doi: 10.1128/jvi.74.2.627-643.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunel FM, Zwick MB, Cardoso RM, Nelson JD, Wilson IA, Burton DR, Dawson PE. Structure-function analysis of the epitope for 4E10, a broadly neutralizing human immunodeficiency virus type 1 antibody. Journal of virology. 2006;80:1680–1687. doi: 10.1128/JVI.80.4.1680-1687.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardoso RM, Zwick MB, Stanfield RL, Kunert R, Binley JM, Katinger H, Burton DR, Wilson IA. Broadly neutralizing anti-HIV antibody 4E10 recognizes a helical conformation of a highly conserved fusion-associated motif in gp41. Immunity. 2005;22:163–173. doi: 10.1016/j.immuni.2004.12.011. [DOI] [PubMed] [Google Scholar]
- Eisenberg D. Three-dimensional structure of membrane and surface proteins. Annual review of biochemistry. 1984;53:595–623. doi: 10.1146/annurev.bi.53.070184.003115. [DOI] [PubMed] [Google Scholar]
- Fugmann SD, Lee AI, Shockett PE, Villey IJ, Schatz DG. The RAG proteins and V(D)J recombination: complexes, ends, and transposition. Annual review of immunology. 2000;18:495–527. doi: 10.1146/annurev.immunol.18.1.495. [DOI] [PubMed] [Google Scholar]
- Gu H, Forster I, Rajewsky K. Sequence homologies, N sequence insertion and JH gene utilization in VH-D-JH joining: implications for the joining mechanism and the ontogenetic timing of Ly1 B cell and B-CLL progenitor generation. EMBO Journal. 1990;9:2133–2140. doi: 10.1002/j.1460-2075.1990.tb07382.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris A, Borgnia MJ, Shi D, Bartesaghi A, He H, Pejchal R, Kang YK, Depetris R, Marozsan AJ, Sanders RW, et al. Trimeric HIV-1 glycoprotein gp140 immunogens and native HIV-1 envelope glycoproteins display the same closed and open quaternary molecular architectures. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:11440–11445. doi: 10.1073/pnas.1101414108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heilkenbrinker U, Dietrich R, Didier A, Zhu K, Lindback T, Granum PE, Martlbauer E. Complex formation between NheB and NheC is necessary to induce cytotoxic activity by the three-component Bacillus cereus Nhe enterotoxin. PloS one. 2013;8:e63104. doi: 10.1371/journal.pone.0063104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ippolito GC, Schelonka RL, Zemlin M, Ivanov II, Kobayashi R, Zemlin C, Gartland GL, Nitschke L, Pelkonen J, Fujihashi K, et al. Forced usage of positively charged amino acids in immunoglobulin CDR-H3 impairs B cell development and antibody production. The Journal of experimental medicine. 2006;203:1567–1578. doi: 10.1084/jem.20052217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov II, Schelonka RL, Zhuang Y, Gartland GL, Zemlin M, Schroeder HW., Jr Development of the expressed Ig CDR-H3 repertoire is marked by focusing of constraints in length, amino acid use, and charge that are first established in early B cell progenitors. Journal of immunology. 2005;174:7773–7780. doi: 10.4049/jimmunol.174.12.7773. [DOI] [PubMed] [Google Scholar]
- Ivanov II, Link JM, Ippolito GC, Schroeder HW., Jr . Constraints on hydropathicity and sequence composition of HCDR3 are conserved across evolution. In: Zanetti M, Capra JD, editors. The Antibodies. London: Taylor and Francis Group; 2002. pp. 43–67. [Google Scholar]
- Kunert R, Wolbank S, Stiegler G, Weik R, Katinger H. Characterization of molecular features, antigen-binding, and in vitro properties of IgG and IgM variants of 4E10, an anti-HIV type 1 neutralizing monoclonal antibody. AIDS Res Hum Retroviruses. 2004;20:755–762. doi: 10.1089/0889222041524571. [DOI] [PubMed] [Google Scholar]
- Kyte J, Doolittle RF. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- LeBien TW, Tedder TF. B lymphocytes: how they develop and function. Blood. 2008;112:1570–1580. doi: 10.1182/blood-2008-02-078071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao HX, Bonsignori M, Alam SM, McLellan JS, Tomaras GD, Moody MA, Kozink DM, Hwang KK, Chen X, Tsao CY, et al. Vaccine induction of antibodies against a structurally heterogeneous site of immune pressure within HIV-1 envelope protein variable regions 1 and 2. Immunity. 2013;38:176–186. doi: 10.1016/j.immuni.2012.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao HX, Sutherland LL, Xia SM, Brock ME, Scearce RM, Vanleeuwen S, Alam SM, McAdams M, Weaver EA, Camacho Z, et al. A group M consensus envelope glycoprotein induces antibodies that neutralize subsets of subtype B and C HIV-1 primary viruses. Virology. 