Abstract
Background
Several host genetic factors are thought to affect susceptibility to Helicobacter pylori infection-related diseases, including tumor necrosis factor (TNF)-α. Previous studies have evaluated the association between TNFA gene polymorphisms and H. pylori infection, but the results were inconclusive. We conducted this meta-analysis to clarify the association between TNFA polymorphisms and H. pylori infection.
Methods
Published literature within PubMed, Embase, and the Cochrane Library were used in our meta-analysis. Data were analyzed with the Stata13.1 software package using pooled odds ratios (ORs) with 95% confidence intervals (CI).
Results
A total of 24 studies were included in our study. The TNFA -308G>A polymorphism was associated with decreasing H. pylori infection (AA vs. AG+GG, OR = 0.64, 95% CI = 0.43–0.97; AA vs. GG, OR = 0.64, 95% CI = 0.43–0.97). A significantly decreased risk was also found for -1031T>C polymorphism (CC vs. CT+TT, OR = 0.61, 95% CI = 0.44–0.84). -863C>A polymorphism was associated with increasing risk of H. pylori infection (AA+AC vs. CC, OR = 1.47, 95% CI = 1.16–1.86; A allele vs. C allele, OR = 1.40, 95% CI = 1.14–1.72). There was no significant association between -857C>T polymorphism and H. pylori infection. When stratified analysis was conducted on H. pylori infection detection methods, -857C>T and -863C>A polymorphisms were associated with H. pylori infection for the non-ELISA subgroup. When stratified for ethnicity or study design, -863C>A significantly increased the risk and -1031T>C decreased the risk for the Asian subgroup and hospital-based subgroup.
Conclusion
Results of our meta-analysis demonstrate that TNFA -308G>A and -1031 T>C polymorphisms may be protective factors against H. pylori infection, and -863C>A may be a risk factor, especially in Asian populations. Further studies with larger sample sizes are required to validate these results.
Introduction
Helicobacter pylori, one of the most common pathogens worldwide, has proven to be associated with gastritis, peptic ulcers, gastric cancer, and mucosa-associated lymphoid tissue (MALT) lymphoma [1]. Some individuals when exposed to H. pylori may escape from persistent infection, even if they live in regions where H. pylori infection is highly prevalent. Previous studies indicate that host factors may play an important role during H. pylori infection [2]. Host cytokines and their gene polymorphisms may be host factors that affect an individual’s susceptibility to H. pylori-related diseases [3, 4]. H. pylori infection can induce production of some cytokines, including interleukin (IL)-1, -2, -4, -6, -8, -10, -17, interferon (IFN)-β, and tumor necrosis factor (TNF)-α [5]. These host cytokines affect the occurrence and development of the gastric mucosal inflammatory response, which is a key event of H. pylori infection [6].
TNF-α, a host cytokine induced by H. pylori in gastric mucosal, is supposed to be involved in H. pylori infection [7]. TNF-α is encoded by the TNFA gene, which is clustered on the short arm of human chromosome 6 (6p21.3), between HLA-B and HLA-DR [8]. The TNFA gene is known to have four single nucleotide polymorphisms in the regulatory sequences that may affect its expression: -308G>A, -857C>T, -863 C>A, and -1031T>C. TNF-α can inhibit gastric acid secretion and influence the immune response, which may be associated with persistent H. pylori infection [9].
A number of studies have focused on the association between TNFA gene polymorphisms and H. pylori-related diseases [10–12]. Previous meta-analysis have demonstrated that TNFA gene polymorphisms are associated with gastric cancer and have no association with peptic ulcers [13, 14]. Many studies conducted on gastric diseases have investigated the relationship between TNFA gene polymorphisms and H. pylori infection simultaneously; however, results from these studies are inconclusive. Therefore, we performed this meta-analysis to clarify the association between TNFA gene polymorphisms and H. pylori infection.
Materials and Methods
Search strategy
Pubmed, Embase and Cochrane Library databases were searched up to August 2015. The following terms were used for searching: (TNF-α OR tumor necrosis factor-α OR TNF-A OR tumor necrosis factor-A OR TNF-alpha OR tumor necrosis factor-alpha) AND (polymorphism OR polymorphisms OR SNP) AND (Helicobacter pylori OR H. pylori OR HP). Searches were restricted to English. In order to identify potentially relevant studies, the reference lists of retrieved articles were also examined. In addition, the related citations of results in Pubmed were searched. We also contacted the authors to get more data as possible as we can. When more than one of the same case series was involved in several studies, only the study with the largest sample sizes was selected in our meta-analysis.
Selection criteria
Studies were included if the following conditions were met: (1) A relationship between the TNFA gene polymorphisms and H. pylori infection was described; (2) Case-control designed; (3) Objective H. pylori infection detection methods were used; (4) Sufficient genotype data to calculate the odd ratios (ORs) with a 95% confidence interval (CI) was available.
Data extraction and quality appraisal
The following data were collected from each study: first author’s name; year of publication; ethnicity; country; study design; number of cases and controls; H. pylori infection detection methods; and genotyping method. The Newcastle-Ottawa scale (NOS) [15] was used to assess the quality of studies included, according to three main criteria: selection of cases and controls; comparability of cases and controls; and exposure to risk factors. NOS scores ranged between 0 and 9 stars. Studies with a score of seven stars or greater were considered to be of high quality, while those that scored five stars or less were considered low quality. Two authors (XDS and YYX) of this meta-analysis independently extracted all information and conducted the quality appraisal. Disagreements were resolved by discussion with other authors.
Statistical analysis
Statistical analysis was performed using STATA 13.1 (STATA Corp, College Station, TX, USA). Pooled OR and corresponding 95% CI was used to measure the strength of associations between TNFA gene polymorphisms and H. pylori infection. Heterogeneity among studies was assessed by the Q-test and I2 statistics. P < 0.10 or I2 > 50% indicated significant heterogeneity [16]. If significant heterogeneity exists, the ORs were pooled with a random effect model. Otherwise, a fixed effect model was selected. Subgroup analyses were conducted based on H. pylori infection detection methods (ELISA or non-ELISA methods (including bacterial culture, rapid urease test (RUT), polymerase chain reaction (PCR), urea breath test (UBT), Helicobacter pylori stool antigen test (HpSAT) and histological examination)), study designs (hospital-based (HB) or population-based (PB)) and ethnicity (Asian or Caucasian). Publication bias was examined using a Begg’s funnel plot or Egger’s plot, and the significance level was set at 0.05 for both. Hardy-Weinberg equilibrium was assessed by the χ2 test for goodness of fit, with a P-value less than 0.05 considered a significant deviation.
