Abstract
Bacteriophages are the viruses of bacteria, which have contributed extensively to our understanding of life and modern biology. The phage-mediated bacterial growth inhibition represents immense untapped source for novel antimicrobials. Insights into the interaction between mycobacteriophage and Mycobacterium host will inform better utilizing of mycobacteriophage. In this study, RNA sequencing technology (RNA-seq) was used to explore the global response of Mycobacterium smegmatis mc2155 at an early phase of infection with mycobacteriophage SWU1, key host metabolic processes of M. smegmatis mc2155 shut off by SWU1, and the responsible phage proteins. The results of RNA-seq were confirmed by Real-time PCR and functional assay. 1174 genes of M. smegmatis mc2155 (16.9% of the entire encoding capacity) were differentially regulated by phage infection. These genes belong to six functional categories: (i) signal transduction, (ii) cell energetics, (iii) cell wall biosynthesis, (iv) DNA, RNA, and protein biosynthesis, (v) iron uptake, (vi) central metabolism. The transcription patterns of phage SWU1 were also characterized. This study provided the first global glimpse of the reciprocal reprogramming between the mycobacteriophage and Mycobacterium host.
Keywords: transcriptome, mycobacteriophage, mycobacterium, interaction, ion channel
Introduction
Bacteriophages, the viruses of bacteria with an estimated size of 1031 in the biosphere (Whitman et al., 1998), represent an enormous resource for biomedicine and biotechnology, as seen by the growing interest in their therapeutic and food safety applications (García et al., 2008; Monk et al., 2010). Insights into the evolutionary arms race between phage and bacterium have revealed many new molecular machineries such as the widespread bacterial defense system called CRISPR/Cas, which in turn has inspired revolutionary genome editing tools (Barrangou et al., 2007), and exciting novel approaches for antimicrobials discovery (Liu et al., 2004; Samaddar et al., 2016). Well-characterized bacteria hosts,including Escherichia coli, Salmonella typhimirium, and Bacillus subtilis, have made significant contribution to the understanding of factors essential for phage replication, assembly, and lysis (Young, 2013; Sun et al., 2014).
New methodologies are increasingly used to characterize the interaction between phage and its host, such as the microarrays for profiling phage transcription throughout the T4-E. coli infection cycle, and the host during phage infection being profiled (Poranen et al., 2006; Ravantti et al., 2008; Fallico et al., 2011). RNA sequencing (RNA-seq) offers unique opportunity for in-depth interrogating reciprocal reprogramming of phage and bacterium during the phage infection cycle (Garber et al., 2011). However, clinical relevant phage–bacteria interactions are scarce and urgently needed. Mycobacterium includes notorious pathogens causing serious diseases in mammals, such as tuberculosis (Mycobacterium tuberculosis) and leprosy (M. leprae). More than 5850 mycobacteriophages, bacteriophage known to infect mycobacteria, have been isolated using a single host strain, Mycobacterium smegmatis mc2155, over 600 of which have been completely sequenced (http://phagesdb.org/). Most mycobacteriophages are isolated from America. We have isolated and characterized the first mycobacteriophage SWU1, a lytic phage, from China soil (Fan et al., 2012). In this study, we used RNA-seq and functional assay to characterize the reciprocal reprogramming between SWU1 and Mycobacterium, with special aim to unveil how mycobacteriophages usurp the biomolecular machinery of their mycobacterial host.
Materials and methods
Bacterial strains, phage, and media
M. smegmatis mc2155 and mycobacteriophage SWU1 were used for profiling the global gene expression dynamics of the host response to phage infection. M. smegmatis mc2155 and SWU1 were grown as described previously (Fan et al., 2012; Chen et al., 2013).
