Abstract
Objective
The purpose of this study was to assess differences in gene expression between cementoblasts and osteoblasts using gene profiling of cell populations isolated directly from osteocalcin-GFP transgenic mice.
Background
Cementum and bone are similar mineralized tissues, but cementum accumulates much more slowly, does not have vasculature or innervation or undergo remodeling. Despite these differences, there are no well-established markers to distinguish cementoblasts from other mature mineralizing cells such as osteoblasts and odontoblasts.
Methods
Osteocalcin-GFP (OC-GFP) reporter mice were used as they show labeling of cementoblasts, osteoblasts and odontoblasts, but not periodontal ligament fibroblasts, within the periodontium. We sorted cells digested from the molar root surface to isolate OC-GFP+ cementoblasts. Osteoblasts were isolated from calvarial digests. Microarray analysis was performed, and selected results were confirmed by real time PCR and immunostaining or in situ hybridization.
Results
Microarray analysis identified 95 genes that were expressed at least 2-fold higher in cementoblasts than osteoblasts. Our analysis indicated that the Wnt signaling pathway was differentially regulated, as were genes related to skeletal development. Real time PCR confirmed that expression of Wnt inhibitors Wif1 and Sfrp1 was elevated in cementoblasts compared to osteoblasts, and Wif1 expression was localized to the apical root region. In addition, the transcription factor Barx1 was expressed at higher levels in cementoblasts, and immunohistochemistry indicated that BARX1 expression was detectable in apical cementoblasts and cementocytes, but was not present in osteoblasts or odontoblasts.
Conclusion
The OC-GFP mouse provides a good model for selectively isolating cementoblasts, and allowed for identification of differentially expressed genes between cementoblasts and osteoblasts.
Keywords: Cementoblast, osteoblast, osteocalcin, gene expression, Wnt signaling pathway
Introduction
Advanced periodontal disease results in clinically significant loss of periodontal tissues including cementum and alveolar bone (1). Effective treatment of this condition relies on the ability to address the underlying infection or pathology leading to tissue loss, then to regenerate replacement tissues. This may be achieved by promoting differentiation of tissue resident stem/progenitor cells using a combination of biomaterials and growth factors (2). However, this requires an understanding of the differentiation of periodontal cell lineages, specifically cementoblasts (CB), alveolar bone osteoblasts, and periodontal ligament (PDL) fibroblasts. While research on osteoprogenitor cell commitment from mesenchymal stem cells has been extensively studied, cementoblast differentiation is not well understood. Study of cementoblasts has been hampered by difficulties in selectively labeling and isolating this cell population. Isolation of cementoblasts relies on distinguishing them from the adjacent PDL fibroblasts, and osteoblasts in the alveolar bone, which are also mesenchymal cells that produce collagenous extracellular matrix. There is still debate regarding whether cementoblasts genuinely represent a cell type distinct from osteoblasts (3). Currently, CBs are defined by their location, lining the tooth root surface, and morphology similar to osteoblasts. Cementum is similar to bone, consisting of a mineralized collagen matrix, and containing non-collagenous proteins including bone sialoprotein, osteocalcin (OC) and osteopontin, however it lacks vasculature and innervation (4). At least two different types of cementum are found in healthy teeth, acellular cementum which is critical for attachment of the tooth to the surrounding PDL, and cellular cementum, which appears to play a role in occlusional positioning (4). Studies directed at identifying CB specific genes, have determined a few genes that show some specificity to cementoblasts such as cementum attachment protein (CAP, Ptpla) and glucose transporter GLUT1 (Slc2a1) (5, 6).
There have been a number of studies examining gene and protein expression in cementum or cementoblasts in comparison to bone or other tooth components (6–8). These studies utilized cementoblast cell lines or a heterogenous population dissected by laser capture, or evaluated proteins deposited within matrix. However, despite identification of a few genes that are potentially cementoblast selective, there are still no validated genetic models to specifically study the cementoblast lineage. Identification of selective markers of cementoblasts would allow for studies targeting this cell population by overexpression or gene deletion approaches. We therefore sought to isolate cementoblasts directly and compare their gene expression to osteoblasts. To achieve this we have used an Osteocalcin-GFP (OC-GFP) reporter mouse that specifically identifies cementoblasts and osteoblasts (9). Microarray gene expression profiles of these two cell populations have been analyzed.
Materials & Methods
Mice
Animal protocols were approved by the Institutional Animal Care and Use Committee. OC-GFP mice were previously described (9).
Histology and immunohistochemistry
Jaws, teeth or calvaria were fixed, decalcified and frozen sections were prepared and imaged as previously described (10). Barx1 immunohistochemistry was performed on paraffin sections. Antigen retrieval was done with citrate-based Antigen Unmasking Solution (Vector Laboratories, Burlingame, CA) at 55°C overnight, prior to blocking and overnight incubation with anti-Barx1 (1:250) (Sigma Aldrich, St. Louis, MO), followed by secondary antibody and development with Vectastain ABC kit, and diaminobenzidine (DAB) kit (Vector Laboratories).
RNAscope in situ hybridization
Frozen sections (7µm) were obtained using the CryoJane system (Leica Biosystems, Nussloch, Germany). RNAscope assay was conducted with RNAscope BROWN kit according to the manufacturer’s instructions (Advanced Cell Diagnostics, Hayward, CA). The negative and positive controls were performed on the same samples using the negative and positive probes from the kit. Briefly, the endogenous peroxidase was blocked with the provided pretreatment 1 solution for 10 min at RT, followed by epitope retrieval by boiling in the provided buffer for 5 min, protease digestion for 30 min at 40°C in a hybridization oven (HybEZ Oven, Advanced Cell Diagnostics, Hayward, CA) and target hybridization for 2 h at 40°C followed by six sequential amplification steps. Sections were counterstained with Hematoxylin. Brown punctuate dots in the section are considered positive staining.
