Abstract
Phosphatidylinositol 4,5-bisphosphate (PI4,5P2) is a lipid messenger that regulates a wide variety of cellular functions. The majority of cellular PI4,5P2 is generated by isoforms of the type I phosphatidylinositol phosphate kinases (PIPKI) that are generated from three genes, and each PIPKI isoform has a unique distribution and function in cells. It has been shown that the signaling specificity of PI4,5P2 can be determined by a physical association of PIPKs with PI4,5P2 effectors. IQGAP1 is newly identified as an interactor of multiple isoforms of PIPKs. Considering the versatile roles of IQGAP1 in cellular signaling pathways, IQGAP1 may confer isoform-specific roles of PIPKs in distinct cellular locations. In this mini review, the emerging roles of PIPKs that are regulated by an association with IQGAP1 will be summarized. Focuses will be on cell migration, vesicle trafficking, cell signaling, and nuclear events.
Keywords: Phosphoinositide, Phosphatidylinositol phosphate kinase, IQGAP, Cell signaling
Introduction
Phosphoinositides (phosphorylated phosphatidylinositol (PI) at the 3, 4 and 5 hydroxyl) are key lipid messengers regulating almost every aspect of eukaryotic cell physiology (Lemmon, 2008). Among the 7 isomers, PI 4,5-bisphosphate (PI4,5P2) has a central role in generating other phosphoinositide species and other lipid messengers (Balla, 2013; Di Paolo and De Camilli, 2006). Also, PI4,5P2 directly interacts with vast array of proteins called PI4,5P2 effectors and regulates their functions in the vicinity of various cellular membranes and in the nucleus (Choi et al., 2015; Irvine, 2003; Yin and Janmey, 2003). In most eukaryotic cells, PI4,5P2 is present at a higher concentration than other phosphoinositide isomers, and its overall concentration largely remains unchanged in response to extracellular stimuli (Insall and Weiner, 2001; Lemmon, 2008), recapitulating its housekeeping role in cellular physiology. However, its local concentration at a specific time and location is dramatically altered by the stimuli in order to efficiently regulate stimuli-directed functions. For example, in response to chemokines, PI4,5P2 and PI3,4,5P3 accumulate at the leading edges of migrating cells and regulate targeting and activity of numerous cytoskeleton regulatory proteins (Choi et al., 2013; Franca-Koh et al., 2007).
Changes in local PI4,5P2 concentration are controlled by redistribution and activation of PI4,5P2 generating enzymes in response to the stimuli (Balla, 2013; Divecha, 2010). In higher eukaryotes, the majority of PI4,5P2 is generated by type I and II phosphatidylinositol phosphate kinases (PIPKIs and PIPKIIs) by phosphorylating PI4P and PI5P, respectively (Heck et al., 2007). For both PIPKI and PIPKII there are three genes, (α, β and γ) and splice variants are found in higher eukaryotes (van den Bout and Divecha, 2009). Advances in the phosphoinositide signaling have revealed that many PI4,5P2 effectors are physically associated with PI4,5P2 generating enzymes and the association is closely regulated by the stimuli. In this mechanism, PI4,5P2 generation is tightly linked to effector activation. We and others have discovered PI4,5P2 effectors whose functions are directly regulated by association with PIPKI and PIPKII isoforms. These include actin regulatory proteins, regulators of membrane trafficking, enzymes, adaptors and transcription factors (Barlow et al., 2012; Choi et al., 2015; Ling et al., 2006; Shah et al., 2013).
Recently, the scaffold protein IQ motif containing GTPase activating protein 1 (IQGAP1) has been shown to bind phosphoinositides and to multiple PIPKI and PIPKII members (Choi et al., 2013). Interestingly, the functions that are regulated by IQGAP1 or the PIPKI and PIPKII isoforms often overlap, and they also share many common binding partners (Choi et al., 2015; Hedman et al., 2015). This suggests that IQGAP1 is a key regulator of phosphoinositide signaling. In this mini review, we will summarize the cellular functions of PIPKI and PIPKII isoforms that are regulated by IQGAP1 and how phosphoinositides may regulate IQGAPs.
Phosphatidylinositol phosphate kinase interaction with IQGAP1
The phosphatidylinositol phosphate kinase (PIPKs) family is distinct from other lipid kinases and protein kinases as no significant homology is found (Boronenkov et al., 1998; Heck et al., 2007; Sasaki et al., 2009). Based on sequence homology, domain structure and substrate preference, PIPKs can be classified into three distinct types (type I, II and III). In vivo, type I and II enzymes generate PI4,5P2 by phosphorylating PI4P and PI5P, respectively. Type III enzymes preferentially use PI3P as a substrate to generate PI3,5P2 in vivo (Heck et al., 2007). Type I and II PIPKs have three isoforms (α, β and γ) and multiple splicing variants of each isoform are also reported (van den Bout and Divecha, 2009). Only one type III enzyme is found in mammals, and it has additional domains (FYVE, DEP and TCP-1 domains) that are not found in other PIPKs (Sasaki et al., 2009). Interestingly, each member of the PIPKs shows unique subcellular localization, and it has been hypothesized that protein-protein interactions mediated by variable regions of PIPKs contribute site-specific function of PIPKs (Choi et al., 2015; Irvine, 2003; Yin and Janmey, 2003). For example, at least 6 splice variants of the γ isoform of type I PIPK (PIPKIγi1 to i6) are found, and they only differ in the C-terminal extensions (Schill and Anderson, 2009). PIPKIγi2 localizes at focal adhesion by interaction with talin1 (Di Paolo et al., 2002; Ling et al., 2002). PIPKIγi5 targets to endosomes by sorting nexins and LAPTM4B (Schill et al., 2014; Tan et al., 2015), and PIPKIγi4 localizes to the nucleus likely by β-catenin (Schramp et al., 2011). IQGAP1 is shown to interact with PIPKIα, PIPKIγi1, PIPKIγi5, PIPKIIα and PIPKIIβ in vitro and in vivo (Choi et al., 2013). IQGAP1 interaction with multiple PIPKs suggests that the highly conserved kinase core domain likely mediates the interaction.