Results
Study characteristics
A total of 230 articles were retrieved from the initial search. From these, 164 articles were assessed for ineligibility after reading titles and abstracts, and 47 articles with insufficient data were excluded after reading the full texts. In addition, 5 papers were included through references. According to our inclusion and exclusion criteria, 24 articles were used for this meta-analysis finally [17–40]. The study selection process is summarized in Fig 1. Of the studies included, 17 concerned -308 G>A, nine concerned -857C>T, four concerned -863C>A, 10 concerned -1031T>C; 14 were on Asians, five were on Caucasians, one was on Africans, four were on mixed ethnicity (Table 1).
Fig 1. Flow diagram of the study selection process.
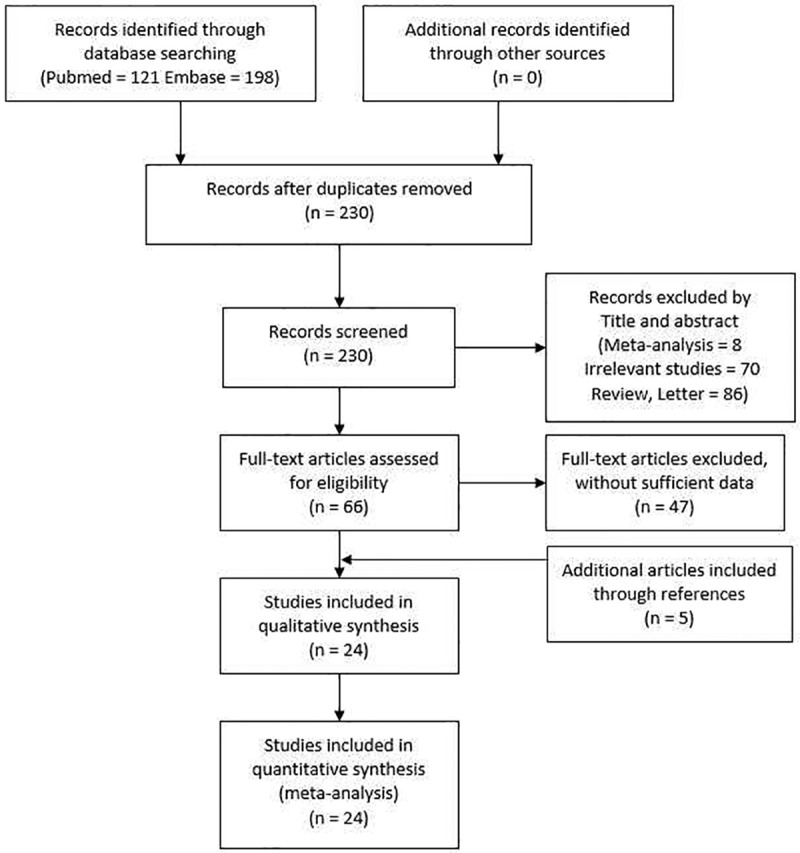
Table 1. Main characteristics of studies included in meta-analysis.
| Author | Year | Country | Ethnicity | Study design | Cases (Hp+) | Controls (Hp-) | Detection of Hp | Genotyping | NOS(score) | HWE(P) | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1/1 | 1/2 | 2/2 | 1/1 | 1/2 | 2/2 | |||||||||
| -308G>A | ||||||||||||||
| Kunstmann | 1999 | Germany | Caucasian | HB | 10 | 50 | 145 | 8 | 39 | 132 | RUT,HE,BC | ASH | 5 | 0.055 |
| Li | 2005 | China | Asian | PB | 3 | 37 | 314 | 0 | 24 | 96 | ELISA | RFLP | 7 | 0.102 |
| Lu | 2005 | China | Asian | HB | 3 | 59 | 351 | 1 | 15 | 84 | HE,BC | SSOP | 6 | 0.989 |
| Kim | 2006 | Korea | Asian | PB | 9 | 172 | 982 | 3 | 23 | 159 | ELISA,RUT,HE,BC | Taqman, RFLP | 6 | 0.153 |
| Sugimoto | 2007 | Japan | Asian | HB | 0 | 11 | 462 | 0 | 3 | 169 | ELISA,RUT | RFLP | 6 | 1.000 |
| Chakravorty | 2008 | India | Asian | HB | 3 | 33 | 117 | 6 | 36 | 115 | RUT,HE,BC | RFLP | 5 | 0.174 |
| Szoke | 2008 | Hungary | Caucasian | HB | 1 | 15 | 59 | 5 | 32 | 106 | HE | RFLP | 4 | 0.322 |
| Gao | 2009 | Germany | Caucasian | PB | 9 | 96 | 291 | 4 | 29 | 98 | ELISA | Pyrosequencing | 8 | 0.492 |
| GonzÂlez | 2009 | Spain | Caucasian | HB | 1 | 12 | 44 | 0 | 4 | 20 | RUT,HE | RFLP | 5 | 1.000 |
| Queiroz | 2009 | Brazil | Mixed | PB | 6 | 81 | 282 | 7 | 42 | 121 | ELISA,UBT | RFLP | 7 | 0.274 |
| Cheng | 2010 | China | Asian | HB | 4 | 61 | 300 | 6 | 73 | 360 | RUT,HE,BC | RFLP | 6 | 0.434 |
| Kang | 2012 | Korea | Asian | HB | 2 | 37 | 245 | 1 | 19 | 96 | RUT,HE,BC | RFLP | 7 | 1.000 |
| Kimang'a | 2012 | Kenya | African | HB | 0 | 97 | 54 | 2 | 81 | 36 | RUT,HE,HpSAT,PCR | RFLP | 4 | 0 |
| Santos | 2012 | Brazil | Mixed | HB | 3 | 50 | 122 | 0 | 4 | 22 | RUT,HE,PCR | RFLP | 5 | 1.000 |
| Queiroz | 2013 | Brazil | Mixed | HB | 0 | 15 | 32 | 0 | 23 | 55 | RUT,UBT,HE,BC | RFLP | 6 | 0.255 |
| Kulmambetova | 2014 | Kazakhstan | Asian | PB | 1 | 26 | 115 | 7 | 90 | 326 | HE | Taqman | 6 | 0.784 |
| Salagacka | 2014 | Poland | Caucasian | HB | 1 | 32 | 68 | 2 | 30 | 69 | RUT | RFLP | 5 | 0.751 |
| -857C>T | ||||||||||||||
| Hamajima | 2003 | Japan | Asian | HB | 28 | 209 | 507 | 14 | 164 | 424 | ELISA | CTPP | 7 | 0.691 |
| Lu | 2005 | China | Asian | HB | 6 | 100 | 315 | 4 | 26 | 70 | HE,BC | SSOP | 6 | 0.625 |
| Atsuta | 2006 | Brazil | Asian | PB | 19 | 146 | 287 | 17 | 155 | 326 | ELISA | CTPP | 8 | 0.786 |
| Tseng | 2006 | Jamaica | Mixed | PB | 0 | 2 | 34 | 0 | 8 | 142 | ELISA | Taqman | 8 | 1.000 |
| Saijo | 2007 | Japan | Asian | PB | 3 | 64 | 170 | 11 | 47 | 115 | ELISA | Taqman | 7 | 0.498 |