Isolation of cementoblast and osteoblast RNA
In order to obtain suitable cell numbers for sorting, tissues from multiple animals were pooled prior to digestion. Calvaria and jaws were isolated from 6–10 OC-GFP animals aged 5–7 weeks. Calvaria were dissected, cut into 2mm wide strips and digested at 37°C in PBS containing 0.2% collagenase A (Roche, Mannheim, Germany), 0.25% trypsin (Life Technologies, Carlsbad, CA) with agitation. The cells from the first 20 minute digest were discarded, and the remaining tissue digested for 1h and the cells collected (11). Eight molars per mouse were extracted and separated from alveolar bone under a dissecting microscope (total 48–80 molars). A single digestion was performed for 1h. OC-GFP+ cells were isolated using a FACSAria II (BD Biosciences, San Jose, CA). Gates were determined using cells isolated from non-transgenic animals. Cells were sorted into chilled culture medium containing 20% fetal bovine serum. Following sorting, the cell pellets were resuspended in Trizol reagent (Life Technologies), and after addition of 20µg glycogen (Roche), RNA was purified according to the manufacturer's instructions and 10–30ng of RNA were obtained from molars, while 100–250ng were generated from calvarial samples.
Gene expression analysis
RNA quality was verified on a Bioanalyzer RNA Nano Chip (Agilent Technologies, Santa Clara, CA), and biotinylated cRNA was generated from 10–250ng RNA using the MessageAmp Premier RNA Amplification Kit (Life Technologies) according to the manufacturer’s instructions. Biotinylated cRNA was hybridized to MouseRef-8 BeadChips (Illumina, San Diego, CA) for 16 hours at 58°C. BeadChips were then washed, stained with streptavidin-Cy3, and scanned on the Illumina BeadArray Reader. Microarray data were processed with the Bioconductor lumi package using a variance-stabilizing transformation algorithm for data normalization, then analyzed using the PBC pattern-based clustering program (12, 13). Gene ontology analysis was performed using the GOStat package on genes showing greater than 2-fold change in one direction between two selected groups. Lists were filtered to only include categories with >10 or <500 total genes and p<10−2, then categories containing very similar genes were grouped and a representative group with the lowest p value was included in the table presented. Microarray data were confirmed using reverse transcribed amplified cRNA using the Improm-II reverse transcription system (Promega, Madison, WI). SYBR green-based assays were performed using SYBR Select Master Mix, and 1µM final primer concentration in an Applied Biosystems 7900HT instrument. Primer sequences for the genes examined are shown in Supplemental Table 1. All assays included a dissociation curve step and a no-template control to ensure specificity of signal. GAPDH was used as a calibrator gene to normalize expression, and the ΔCt method was used to analyze data.
Results
Isolation of cementoblasts and osteoblasts
In order to specifically isolate cementoblasts, it is critical to separate them from adjacent PDL fibroblasts. Our group has previously developed mice with reporters that label various stages of the osteogenic lineage, and some of these were evaluated for selective expression in cementoblasts. Col2.3-GFP is expressed in cementoblasts as well as a subset of PDL fibroblasts (10) making it unsuitable for selective isolation of cementoblasts. We therefore evaluated OC-GFP expression in the periodontium, as OC expression has previously been shown by a number of investigators to be expressed in cementoblasts but not by PDL fibroblasts (3, 14). OC-GFP labels osteoblasts lining the calvarial bone surface (Fig. 1A). Within the periodontium it labels a single layer of cells on the surface of the root, consistent with a cementoblast phenotype in both the cervical (acellular) and apical regions (Fig. 1B–C). In addition, the reporter labels osteoblasts lining the alveolar bone and odontoblasts (Fig. 1B). To specifically isolate cementoblasts, teeth were extracted to retain cementoblasts and partial PDL, but to remove alveolar bone (Fig. 1D–E). Surface cells were obtained by enzymatic digestion (Fig. 1F–G). Cementocytes also express GFP (Fig. 1C), and we cannot exclude the possibility that some of the cementocytes located in close proximity to the cementum surface are also collected during enzymatic digestion. OC-GFP+ cells were then isolated by FACS as indicated in Figure 2. Periodontal digests yielded around 3000 GFP+ cells. Calvarial osteoblasts were chosen for comparison, as they are a readily accessible craniofacial osteoblast population unlikely to be contaminated by other mineralizing cell types such as cementoblasts. We were able to sort around 20,000 GFP+ cells from calvarial digests. RQI values for the RNA samples were between 6.8 and 8.6.
