Abstract
A PCR-based assay was developed for more accurate identification of Vibrio parahaemolyticus through targeting the blaCARB-17 like element, an intrinsic β-lactamase gene that may also be regarded as a novel species-specific genetic marker of this organism. Homologous analysis showed that blaCARB-17 like genes were more conservative than the tlh, toxR and atpA genes, the genetic markers commonly used as detection targets in identification of V. parahaemolyticus. Our data showed that this blaCARB-17-specific PCR-based detection approach consistently achieved 100% specificity, whereas PCR targeting the tlh and atpA genes occasionally produced false positive results. Furthermore, a positive result of this test is consistently associated with an intrinsic ampicillin resistance phenotype of the test organism, presumably conferred by the products of blaCARB-17 like genes. We envision that combined analysis of the unique genetic and phenotypic characteristics conferred by blaCARB-17 shall further enhance the detection specificity of this novel yet easy-to-use detection approach to a level superior to the conventional methods used in V. parahaemolyticus detection and identification.
Keywords: Vibrio parahaemolyticus, blaCARB-17, molecular detection, PCR
Introduction
Vibrio sp. are gram-negative and halophilic bacteria that inhabit the estuarine and marine environment and some species can cause gastrointestinal diseases in human (Austin, 2010; Scallan et al., 2011; Letchumanan et al., 2014). Infections caused by the pathogenetic Vibrio sp. are often due to consumption of raw or undercooked seafood, with V. parahaemolyticus being one of the most important foodborne pathogens worldwide (Su and Liu, 2007; Gonzalez-Escalona et al., 2011; Letchumanan et al., 2015). Although infections caused by V. parahaemolyticus are always self-limiting, they can be life-threatening in patients who suffer from liver dysfunction or suppressed immunity (Ottaviani et al., 2012).
Identification of V. parahaemolyticus is conventionally conducted by performing biochemical tests upon isolation of the organisms from selective agar plates (Di Pinto et al., 2011). However, identification of V. parahaemolyticus by phenotypic approaches has some drawbacks such as being labor-intensive, time-consuming and not very effective in terms of detection specificity (Di Pinto et al., 2011; Izumiya et al., 2011). To cope with the problems caused by conventional microbiological culture method, some rapid detection techniques based on genus or species-specific genotypic features have been developed recently (Bej et al., 1999; Ward and Bej, 2006; Bauer and Rorvik, 2007; Neogi et al., 2010; Izumiya et al., 2011; Liu et al., 2012; Vinothkumar et al., 2013). Many of the targeting genes used in these approaches are phylogenetic markers or those involved in virulence (tlh, toxR, atpA etc.), yet some of which are not highly species-specific as different Vibrio species may share similar sequences, thus reducing the accuracy and specificity of such detection methods. V. parahaemolyticus is a member of the V. harveyi group, which comprises V. alginolyticus, V. harveyi, and V. campbellii etc. These species exhibited a high degree of genetic relatedness in phylogenetic analysis (Thompson et al., 2007). However, in our routine identification of V. parahaemolyticus, we noticed that PCR targeting tlh often could not differentiate organisms in the V. harveyi group, especially V. parahaemolyticus and V. alginolyticus. This phenomena is in agreement with previous findings that tlh was distributed among V. alginolyticus (Xie et al., 2005), and that similar virulence-related genes in V. parahaemolyticus also existed in other Vibrionaceae species(Klein et al., 2014). However, no investigation has been done to compare the specificity of these genetic markers in detecting V. parahamolyticus.
Our laboratory recently identified a β-lactamase that contributed to intrinsic ampicillin resistance in V. parahaemolyticus (Chiou et al., 2015). The gene encoding this enzyme is an intrinsic gene in V. parahaemolyticus and is more conserved in this species compared to other gene markers. It bears all the hallmarks of a unique marker suitable for V. parahaemolyticus detection and identification. In recent years, species-specific β-lactamase genes are being explored as targets for development of combined genetic and phenotypic bacteria identification approaches. An excellent example being the PCR detection method targeting the intrinsic β-lactamase gene, blaOXA-51 like gene, which can be applied for Acinetobacter baumannii detection (Turton et al., 2006). In this study, we attempted to develop a reliable and simple PCR assay targeting blaCARB-17 for detection and identification of V. parahaemolyticus.