IQGAP1 is a member of IQGAP protein family, and in higher eukaryotes three isoforms are expressed (IQGAP1, 2 and 3) (Smith et al., 2015). IQGAP1 scaffolds many distinct signaling pathways by interacting with numerous proteins. More than a hundred IQGAP1 interacting proteins have been discovered, and many of these directly or indirectly bind to IQGAP1 through the calponin homology domain, coiled-coil domain, WW domain, IQ domain, GTPase activating protein (GAP)-related domain, (GRD) and Ras GAP C-terminal domain (RGCT) (Hedman et al., 2015; Smith et al., 2015). For example, PIPKIs are shown to interact on the IQ domain of IQGAP1 (Choi et al., 2013). This raises the possibility that different PIPK isoforms compete for interaction with IQGAP1 by binding to similar regions of IQGAP1. However, overexpression or knockdown of a single PIPKI isoform has no impact on the interaction of other isoforms with IQGAP1 (Choi and Anderson, unpublished data), indicating that there are distinct PIPKI-IQGAP1 complexes in a cell. A recent unbiased proteomic analysis revealed that the protein copy number of IQGAP1 is at least two orders of magnitude higher than that of most interacting proteins (Schwanhausser et al., 2011), supporting the idea that IQGAP1 forms complexes with different PIPKI isoforms. Also, IQGAP1 is found in various locations in cells including leading edge membranes, cell-cell contact sites, intracellular membranes, cell-extracellular matrix (ECM) contact sites, cytoplasm and the nucleus where PIPKs are also found (Hedman et al., 2015; Smith et al., 2015).
IQGAP1 interaction with PIPKIγ regulates cell motility
In vitro, IQGAP1 binds to PIPKIγ with a moderate binding affinity (Choi et al., 2013). The binding is increased by stimuli for cell migration such as growth factors and ECM, suggesting their roles in cell migration. Biochemical and cell biological analyses revealed that PIPKIγ and IQGAP1 association with membrane fraction are increased in migrating cells (Choi et al., 2013). As the product of PIPKI, PI4,5P2, is a key mediator of cytoskeleton regulator protein binding to membrane, one could speculate that PI4,5P2 might modulate IQGAP1 membrane targeting. However, with a PI4,5P2-binding defective mutant, membrane binding remains largely unchanged, ruling out this possibility. Instead, IQGAP1 membrane recruitment is regulated by Rac1, Cdc42, Dia1 and PIPKIγ (Brandt et al., 2007; Choi et al., 2013; Fukata et al., 2002; Watanabe et al., 2004).
In unstimulated conditions, IQGAP1 is maintained in an inactive conformation that is mediated by at least two points of intramolecular interactions (Figure 1). Interaction of N- and C-terminal halves is relieved by phosphorylation at the serine1443 residue, allowing binding of PIPKIγ (Choi et al., 2013; Li et al., 2005). Rac1 and Cdc42 binding on the GRD domain or PI4,5P2 binding on the RGCT domain resolves the secondary interaction between the GRD and RGCT domains (Choi et al., 2013; Fukata et al., 2002; Watanabe et al., 2004). In turn, the unmasked RGCT domain recruits regulators of the actin cytoskeleton including the N-WASP and Arp2/3 complex, leading to de novo actin polymerization at the leading edge (Le Clainche et al., 2007; Rohatgi et al., 2000). Microtubule plus end organizers APC and CLIP-170 are also recruited to the RGCT domain and further support directional cell migration by regulating polarized vesicle trafficking to the leading edge (Fukata et al., 2002; Hedman et al., 2015; Watanabe et al., 2004).
Figure 1. A proposed mechanism of IQGAP1 activation.
IQGAP1 folds into an inactive conformation by intramolecular interactions. Phosphorylation at serine 1443 residue relieves the N- and C-termini interaction. This phosphorylation also allows PIPKIγ binding on the IQ domain. Small GTPases Rac1 and Cdc42 binding on the GRD domain or PI4,5P2 binding on RGCT domain further opens up IQGAP1 structure. The relieved RGCT domain recruits downstream effector proteins such as N-WASP, CLIP-170 and the exocyst complex.
Invadopodia are podosome-like protrusions of the plasma membrane that are found in invading cancer cells. Invadopodia are rich in cytoskeleton, its regulatory proteins, and matrix degrading proteases (Linder, 2007; Ridley, 2011). IQGAP1 controls cancer cell invasion into ECM by regulating invadopodia formation. At the tip of invadopodia, IQGAP1 recruits the exocyst, an octameric protein complex that mediates tethering of post-Golgi vesicles to the plasma membrane, and regulates deposition of invadopodium components (Sakurai-Yageta et al., 2008). Interestingly, the exocyst binding region is on the RGCT domain of IQGAP1 suggesting that invadopodia-targeted IQGAP1 might be activated by PIPKIγ and PI4,5P2. Consistent with this possibility, PIPKIγi2 is reported to interact with the exocyst and regulate β1-integrin deposition to the leading edge membrane (Thapa et al., 2012). Also, depletion of PIPKIγ reduces invadopodia formation (Choi and Anderson, unpublished data).
Consistent with cellular roles of PIPKIs and IQGAP1 in cell migration, PIPKIγ knockout mice are lethal beyond embryonic day 11.5 due to defects in cardiovascular development. In cells derived from the PIPKIγ knockout mice PI4,5P2 levels and cell motility are reduced (Wang et al., 2007). IQGAP1 knockout mice develop normally, but endothelial and vascular smooth muscle cells derived from the knockout mice show retarded motility and proliferation (Ikeda et al., 2005; Kohno et al., 2013). Also, IQGAP1 regulates hypertrophy and survival of cardiomyocytes upon pressure overload (Sbroggio et al., 2011a; Sbroggio et al., 2011b). These studies indicate that IQGAP1 and PIPKIγ knockout mice display defects in the cardiovascular system due to retarded cell motility. However, it remains to be tested if the interaction of IQGAP1 with PIPKIγ is required for the cardiovascular function in mice.