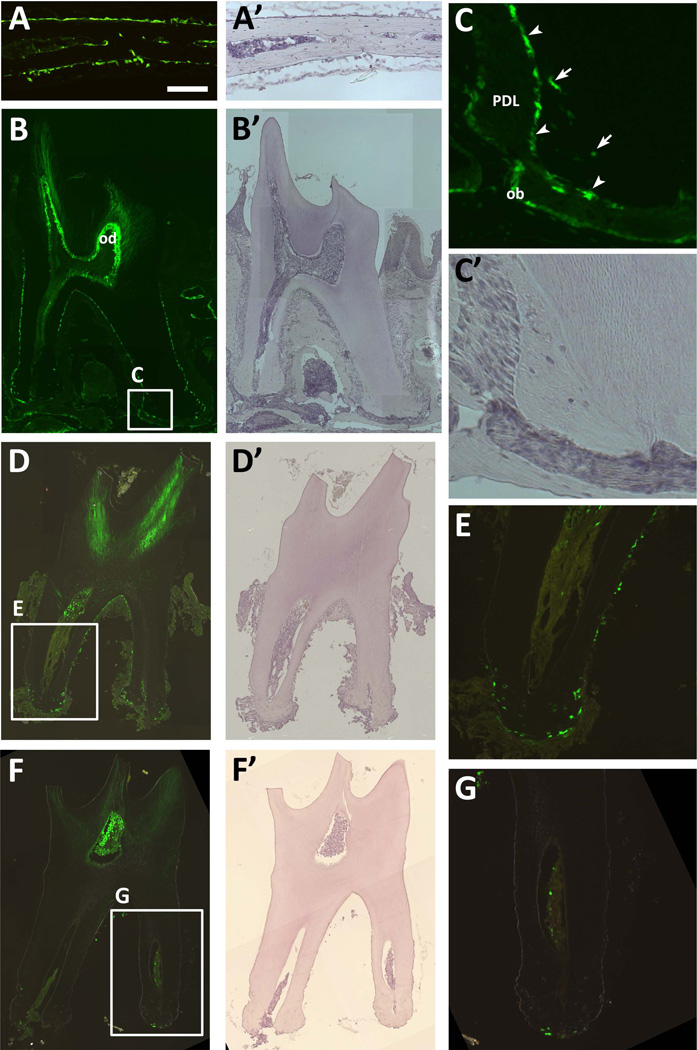
Figure 1. OC-GFP labels osteoblasts and cementoblasts.
(A) Expression of OC-GFP is restricted to osteoblasts in the calvaria. (B–C) A section of the second molar indicates that OC-GFP is present in cementoblasts (arrowheads) and cementocytes (arrows), as well as odontoblasts (od) and osteoblasts (ob) lining the alveolar bone, but not cells within the periodontal ligament (PDL). (D–E) After tooth extraction, cementoblasts remain on the tooth surface, but osteoblasts associated with alveolar bone are absent. (F–G) Following digestion, cells are no longer present on the surface of the tooth, including OC-GFP+ cementoblasts. A 100µm scale bar for the higher magnification images (A, C, E, G) is shown in A. A’-D’ and F’ show hematoxylin staining for the corresponding fluorescent image.
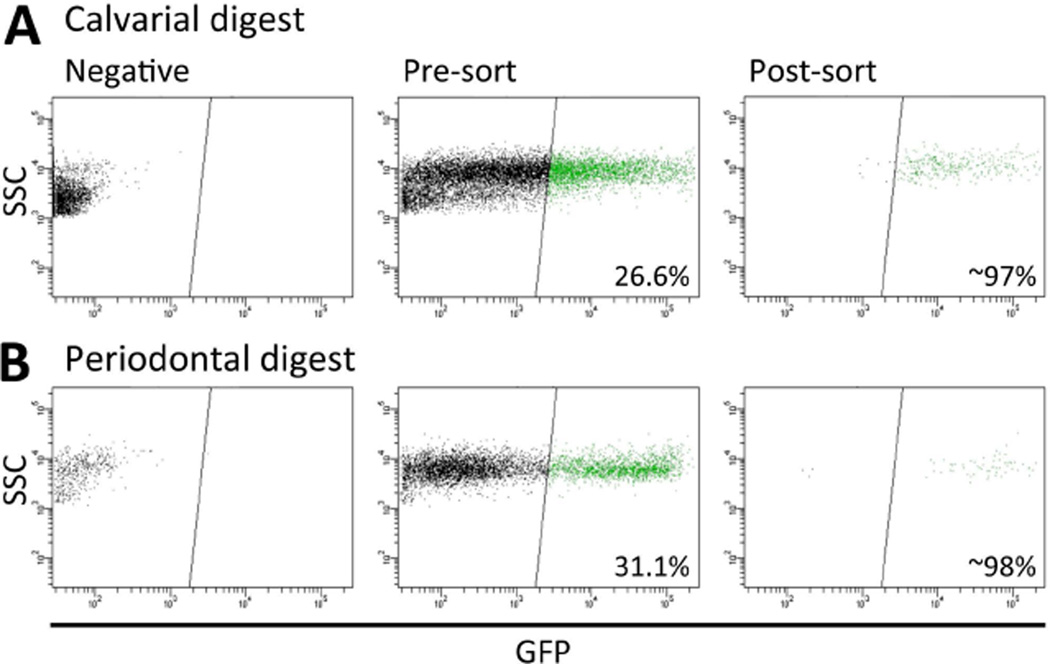
Figure 2. OC-GFP positive cells were sorted from calvarial and periodontal cells.
After dissection and digestion cells from (A) calvaria, and (B) periodontium attached to extracted teeth were sorted from OC-GFP mice and GFP+ cells isolated. Reanalysis indicated that over 95% of the sorted cells were GFP+.