Sample collection for illumina RNA deep sequencing (RNA-seq) technology
An overnight culture was diluted with fresh medium (200 ml) and grown at 37°C until the optical density at 600 nm (OD600) up to 2. The culture was harvested by centrifugation, washed twice to remove Tween-80 using fresh 7H9 (without Tween-80), resuspended using MP buffer, and halved (about 1011 cells per culture). One portion of the cell culture was infected with SWU1 with a multiplicity of infection of 10 (Ravantti et al., 2008); the other portion was a non-infected control. Cultures were maintained at 37°C. Samples for RNA isolation were taken from infected cultures at time points of 30, 180, and 315 min p.i (named inf_30, inf_180, and inf_315). Sample for RNA isolation was also taken from uninfected cultures at time points of 30 min p.i (named uninf_30). The RNA of inf_30, inf_180, and inf_315 was used to study early, middle, and late transcriptional feature during the SWU1 replicative cycle; The RNA of inf_30, and uninf_30 was used to profile the M. smegmatis mc2155 response at the early infection by Mycobacteriophage SWU1.
RNA extraction
For every sample, the cultures were centrifuged at 12,000 r.p.m. for 10 min at 4°C. Cell pellets were snap frozen in liquid nitrogen and stored for subsequent RNA isolation. Total RNA was extracted using Trizol (Liao et al., 2013). The concentration, quality and integrity were determined using a NanoDrop spectrophotometer and an Agilent 2100 Bioanalyzer. The RNA integrity (RIN) value of samples were 6.9 (uninf_30), 8.6 (inf_30), 8.6 (inf_180), and 8.8 (inf_315) (Figure S1).
cDNA library construction, illumina sequencing, data processing, and analysis
Ribo-Zero™rRNA Removal Kit (Bacteria) was used to remove rRNAs and enrich mRNA. The mRNA was fragmented and used as template to synthesize first-stranded cDNA with reverse transcriptase and random hexamer-primers. Second-stranded cDNA was synthesized using RNase H and DNA polymerase I. These double-stranded cDNA fragments underwent process of end repair, addition of a single “A” base and ligation of adapters (TruSeq™ RNA Sample Prep Kit, Illumina). Adaptor modified fragments were selected by AMPure XP beads and amplified through PCR to create the final cDNA library. Transcriptome sequencing was carried out on an Illumina HiSeqTM2000 platform using 2 × 100 bp reads at the Beijing Novogene company (Beijing, China).
Raw data (raw reads) of fastq format were firstly processed through in-house PERL scripts. In this step, clean data (clean reads) were obtained by removing reads containing adapter, reads containing ploy-N (N > 10%) and low quality reads from raw data. At the same time, Q20, Q30, and GC content sequence of the clean data were calculated. Based on high quality clean data, all the downstream analyses were carried out.
The high quality reads were mapped to the genome of M. smegmatis mc2155 and SWU1. For differential expression analysis, the read counts were adjusted by edger program package through one scaling normalized factor (Robinson et al., 2010). Differential expression analysis of two conditions was performed using the DEGSeq R package (1.12.0). The P-values were adjusted using the Benjamini and Hochberg method. Corrected P-value of 0.005 and log2 (Fold change) of 1 were set as the threshold for significantly differential expression. Different expression genes between infected 30 min and uninfected 30 min can be found as Table S1 online.
KEGG is a database resource for understanding high-level functions and utilities of the biological system (Kanehisa and Goto, 2000), such as the cell, the organism, and the ecosystem, from molecular-level information, especially large-scale molecular datasets generated by genome sequencing and other high-throughput experimental technologies (http://www.genome.jp/kegg/). We used KOBAS software (Xie et al., 2011) to test the statistical enrichment of differential expression genes in KEGG pathways.
Measurements of ion fluxes
For the ion flux experiments, we harvested a log-phase culture of M. smegmatis using centrifugation and washed them with MP buffer to remove Tween-80. This cell suspension was kept on 4°C until used. SWU1 was added into the cell suspension (MOI = 10). The same volume of MP buffer was added as negative control. The concentrations of ions (potassium, sodium, calcium, magnesium, ferrum, manganese, barium, cesium, strontium, aluminum) in the solution were detected by OPTIMA ICP-OES 2100DV (PerkinElmer) in 0 and 30 min using published protocol. Two independent experiments were performed for all measurements (Xiang et al., 2011).