| Sugimoto | 2007 | Japan | Asian | HB | 33 | 125 | 315 | 7 | 40 | 125 | ELISA,RUT | RFLP | 6 | 0.179 |
| Chakravorty | 2008 | India | Asian | HB | 1 | 10 | 142 | 1 | 16 | 140 | RUT,HE,BC | RFLP | 5 | 0.91 |
| Abdiev | 2010 | Uzbeks | Asian | HB | 1 | 44 | 79 | 2 | 9 | 31 | ELISA | CTPP | 5 | 0.501 |
| Salagacka | 2014 | Poland | Caucasian | HB | 2 | 17 | 83 | 3 | 23 | 75 | RUT | RFLP | 5 | 0.691 |
| -863C>A | ||||||||||||||
| Lu | 2005 | China | Asian | HB | 12 | 118 | 293 | 3 | 23 | 74 | HE,BC | SSOP | 6 | 0.703 |
| Sugimoto | 2007 | Japan | Asian | HB | 12 | 153 | 308 | 3 | 44 | 125 | ELISA,RUT | RFLP | 6 | 0.890 |
| Chakravorty | 2008 | India | Asian | HB | 11 | 56 | 86 | 6 | 42 | 109 | RUT,HE,BC | RFLP | 5 | 0.600 |
| Salagacka | 2014 | Poland | Caucasian | HB | 3 | 28 | 71 | 1 | 22 | 78 | RUT | RFLP | 5 | 0.939 |
| -1031T>C | ||||||||||||||
| Hamajima | 2003 | Japan | Asian | HB | 13 | 208 | 540 | 21 | 177 | 412 | ELISA | CTPP | 7 | 0.714 |
| Lu | 2005 | China | Asian | HB | 5 | 107 | 309 | 2 | 20 | 78 | HE,BC | SSOP | 6 | 0.885 |
| Ando | 2006 | Japan | Asian | HB | 0 | 49 | 141 | 3 | 22 | 32 | ELISA,UBT,HE | CTPP | 5 | 0.935 |
| Atsuta | 2006 | Brazil | Asian | PB | 14 | 120 | 322 | 17 | 149 | 326 | ELISA | CTPP | 8 | 0.996 |
| Saijo | 2007 | Japan | Asian | PB | 5 | 80 | 152 | 7 | 51 | 115 | ELISA | Taqman | 7 | 0.656 |
| Sugimoto | 2007 | Japan | Asian | HB | 12 | 158 | 303 | 2 | 46 | 124 | ELISA,RUT | RFLP | 6 | 0.461 |
| Chakravorty | 2008 | India | Asian | HB | 14 | 60 | 79 | 30 | 55 | 70 | RUT,HE,BC | RFLP | 5 | 0.003 |
| Abdiev | 2010 | Uzbeks | Asian | HB | 1 | 41 | 82 | 3 | 6 | 33 | ELISA | CTPP | 5 | 0.031 |
| Zhao | 2013 | Indonesia | Asian | PB | 6 | 16 | 11 | 36 | 105 | 120 | UBT | CTPP | 7 | 0.098 |
| Salagacka | 2014 | Poland | Caucasian | HB | 1 | 31 | 54 | 2 | 31 | 59 | RUT | RFLP | 5 | 0.537 |
Hp: H. pylori; +: positive; -: negative; 1/1: variant homozygote; 1/2: heterozygote; 2/2: wild type homozygote; PB: population-based; HB: hospital-based; ELISA: enzyme-linked immunosorbent assay; RUT: rapid urease test; UBT: urease breath test; HpSAT: Helicobacter pylori stool antigen test; HE: histological examination; BC: bacterial culture; ASH: allele specific hybridization; RFLP: restriction fragment length polymorphism; CTPP: confronting two-pair primers; SSOP: sequence-specific oligonucleotide probe.
Meta-analysis results
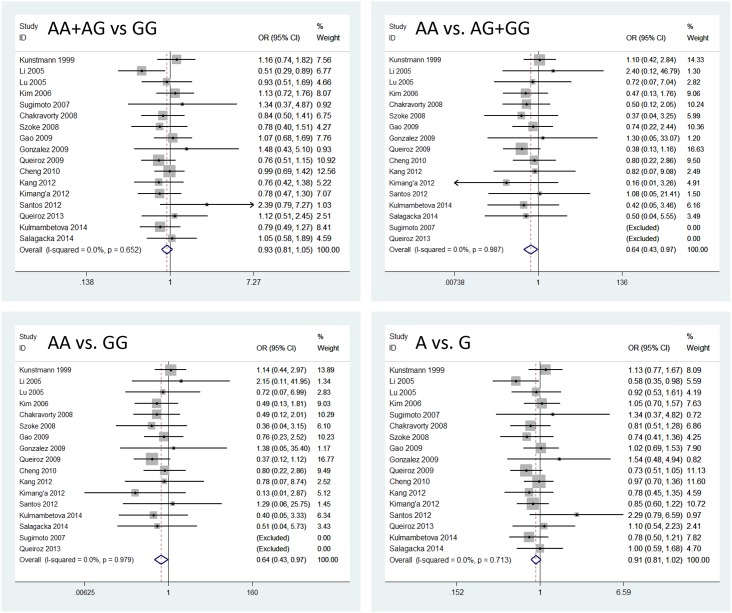
The TNFA gene -308G>A polymorphism was associated with decreasing H. pylori infection in recessive and homozygote models (AA+AG vs. GG, OR = 0.93, 95% CI = 0.81–1.05; AA vs. AG+GG, OR = 0.64, 95% CI = 0.43–0.97; AA vs. GG, OR = 0.64, 95% CI = 0.43–0.97; A allele vs. G allele, OR = 0.91, 95% CI = 0.81–1.02) (Fig 2). For the -1031T>C polymorphism, a significantly decreased risk was also found in recessive model (CC+CT vs. TT, OR = 1.00, 95% CI = 0.81–1.23; CC vs. CT+TT, OR = 0.61, 95% CI = 0.44–0.84; CC vs. TT, OR = 0.63, 95% CI = 0.39–1.03; C allele vs. T allele, OR = 0.94, 95% CI = 0.78–1.13). In contrast, the -863C>A polymorphism was associated with an increasing risk of H. pylori infection in dominant and allelic models (AA+AC vs. CC, OR = 1.47, 95% CI = 1.16–1.86; AA vs. AC+CC, OR = 1.58, 95% CI = 0.82–3.03; AA vs. CC, OR = 1.77, 95% CI = 0.92–3.43; A allele vs. C allele, OR = 1.40, 95% CI = 1.14–1.72). There was no significant association between the -857C>T polymorphism and H. pylori infection (Table 2).