IQGAP1 interaction with PIPKIγ regulates phosphoinositide signaling
PIPKIs generate PI4,5P2 and PI4,5P2 can be further phosphorylated by class I phosphoinositide 3-kinases (PI3Ks) to generate PI3,4,5P3. Several key signaling proteins such as phosphoinositide-dependent kinase 1(PDK1) and Akt have PI3,4,5P3 binding modules and PI3,4,5P3 binding regulates their activities and targeting to membranes (Fayard et al., 2005). As expected, PIPKI isoforms are required for Akt activation. In Dictyostelium, depletion of a PIPKI isoform reduces approximately 90% of PI4,5P2 levels, leading to attenuation of Akt phosphorylation (Fets et al., 2014). In human keratinocytes, knockdown of PIPKIα reduces extracellular, calcium-induced Akt activation (Xie et al., 2009). In advanced human prostate cancer biopsies, PIPKIα expression is correlated with elevated PI3,4,5P3 levels and Akt activation (Semenas et al., 2014). In human breast cancer cell lines, depletion of PIPKIγi2 reduces serum and ECM-stimulated Akt phosphorylation (Thapa et al., 2015). In this study, PIPKIγi2 is coimmunoprecipitated with class I PI3Ks.
IQGAP1 also has a clear role in Akt activation. In the heart of IQGAP1-null mice, Akt phosphorylation is reduced in response to prolonged pressure overload, and IQGAP1 is coimmunoprecipitated with Akt (Sbroggio et al., 2011b). In hepatocellular carcinoma, IQGAP1 overexpression is correlated with Akt activation (Chen et al., 2010). Also, IQGAP1 regulates activity of Akt downstream target TOR complex 1 (Tekletsadik et al., 2012). However, the detailed mechanism of how IQGAP1 regulates Akt activation remains to be defined. As IQGAP1 interacts with PIPKIγ and Akt is shown to form a complex with IQGAP1 and PIPKIs separately, a possible mechanism is by forming an IQGAP1-PIPKIs-Akt ternary complex. In the complex, locally generated PI4,5P2 by PIPKIγ or other PIPKIs could be converted to PI3,4,5P3 by the action of class I PI3Ks and then PI3,4,5P3 will recruit and activate Akt. Consistent with this possibility, class I PI3Ks are shown to interact with PIPKIγi2, (Thapa et al., 2015) and, in a proteomic analysis, IQGAP1 has been shown to bind to an isoform of class I PI3K catalytic subunit (Brehme et al., 2009). Another possibility comes from the finding that IQGAP1 binds to both PI4,5P2 and PI3,4,5P3 (Dixon et al., 2011; Dixon et al., 2012). In this scenario, PI4,5P2 and PI3,4,5P3 could be bound to IQGAP1 and passed to class I PI3Ks and Akt for direct activation. Indeed, several PI4,5P2 sequestering proteins function as PI4,5P2 reservoirs, and the stored PI4,5P2 can be used by PI4,5P2 effectors in response to stimuli (McLaughlin and Murray, 2005). The switching stimuli include activation of membrane receptors, and both IQGAP1 and PIPKIγ are known to interact and regulate membrane receptors such as integrins and growth factor receptors (Hedman et al., 2015; Thapa and Anderson, 2013; Thapa et al., 2015). Additionally, IQGAP1 interacts with phosphatase and tensin homolog (PTEN) which reverts PI3,4,5P3 to PI4,5P2 (Gunaratne et al., 2011), suggesting that the IQGAP1-PIPKIs nexus functions as a master regulator of phosphoinositide signaling by fine-tuning its activity spatiotemporally.
Could IQGAP1 and PIP kinases regulate nuclear signaling?
Four isoforms of PIPKs are shown to localize in the nucleus (PIPKIα, PIPKIγi4, PIPKIIα and PIPKIIβ) and all of these isoforms potentially interact with IQGAP1 (Barlow et al., 2012; Choi et al., 2013; Shah et al., 2013). Consistently, roles of IQGAP1 in the nucleus are emerging. IQGAP1 accumulates in the nucleus in early S phase and regulates cell cycle progression from S to G2/M phase (Johnson et al., 2011). Also, IQGAP1 interacts with many modulators of Wnt signaling pathway and, importantly, IQGAP1 regulates nuclear translocation and transcriptional activity of β-catenin (Carmon et al., 2014; Goto et al., 2013a; Goto et al., 2013b). We have shown that PIPKIγi4 also controls transcriptional activity of β-catenin in the nucleus (Schramp et al., 2011), suggesting that IQGAP1 and PIPKIγ may work together to regulate β-catenin. However, how IQGAP1 and PIPKIγ work together to control β-catenin is not defined.
Many nuclear proteins such as transcription regulatory proteins, RNA polymerases, kinases and translation regulators have been identified and validated to bind to phosphoinositides; phosphoinositide binding controls their activities (Choi et al., 2015; Lewis et al., 2011). The nuclear receptor SF-1 is one of the best characterized (Blind, 2014). Hydrophobic acyl chains of phosphoinositides (PI4,5P2 and PI3,4,5P3) are sequestered in the ligand-binding domain of SF-1. However, the structure shows that the phosphoinositide head groups are fully exposed to solvent (Blind et al., 2014). Interestingly, the solvent-exposed head groups can be further modified by kinases and phosphatases. SF-1 bound PI4,5P2 is phosphorylated by the lipid kinase IMPK, and PTEN dephosphorylates PI3,4,5P3 (Blind et al., 2012). Functionally, the PI3,4,5P3 binding stabilizes co-activator binding and increases transcriptional activity of SF-1 (Blind et al., 2014; Lin et al., 2009). We speculate that PIPKIγi4 or other PIPK isoforms regulate β-catenin in a similar manner. PI4,5P2 generated by PIPKIγi4 might bind and control β-catenin directly, or its regulator proteins and IQGAP1 might function as a scaffold that brings PIPKIγi4 to β-catenin.