Materials and Methods
Bacterial Strains
A total of 120 V. parahaemolyticus strains and 109 non- V. parahaemolyticus strains were included in this study. All strains were identified using 16S rRNA sequencing, API 20E test strips and Vitek 2 Compact system (bioMerieux, Inc.). Genomic DNA extraction was conducted by the boiling method as previously described (Pathmanathan et al., 2003). Briefly, 1 ml of overnight culture was centrifuged and the pellet was suspended in 400 μl of ddH2O. The bacterial suspension was boiled for 5 min and centrifuged at 11,000 g for 6 min. The supernatant was used as DNA template for PCR assay.
Phylogenetic Analysis of Different Genetic Markers within the Vibrio sp.
Homology analysis of different atpA, tlh, toxR and blaCARB-17 variants was performed with the DNAMAN software (Lynnon Biosoft Corporation, USA1) by quick alignment method and default parameters were used. Sequences available at NCBI nucleotide collection (nr) and whole genome shotgun(wgs) databases were employed to retrieve atpA, tlh, toxR and blaCARB-17-like sequences. The four genes were almost identical among the strains from the same species, therefore only one representative sequence per specie was used to build the homology tree. The sequences used to construct the homology trees were displayed in Supplementary Materials.
Development of a PCR Method Targeting blaCARB–17 Like Genes in V. parahaemolyticus
In order to design specific primers for blaCARB-17 gene detection in V. parahaemolyticus, the conserved regions of this gene in the V. parahaemolyticus genome was screened for selection of target regions, followed by development of a PCR-based mismatch amplification mutation assay. Upon sequence alignment, two regions (550–565, 834–852) that corresponded to the location of blaCARB-17 (KJ934265) were selected for primer design. Eventually, two degenerate primers (Table 1), named CARB-VP-F (ACYTTGATGGAAGATA) and CARB-VP-R (YTAACTTTCTTTGTAGTGM) respectively, were generated. Primer-Blast was used to check primer pair specificity2. The result showed that this primer set did not exhibit significant sequence homology to other DNA fragments in the NCBI nr database.
Table 1.
Primers used in comparison of detection specificity of different V. parahaemolyticus detection methods.
| Primer names | Primer sequences | Product length | Target genes | References |
|---|---|---|---|---|
| CARB-F | ACC(T)TTGATGGAAGATA | 303 bp | blaCARB-17 | This study |
| CARB-R | T(C)TAACTTTCTTTGTAGTGC(A) | |||
| TLH-F | AAAGCGGATTATGCAGAAGCACTG | 450 bp | tlh | Dickinson et al., 2013 |
| TLF-R | GCTACTTTCTAGCATTTTCTCTGC | |||
| atpA-VP-F | TACTAGGCCGCGTAGTA | 794 bp | atpA | Izumiya et al., 2011 |
| atpA-VP-R | CGCTGGACGTACACCT | |||
| toxR-VP-F | GTCTTCTGACGCAATCGTTG | 350 bp | toxR | Kim et al., 1999 |
| toxR-VP-R | ATACGAGTGGTTGCTGTCATG |
PCR reactions with the designed primers were optimized by testing different annealing temperatures, primer concentrations and extension times. Each reaction mixture (20 μl) contained 10 μl of Premix Ex TaqTM (TaKaRa, Japan), 0.5 μl of DNA template, 1 μl of forward and reverse primers(10 pM) respectively, and 7.5 μl of nuclease-free water. The PCR amplification program was as follows: 95°C for 5 min, followed by 30 cycles of 95°C for 30 s, 50°C for 30 s, and 72°C for 30 s, and final elongation at 72°C for 5 min. The annealing temperature 50°C was obtained through the comparison of three different temperatures, 45, 50, and 55°C. The PCR products were differentiated on 1.5% agarose and visualized by the Gel Doc System (Biorad). The specificity of the PCR method was tested with 120 V. parahaemolyticus strains and 109 non- V. parahaemolyticus strains (Table 2).