Fig 2. Forest plots for all models to show an association between the TNFA -308G>A polymorphism and H. pylori infection.
Table 2. Meta-analysis of the association between TNFA polymorphisms and H. pylori infection.
| Study Group | Study(n) | Dominant model | Recessive model | Homozygote model | Allelic model | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| OR | 95%CI | I2 | OR | 95%CI | I2 | OR | 95%CI | I2 | OR | 95%CI | I2 | ||
| -308G>A | |||||||||||||
| Total | 17 | 0.93 | 0.81–1.05 | 0% | 0.64 | 0.43–0.97 | 0% | 0.64 | 0.43–0.97 | 0% | 0.91 | 0.81–1.02 | 0% |
| ELISA | 5 | 0.88 | 0.71–1.10 | 38.8% | 0.57 | 0.30–1.11 | 0% | 0.57 | 0.29–1.09 | 0% | 0.86 | 0.70–1.05 | 19.9% |
| Non-ELISA | 12 | 0.95 | 0.81–1.11 | 0% | 0.69 | 0.41–1.15 | 0% | 0.69 | 0.41–1.15 | 0% | 0.94 | 0.81–1.08 | 0% |
| Asian | 8 | 0.88 | 0.73–1.05 | 0% | 0.65 | 0.34–1.22 | 0% | 0.63 | 0.34–1.19 | 0% | 0.87 | 0.74–1.03 | 0% |
| Caucasian | 5 | 1.06 | 0.82–1.36 | 0% | 0.82 | 0.43–1.56 | 0% | 0.84 | 0.44–1.60 | 0% | 1.02 | 0.82–1.28 | 0% |
| HB | 12 | 0.98 | 0.82–1.16 | 0% | 0.72 | 0.42–1.22 | 0% | 0.72 | 0.42–1.23 | 0% | 0.96 | 0.83–1.11 | 0% |
| PB | 5 | 0.85 | 0.70–1.05 | 35.4% | 0.55 | 0.30–1.03 | 0% | 0.54 | 0.29–1.02 | 0% | 0.84 | 0.70–1.01 | 13.2% |
| -857C>T | |||||||||||||
| Total | 9 | 1.04 | 0.91–1.19 | 16% | 0.81 | 0.44–1.49 | 55.5% | 0.81 | 0.43–1.52 | 56.7% | 0.98 | 0.82–1.17 | 42.5% |
| ELISA | 6 | 1.10 | 0.95–1.28 | 0% | 0.91 | 0.43–1.93 | 67.3% | 0.94 | 0.45–2.00 | 67.3% | 1.09 | 0.96–1.24 | 36.2% |
| Non-ELISA | 3 | 0.72 | 0.51–1.02 | 0% | 0.50 | 0.18–1.36 | 0% | 0.47 | 0.17–1.29 | 0% | 0.72 | 0.52–0.98 | 0% |
| Asian | 7 | 1.06 | 0.92–1.21 | 22.3% | 0.81 | 0.41–1.58 | 61.1% | 0.82 | 0.42–1.62 | 61.9% | 1.00 | 0.83–1.21 | 50.3% |
| HB | 6 | 1.06 | 0.90–1.26 | 34.4% | 1.25 | 0.81–1.92 | 40.4% | 1.27 | 0.82–1.97 | 41.6% | 1.07 | 0.93–1.24 | 44.4% |
| PB | 3 | 0.99 | 0.79–1.24 | 0% | 0.53 | 0.08–3.34 | 84.6% | 0.53 | 0.08–3.49 | 85.2% | 0.91 | 0.63–1.31 | 52.5% |
| -863C>A | |||||||||||||
| Total (HB) | 4 | 1.47 | 1.16–1.86 | 0% | 1.58 | 0.82–3.03 | 0% | 1.77 | 0.92–3.43 | 0% | 1.40 | 1.14–1.72 | 0% |
| Non-ELISA | 3 | 1.50 | 1.11–2.02 | 0% | 1.62 | 0.76–3.46 | 0% | 1.83 | 0.85–3.94 | 0% | 1.43 | 1.11–1.86 | 0% |
| Asian | 3 | 1.47 | 1.14–1.90 | 0% | 1.48 | 0.75–2.93 | 0% | 1.67 | 0.84–3.32 | 0% | 1.39 | 1.11–1.74 | 0% |
| -1031T>C | |||||||||||||
| Total | 10 | 1.00 | 0.81–1.23 | 55.4% | 0.61 | 0.44–0.84 | 34.7% | 0.63 | 0.39–1.03 | 40.3% | 0.94 | 0.78–1.13 | 59.7% |
| ELISA | 6 | 0.96 | 0.72–1.27 | 67.9% | 0.57 | 0.28–1.13 | 48.6% | 0.57 | 0.28–1.15 | 49.3% | 0.91 | 0.72–1.16 | 67.2% |
| Non-ELISA | 4 | 1.06 | 0.81–1.39 | 23.8% | 0.60 | 0.36–1.01 | 26.0% | 0.63 | 0.37–1.08 | 42.1% | 1.01 | 0.71–1.42 | 56.7% |
| Asian | 9 | 1.00 | 0.79–1.25 | 60.1% | 0.62 | 0.38–1.00 | 41.9% | 0.63 | 0.38–1.06 | 46.9% | 0.94 | 0.77–1.15 | 63.9% |
| HB | 7 | 0.99 | 0.74–1.32 | 63.1% | 0.48 | 0.32–0.72 | 29.1% | 0.48 | 0.32–0.73 | 33.7% | 0.91 | 0.70–1.18 | 67.7% |
| PB | 3 | 0.95 | 0.76–1.18 | 49.1% | 0.90 | 0.53–1.51 | 0% | 0.91 | 0.54–1.56 | 18.2% | 0.95 | 0.79–1.14 | 41.6% |
Significant results were shown in bold.