The concentration of PI4P is at least 20 times higher than that of PI5P in cells (Lemmon, 2008). Thus, it has been believed that most PI4,5P2 is generated by PIPKI isoforms and a smaller pool by PIPKII isoform. Although the relative concentration of phosphoinositide in the nucleus remains to be determined, the roles of PIPKII isoforms in the nucleus appear to regulate the levels of PI5P, and this could be more important than generating PI4,5P2 (Shah et al., 2013). Consistently, nuclear PIPKII isoforms have been implicated in regulation of PI5P-binding proteins. PI5P binds to a set of plant homeodomain (PHD) finger-containing nuclear proteins, and PI5P binding regulates their targeting and activities. For example, a modulator of histone acetyltransferase, inhibitor of growth 2 (ING2), binds to PI5P by the PHD domain, and PI5P binding regulates nuclear localization of ING2. Depletion of nuclear PI5P by PIPKIIβ overexpression reduces nuclear localization of ING2 and acetylation of p53 by ING2 (Gozani et al., 2003). Recently, PI5P was shown to bind to another PHD finger protein (the transcription complex component TAF3) and regulate basal transcription activity (Stijf-Bultsma et al., 2015). The roles of IQGAP1 in association with PIPKIIs are totally unknown. Interestingly, IQGAP1 is predicted to bind to ING1 and p53 (http://dcv.uhnres.utoronto.ca/FPCLASS/ppis/). We hypothesize that IQGAP1 and PIPKIIs might function together to control PI5P availability and regulate PI5P binding proteins in the nucleus. Alternatively, as IQGAP1 is shown to interact with PI5P (Choi et al., 2013), IQGAP1 might function as a PI5P reservoir in the nucleus.
IQGAP1 and IQGAP2 function differently in regulation of cell signaling
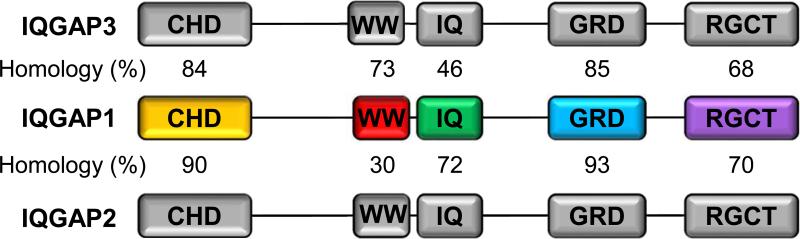
IQGAP2 is homologous to IQGAP1. At the amino acid sequence level, IQGAP2 is 62% identical to IQGAP1. Furthermore, IQGAP2 has five domains that are also found in IQGAP1 with varying identity to IQGAP1 (Figure 2). However, these two similar proteins function oppositely in cells. IQGAP1 is implicated in many human cancers, and its oncogenic functions are validated in diverse cancer models, whereas IQGAP2 functions unequivocally as a tumor suppressor in multiple studies (Brown and Sacks, 2006; Hedman et al., 2015; Smith et al., 2015; White et al., 2009). Until now, how IQGAP1 and IQGAP2 function differently remained largely unknown. Homology between the two proteins raises the possibility that IQGAP2 might compete for binding to IQGAP1-interacting proteins that attenuate IQGAP1's cellular activities. In support of this possibility, IQGAP2-deficient mice develop late onset of hepatocellular carcinoma, which is dependent on IQGAP1 expression (Schmidt et al., 2008). Also, IQGAP2 interacts with many IQGAP1 binding partners including calmodulin, Arp2/3 complex, β-catenin, Rac1, Cdc42 and PI3,4,5P3 (Hedman et al., 2015). However, it remains to be tested whether IQGAP2 competes with IQGAP1 for binding to these molecules. PIPKIs interact with IQGAP1 on the IQ domain that has 72% identity between IQGAP1 and IQGAP2 (Figure 2). Thus, it is likely that PIPKIs also interact with IQGAP2 on the IQ domain. Importantly, in our preliminary data, IQGAP1 depletion increases IQGAP2 interaction with a PIPKI isoform, supporting the competitive binding of IQGAP1 and IQGAP2 (Choi and Anderson, unpublished data).
Figure 2. Homology between IQGAP1 and IQGAP2.
Both IQGAP1 and IQGAP2 contain five domains and each domain has different homology. Percent of identity is shown in the middle.
The intramolecular interactions of IQGAP1 block the binding to downstream effector molecules (Figure 1). IQGAP2 possibly forms a complex with IQGAP1 that blocks IQGAP1 interactions with its effectors. For example, the relieving of the interaction between GRD and RGCT domains is required for the exocyst complex, CLIP-170 and N-WASP binding to the RGCT domain (Fukata et al., 2002; Grohmanova et al., 2004; Le Clainche et al., 2007; Watanabe et al., 2004). The GRD domain of IQGAP2 might bind to the RGCT domain of IQGAP1 to inhibit the effector recruitment. Consistently, the GRD domain of IQGAP2 is highly homologous to IQGAP1 (93% identical) and, importantly, IQGAP2 is coimmunoprecipitated with IQGAP1 (Schmidt et al., 2008).
An proteomic study reveals that IQGAP1 is much more abundant than IQGAP2 (Schwanhausser et al., 2011), suggesting that IQGAP2 inhibits IQGAP1 functions not only by competing for binding partners. Indeed, minor alteration of IQGAP2 expression efficiently modulates signaling pathways that are regulated by IQGAP1 (White et al., 2010; Xie et al., 2012). In this regard, IQGAP2 may directly regulate IQGAP1 activity by inhibiting activators of IQGAP1 such as Rac1, Cdc42 and phosphoinositides. IQGAP2 is reported to bind to PI3,4,5P3, (Dixon et al., 2012) and the PI4,5P2 binding region on IQGAP1 is highly homologous to IQGAP2 (Choi et al., 2013), suggesting that IQGAP2 may also bind to PI4,5P2. In this scenario, IQGAP2 sequesters PI4,5P2 that is generated by PIPKIs by interacting with PIPKIs leading to inactivation of IQGAP1.