2006a;353:268–282. doi: 10.1016/j.virol.2006.04.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao HX, Sutherland LL, Xia SM, Brock ME, Scearce RM, Vanleeuwen S, Alam SM, McAdams M, Weaver EA, Camacho Z, et al. A group M consensus envelope glycoprotein induces antibodies that neutralize subsets of subtype B and C HIV-1 primary viruses. Virology. 2006b;353:268–282. doi: 10.1016/j.virol.2006.04.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutje Hulsik D, Liu YY, Strokappe NM, Battella S, El Khattabi M, McCoy LE, Sabin C, Hinz A, Hock M, Macheboeuf P, et al. A gp41 MPER-specific llama VHH requires a hydrophobic CDR3 for neutralization but not for antigen recognition. PLoS Pathog. 2013;9:e1003202. doi: 10.1371/journal.ppat.1003202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahmoud TI, Schroeder HW, Jr, Kearney JF. Limiting CDR-H3 diversity abrogates the antibody response to the bacterial polysaccharide alpha 1-->3 dextran. Journal of immunology. 2011;187:879–886. doi: 10.4049/jimmunol.1100957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLellan JS, Pancera M, Carrico C, Gorman J, Julien JP, Khayat R, Louder R, Pejchal R, Sastry M, Dai K, et al. Structure of HIV-1 gp120 V1/V2 domain with broadly neutralizing antibody PG9. Nature. 2011;480:336–343. doi: 10.1038/nature10696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meffre E, Milili M, Blanco-Betancourt C, Antunes H, Nussenzweig MC, Schiff C. Immunoglobulin heavy chain expression shapes the B cell receptor repertoire in human B cell development. The Journal of clinical investigation. 2001;108:879–886. doi: 10.1172/JCI13051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadel B, Feeney AJ. Influence of coding-end sequence on coding-end processing in V(D)J recombination. Journal of Immunology. 1995;155:4322–4329. [PubMed] [Google Scholar]
- Nguyen HH, Zemlin M, Ivanov II, Andrasi J, Zemlin C, Vu HL, Schelonka R, Schroeder HW, Jr, Mestecky J. Heterosubtypic immunity to influenza A virus infection requires a properly diversified antibody repertoire. Journal of virology. 2007;81:9331–9338. doi: 10.1128/JVI.00751-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nitschke L, Kestler J, Tallone T, Pelkonen S, Pelkonen J. Deletion of the DQ52 element within the Ig heavy chain locus leads to a selective reduction in VDJ recombination and altered D gene usage. Journal of Immunology. 2001;166:2540–2552. doi: 10.4049/jimmunol.166.4.2540. [DOI] [PubMed] [Google Scholar]
- Ofek G, McKee K, Yang Y, Yang ZY, Skinner J, Guenaga FJ, Wyatt R, Zwick MB, Nabel GJ, Mascola JR, et al. Relationship between antibody 2F5 neutralization of HIV-1 and hydrophobicity of its heavy chain third complementarity-determining region. Journal of virology. 2010;84:2955–2962. doi: 10.1128/JVI.02257-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schelonka RL, Ivanov II, Jung DH, Ippolito GC, Nitschke L, Zhuang Y, Gartland GL, Pelkonen J, Alt FW, Rajewsky K, et al. A single DH gene segment creates its own unique CDR-H3 repertoire and is sufficient for B cell development and immune function. Journal of immunology. 2005;175:6624–6632. doi: 10.4049/jimmunol.175.10.6624. [DOI] [PubMed] [Google Scholar]
- Schelonka RL, Zemlin M, Kobayashi R, Ippolito GC, Zhuang Y, Gartland GL, Szalai A, Fujihashi K, Rajewsky K, Schroeder HW., Jr Preferential use of DH reading frame 2 alters B cell development and antigen-specific antibody production. Journal of immunology. 2008;181:8409–8415. doi: 10.4049/jimmunol.181.12.8409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder HW, Jr, Zemlin M, Khass M, Nguyen HH, Schelonka RL. Genetic control of DH reading frame and its effect on B-cell development and antigen-specifc antibody production. Crit Rev Immunol. 2010a;30:327–344. doi: 10.1615/critrevimmunol.v30.i4.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder HW, Jr, Zemlin M, Khass M, Nguyen HH, Schelonka RL. Genetic control of DH reading frame and its effect on B-cell development and antigen-specifc antibody production. Critical reviews in immunology. 2010b;30:327–344. doi: 10.1615/critrevimmunol.v30.i4.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shukla NM, Salunke DB, Balakrishna R, Mutz CA, Malladi SS, David SA. Potent adjuvanticity of a pure TLR7-agonistic imidazoquinoline dendrimer. PloS one. 2012;7:e43612. doi: 10.1371/journal.pone.0043612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatatos L, Lim M, Cheng-Mayer C. Generation and structural analysis of soluble oligomeric gp140 envelope proteins derived from neutralization-resistant and neutralization-susceptible primary HIV type 1 isolates. AIDS Res Hum Retroviruses. 2000;16:981–994. doi: 10.1089/08892220050058407. [DOI] [PubMed] [Google Scholar]