Variable H. pylori infection detection methods were used in the studies included in this meta-analysis (Table 1). These methods were different in sensitivity and specificity, and various methods could cause various results of diagnosing H. pylori infection. ELISA method had special features during H. pylori epidemiological survey, so we performed a subgroup analysis for ELISA and non-ELISA methods. The TNFA -308G>A and -1031T>C polymorphisms had no association with H. pylori infection for ELISA or non-ELISA subgroups. -857C>T polymorphism significantly decreased the risk of H. pylori infection in allelic model for the non-ELISA subgroup, and -863C>A polymorphism increased the risk in dominant and allelic models for the non-ELISA subgroup. We also conducted a subgroup analysis on ethnicity. The results showed that the -863C>A polymorphism had a significant association with H. pylori infection in dominant and allelic models for the Asian subgroup, and -1031T>C polymorphism was associated with H. pylori infection in recessive model for the Asian subgroup too. -308G>A and -857C>T polymorphisms did not have significant association with H. pylori infection for Asian or Caucasian subgroups. A stratified analysis on study design was also performed, and the results indicated that -863C>A significantly increased the risk and -1031T>C decreased the risk for HB subgroups. All results of the meta-analysis are shown in Table 2.
Heterogeneity and sensitivity analysis
Significant heterogeneity was observed in the TNFA -857C>T and -1031T>C polymorphism results. We then conducted sensitivity analysis to identify the results by omitting one study in turn. Heterogeneity decreased when a study by Saijo et al. [34] was excluded for the -857C>T polymorphism and a study by Ando et al. [21] was excluded for the -1031T>C polymorphism. The pooled ORs were not significantly altered in all investigated SNPs by sequential omission of included studies.
Publication bias
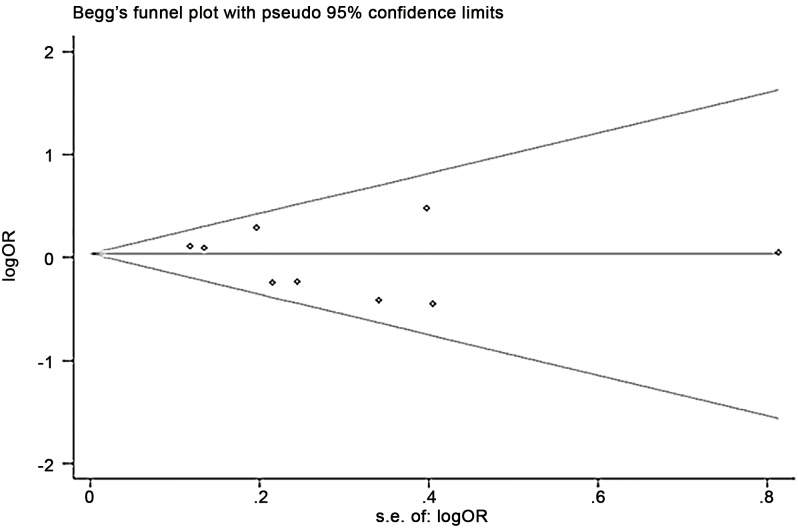
Begg’s funnel plot of SNPs did not reveal any evidence of significant publication bias (Fig 3). Begg’s or Egger’s tests also showed no statistical significance for examining publication bias in the dominant model (-308G>A, Begg’s test P = 0.27, Egger’s test P = 0.26; -857C>T, Begg’s test P = 0.60, Egger’s test P = 0.35; -863C>A, Begg’s test P = 1.00, Egger’s test P = 0.98; and -1031T>C, Begg’s test P = 0.37, Egger’s test P = 0.28).
Fig 3. Begg’s funnel plot of all studies included in the meta-analysis for -857C>T polymorphism.
Se: standard error.
Discussion
Results of our meta-analysis indicate that TNFA -308G>A and -1031T>C polymorphisms might be associated with a decreasing risk of H. pylori infection, while the -863C>A polymorphism could increase the risk of H. pylori infection. When stratified analysis was conducted on ethnicity in our meta-analysis, only -863C>A and -1031T>C polymorphisms had significant association with H. pylori infection in Asian population. -308G>A and -857C>T polymorphisms had no significant association with H. pylori infection in Asian or Caucasian population. TNFA polymorphisms did not show up in a genome wide association study in Europeans [41], which was consistent with the results of our meta-analysis in Caucasian subgroup. The association between TNFA polymorphisms and H. pylori infection may be more meaningful in Asian population. When stratified for study design, -863C>A significantly increased the risk and -1031T>C decreased the risk for the HB subgroups.
Significant heterogeneity existed in meta-analysis results of -857C>T and -1031T>C polymorphisms, and heterogeneity decreased after excluding the study of Saijo et al. [34] for -857C>T polymorphism and the study of Ando et al. [21] for -1031T>C polymorphism, which suggests that the above two studies might be the source of heterogeneity. Subjects of the study by Saijo et al. were all healthy Japanese transit company employees whose ages ranged from 35–60 years, including 413 men and only 5 women. Specific gender, age and occupational composition of the subjects might lead to the difference between the study by Saijo et al. and other including studies. 41% of the subjects of the study by Ando et al. suffered from gastro-oesophageal reflux disease, which might be the source of heterogeneity between the study by Ando et al. and other including studies. No significant difference with pooled ORs was shown in the sensitivity analysis. In our study selection process, two studies on -238G>A, one study on -555G>A, and one study on -806C>T investigated the association with H. pylori infection, and all reported no significant association. We did not conduct a meta-analysis in three TNFA SNPs [20, 27, 40].
Numerous methods have been developed for diagnosing H. pylori infection, such as bacterial culture, RUT, PCR, UBT, histological examination and serum antibody detection [42]. Bacterial culture, RUT, UBT and histological examination can be affected by biopsy location, bacterial density and morphology, fastidious growth requirements, and so on [43]. Serology could not distinguish between current and past H. pylori infection, but an IgG-positive sample can show that the host is susceptible to H. pylori [44]. Since variable H. pylori infection detection methods were used in studies included in our meta-analysis, which could cause different results of diagnosing H. pylori infection, we conducted subgroup analyses (ELISA vs. non-ELISA methods) to verify the association between TNFA polymorphisms and H. pylori infection. A significant association was found between the TNFA -863C>A polymorphism and H. pylori infection for the non-ELISA subgroup in dominant and allelic models, and between -857C>T and H. pylori infection for the non-ELISA subgroup in allelic model. -308G>A and -1031T>C polymorphisms had no association with H. pylori infection for ELISA or no-ELISA subgroups.