Gene expression of cementoblasts versus osteoblasts
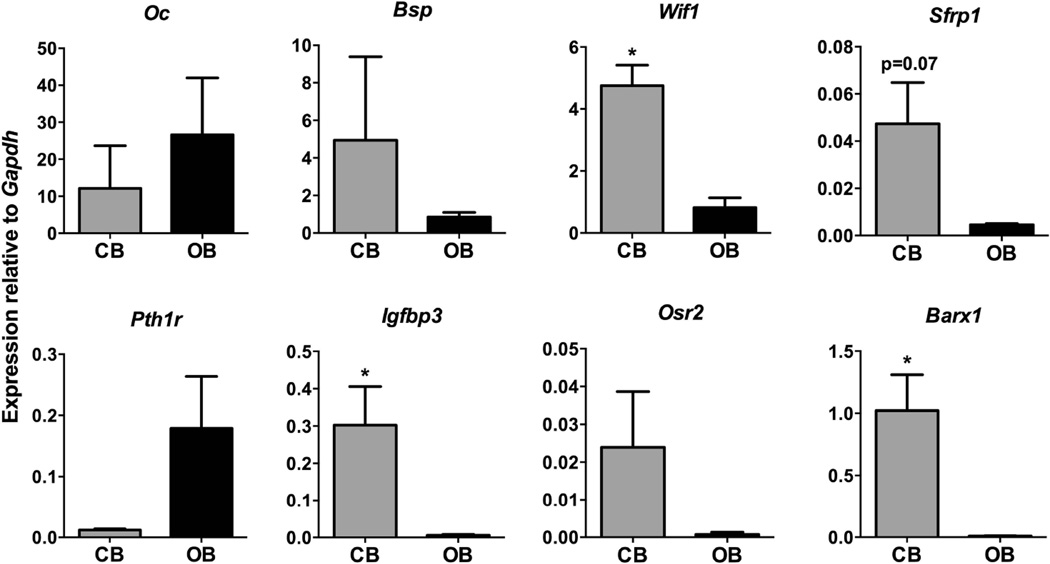
Microarray analysis indicated that 95 genes were at least 2-fold higher in cementoblasts compared to osteoblasts, while 118 genes were elevated in osteoblasts. The full dataset can be found in the GEO repository under accession GSE60394. Gene ontology analysis of the differentially regulated genes indicated that in addition to enrichment of a number of developmental categories, cementoblasts also showed enrichment of Wnt signaling pathway molecules (Table 1, Supplemental Table 2). We observed a number of genes associated with skeletal development both in the cementoblast-enriched, and osteoblast-enriched gene lists (Supplemental Table 3), suggesting that although many marker genes are expressed in both tissues, they may be expressed at different levels. The cell adhesion category, that also shows significant changes, also contains many matrix genes, including a number of collagens. We also noted enrichment of epithelial genes, particularly keratins (Supplemental Table 4). Upon further examination, we observed that cementoblasts were negative for keratin 14 promoter driven GFP and keratin 14 immunostaining (data not shown), consistent with a previous study, (15) however it was strongly expressed in the gingiva. We therefore believe that some of the cementoblast populations contained enough contamination from cells of the gingiva to result in enrichment of some epithelial genes such as keratins. We confirmed expression levels of a number of genes of interest using real time PCR (Fig. 3). As expected, osteocalcin was expressed at high levels in both osteoblasts and cementoblasts, and there was no difference between groups. Similar results were observed for bone sialoprotein expression. Since Wnt signaling has been identified as an important pathway in both tooth and bone development, and bone homeostasis, we sought to confirm changes in regulators of Wnt signaling. Two inhibitors of Wnt signaling, Wif1 and Sfrp1 were expressed at higher levels in cementoblasts, and this was confirmed by real time PCR. The change in Sfrp1 did not reach statistical significance, but levels were consistently higher in cementoblasts. In situ hybridization for Wif1 confirmed high expression in cementoblasts in the apical root area (Fig. 4). Lower expression was also seen in some PDL and pulp cells, and a very small proportion of osteoblasts on the alveolar bone surface. Expression of the PTH/PTHrP receptor Pth1r was consistently elevated in osteoblasts compared to cementoblasts, in line with results of a previous study (16). We also confirmed elevated expression of the insulin-like growth factor binding protein Igfbp3 and transcription factors Osr2 and Barx1 in cementoblasts compared to osteoblasts. Both transcription factors play roles in murine tooth development, (17, 18) and this data suggests they may also be important postnatally.
Table 1.
Gene ontology analysis of microarray data
| GO Id | P value | Odds ratio |
Expected gene count |
Actual gene count |
Size | GO Term |
|---|---|---|---|---|---|---|
| Higher in cementoblasts | ||||||
| GO:0009888 | 3.21E-09 | 10.91 | 1.57 | 13 | 259 | tissue development |
| GO:0016055 | 2.22E-07 | 15.25 | 0.65 | 8 | 107 | Wnt receptor signaling pathway |
| GO:0007155 | 5.55E-07 | 6.31 | 2.85 | 14 | 472 | cell adhesion |
| GO:0002009 | 2.41E-06 | 13.64 | 0.62 | 7 | 102 | morphogenesis of an epithelium |
| GO:0001501 | 5.45E-06 | 9.68 | 0.99 | 8 | 163 | skeletal development |
| GO:0006817 | 3.11E-05 | 15.97 | 0.37 | 5 | 61 | phosphate transport |
| GO:0009887 | 7.01E-05 | 4.81 | 2.74 | 11 | 453 | organ morphogenesis |
| GO:0045104 | 1.34E-04 | 37.09 | 0.1 | 3 | 17 | intermediate filament cytoskeleton organization and biogenesis |
| GO:0043062 | 1.37E-04 | 11.44 | 0.5 | 5 | 83 | extracellular structure organization and biogenesis |
| GO:0000902 | 3.80E-03 | 3.85 | 2.03 | 7 | 335 | cell morphogenesis |
| GO:0035295 | 4.00E-03 | 5.2 | 1.06 | 5 | 175 | tube development |
| GO:0009790 | 4.32E-04 | 4.51 | 2.31 | 9 | 382 | embryonic development |
| GO:0030334 | 5.10E-03 | 9.4 | 0.35 | 3 | 58 | regulation of cell migration |
| GO:0031324 | 8.70E-03 | 3.66 | 1.8 | 6 | 297 | negative regulation of cellular metabolic process |
| Higher in osteoblasts | ||||||
| GO:0001501 | 7.29E-04 | 6.17 | 1.09 | 6 | 163 | skeletal development |
| GO:0046849 | 3.90E-03 | 6.76 | 0.65 | 4 | 97 | bone remodeling |
| GO:0048747 | 6.70E-03 | 8.47 | 0.39 | 3 | 58 | muscle fiber development |
Figure 3. Confirmation of differentially expressed genes by real time PCR.