Table 2.
Results of the specificity of PCR methods targeting different genes in Vibrio parahaemolyticus and non- Vibrio parahaemolyticus strains.
| Species | Source | No. of strains | Positive rate (No. of positive strains) |
|||
|---|---|---|---|---|---|---|
| blaCARB-17 | tlh | atpA | toxR | |||
| Vibrio parahaemolyticus | Food, Clinical | 120 | 100% | 100% | 100% | 100% |
| Vibrio cholerae | Food | 26 | 0 | 0 | 89% (23) | 0 |
| Vibrio vulnificus | Food, Clinical | 4 | 0 | 0 | 0 | 0 |
| Vibrio alginolyticus | Food | 35 | 0 | 20% (7) | 2.8% (1) | 0 |
| Vibrio metschnikovii | Food | 1 | 0 | 0 | 0 | 0 |
| Vibrio fluvialis ATCC33809 | ATCC | 1 | 0 | 0 | 0 | 0 |
| Vibrio harveyi ATCC33842 | ATCC | 1 | 0 | 0 | 0 | 0 |
| Vibrio mimicus ATCC 33653 | ATCC | 1 | 0 | 0 | 0 | 0 |
| Vibrio campbellii ATCC 33865 | ATCC | 1 | 0 | 0 | 0 | 0 |
| Vibrio natriegens ATCC 14048 | Food | 1 | 0 | 0 | 0 | 0 |
| Aeromonas sp. | Food | 7 | 0 | 0 | 0 | 0 |
| Escherichia coli | Food, clinical | 10 | 0 | 0 | 0 | 0 |
| Salmonella sp. | Food, clinical | 10 | 0 | 0 | 0 | 0 |
| Enterobacter sp. | Clinical | 2 | 0 | 0 | 0 | 0 |
| Pseudomonas aeruginosa PAO1 | Clinical | 1 | 0 | 0 | 0 | 0 |
| Citrobacter freundii | Clinical | 2 | 0 | 0 | 0 | 0 |
| Klebsiella pneumonia | Clinical | 1 | 0 | 0 | 0 | 0 |
| Proteus mirabilis | Food | 2 | 0 | 0 | 0 | 0 |
| Myroides odoratimimus | Food | 2 | 0 | 0 | 0 | 0 |
| Staphylococcus aureus | Food | 1 | 0 | 0 | 0 | 0 |
Comparison of blaCARB Detection with Other Reported PCR Detection Methods
Other reported PCR detection methods targeting tlh, atpA and toxR genes were included in this study to compare the specificity between these methods (Kim et al., 1999; Izumiya et al., 2011; Dickinson et al., 2013). The primers used were listed in Table 1. PCR reactions were conducted according to those conditions previously reported.
Result and Discussion
With thorough bioinformatics analysis of the putative β-lactamase gene, we identified 32 CARB-like variants among the 293 available V. parahaemolyticus whole genome sequences in NCBI WGS database as of October 1, 2014. Apart from V. parahaemolyticus, CARB-like genes were found to distribute among several other Vibrio sp., such as V. alginolyticus, V. harveyi, V. campbellii, V. jasicida, V. natriegens, V. owensii, and V. rotiferianus after conducting the BLAST with blaCARB-17 gene in nucleotide collection (nr) database. Upon phylogenetic analysis, we found that the blaCARB-17 like genes in V. parahaemolyticus exhibited the lowest degree of similarity (78% homology) with that in V. alginolyticus (Figure 1). In order to compare the uniqueness of this gene with other genetic markers used to detect V. parahaemolyticus, we selected tlh, atpA and toxR genes within the Vibrio sp. and compared their genetic relatedness. The results showed that the degree of homology between V. parahaemolyticus and V. alginolyticus were respectively 85, 97, and 86% for the tlh, atpA and toxR genes; these values were higher than that of the blaCARB-17 gene, indicating that the blaCARB-17 like gene is the most divergent among these genes in Vibrio sp., therefore offering the highest specificity for detection.