Gastric acid secretion is supposed to be inhibited by TNF-α, which was produced by macrophages in the gastric submucosa [45]. Since the TNFA -863A polymorphism is related to high transcriptional promoter activity [46], carriers of the TNFA -863A polymorphism may have a significantly higher level of TNF-α than those with the C allele. High concentrations of TNF-α could directly suppress gastric acid secreted by parietal cells, and simultaneously inhibit the functions of gastrin-stimulated enterochromaffin-like cells to decrease the secretion of histamine, which can elevate gastric secretion [46]. In addition, a high level of TNF-α could amplify inflammatory responses by activating neutrophils, T cells, and B cells. Low levels of gastric acid, and an aggressive inflammatory response, can facilitate the colonization of the gastric mucosa with H. pylori from the gastric antrum to the corpus [9]. This might increase the risk of developing atrophic gastritis, or even gastric cancer.
Although there are papers reporting that -308G>A and -1031T>C polymorphisms are also related to high transcriptional promoter activity [47–49], our meta-analysis revealed that -308G>A and -1031T>C polymorphisms could decrease the risk of H. pylori infection. This difference may be linked with the sample size and ethnicity. Moreover, TNF-α possibly regulates H. pylori infection through other mechanisms. Further studies are needed to confirm the mechanisms.
There were some limitations to this study. Firstly, most of the studies included for -857C>T, -863C>A, and -1031T>C polymorphisms were conducted on Asian populations, so further research with other ethnic populations are needed. Secondly, only a low number of studies were included. Therefore, more studies involving much larger sampling sizes should be conducted. Thirdly, it is also possible that language bias might exist, as our meta-analysis only included articles published in English.
Conclusions
This meta-analysis is the first to investigate the association between TNFA polymorphisms and H. pylori infection. Our conclusion suggests that TNFA -308G>A and -1031T>C polymorphisms may be associated with a decreasing risk of H. pylori infection, and the -863C>A polymorphism may be associated with an increased risk of H. pylori infection. There was no significant association between -308G>A and H. pylori infection for Asian or Caucasian subgroups. TNFA -863C>A and -1031T>C polymorphisms had significant associations with H. pylori infection for Asian and HB subgroups, and -857C>T and -863C>A polymorphisms had significant associations with H. pylori infection for non-ELISA subgroup. Further studies with different ethnicities and larger sample size are required to validate our results.
Supporting Information
(DOC)
(DOC)
(DOCX)
(DOCX)
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This research was supported by the open fund of Key Laboratory for Gastrointestinal Diseases of Gansu Province (gswcky-2013-002). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Björkholm B, Falk P, Engstrand L, Nyrén O. Helicobacter pylori: Resurrection of the cancer link. Journal of Internal Medicine. 2003;253(2):102–19. [DOI] [PubMed] [Google Scholar]
- 2.Axon A. Helicobacter pylori and public health. Helicobacter. 2014;19 Suppl 1(1523–5378 (Electronic)):68–73. 10.1111/hel.12155 . [DOI] [PubMed] [Google Scholar]
- 3.Ryan BM, Murphy G, O'Morain CA. Host cytokine responses to Helicobacter pylori: An important determinant of clinical outcome. Irish Journal of Medical Science. 2001;170(2):90–1. [DOI] [PubMed] [Google Scholar]
- 4.Basso D, Plebani M. H. pylori infection: bacterial virulence factors and cytokine gene polymorphisms as determinants of infection outcome. Crit Rev Clin Lab Sci. 2004;41(3):313–37. Epub 2004/08/17. 10.1080/10408360490472804 . [DOI] [PubMed] [Google Scholar]
- 5.Goto H. Helicobacter pylori and gastric diseases. Nagoya J Med Sci. 2003;66(3–4):77–85. Epub 2004/01/20. . [PubMed] [Google Scholar]
- 6.Roberts-Thomson IC, Butler WJ. Polymorphism and gastric cancer. Journal of Gastroenterology and Hepatology (Australia). 2005;20(5):793–4. [DOI] [PubMed] [Google Scholar]
- 7.Xiang Y, Yang ZB, Chen P. Relationship between IL-1B and TNF-α gene polymorphisms and susceptibilities to gastric ulcer and cancer. Chinese Journal of Biologicals. 2009;22(10):1010–4. [Google Scholar]
- 8.Hamajima N. Persistent Helicobacter pylori infection and genetic polymorphisms of the host. Nagoya J Med Sci. 2003;66(3–4):103–17. Epub 2004/01/20. . [PubMed] [Google Scholar]
- 9.Hamajima N, Hishida A. Genetic traits for the persistence of Helicobacter pylori infection. Personalized Medicine. 2010;7(3):249–62. [DOI] [PubMed] [Google Scholar]
- 10.Zambon CF, Basso D, Navaglia F, Belluco C, Falda A, Fogar P, et al. Pro- and anti-inflammatory cytokines gene polymorphisms and Helicobacter pylori infection: interactions influence outcome. Cytokine. 2005;29(4):141–52. Epub 2005/01/18. 10.1016/j.cyto.2004.10.013 . [DOI] [PubMed] [Google Scholar]
- 11.Garcia-Gonzalez MA, Savelkoul PH, Benito R, Santolaria S, Crusius JB, Pena AS, et al. No allelic variant associations of the IL-1 and TNF gene polymorphisms in the susceptibility to duodenal ulcer disease. Int J Immunogenet. 2005;32(5):299–306. Epub 2005/09/17. 10.1111/j.1744-313X.2005.00528.x . [DOI] [PubMed] [Google Scholar]
- 12.Salagacka A, Zebrowska M, Jelen A, Mirowski M, Balcerczak E. Investigation of -308G>A and -1031T>C polymorphisms in the TNFA promoter region in Polish peptic ulcer patients. Gut Liver. 2014;8(6):632–6. Epub 2014/11/05. 10.5009/gnl13224 ; PubMed Central PMCID: PMCPmc4215449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang BB, Liu XZ, Sun J, Yin YW, Sun QQ. Association between TNF alpha gene polymorphisms and the risk of duodenal ulcer: a meta-analysis. PLoS One. 2013;8(2):e57167 Epub 2013/03/02. 10.1371/journal.pone.0057167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guo XF, Wang J, Yu SJ, Song J, Ji MY, Cao Z, et al. TNF-alpha-308 polymorphism and risk of digestive system cancers: a meta-analysis. World journal of gastroenterology: WJG. 2013;19(48):9461–71. Epub 2014/01/11. 10.3748/wjg.v19.i48.9461 ; PubMed Central PMCID: PMCPmc3882423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.GA Wells BS, D O'Connell, J Peterson, V Welch, M Losos, P Tugwell. The Newcastle-Ottawa Scale (NOS) for assessing the quality of nonrandomised studies in meta-analyses Ottawa Health Research Institute2011. Available: http://www.ohri.ca/programs/clinical_epidemiology/oxford.asp.