Expression of the genes indicated was quantified using real time PCR in cementoblast (CB) and osteoblast (OB) samples, and expression was normalized to Gapdh expression. Statistical significance was calculated using a Student’s t test. * p<0.05.
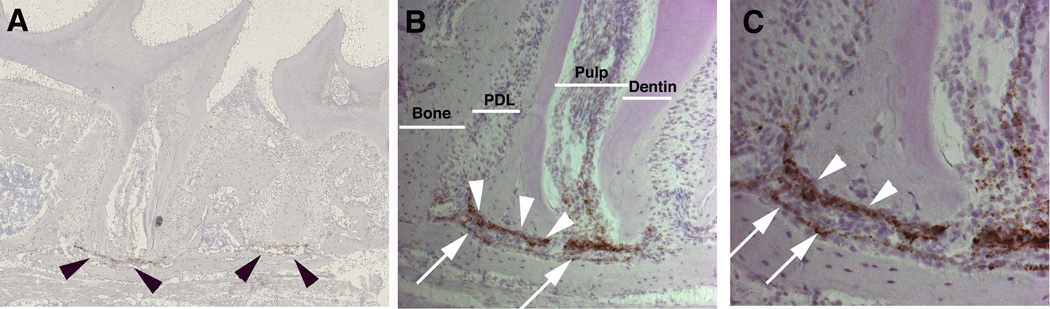
Figure 4. Wif1 mRNA expression in the periodontium.
Wif1 expression was identified in molar sections using RNAScope in situ hybridization. Strong brown signal was observed in cementoblasts lining the apical region of the root (arrowheads). The majority of osteoblasts lining alveolar bone were negative (arrows). Images of a representative section are shown as a composite scan (A), and at 10× (B) and 20× (C) magnification.
We confirmed the difference in expression of Barx1 at the protein level using immunohistochemistry. Cementoblasts in the apical root region, and many cementocytes expressed BARX1 (Fig. 5). In contrast, cells in the PDL did not stain for BARX1, nor did cells on the alveolar bone surface. However we also noted staining in a subset of osteocytes indicating that BARX1 expression is not completely cementoblast-selective within the periodontium.
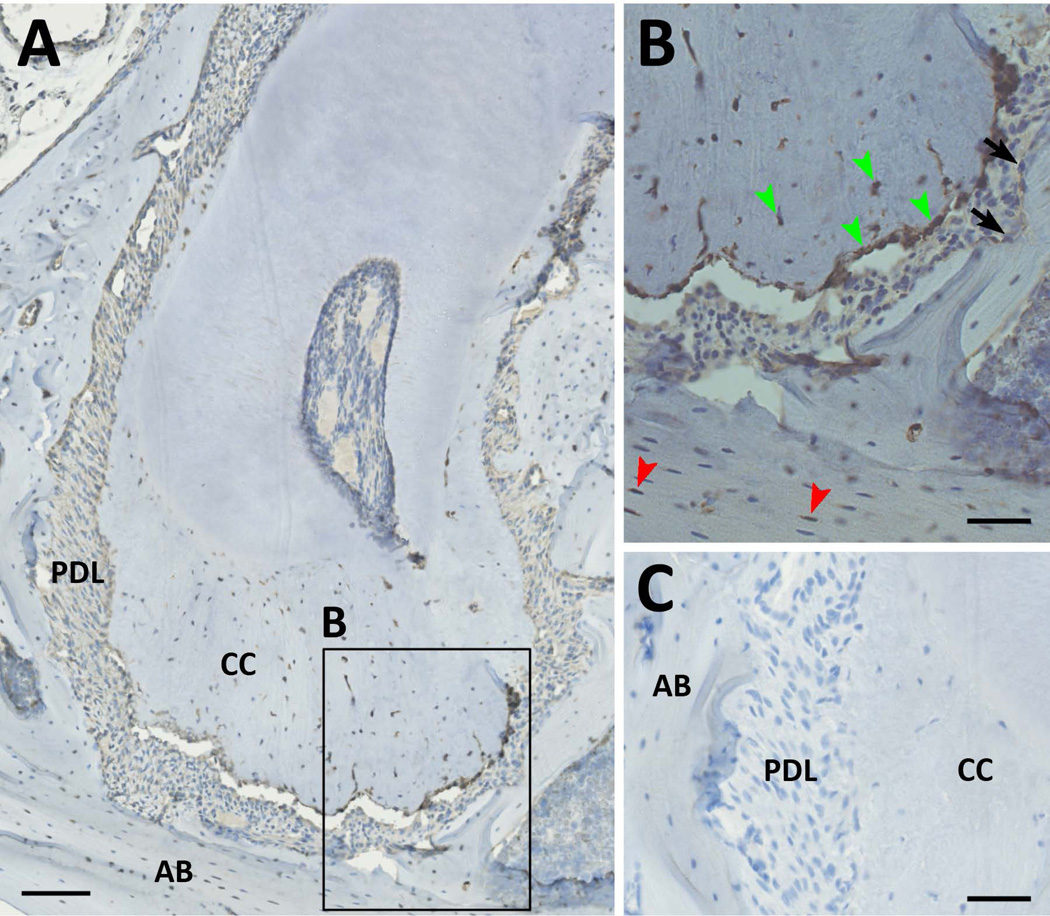
Figure 5. Barx1 protein expression in the periodontium.