Summary and future prospects
PIPKs generate a lipid messenger PI4,5P2, and PI4,5P2 signaling specificity is determined by PIPK association with PI4,5P2 effectors (Choi et al., 2015). IQGAP1 interacts with multiple PIPK isoforms and itself is PI4,5P2 effector in regulation of cell migration (Choi et al., 2013). IQGAP1 assembles with PIPKs in various cellular locations to regulate other cellular processes by association with other PI4,5P2 effectors. This suggests that IQGAP1 functions as a signaling platform in regulation of phosphoinositide signaling. Cellular events that are regulated by PIPK association with IQGAP1 include cell migration, membrane trafficking, Akt signaling, and nuclear signaling. Further studies are needed to investigate the roles of IQGAP1 with PIPKs in the nucleus. Clearly, nonmembrane pools of phosphoinositides exist and regulate a variety of nuclear proteins (Shah et al., 2013), while the mechanism is less clear. We have shown that the activity of a poly(A) polymerase is directly regulated by PI4,5P2 that is generated by the associated PIPKIα (Laishram and Anderson, 2010; Li and Anderson, 2014; Li et al., 2012; Mellman et al., 2008). Study of the roles of IQGAP1 in this pathway would be interesting. Also, a direct contribution of PI4,5P2-generating enzymes in PI3,4,5P3 generation by PI3Ks is has long been suspected. As both IQGAP1 and PIPKIs have validated roles in Akt signaling, it would be worthwhile to investigate how IQGAP1 and PIPKIs regulate PI3K signaling. Finally, the roles of IQGAP2 in control of PIPKs-associated IQGAP1 would be of great interest.
Acknowledgements
Due to space constraints some relevant studies may not have been referenced. This work was supported by NIH grants to RAA and American Heart Association fellowships to SC.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
The authors have declared no conflict of interest.
References
- Balla T. Phosphoinositides: tiny lipids with giant impact on cell regulation. Physiol Rev. 2013;93:1019–1137. doi: 10.1152/physrev.00028.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barlow CA, Laishram RS, Anderson RA. Nuclear phosphoinositides: a signaling enigma wrapped in a compartmental conundrum. Trends Cell Biol. 2012;20:25–35. doi: 10.1016/j.tcb.2009.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blind RD. Disentangling biological signaling networks by dynamic coupling of signaling lipids to modifying enzymes. Adv Biol Regul. 2014;54:25–38. doi: 10.1016/j.jbior.2013.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blind RD, Sablin EP, Kuchenbecker KM, Chiu HJ, Deacon AM, Das D, Fletterick RJ, Ingraham HA. The signaling phospholipid PIP3 creates a new interaction surface on the nuclear receptor SF-1. Proc Natl Acad Sci U S A. 2014;111:15054–15059. doi: 10.1073/pnas.1416740111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blind RD, Suzawa M, Ingraham HA. Direct modification and activation of a nuclear receptor-PIP(2) complex by the inositol lipid kinase IPMK. Sci Signal. 2012;5:ra44. doi: 10.1126/scisignal.2003111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boronenkov IV, C. LJ, Umeda M, Anderson RA. Phosphoinositide signaling pathways in nuclei are associated with nuclear speckles containing pre-mRNA processing factors. Mol Biol Cell. 1998;9:3547–3560. doi: 10.1091/mbc.9.12.3547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandt DT, Marion S, Griffiths G, Watanabe T, Kaibuchi K, Grosse R. Dia1 and IQGAP1 interact in cell migration and phagocytic cup formation. J Cell Biol. 2007;178:193–200. doi: 10.1083/jcb.200612071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brehme M, Hantschel O, Colinge J, Kaupe I, Planyavsky M, Kocher T, Mechtler K, Bennett KL, Superti-Furga G. Charting the molecular network of the drug target Bcr-Abl. Proc Natl Acad Sci U S A. 2009;106:7414–7419. doi: 10.1073/pnas.0900653106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown MD, Sacks DB. IQGAP1 in cellular signaling: bridging the GAP. Trends Cell Biol. 2006;16:242–249. doi: 10.1016/j.tcb.2006.03.002. [DOI] [PubMed] [Google Scholar]
- Carmon KS, Gong X, Yi J, Thomas A, Liu Q. RSPO-LGR4 functions via IQGAP1 to potentiate Wnt signaling. Proc Natl Acad Sci U S A. 2014;111:E1221–1229. doi: 10.1073/pnas.1323106111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F, Zhu HH, Zhou LF, Wu SS, Wang J, Chen Z. IQGAP1 is overexpressed in hepatocellular carcinoma and promotes cell proliferation by Akt activation. Exp Mol Med. 2010;42:477–483. doi: 10.3858/emm.2010.42.7.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi S, Thapa N, Hedman AC, Li Z, Sacks DB, Anderson RA. IQGAP1 is a novel phosphatidylinositol 4,5 bisphosphate effector in regulation of directional cell migration. EMBO J. 2013;32:2617–2630. doi: 10.1038/emboj.2013.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi S, Thapa N, Tan X, Hedman AC, Anderson RA. PIP kinases define PI4,5P(2)signaling specificity by association with effectors. Biochim Biophys Acta. 2015;1851:711–723. doi: 10.1016/j.bbalip.2015.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Paolo G, De Camilli P. Phosphoinositides in cell regulation and membrane dynamics. Nature. 2006;443:651–657. doi: 10.1038/nature05185. [DOI] [PubMed] [Google Scholar]