- 16.Higgins JP, Thompson SG. Quantifying heterogeneity in a meta-analysis. Statistics in medicine. 2002;21(11):1539–58. 10.1002/sim.1186 . [DOI] [PubMed] [Google Scholar]
- 17.Kunstmann E, Epplen C, Elitok E, Harder M, Suerbaum S, Peitz U, et al. Helicobacter pylori infection and polymorphisms in the tumor necrosis factor region. Electrophoresis. 1999;20(8):1756–61. Epub 1999/08/06. . [DOI] [PubMed] [Google Scholar]
- 18.Hamajima N, Shibata A, Katsuda N, Matsuo K, Ito H, Saito T, et al. Subjects with TNF-A-857TT and -1031TT genotypes showed the highest Helicobacter pylori seropositive rate compared with those with other genotypes. Gastric Cancer. 2003;6(4):230–6. Epub 2004/01/13. 10.1007/s10120-003-0258-z . [DOI] [PubMed] [Google Scholar]
- 19.Li C, Xia B, Yang Y, Li J, Xia HH. TNF gene polymorphisms and Helicobacter Pylori infection in gastric carcinogenesis in Chinese population. The American journal of gastroenterology. 2005;100(2):290–4. Epub 2005/01/26. 10.1111/j.1572-0241.2005.40806.x . [DOI] [PubMed] [Google Scholar]
- 20.Lu CC, Sheu BS, Chen TW, Yang HB, Hung KH, Kao AW, et al. Host TNF-alpha-1031 and -863 promoter single nucleotide polymorphisms determine the risk of benign ulceration after H. pylori infection. The American journal of gastroenterology. 2005;100(6):1274–82. Epub 2005/06/03. 10.1111/j.1572-0241.2005.40852.x . [DOI] [PubMed] [Google Scholar]
- 21.Ando T, El-Omar EM, Goto Y, Nobata K, Watanabe O, Maeda O, et al. Interleukin 1B proinflammatory genotypes protect against gastro-oesophageal reflux disease through induction of corpus atrophy. Gut. 2006;55(2):158–64. Epub 2005/08/27. 10.1136/gut.2005.072942 ; PubMed Central PMCID: PMCPmc1856489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Atsuta Y, Ito LS, Oba-Shinjo SM, Uno M, Shinjo SK, Marie SKN, et al. Associations of TNF-A-1031TT and -857TT genotypes with Helicobacter pylori seropositivity and gastric atrophy among Japanese Brazilians. International Journal of Clinical Oncology. 2006;11(2):140–5. [DOI] [PubMed] [Google Scholar]
- 23.Kim N, Cho SI, Yim JY, Kim JM, Lee DH, Park JH, et al. The effects of genetic polymorphisms of IL-1 and TNF-A on Helicobacter pylori-induced gastroduodenal diseases in Korea. Helicobacter. 2006;11(2):105–12. Epub 2006/04/04. 10.1111/j.1523-5378.2006.00384.x . [DOI] [PubMed] [Google Scholar]
- 24.Sugimoto M, Furuta T, Shirai N, Nakamura A, Xiao F, Kajimura M, et al. Different effects of polymorphisms of tumor necrosis factor-alpha and interleukin-1 beta on development of peptic ulcer and gastric cancer. Journal of gastroenterology and hepatology. 2007;22(1):51–9. Epub 2007/01/05. 10.1111/j.1440-1746.2006.04442.x . [DOI] [PubMed] [Google Scholar]
- 25.Chakravorty M, Datta de D, Choudhury A, Santra A, Roychoudhury S. Association of specific haplotype of TNFα with Helicobacter pylori-mediated duodenal ulcer in eastern Indian population. Journal of Genetics. 2008;87(3):299–304. [DOI] [PubMed] [Google Scholar]
- 26.Szoke D, Molnar B, Solymosi N, Klausz G, Gyulai Z, Toth B, et al. T-251A polymorphism of IL-8 relating to the development of histological gastritis and G-308A polymorphism of TNF-α relating to the development of macroscopic erosion. European Journal of Gastroenterology and Hepatology. 2008;20(3):191–5. 10.1097/MEG.0b013e3282f1d29f [DOI] [PubMed] [Google Scholar]
- 27.García-González MA, Aísa MAP, Strunk M, Benito R, Piazuelo E, Jiménez P, et al. Relevance of IL-1 and TNF gene polymorphisms on interleukin-1β and tumor necrosis factor-α gastric mucosal production. Human Immunology. 2009;70(11):935–45. 10.1016/j.humimm.2009.07.024 [DOI] [PubMed] [Google Scholar]
- 28.Abdiev S, Ahn KS, Khadjibaev A, Malikov Y, Bahramov S, Rakhimov B, et al. Helicobacter pylori infection and cytokine gene polymorphisms in Uzbeks. Nagoya J Med Sci. 2010;72(3–4):167–72. Epub 2010/10/15. . [PMC free article] [PubMed] [Google Scholar]
- 29.Kimang'a AN. IL-1B-511 Allele T and IL-1RN-L/L Play a Pathological Role in Helicobacter Pylori (H. Pylori) Disease Outcome in the African Population. Ethiopian journal of health sciences. 2012;22(3):163–9. Epub 2012/12/05. ; PubMed Central PMCID: PMCPmc3511894. [PMC free article] [PubMed] [Google Scholar]
- 30.Santos JC, Ladeira MS, Pedrazzoli J Jr., Ribeiro ML. Relationship of IL-1 and TNF-alpha polymorphisms with Helicobacter pylori in gastric diseases in a Brazilian population. Brazilian journal of medical and biological research = Revista brasileira de pesquisas medicas e biologicas / Sociedade Brasileira de Biofisica [et al]. 2012;45(9):811–7. Epub 2012/06/21. ; PubMed Central PMCID: PMCPmc3854325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Queiroz DM, Rocha AM, Melo FF, Rocha GA, Teixeira KN, Carvalho SD, et al. Increased gastric IL-1beta concentration and iron deficiency parameters in H. pylori infected children. PLoS One. 2013;8(2):e57420 Epub 2013/03/02. 10.1371/journal.pone.0057420 ; PubMed Central PMCID: PMCPmc3581450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhao Y, Wang JW, Tanaka T, Hosono A, Ando R, Tokudome S, et al. Association between TNF-alpha and IL-1beta genotypes vs Helicobacter pylori infection in Indonesia. World journal of gastroenterology: WJG. 2013;19(46):8758–63. Epub 2014/01/01. 10.3748/wjg.v19.i46.8758 ; PubMed Central PMCID: PMCPmc3870525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Salagacka A, Zebrowska M, Jelen A, Mirowski M, Balcerczak E. Haplotype analysis of TNFA gene in peptic ulcer patients. International Journal of Human Genetics. 2014;14(1):9–15. [Google Scholar]