Validation of transcript levels using real-time PCR
Total RNA samples for real-time PCR were isolated and purified from those samples (uninf_30 and inf_30) by the method described above. Used purified RNA as templates, cDNA production was synthesized using the Transcriptor First Strand cDNA Synthesis Kit (Roche) and was used as templates for real-time PCR. Table S2 lists gene targets and oligonucleotide primers used for Real-time PCR. Real-time PCR was carried out using the SsoAdvanced™ SYBR® Green supermix (BIO-RAD) in Bio-Rad PTC-200 thermal cyclers. 16s rRNA gene was included as control. Three independent experiments were performed.
Statistical methods
All analyses were carried out by SPSS software (version 13.0 for Windows). To analyze the differences between groups, we used a one-way ANOVA test. A p-value of < 0.05 was defined statistically significant.
Results
Waves of transcription of mycobacteriophage SWU1 genome
There are a great number of mycobacteriophages sequenced by Graham Hatfulls group (Pope et al., 2015). Using previously reported approach (Hatfull et al., 2006), SWU1 (GenBank accession number: JF946695) belongs to mycobacteriophage A2 Subcluster of Cluster A. The number of Subcluster A2 phages is 41 before 22 December 2015 (http://phagesdb.org). Comparative genomics analysis shows that all Subcluster A2 phages are similar to SWU1 (Figure S2). SWU1 can be a representative of the Subcluster A2 mycobacteriophages.
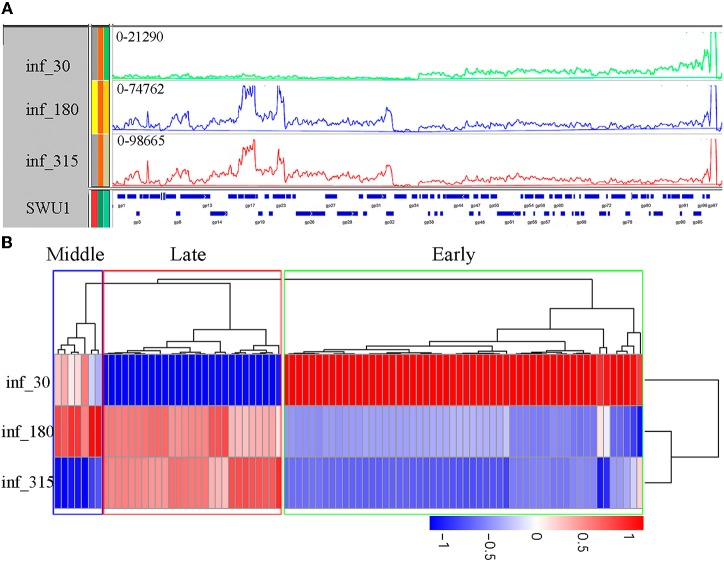
One-step growth analysis as described in prior report (Fan et al., 2015) revealed that the eclipse period of SWU1 was about 30 min, and burst period about 270 min. To define the gene expression of SWU1 during infection, we used RNA-seq to determine transcription profiles (Figure 1A) in early, middle, and late phage of SWU1 infections, namely 30, 180, and 315 min after absorption, respectively, according to the one-step growth curve. Multiplicity of infection (MOI) of 10 which can infect most hosts (Ravantti et al., 2008), was used in the experiment of SWU1 infection. Clustering of differentially expressed genes (Figure 1B) showed that some highly expressed genes located on the right arm of SWU1 in early phase of SWU1 infection, while other highly expressed genes located on the left arm in middle and late phase. A list of genes displaying three different expression profiles can be found as Table S3 online. Early proteins involved in the bacteriophage–host interaction might be of great value for further study. A promising early protein is SWU1 gp64 that can inhibit the growth of M. smegmatis and E. coli (our unpublished data).
Figure 1.