FIGURE 1.
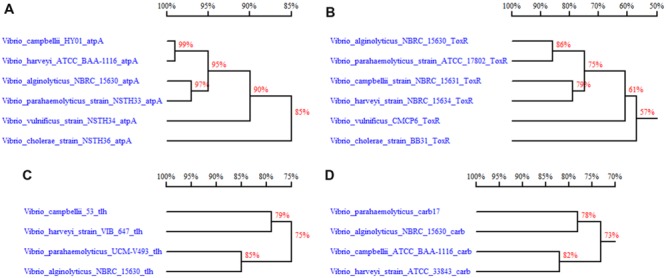
Comparison of sequence homology comparison between related Vibrio sp. for four different genes used as Vibrio parahaemolyticus detection targets. (A) atpA gene homology comparison between Vibrio sp. (NCBI accession no. AAWP01000035, CP006605.1, BATK01000022.1, KF886608.1, KF886609, KF886611); (B) toxR gene homology comparison between Vibrio sp.; (C) tlh gene homology comparison between species in the V. harveyi group (AB271112, DQ224369.1, CP007005.1, CP006719.1); (D) blaCARB-17 gene homology comparison between species in the V. harveyi group (KJ934265.1, CP006719.1, JPTG01000491, CP009468). Note: Some accession numbers represent the genome sequences, which harbor the analyzed genes. For the accurate gene sequences, refer to the Supplementary Sequences.
The PCR assay designed in this study for detection of the blaCARB-17 like gene in V. parahaemolyticus yielded an amplified fragment of 303bp (Supplementary Figure 1). The optimal annealing temperature was determined to be 50°C after optimization (Supplementary Figure 2). The specificity of the developed PCR in this study and other published methods (Izumiya et al., 2011; Dickinson et al., 2013) were verified in parallel with different strains (Table 2), with results showing that PCR method based on blaCARB-17 yielded 100% specificity for V. parahaemolyticus, while the methods based on detecting atpA and tlh were less specific and occasionally produced false positive result. This will undoubtedly reduce the accuracy of V. parahaemolyticus identification and may result in incorrect clinical diagnosis. The PCR method based on toxR gene detection displayed similar specificity as that targeting to blaCARB-17 in this study (Kim et al., 1999). Primers targeting the atpA gene exhibited very high false positive rate (89%) for V. cholerae and 2.8% false positive rate for V. alginolyticus, whereas primers targeting the tlh gene yield 20% false positive rate for V. alginolyticus (Table 2). This indicates that the choice of these two targets is not rigorous enough in terms of detection specificity. Some of the PCR results have been displayed in Supplementary Figure 3. The different tlh gene variants in V. alginolyticus and V. parahaemolyticus were detrimental to the specificity of the primers. All the tlh genes in WGS database from V. alginolyticus and V. parahaemolyticus were included in Supplementary Sequences. In contrast, the use of blaCARB-17 specific primers did not result in any false positive detection for all the bacteria tested, and consistently maintained 100% detection specificity for V. parahaemolyticus. Although many molecular detection methods have been developed to identify V. parahaemolyticus rapidly, some do not have a satisfactory level of specificity, hindering extensive application in routine laboratory tests (Xie et al., 2005; Klein et al., 2014). In this work, we showed that the blaCARB-17 gene is a V. harveyi clade (including V. parahaemolyticus) specific gene that can be used as a novel target for identification of V. parahaemolyticus by using degenerated primers. Combined with other specific target genes in other Vibrio sp., this novel target gene may be used to design multiplex-PCR approaches to detect food contamination by V. parahaemolyticus rapidly. With the increasing amount data of genome sequences, more species-specific genetic markers could be mined in silico through bioinformatics techniques, and relieve the laborious works required for specificity testing.