- Di Paolo G, Pellegrini L, Letinic K, Cestra G, Zoncu R, Voronov S, Chang S, Guo J, Wenk MR, De Camilli P. Recruitment and regulation of phosphatidylinositol phosphate kinase type 1 gamma by the FERM domain of talin. Nature. 2002;420:85–89. doi: 10.1038/nature01147. [DOI] [PubMed] [Google Scholar]
- Divecha N. Lipid kinases: charging PtdIns(4,5)P2 synthesis. Current biology : CB. 2010;20:R154–157. doi: 10.1016/j.cub.2010.01.016. [DOI] [PubMed] [Google Scholar]
- Dixon MJ, Gray A, Boisvert FM, Agacan M, Morrice NA, Gourlay R, Leslie NR, Downes CP, Batty IH. A screen for novel phosphoinositide 3-kinase effector proteins. Mol Cell Proteomics. 2011;10:M110 003178. doi: 10.1074/mcp.M110.003178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon MJ, Gray A, Schenning M, Agacan M, Tempel W, Tong Y, Nedyalkova L, Park HW, Leslie NR, van Aalten DM, et al. IQGAP Proteins Reveal an Atypical Phosphoinositide (aPI) Binding Domain with a Pseudo C2 Domain Fold. J Biol Chem. 2012;287:22483–22496. doi: 10.1074/jbc.M112.352773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fayard E, Tintignac LA, Baudry A, Hemmings BA. Protein kinase B/Akt at a glance. J Cell Sci. 2005;118:5675–5678. doi: 10.1242/jcs.02724. [DOI] [PubMed] [Google Scholar]
- Fets L, Nichols JM, Kay RR. A PIP5 kinase essential for efficient chemotactic signaling. Curr Biol. 2014;24:415–421. doi: 10.1016/j.cub.2013.12.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franca-Koh J, Kamimura Y, Devreotes PN. Leading-edge research: PtdIns(3,4,5)P3 and directed migration. Nature cell biology. 2007;9:15–17. doi: 10.1038/ncb0107-15. [DOI] [PubMed] [Google Scholar]
- Fukata M, Watanabe T, Noritake J, Nakagawa M, Yamaga M, Kuroda S, Matsuura Y, Iwamatsu A, Perez F, Kaibuchi K. Rac1 and Cdc42 capture microtubules through IQGAP1 and CLIP-170. Cell. 2002;109:873–885. doi: 10.1016/s0092-8674(02)00800-0. [DOI] [PubMed] [Google Scholar]
- Goto T, Sato A, Adachi S, Iemura S, Natsume T, Shibuya H. IQGAP1 protein regulates nuclear localization of beta-catenin via importin-beta5 protein in Wnt signaling. J Biol Chem. 2013a;288:36351–36360. doi: 10.1074/jbc.M113.520528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goto T, Sato A, Shimizu M, Adachi S, Satoh K, Iemura S, Natsume T, Shibuya H. IQGAP1 functions as a modulator of dishevelled nuclear localization in Wnt signaling. PLoS One. 2013b;8:e60865. doi: 10.1371/journal.pone.0060865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gozani O, Karuman P, Jones DR, Ivanov D, Cha J, Lugovskoy AA, Baird CL, Zhu H, Field SJ, Lessnick SL, et al. The PHD finger of the chromatin-associated protein ING2 functions as a nuclear phosphoinositide receptor. Cell. 2003;114:99–111. doi: 10.1016/s0092-8674(03)00480-x. [DOI] [PubMed] [Google Scholar]
- Grohmanova K, Schlaepfer D, Hess D, Gutierrez P, Beck M, Kroschewski R. Phosphorylation of IQGAP1 modulates its binding to Cdc42, revealing a new type of rho-GTPase regulator. J Biol Chem. 2004;279:48495–48504. doi: 10.1074/jbc.M408113200. [DOI] [PubMed] [Google Scholar]
- Gunaratne J, Goh MX, Swa HL, Lee FY, Sanford E, Wong LM, Hogue KA, Blackstock WP, Okumura K. Protein interactions of phosphatase and tensin homologue (PTEN) and its cancer-associated G20E mutant compared by using stable isotope labeling by amino acids in cell culture-based parallel affinity purification. J Biol Chem. 2011;286:18093–18103. doi: 10.1074/jbc.M111.221184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heck JN, Mellman DL, Ling K, Sun Y, Wagoner MP, Schill NJ, Anderson RA. A conspicuous connection: structure defines function for the phosphatidylinositol-phosphate kinase family. Crit Rev Biochem Mol Biol. 2007;42:15–39. doi: 10.1080/10409230601162752. [DOI] [PubMed] [Google Scholar]
- Hedman AC, Smith JM, Sacks DB. The biology of IQGAP proteins: beyond the cytoskeleton. EMBO Rep. 2015;16:427–446. doi: 10.15252/embr.201439834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda S, Yamaoka-Tojo M, Hilenski L, Patrushev NA, Anwar GM, Quinn MT, Ushio-Fukai M. IQGAP1 regulates reactive oxygen species-dependent endothelial cell migration through interacting with Nox2. Arterioscler Thromb Vasc Biol. 2005;25:2295–2300. doi: 10.1161/01.ATV.0000187472.55437.af. [DOI] [PubMed] [Google Scholar]
- Insall RH, Weiner OD. PIP3, PIP2, and cell movement--similar messages, different meanings? Dev Cell. 2001;1:743–747. doi: 10.1016/s1534-5807(01)00086-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irvine RF. Nuclear lipid signalling. Nat Rev Mol Cell Biol. 2003;4:349–360. doi: 10.1038/nrm1100. [DOI] [PubMed] [Google Scholar]
- Johnson M, Sharma M, Brocardo MG, Henderson BR. IQGAP1 translocates to the nucleus in early S-phase and contributes to cell cycle progression after DNA replication arrest. Int J Biochem Cell Biol. 2011;43:65–73. doi: 10.1016/j.biocel.2010.09.014. [DOI] [PubMed] [Google Scholar]