(A–B) Immunohistochemistry for Barx1 in a molar section is shown. Expression is present in cementoblasts in the apical region and many of the cementocytes (green arrowheads). Expression is also detectable in selected osteocytes within the alveolar bone (red arrowheads). In contrast, staining is not visible in alveolar osteoblasts (black arrows). (C) The negative control shows no DAB staining. Scale bars represent 100µm (A) and 50µm (B–C). AB, alveolar bone; CC, cellular cementum; PDL, periodontal ligament.
Discussion
Cementoblasts comprise the cellular component of cementum, a tissue that is critical for anchoring the tooth within the periodontium. Cementum has a similar composition to bone, and there are many similarities between osteoblasts and cementoblasts. Osteocalcin is a marker of both osteoblasts and cementoblasts, and in the periodontium distinguishes these cells from PDL fibroblasts. We took advantage of the OC-GFP mouse to specifically isolate both cell types in order to further define the cementoblast phenotype using gene expression profiling. There have been a number of previous studies examining differences in gene and protein expression between various craniofacial tissue elements. A recent study utilized laser capture microdissection to profile differences between cementoblasts and odontoblasts (8). Given that both these cell types reside in a single layer in direct contact with other types of cells, it is difficult to avoid contamination from adjacent cell populations using this technique. Koike et al. compared gene expression profiles of cultured human cementoblasts with bone marrow stromal cells and found a small number of differentially expressed transcripts (6). Interestingly, in our study, GLUT1 (Slc2a1) is elevated 2.4× in cementoblasts compared to osteoblasts in line with their results, although the other genes reported do not appear to be altered in the current study. CAP has also been proposed as a cementoblast marker, however in this study we found no change in Ptpla expression, consistent with other reports that have questioned the specificity of this marker (4, 5). Finally, a study comparing the proteome of human cementum with bone identified 83 proteins that were detectable in cementum but not bone, 105 in bone only, and numerous others that were expressed at different levels (7). In our study the majority of these proteins that could be identified in the microarray were either undetectable (35%) or did not show any change in expression (53%). This lack of agreement is probably due to a combination of factors, including the difference in species, bias involved in protein extraction and detection, and the inclusion of matrix, which may include proteins produced by other cells types and proteins no longer expressed by the cells. It is also possible that some of the changes identified in this study are specific to the young adult animals used, and may not be present during tooth development, or in fully mature adult animals. Further studies will be required to determine if there are age-related changes in cementoblast gene expression.
Histologically, OC-GFP is specific to the cementoblast layer and cementocytes in extracted teeth, however our microarray data suggest that the sorting may not have produced a completely pure cementoblast population in all cases. Two of the cementoblast samples show high expression of a number of epithelial markers such as Krt14, however lineage tracing studies have shown the Krt14-expressing cells make a very minimal contribution to the cementoblast layer, and K14 is absent from cementoblasts in both the apical and cervical regions of the root (15). We therefore suspect contamination from gingival cells in these samples, however, they are included in the analyses presented. In contrast, we do not see expression of Dspp, suggesting that we do not have odontoblast contamination using this methodology, although odontoblasts are also OC-GFP+. To clearly present this point raw data from all the samples has been submitted to GEO repository, which will allow other investigators to obtain detailed comparisons of the experiments.
One of the interesting findings in this study, that has not been noted in previous expression profiling studies, is alteration in components of the Wnt signaling pathway. We found that two Wnt inhibitors, Wif1 and Sfrp1 were elevated in cementoblasts, as well as Barx1, which has been shown to function as a Wnt antagonist by directing Sfrp expression (19). Wif1 and Barx1 expression were both localized to the apical root area. Wnt signaling is critical for bone development and growth, and remains important in adult bone homeostasis (20). A number of recent studies have demonstrated that appropriate levels of Wnt signaling are also necessary for tooth development. Postnatal knockout of β-catenin in the odontoblast and cementoblast lineages using OC-Cre prevents development of molar roots (21), and in two models of postnatal Wnt inhibition, cementoblasts on the surface of acellular cementum are largely replaced by osteoclasts, (22, 23) suggesting that Wnt signaling is critical for cementoblast differentiation. Constitutive activation of β-catenin using OC-Cre results in shortened roots, delayed eruption and excessive production of both dentin and cementum (24). Wnt signaling is also active in adult cementoblasts, as indicated by Axin2-driven LacZ activity, (25) but once dentition is complete its function is unclear. Restricting Wnt signalling in mature cementoblasts should be evaluated as a potential mechanism for maintaining much slower matrix production by cementoblasts than osteoblasts. Regulation of canonical Wnt signaling is critical for normal bone remodeling that is under constant mechanical stimulation. In adult bone, expression of Wnt inhibitors such as Sost decreases in response to mechanical stimulation leading to increased Wnt signaling and bone formation (20). As constant mechanical pressure is present on the apical area of the root one could speculate that expression of Wnt inhibitors in this area is necessary to prevent non-physiological cementum expansion. This situation has to be explored when increased mechanical forces are applied to teeth during orthodontic tooth movement, and subsequent remodeling of bone and cementum is induced (resorption of compression and formation on tension side).