Conclusion
In this report, we used the available genome sequences in NCBI to identify a resistance gene known as blaCARB-17 like gene, which is intrinsic to V. parahaemolyticus. Based on the DNA sequences, a set of degenerated primers were designed to detect this major foodborne pathogen. The blaCARB-17 gene can be used as a novel V. parahaemolyticus detection marker, or used in combination with other markers to detect different Vibrio sp. simultaneously and rapidly. The specificity of blaCARB-17 gene together with its ampicillin resistance phenotype offer this detection method higher accuracy and specificity than other previously reported methods and will be of great benefit for food safety and clinical diagnosis.
Author Contributions
RL designed and conducted the experiments and wrote the manuscript, JC initiated the project, EC designed the experiment and edited the manuscript, SC designed the experiment, supervised the project and edited the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by the Chinese National Key Basic Research and Development Program (2013CB127200) and the Research Fund for the Control of Infectious Diseases of the Food and Health Bureau, the Government of the Hong Kong SAR (13121422 to SC).
Footnotes
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2016.00044
References
- Austin B. (2010). Vibrios as causal agents of zoonoses. Veter. Microbiol. 140 310–317. 10.1016/j.vetmic.2009.03.015 [DOI] [PubMed] [Google Scholar]
- Bauer A., Rorvik L. M. (2007). A novel multiplex PCR for the identification of Vibrio parahaemolyticus, Vibrio cholerae and Vibrio vulnificus. Lett. Appl. Microbiol. 45 371–375. 10.1111/j.1472-765X.2007.02195.x [DOI] [PubMed] [Google Scholar]
- Bej A. K., Patterson D. P., Brasher C. W., Vickery M. C. L., Jones D. D., Kaysner C. A. (1999). Detection of total and hemolysin-producing Vibrio parahaemolyticus in shellfish using multiplex PCR amplification of tl, tdh and trh. J. Microbiol. Methods 36 215–225. 10.1016/S0167-7012(99)00037-8 [DOI] [PubMed] [Google Scholar]
- Chiou J., Li R., Chen S. (2015). CARB-17 family of beta-lactamases mediated intrinsic resistance to penicillins in Vibrio parahaemolyticus. Antimicrob. Agents Chemother. 59 3593–3595. 10.1128/AAC.00047-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Pinto A., Terio V., Novello L., Tantillo G. (2011). Comparison between thiosulphate-citrate-bile salt sucrose (TCBS) agar and CHROMagar Vibrio for isolating Vibrio parahaemolyticus. Food Control 22 124–127. 10.1016/j.foodcont.2010.06.013 [DOI] [Google Scholar]
- Dickinson G., Lim K. Y., Jiang S. C. (2013). Quantitative microbial risk assessment of pathogenic Vibrios in marine recreational waters of Southern California. Appl. Environ. Microbiol. 79 294–302. 10.1128/Aem.02674-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Escalona N., Strain E. A., De Jesus A. J., Jones J. L., DePaola A. (2011). Genome sequence of the clinical O4:K12 serotype Vibrio parahaemolyticus strain 10329. J. Bacteriol. 193 3405–3406. 10.1128/Jb.05044-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izumiya H., Matsumoto K., Yahiro S., Lee J., Morita M., Yamamoto S., et al. (2011). Multiplex PCR assay for identification of three major pathogenic Vibrio spp., Vibrio cholerae, Vibrio parahaemolyticus, and Vibrio vulnificus. Mol. Cell. Probes 25 174–176. 10.1016/j.mcp.2011.04.004 [DOI] [PubMed] [Google Scholar]