- Kohno T, Urao N, Ashino T, Sudhahar V, Inomata H, Yamaoka-Tojo M, McKinney RD, Fukai T, Ushio-Fukai M. IQGAP1 links PDGF receptor-beta signal to focal adhesions involved in vascular smooth muscle cell migration: role in neointimal formation after vascular injury. Am J Physiol Cell Physiol. 2013;305:C591–600. doi: 10.1152/ajpcell.00011.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laishram RS, Anderson RA. The poly A polymerase Star-PAP controls 3′-end cleavage by promoting CPSF interaction and specificity toward the pre-mRNA. The EMBO Journal. 2010;29:4132–4145. doi: 10.1038/emboj.2010.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Clainche C, Schlaepfer D, Ferrari A, Klingauf M, Grohmanova K, Veligodskiy A, Didry D, Le D, Egile C, Carlier MF, et al. IQGAP1 stimulates actin assembly through the N-WASP-Arp2/3 pathway. J Biol Chem. 2007;282:426–435. doi: 10.1074/jbc.M607711200. [DOI] [PubMed] [Google Scholar]
- Lemmon MA. Membrane recognition by phospholipid-binding domains. Nat Rev Mol Cell Biol. 2008;9:99–111. doi: 10.1038/nrm2328. [DOI] [PubMed] [Google Scholar]
- Lewis AE, Sommer L, Arntzen MO, Strahm Y, Morrice NA, Divecha N, D'Santos CS. Identification of nuclear phosphatidylinositol 4,5-bisphosphate-interacting proteins by neomycin extraction. Mol Cell Proteomics. 2011;10:M110 003376. doi: 10.1074/mcp.M110.003376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Anderson RA. Star-PAP controls HPV E6 regulation of p53 and sensitizes cells to VP-16. Oncogene. 2014;33:928–932. doi: 10.1038/onc.2013.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Laishram RS, Ji Z, Barlow CA, Tian B, Anderson RA. Star-PAP Control of BIK Expression and Apoptosis Is Regulated by Nuclear PIPKIalpha and PKCdelta Signaling. Molecular Cell. 2012;45:25–37. doi: 10.1016/j.molcel.2011.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, McNulty DE, Marler KJ, Lim L, Hall C, Annan RS, Sacks DB. IQGAP1 promotes neurite outgrowth in a phosphorylation-dependent manner. J Biol Chem. 2005;280:13871–13878. doi: 10.1074/jbc.M413482200. [DOI] [PubMed] [Google Scholar]
- Lin BC, Suzawa M, Blind RD, Tobias SC, Bulun SE, Scanlan TS, Ingraham HA. Stimulating the GPR30 estrogen receptor with a novel tamoxifen analogue activates SF-1 and promotes endometrial cell proliferation. Cancer Res. 2009;69:5415–5423. doi: 10.1158/0008-5472.CAN-08-1622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linder S. The matrix corroded: podosomes and invadopodia in extracellular matrix degradation. Trends Cell Biol. 2007;17:107–117. doi: 10.1016/j.tcb.2007.01.002. [DOI] [PubMed] [Google Scholar]
- Ling K, Doughman RL, Firestone AJ, Bunce MW, Anderson RA. Type I gamma phosphatidylinositol phosphate kinase targets and regulates focal adhesions. Nature. 2002;420:89–93. doi: 10.1038/nature01082. [DOI] [PubMed] [Google Scholar]
- Ling K, Schill NJ, Wagoner MP, Sun Y, Anderson RA. Movin’ on up: the role of PtdIns(4,5)P(2) in cell migration. Trends Cell Biol. 2006;16:276–284. doi: 10.1016/j.tcb.2006.03.007. [DOI] [PubMed] [Google Scholar]
- McLaughlin S, Murray D. Plasma membrane phosphoinositide organization by protein electrostatics. Nature. 2005;438:605–611. doi: 10.1038/nature04398. [DOI] [PubMed] [Google Scholar]
- Mellman D, Gonzales M, Song C, Barlow C, Wang P, Kendziorski C, Anderson R. A PtdIns4,5P2-regulated nuclear poly(A) polymerase controls expression of select mRNAs. Nature. 2008;451:1013–1017. doi: 10.1038/nature06666. [DOI] [PubMed] [Google Scholar]
- Ridley AJ. Life at the leading edge. Cell. 2011;145:1012–1022. doi: 10.1016/j.cell.2011.06.010. [DOI] [PubMed] [Google Scholar]
- Rohatgi R, Ho HY, Kirschner MW. Mechanism of N-WASP activation by CDC42 and phosphatidylinositol 4, 5-bisphosphate. J Cell Biol. 2000;150:1299–1310. doi: 10.1083/jcb.150.6.1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakurai-Yageta M, Recchi C, Le Dez G, Sibarita JB, Daviet L, Camonis J, D'Souza-Schorey C, Chavrier P. The interaction of IQGAP1 with the exocyst complex is required for tumor cell invasion downstream of Cdc42 and RhoA. J Cell Biol. 2008;181:985–998. doi: 10.1083/jcb.200709076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki T, Takasuga S, Sasaki J, Kofuji S, Eguchi S, Yamazaki M, Suzuki A. Mammalian phosphoinositide kinases and phosphatases. Prog Lipid Res. 2009;48:307–343. doi: 10.1016/j.plipres.2009.06.001. [DOI] [PubMed] [Google Scholar]
- Sbroggio M, Bertero A, Velasco S, Fusella F, De Blasio E, Bahou WF, Silengo L, Turco E, Brancaccio M, Tarone G. ERK1/2 activation in heart is controlled by melusin, focal adhesion kinase and the scaffold protein IQGAP1. J Cell Sci. 2011a;124:3515–3524. doi: 10.1242/jcs.091140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sbroggio M, Carnevale D, Bertero A, Cifelli G, De Blasio E, Mascio G, Hirsch E, Bahou WF, Turco E, Silengo L, et al. IQGAP1 regulates ERK1/2 and AKT signalling in the heart and sustains functional remodelling upon pressure overload. Cardiovasc Res. 2011b;91:456–464. doi: 10.1093/cvr/cvr103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schill NJ, Anderson RA. Two novel phosphatidylinositol-4-phosphate 5-kinase type Igamma splice variants expressed in human cells display distinctive cellular targeting. Biochem J. 2009;422:473–482. doi: 10.1042/BJ20090638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schill NJ, Hedman AC, Choi S, Anderson RA. Isoform 5 of PIPKIgamma regulates the endosomal trafficking and degradation of E-cadherin. J Cell Sci. 2014;127:2189–2203. doi: 10.1242/jcs.132423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt VA, Chiariello CS, Capilla E, Miller F, Bahou WF. Development of hepatocellular carcinoma in Iqgap2-deficient mice is IQGAP1 dependent. Mol Cell Biol. 2008;28:1489–1502. doi: 10.1128/MCB.01090-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schramp M, Thapa N, Heck J, Anderson R. PIPKIgamma regulates beta-catenin transcriptional activity downstream of growth factor receptor signaling. Cancer Res. 2011;71:1282–1291. doi: 10.1158/0008-5472.CAN-10-2480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwanhausser B, Busse D, Li N, Dittmar G, Schuchhardt J, Wolf J, Chen W, Selbach M. Global quantification of mammalian gene expression control. Nature. 