- 34.Saijo Y, Yoshioka E, Fukui T, Kawaharada M, Sata F, Sato H, et al. H pylori seropositivity and cytokine gene polymorphisms. World Journal of Gastroenterology. 2007;13(33):4445–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gao L, Weck MN, Nieters A, Brenner H. Inverse association between a pro-inflammatory genetic profile and Helicobacter pylori seropositivity among patients with chronic atrophic gastritis: Enhanced elimination of the infection during disease progression? European Journal of Cancer. 2009;45(16):2860–6. 10.1016/j.ejca.2009.04.015 [DOI] [PubMed] [Google Scholar]
- 36.Cheng HH, Chang CS, Wang HJ, Wang WC. Interleukin-1beta and -10 polymorphisms influence erosive reflux esophagitis and gastritis in Taiwanese patients. Journal of gastroenterology and hepatology. 2010;25(8):1443–51. Epub 2010/07/28. 10.1111/j.1440-1746.2010.06310.x . [DOI] [PubMed] [Google Scholar]
- 37.Kang JM, Kim N, Shin CM, Lee HS, Lee DH, Jung HC, et al. Predictive factors for improvement of atrophic gastritis and intestinal metaplasia after Helicobacter pylori eradication: a three-year follow-up study in Korea. Helicobacter. 2012;17(2):86–95. Epub 2012/03/13. 10.1111/j.1523-5378.2011.00918.x . [DOI] [PubMed] [Google Scholar]
- 38.Kulmambetova GN, Imanbekova MK, Logvinenko AA, Sukashev AT, Filipenko ML, Ramanculov EM. Association of cytokine gene polymorphisms with gastritis in a Kazakh population. Asian Pacific journal of cancer prevention: APJCP. 2014;15(18):7763–8. Epub 2014/10/09. . [PubMed] [Google Scholar]
- 39.Queiroz DM, Saraiva IE, Rocha GA, Rocha AM, Gomes LI, Melo FF, et al. IL2-330G polymorphic allele is associated with decreased risk of Helicobacter pylori infection in adulthood. Microbes and infection / Institut Pasteur. 2009;11(12):980–7. Epub 2009/07/30. 10.1016/j.micinf.2009.07.008 . [DOI] [PubMed] [Google Scholar]
- 40.Tseng FC, Brown EE, Maiese EM, Yeager M, Welch R, Gold BD, et al. Polymorphisms in cytokine genes and risk of Helicobacter pylori infection among Jamaican children. Helicobacter. 2006;11(5):425–30. Epub 2006/09/12. 10.1111/j.1523-5378.2006.00433.x . [DOI] [PubMed] [Google Scholar]
- 41.Mayerle J, den Hoed CM, Schurmann C, Stolk L, Homuth G, Peters MJ, et al. Identification of genetic loci associated with Helicobacter pylori serologic status. Jama. 2013;309(18):1912–20. Epub 2013/05/09. 10.1001/jama.2013.4350 . [DOI] [PubMed] [Google Scholar]
- 42.Patel SK, Pratap CB, Jain AK, Gulati AK, Nath G. Diagnosis of Helicobacter pylori: what should be the gold standard? World J Gastroentero. 2014;20(2219–2840 (Electronic)):12847–59. D—NLM: PMC4177467 OTO—NOTNLM. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McNulty CA, Lehours P Fau—Megraud F, Megraud F. Diagnosis of Helicobacter pylori Infection. Helicobacter. 2011;16 Suppl 1(1523–5378 (Electronic)):10–8. [DOI] [PubMed] [Google Scholar]
- 44.Sun X, Xu Y, Zhang F, Jing T, Han J, Zhang J. Association between the IL1B -31C > T polymorphism and Helicobacter pylori infection in Asian and Latin American population: A meta-analysis. Microbial pathogenesis. 2015;86:45–52. Epub 2015/07/19. 10.1016/j.micpath.2015.07.010 . [DOI] [PubMed] [Google Scholar]
- 45.D'Elios MM, Czinn SJ. Immunity, inflammation, and vaccines for Helicobacter pylori. Helicobacter. 2014;19 Suppl 1(1523–5378 (Electronic)):19–26. [DOI] [PubMed] [Google Scholar]
- 46.Beales IL, Calam J. Interleukin 1 beta and tumour necrosis factor alpha inhibit acid secretion in cultured rabbit parietal cells by multiple pathways. Gut. 1998;42(2):227–34. D—NLM: PMC1726991 EDAT- 1998/04/16 MHDA- 1998/04/16 00:01 CRDT- 1998/04/16 00:00 PST—ppublish. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kroeger KM, Carville KS, Abraham LJ. The -308 tumor necrosis factor-alpha promoter polymorphism effects transcription. Molecular immunology. 1997;34(5):391–9. Epub 1997/04/01. . [DOI] [PubMed] [Google Scholar]
- 48.Soga Y, Nishimura F, Ohyama H, Maeda H, Takashiba S, Murayama Y. Tumor necrosis factor-alpha gene (TNF-alpha) -1031/-863, -857 single-nucleotide polymorphisms (SNPs) are associated with severe adult periodontitis in Japanese. Journal of clinical periodontology. 2003;30(6):524–31. Epub 2003/06/11. . [DOI] [PubMed] [Google Scholar]
- 49.Higuchi T, Seki N, Kamizono S, Yamada A, Kimura A, Kato H, et al. Polymorphism of the 5'-flanking region of the human tumor necrosis factor (TNF)-alpha gene in Japanese. Tissue antigens. 1998;51(6):605–12. Epub 1998/08/07. . [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOC)
(DOC)
(DOCX)
(DOCX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.