- Kim Y. B., Okuda J., Matsumoto C., Takahashi N., Hashimoto S., Nishibuchi M. (1999). Identification of Vibrio parahaemolyticus strains at the species level by PCR targeted to the toxR gene. J. Clin. Microbiol. 37 1173–1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein S. L., West C. K. G., Mejia D. M., Lovell C. R. (2014). Genes similar to the Vibrio parahaemolyticus virulence-related genes tdh, tlh, and vscC2 occur in other Vibrionaceae species isolated from a pristine estuary. Appl. Environ. Microbiol. 80 595–602. 10.1128/Aem.02895-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letchumanan V., Chan K. G., Lee L. H. (2014). Vibrio parahaemolyticus: a review on the pathogenesis, prevalence, and advance molecular identification techniques. Front. Microbiol. 5:705 10.3389/fmicb.2014.00705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letchumanan V., Yin W. F., Lee L. H., Chan K. G. (2015). Prevalence and antimicrobial susceptibility of Vibrio parahaemolyticus isolated from retail shrimps in Malaysia. Front. Microbiol. 6:33 10.3389/fmicb.2015.00033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B., He X. H., Chen W. Y., Yu S. J., Shi C. L., Zhou X. J., et al. (2012). Development of a real time PCR assay for rapid detection of Vibrio parahaemolyticus from seafood. Protein Cell 3 204–212. 10.1007/s13238-012-2017-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neogi S. B., Chowdhury N., Asakura M., Hinenoya A., Haldar S., Saidi S. M., et al. (2010). A highly sensitive and specific multiplex PCR assay for simultaneous detection of Vibrio cholerae, Vibrio parahaemolyticus and Vibrio vulnificus. Lett. Appl. Microbiol. 51 293–300. 10.1111/j.1472-765X.2010.02895.x [DOI] [PubMed] [Google Scholar]
- Ottaviani D., Leoni F., Serra R., Serracca L., Decastelli L., Rocchegiani E., et al. (2012). Nontoxigenic Vibrio parahaemolyticus strains causing acute gastroenteritis. J. Clin. Microbiol. 50 4141–4143. 10.1128/Jcm.01993-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pathmanathan S. G., Cardona-Castro N., Sanchez-Jimenez M. M., Correa-Ochoa M. M., Puthucheary S. D., Thong K. L. (2003). Simple and rapid detection of Salmonella strains by direct PCR amplification of the hilA gene. J. Med. Microbiol. 52 773–776. 10.1099/jmm.0.05188-0 [DOI] [PubMed] [Google Scholar]
- Scallan E., Hoekstra R. M., Angulo F. J., Tauxe R. V., Widdowson M. A., Roy S. L., et al. (2011). Foodborne illness acquired in the united states-major pathogens. Emerg. Infect. Dis. 17 7–15. 10.3201/eid1701.P11101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su Y. C., Liu C. C. (2007). Vibrio parahaemolyticus: a concern of seafood safety. Food Microbiol. 24 549–558. 10.1016/J.Fm.2007.01.005 [DOI] [PubMed] [Google Scholar]
- Thompson C. C., Thompson F. L., Vicente A. C. P., Swings J. (2007). Phylogenetic analysis of vibrios and related species by means of atpA gene sequences. Int. J. Syst. Evol. Microbiol. 57 2480–2484. 10.1099/ijs.0.65223-0. [DOI] [PubMed] [Google Scholar]
- Turton J. F., Woodford N., Glover J., Yarde S., Kaufmann M. E., Pitt T. L. (2006). Identification of Acinetobacter baumannii by detection of the blaOXA-51-like carbapenemase gene intrinsic to this species. J. Clin. Microbiol. 44 2974–2976. 10.1128/JCM.01021-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinothkumar K., Bhardwaj A. K., Ramamurthy T., Niyogi S. K. (2013). Triplex PCR assay for the rapid identification of 3 major Vibrio species, Vibrio cholerae, Vibrio parahaemolyticus, and Vibrio fluvialis. Diagn. Microbiol. Infect. Dis. 76 526–528. 10.1016/j.diagmicrobio.2013.04.005 [DOI] [PubMed] [Google Scholar]
- Ward L. N., Bej A. K. (2006). Detection of Vibrio parahaemolyticus in shellfish by use of multiplexed real-time PCR with TaqMan fluorescent probes. Appl. Environ. Microbiol. 72 2031–2042. 10.1128/Aem.72.3.2031-2042.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z. Y., Hu C. Q., Chen C., Zhang L. P., Ren C. H. (2005). Investigation of seven Vibrio virulence genes among Vibrio alginolyticus and Vibrio parahaemolyticus strains from the coastal mariculture systems in Guangdong, China. Lett. Appl. Microbiol. 41 202–207. 10.1111/j.1472-765X.2005.01688.x [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