2011;473:337–342. doi: 10.1038/nature10098. [DOI] [PubMed] [Google Scholar]
- Semenas J, Hedblom A, Miftakhova RR, Sarwar M, Larsson R, Shcherbina L, Johansson ME, Harkonen P, Sterner O, Persson JL. The role of PI3K/AKT-related PIP5K1alpha and the discovery of its selective inhibitor for treatment of advanced prostate cancer. Proc Natl Acad Sci U S A. 2014;111:E3689–3698. doi: 10.1073/pnas.1405801111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah ZH, Jones DR, Sommer L, Foulger R, Bultsma Y, D'Santos C, Divecha N. Nuclear phosphoinositides and their impact on nuclear functions. FEBS Journal. 2013;280:6295–6310. doi: 10.1111/febs.12543. [DOI] [PubMed] [Google Scholar]
- Smith JM, Hedman AC, Sacks DB. IQGAPs choreograph cellular signaling from the membrane to the nucleus. Trends Cell Biol. 2015;25:171–184. doi: 10.1016/j.tcb.2014.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stijf-Bultsma Y, Sommer L, Tauber M, Baalbaki M, Giardoglou P, Jones DR, Gelato KA, van Pelt J, Shah Z, Rahnamoun H, et al. The basal transcription complex component TAF3 transduces changes in nuclear phosphoinositides into transcriptional output. Mol Cell. 2015;58:453–467. doi: 10.1016/j.molcel.2015.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan X, Sun Y, Thapa N, Liao Y, Hedman AC, Anderson RA. LAPTM4B is a PtdIns(4,5)P2 effector that regulates EGFR signaling, lysosomal sorting, and degradation. EMBO J. 2015;34:475–490. doi: 10.15252/embj.201489425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tekletsadik YK, Sonn R, Osman MA. A conserved role of IQGAP1 in regulating TOR complex 1. J Cell Sci. 2012;125:2041–2052. doi: 10.1242/jcs.098947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thapa N, Anderson RA. PIP2 signaling, an integrator of cell polarity and vesicle trafficking in directionally migrating cells. Cell Adh Migr. 2013;6:409–412. doi: 10.4161/cam.21192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thapa N, Choi S, Tan X, Wise T, Anderson RA. Phosphatidylinositol Phosphate 5-Kinase Igamma and Phosphoinositide 3-Kinase/Akt Signaling Couple to Promote Oncogenic Growth. J Biol Chem. 2015;290:18843–18854. doi: 10.1074/jbc.M114.596742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thapa N, Sun Y, Schramp M, Choi S, Ling K, Anderson RA. Phosphoinositide signaling regulates the exocyst complex and polarized integrin trafficking in directionally migrating cells. Dev Cell. 2012;22:116–130. doi: 10.1016/j.devcel.2011.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Bout I, Divecha N. PIP5K-driven PtdIns(4,5)P2 synthesis: regulation and cellular functions. J Cell Sci. 2009;122:3837–3850. doi: 10.1242/jcs.056127. [DOI] [PubMed] [Google Scholar]
- Wang Y, Lian L, Golden JA, Morrisey EE, Abrams CS. PIP5KI gamma is required for cardiovascular and neuronal development. Proc Natl Acad Sci U S A. 2007;104:11748–11753. doi: 10.1073/pnas.0700019104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe T, Wang S, Noritake J, Sato K, Fukata M, Takefuji M, Nakagawa M, Izumi N, Akiyama T, Kaibuchi K. Interaction with IQGAP1 links APC to Rac1, Cdc42, and actin filaments during cell polarization and migration. Dev Cell. 2004;7:871–883. doi: 10.1016/j.devcel.2004.10.017. [DOI] [PubMed] [Google Scholar]
- White CD, Brown MD, Sacks DB. IQGAPs in cancer: a family of scaffold proteins underlying tumorigenesis. FEBS Lett. 2009;583:1817–1824. doi: 10.1016/j.febslet.2009.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White CD, Khurana H, Gnatenko DV, Li Z, Odze RD, Sacks DB, Schmidt VA. IQGAP1 and IQGAP2 are reciprocally altered in hepatocellular carcinoma. BMC Gastroenterol. 2010;10:125. doi: 10.1186/1471-230X-10-125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Y, Yan J, Cutz JC, Rybak AP, He L, Wei F, Kapoor A, Schmidt VA, Tao L, Tang D. IQGAP2, A candidate tumour suppressor of prostate tumorigenesis. Biochim Biophys Acta. 2012;1822:875–884. doi: 10.1016/j.bbadis.2012.02.019. [DOI] [PubMed] [Google Scholar]
- Xie Z, Chang SM, Pennypacker SD, Liao EY, Bikle DD. Phosphatidylinositol-4-phosphate 5-kinase 1alpha mediates extracellular calcium-induced keratinocyte differentiation. Mol Biol Cell. 2009;20:1695–1704. doi: 10.1091/mbc.E08-07-0756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin HL, Janmey PA. Phosphoinositide regulation of the actin cytoskeleton. Annu Rev Physiol. 2003;65:761–789. doi: 10.1146/annurev.physiol.65.092101.142517. [DOI] [PubMed] [Google Scholar]