Transcription of the SWU1 genome determined by RNA-seq. (A) Transcription patterns in the SWU1 genome determined by RNAseq. RNA was isolated during early SWU1 infection (green), middle SWU1 infection (blue), and late SWU1 infection (red). At all time points, one sample was taken for sequencing (Illumina HiSeqTM2000). The Y-axis is the number of RNAseq reads. The X-axis is the map of the SWU1 genome. (B) Cluster analysis of differentially expressed genes of SWU1. Based on log10 (RPKM+1), we cluster the differentially expressed genes in different phase of SWU1 infection. Red stands for high expressed genes; blue stands for low expressed genes. In early phase of SWU1 infection, a lesser number of genes were transcribed. Most of genes transcribed were located on the right arm of SWU1 genome. In middle and late phase of SWU1 infection, most of genes transcribed were located on the left arm of SWU1 genome.
Overview of phage-induced host global gene expression alteration
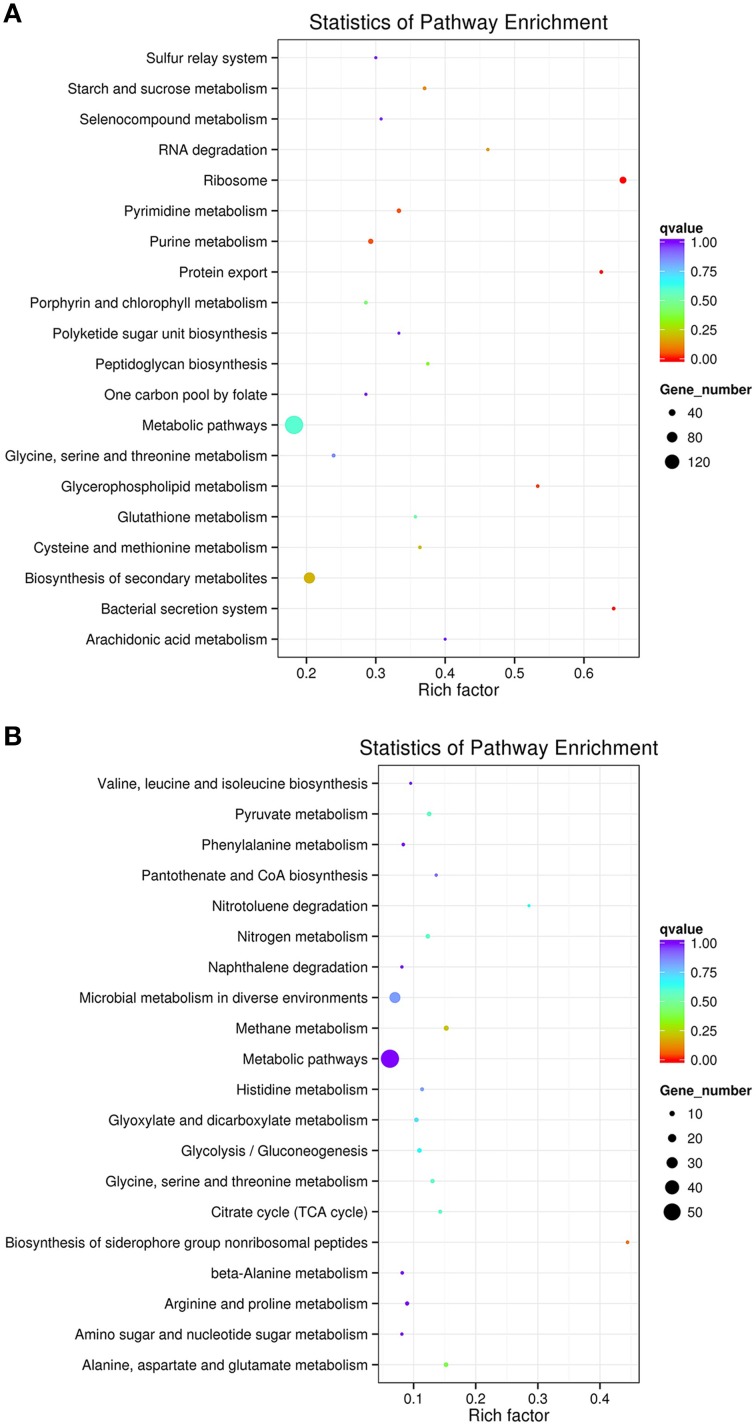
To profile M. smegmatis mc2155 gene expression dynamics induced by SWU1 infections, we used RNA-seq to determine the transcriptome of the cell culture infected with SWU1 and the non-infected control culture at 30 min after absorption. The phage SWU1 infection significantly shifted host gene expression. The expression of 1174 (825 up-regulated genes; 349 down-regulated genes) genes was changed at least two-fold, up to 16.9% of M. smegmatis mc2155 genome encoding capacity. Except 263 function unknown genes, 911 genes can be assigned to different specific pathways using KEGG (Figure 2). Up-regulated pathways include ribosome, protein export, bacterial secretion system, glycerophospholipid metabolism, RNA degradation. Down-regulated pathways include the biosynthesis of siderophore non-ribosomal peptides, nitrotoluene degradation.