Another difference we noted in this study was decreased expression of Pth1r in cementoblasts compared to osteoblasts. This is consistent with the results of Tenorio and Hughes which indicated that within the periodontium, PTH receptor expression is highest in alveolar bone, but is also present in cementoblasts of cellular cementum (16). These data suggest that cementoblasts may be less responsive to PTH than osteoblasts. Osteoblasts respond to PTH in numerous ways that result in increased bone formation, and changes in resorption through modulation of RANKL and OPG expression (26). In contrast, cementum does not undergo constant remodeling. Igfbp3, which was higher in cementoblasts, has complex roles regulating IGF signaling, but generally sequesters IGFs in an inactive form (27). Its expression has previously been identified in human cementoblasts (28).
One of the genes identified in this study, Barx1, may represent a fairly selective marker of cellular cementoblasts and cementocytes. By E16.5 Barx1 is expressed specifically in the molars, in addition to the stomach (29). Knockout mice show a transient delay in molar but not incisor development, however they show perinatal lethality, so its function in cementoblasts is undefined (18). Osr2 is another transcription factor involved in molar development, and Osr2 knockout mice demonstrate supernumerary tooth development (17). These mice also die soon after birth, so while full ectopic molar development appears normal, any effect of Osr2 on adult tooth function has not been defined. With further validation, Barx1 expression may represent a method to target genetic manipulations to the cellular cementum compartment. The use of OC-GFP does not allow for differentiation between cellular and acellular cementum, however these tissues have distinct structures and functions meaning it is not surprising that we have identified a gene that is not expressed in both areas.
The OC-GFP mouse, combined with dissection and cell sorting represents a powerful tool in which to define specific cellular responses in the different periodontal tissues to stimuli. Similar techniques could be used to define the response of cementoblasts in models of periodontal disease or orthodontic tooth movement. Utilization of an inducible Cre mouse driven by the OC promoter may provide an interesting model to investigate the effects of knocking out molecules in the adult tooth, since all of the constitutively expressed Cre models are active prior to root development (30). Some of the differentially expressed genes in this study, such as Barx1, may provide further ways to preferentially target cementoblasts without affecting the majority of nearby mineralizing cell types.
In summary, we have selectively isolated mouse cementoblasts and compared their expression profile to osteoblasts. We find numerous differences between the two cell types, including changes in Wnt signaling components that suggest that this signaling pathway is differently regulated in the two cell types.
Supplementary Material
Acknowledgements
Research reported in this publication was supported by the National Institute of Arthritis and Musculoskeletal and Skin Diseases of the National Institutes of Health under Award Number AR055607-01. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. This work has been supported by by a Croatian Science Foundation fellowship, Zagreb, Croatia, to HR.
We acknowledge Danka Grcevic from the Department of Physiology and Immunology, University of Zagreb for assistance with cell sorting, and Valerie Horsley from Department of Molecular, Cell and Developmental Biology, Yale University for providing us the tissues from Keratin 14-GFP mice.
Supported by: This work has been supported by NIH/NIAMS AR055607 grant to I.K and Croatian Science Foundation fellowship to HR.
Footnotes
The authors indicate no potential conflicts of interest.
References
- 1.Pihlstrom BL, Michalowicz BS, Johnson NW. Periodontal diseases. Lancet. 2005;366:1809–1820. doi: 10.1016/S0140-6736(05)67728-8. [DOI] [PubMed] [Google Scholar]
- 2.Chen FM, Zhang J, Zhang M, An Y, Chen F, Wu ZF. A review on endogenous regenerative technology in periodontal regenerative medicine. Biomaterials. 2010;31:7892–7927. doi: 10.1016/j.biomaterials.2010.07.019. [DOI] [PubMed] [Google Scholar]
- 3.Bosshardt DD. Are cementoblasts a subpopulation of osteoblasts or a unique phenotype? J Dent Res. 2005;84:390–406. doi: 10.1177/154405910508400501. [DOI] [PubMed] [Google Scholar]
- 4.Foster BL, Somerman MJ. Cementum. In: McCauley LK, Somerman MJ, editors. Mineralized Tissues in Oral and Craniofacial Science. Hoboken, NJ, USA: Wiley-Blackwell; 2012. pp. 169–181. [Google Scholar]
- 5.BarKana I, Narayanan AS, Grosskop A, Savion N, Pitaru S. Cementum attachment protein enriches putative cementoblastic populations on root surfaces in vitro. J Dent Res. 2000;79:1482–1488. doi: 10.1177/00220345000790070901. [DOI] [PubMed] [Google Scholar]
- 6.Koike H, Uzawa K, Grzesik WJ, et al. GLUT1 is highly expressed in cementoblasts but not in osteoblasts. Connect Tissue Res. 2005;46:117–124. doi: 10.1080/03008200591008437. [DOI] [PubMed] [Google Scholar]