- Stamatos NM, Mascola JR, Kalyanaraman VS, Louder MK, Frampton LM, Birx DL, VanCott TC. Neutralizing antibodies from the sera of human immunodeficiency virus type 1-infected individuals bind to monomeric gp120 and oligomeric gp140. Journal of virology. 1998;72:9656–9667. doi: 10.1128/jvi.72.12.9656-9667.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson CL, Wilson TJ, Strauch P, Colonna M, Pelanda R, Torres RM. Type I IFN enhances follicular B cell contribution to the T cell-independent antibody response. The Journal of experimental medicine. 2010;207:1485–1500. doi: 10.1084/jem.20092695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiller T, Tsuiji M, Yurasov S, Velinzon K, Nussenzweig MC, Wardemann H. Autoreactivity in human IgG+ memory B cells. Immunity. 2007;26:205–213. doi: 10.1016/j.immuni.2007.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trad A, Tanasa RI, Lange H, Zemlin M, Schroeder HWJ, Lemke H. Clonal progression during the thymus-dependent B cell response to the hapten oxazolone depends on the immunoglobulin DH gene repertoire. 2014 doi: 10.3389/fimmu.2014.00385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vale AM, Kapoor P, Skibinski GA, Elgavish A, Mahmoud TI, Zemlin C, Zemlin M, Burrows PD, Nobrega A, Kearney JF, et al. The link between antibodies to OxLDL and natural protection against pneumococci depends on D(H) gene conservation. The Journal of experimental medicine. 2013;210:875–890. doi: 10.1084/jem.20121861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verkoczy L, Chen Y, Bouton-Verville H, Zhang J, Diaz M, Hutchinson J, Ouyang YB, Alam SM, Holl TM, Hwang KK, et al. Rescue of HIV-1 broad neutralizing antibody-expressing B cells in 2F5 VH x VL knockin mice reveals multiple tolerance controls. Journal of immunology. 2011;187:3785–3797. doi: 10.4049/jimmunol.1101633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verkoczy L, Chen Y, Zhang J, Bouton-Verville H, Newman A, Lockwood B, Scearce RM, Montefiori DC, Dennison SM, Xia SM, et al. Induction of HIV-1 broad neutralizing antibodies in 2F5 knock-in mice: selection against membrane proximal external region-associated autoreactivity limits T-dependent responses. Journal of immunology. 2013;191:2538–2550. doi: 10.4049/jimmunol.1300971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinuesa CG, Chang PP. Innate B cell helpers reveal novel types of antibody responses. Nat Immunol. 2013;14:119–126. doi: 10.1038/ni.2511. [DOI] [PubMed] [Google Scholar]
- Wells TJ, Whitters D, Sevastsyanovich YR, Heath JN, Pravin J, Goodall M, Browning DF, O'Shea MK, Cranston A, De Soyza A, et al. Increased severity of respiratory infections associated with elevated anti-LPS IgG2 which inhibits serum bactericidal killing. J Exp Med. 2014;211:1893–1904. doi: 10.1084/jem.20132444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West AP, Jr, Scharf L, Scheid JF, Klein F, Bjorkman PJ, Nussenzweig MC. Structural insights on the role of antibodies in HIV-1 vaccine and therapy. Cell. 2014;156:633–648. doi: 10.1016/j.cell.2014.01.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yates NL, Liao HX, Fong Y, deCamp A, Vandergrift NA, Williams WT, Alam SM, Ferrari G, Yang ZY, Seaton KE, et al. Vaccine-induced Env V1–V2 IgG3 correlates with lower HIV-1 infection risk and declines soon after vaccination. Science translational medicine. 2014;6:228ra239. doi: 10.1126/scitranslmed.3007730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zemlin M, Schelonka RL, Ippolito GC, Zemlin C, Zhuang Y, Gartland GL, Nitschke L, Pelkonen J, Rajewsky K, Schroeder HW., Jr Regulation of repertoire development through genetic control of DH reading frame preference. Journal of immunology. 2008;181:8416–8424. doi: 10.4049/jimmunol.181.12.8416. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.