Figure 2.
The scatter plot of KEGG pathways enriched for differential expression genes. Panel (A) shows up-regulated pathways. Panel (B) shows down-regulated pathways. The Y-axis is different KEGG pathway. The X-axis is the number of rich factor. The color of circles stands for the q-value of pathway. The range of q value is from 0 to 1. The size of circles stands for gene number. Some up-regulated genes associated to ribosome, protein export, bacterial secretion system, glycerophospholipid metabolism, and RNA degradation were enriched. Some down-regulated genes associated to siderophore non-ribosomal peptides and nitrotoluene degradation were enriched.
SWU1 infection alters the expression of genes involved in signaling
Signaling is essential to organisms. Mycobacterial serine/threonine protein kinases (STPKs) play important roles in growth, pathogenesis, and cell wall metabolism. During SWU1 infection, some M. smegmatis mc2155 genes involved in signaling were altered. They are listed in Table S4. Some genes encoded Ser/Thr protein kinases, pknA, pknB, pknE, pknH, and pknF, were regulated to respond to SWU1 induction (Table S4). M. smegmatis mc2155 also down-regulated sigA, sigF, and the gene encoding anti-sigma F factor antagonist (Table S4). Overexpression of sigF resulted in the up-regulation of many cell wall-associated proteins (Williams et al., 2007). However, among the 14 genes directly regulated by SigF, only the homologous genes of Rv3476c, Rv1270c, Rv2400c, and Rv1281c, were up-regulated during phage infection, implicating selective regulation by some unknown phage molecules. In early phase of SWU1 infection, the genes encoding transcription factors had been significant affected. Up to 28 transcriptional factor genes were affected. Among which, 13 were up-regulated; 15 were down-regulated (Table S4). These include WhiA, several FadR-like regulators, Lrp/AsnC, several MerR family regulators, and RpiR. The altered expression of 28 M. smegmatis mc2155 transcriptional factors showed extensive transcriptional reprogramming in response to SWU1 infection.