- 7.Salmon CR, Tomazela DM, Ruiz KGS, et al. Proteomic analysis of human dental cementum and alveolar bone. J Proteomics. 2013;91:544–555. doi: 10.1016/j.jprot.2013.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sun JX, Horst OV, Bumgarner R, Lakely B, Somerman MJ, Zhang H. Laser capture microdissection enables cellular and molecular studies of tooth root development. Int J Oral Sci. 2012;4:7–13. doi: 10.1038/ijos.2012.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bilic-Curcic I, Kronenberg M, Jiang X, et al. Visualizing levels of osteoblast differentiation by a two-color promoter-GFP strategy: Type I collagen-GFPcyan and osteocalcin-GFPtpz. Genesis. 2005;43:87–98. doi: 10.1002/gene.20156. [DOI] [PubMed] [Google Scholar]
- 10.Roguljic H, Matthews BG, Yang W, Cvija H, Mina M, Kalajzic I. In vivo Identification of periodontal progenitor cells. J Dent Res. 2013;92:709–715. doi: 10.1177/0022034513493434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kalajzic I, Kalajzic Z, Kaliterna M, et al. Use of type I collagen green fluorescent protein transgenes to identify subpopulations of cells at different stages of the osteoblast lineage. J Bone Miner Res. 2002;17:15–25. doi: 10.1359/jbmr.2002.17.1.15. [DOI] [PubMed] [Google Scholar]
- 12.Shin DG, Hong S-H, Joshi P, et al. PBC: A software framework facilitating pattern-based clustering for microarray data analysis IJCBS International Joint Conference on Bioinformatics, Systems Biology and Intelligent Computing. Shangai, China: 2009. pp. 30–36. [Google Scholar]
- 13.Matthews BG, Grcevic D, Wang LP, et al. Analysis of aSMA-labeled progenitor cell commitment identifies Notch signaling as an important pathway in fracture healing. J Bone Miner Res. 2014;29:1283–1294. doi: 10.1002/jbmr.2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tenorio D, Cruchley A, Hughes FJ. Immunocytochemical investigation of the rat cementoblast phenotype. J Periodontal Res. 1993;28:411–419. [PubMed] [Google Scholar]
- 15.Huang XF, Bringas P, Slavkin HC, Chai Y. Fate of HERS during tooth root development. Dev Biol. 2009;334:22–30. doi: 10.1016/j.ydbio.2009.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tenorio D, Hughes FJ. An immunohistochemical investigation of the expression of parathyroid hormone receptors in rat cementoblasts. Arch Oral Biol. 1996;41:299–305. doi: 10.1016/0003-9969(95)00113-1. [DOI] [PubMed] [Google Scholar]
- 17.Zhang Z, Lan Y, Chai Y, Jiang R. Antagonistic actions of Msx1 and Osr2 pattern mammalian teeth into a single row. Science. 2009;323:1232–1234. doi: 10.1126/science.1167418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Miletich I, Yu WY, Zhang RF, et al. Developmental stalling and organ-autonomous regulation of morphogenesis. Proc Natl Acad Sci U S A. 2011;108:19270–19275. doi: 10.1073/pnas.1112801108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim B-M, Buchner G, Miletich I, Sharpe PT, Shivdasani RA. The stomach mesenchymal transcription factor Barx1 specifies gastric epithelial identity through inhibition of transient Wnt signaling. Dev Cell. 2005;8:611–622. doi: 10.1016/j.devcel.2005.01.015. [DOI] [PubMed] [Google Scholar]
- 20.Baron R, Kneissel M. WNT signaling in bone homeostasis and disease: from human mutations to treatments. Nat Med. 2013;19:179–192. doi: 10.1038/nm.3074. [DOI] [PubMed] [Google Scholar]
- 21.Zhang R, Yang G, Wu X, Xie J, Yang X, Li T. Disruption of Wnt/beta-catenin signaling in odontoblasts and cementoblasts arrests tooth root development in postnatal mouse teeth. Int J Biol Sci. 2013;9:228–236. doi: 10.7150/ijbs.5476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Han XL, Liu M, Voisey A, et al. Post-natal effect of overexpressed DKK1 on mandibular molar formation. J Dent Res. 2011;90:1312–1317. doi: 10.1177/0022034511421926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lim WH, Liu B, Hunter DJ, Cheng D, Mah SJ, Helms JA. Downregulation of Wnt causes root resorption. Am J Orthod Dentofacial Orthop. 2014;146:337–345. doi: 10.1016/j.ajodo.2014.05.027. [DOI] [PubMed] [Google Scholar]
- 24.Kim T-H, Lee J-Y, Baek J-A, et al. Constitutive stabilization of beta-catenin in the dental mesenchyme leads to excessive dentin and cementum formation. Biochem Biophys Res Commun. 2011;412:549–555. doi: 10.1016/j.bbrc.2011.07.116. [DOI] [PubMed] [Google Scholar]
- 25.Lim WH, Liu B, Cheng D, et al. Wnt signaling regulates pulp volume and dentin thickness. J Bone Miner Res. 2014;29:892–901. doi: 10.1002/jbmr.2088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jilka RL. Molecular and cellular mechanisms of the anabolic effect of intermittent PTH. Bone. 2007;40:1434–1446. doi: 10.1016/j.bone.2007.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Martin JL, Baxter RC. Signalling pathways of insulin-like growth factors (IGFs) and IGF binding protein-3. Growth Factors. 2011;29:235–244. doi: 10.3109/08977194.2011.614237. [DOI] [PubMed] [Google Scholar]
- 28.Magnucki G, Schenk U, Ahrens S, et al. Expression of the IGF-1, IGFBP-3 and IGF-1 receptors in dental pulp stem cells and impacted third molars. J Oral Sci. 2013;55:319–327. doi: 10.2334/josnusd.55.319. [DOI] [PubMed] [Google Scholar]
- 29.Tissier-Seta JP, Mucchielli ML, Mark M, Mattei MG, Goridis C, Brunet JF. Barx1, a new mouse homeodomain transcription factor expressed in craniofacial ectomesenchyme and the stomach. Mech Dev. 1995;51:3–15. doi: 10.1016/0925-4773(94)00343-l. [DOI] [PubMed] [Google Scholar]
- 30.Yoshikawa Y, Kode A, Xu LL, et al. Genetic Evidence Points to an Osteocalcin-Independent Influence of Osteoblasts on Energy Metabolism. J Bone Miner Res. 2011;26:2012–2025. doi: 10.1002/jbmr.417. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.