Mycobacterium cell energetics and ion fluxes changes in response to phage infection
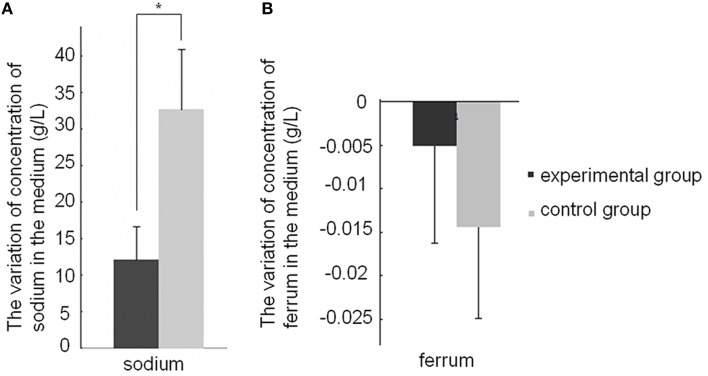
In this study, we showed that SWU1 can up-regulate genes involved in the transfer of K+ and efflux of K+ ions. KdpDE two component system is highly conserved across all bacterial species. Expression of the kdpD (encoding sensor kinase) and the kdpFABC operon, which encodes the high-affinity K+ transporter KdpFABC, was up-regulated early in SWU1 infection. ftsE and ftsX, encoding FtsEX protein complex, an ABC type transporter proteins, were also up-regulated. KdpD senses intracellular K+-limiting signal or extracellular high K+ concentration, and KdpE transmits the signal to the kdpFABC promoter to regulate the expression of kdpFABC (Steyn et al., 2003; Rothenbücher et al., 2006; Laermann et al., 2013). FtsEX protein complex regulates the translocation of K+ ion pump proteins KdpA into inner membrane (Ukai et al., 1998). To study the effect of the SWU1 infection on ions fluxes, we measured the concentration level change of some ions (potassium, sodium, calcium, magnesium, ferrum, manganese, barium, cesium, strontium, aluminum) in the medium 30 min after SWU1 infection, using uninfected bacteria as control. Comparing with the control group, the rising values of concentration of sodium decreased significantly (Figure 3A). That meant the capacity of sodium efflux was suppressed after SWU1 infected. In addition, the iron level is significantly reduced. However, the difference was not statistically significant (Figure 3B; Figure S3). For other ions, there was no difference between control group and experimental group (Figure S3). Quite unexpectedly, the increase of the concentration of K+ ions in the solution was not significant. The subtle change of the efflux of K+ ions beyond the threshold of the measurement might underlie this result.
Figure 3.
Effect of the SWU1 infection on sodium and ferrum fluxes. (A) Positive value stands for the concentration of ions increased after SWU1 infected 30 min. (B) Negative value stands for the concentration of ions decreased after SWU1 infected 30 min. The capacity of sodium efflux was suppressed after SWU1 infected. Iron uptake system was disrupted by SWU1. The data were averaged from two independent experiments ± s.d. Significant differences (*p < 0.05).
Transporter proteins are indispensable for the survival of organism. There are 423 transporter proteins in M. smegmatis mc2155 belonging to five transporter types: ATP-dependent, ion channels, phosphotransferase system (PTS), secondary transporter, and unclassified. Our data showed that after SWU1 infection 119 transporters (28.13%) were regulated (Table S5).
SWU1 infection induces the expression of the cell wall biosynthetic genes
The expression of the genes involved in the biosynthesis of mycolic acids, arabinogalactan (AG), peptidoglycan (PG), phosphatidylinositol mannosides (PIMs), lipomannan (LM), and lipoarabinomannan (LAM) (Belanger et al., 1996; Crick et al., 2001; Brennan, 2003; Takayama et al., 2005; Jankute et al., 2012; Munshi et al., 2013) had been studied. During SWU1 infection, most genes involved in biogenesis, modification, and regulation of the cell envelope were up-regulated (Table S6).
SWU1 hijacked M. smegmatis mc2155 DNA, RNA, and protein synthesis system
Among the 51 M. smegmatis mc2155 genes related to pyrimidine metabolism, 17 were up-regulated, and one of them was down-regulated during the early infection. Among 82 genes related to purine metabolism, 24 of them were up-regulated, and two of them were down-regulated. The data supported active mobilization of host DNA and RNA synthesis capacity by the phage.
The bacterial transcription largely depends on DNA-directed RNA polymerase. We investigated the components of RNA polymerase holoenzyme, and found that the expression of rpoA, rpoB, rpoC, and rpoZ was all up-regulated. Moreover, the transcription of sigA (rpoD) and sigF also was up-regulated. The up-regulation of these components demonstrated that SWU1 has usurped the host RNA synthesis machinery.
Ribosome is the factory for protein synthesis. In M. smegmatis mc2155, there are totally 64 ribosomal proteins, which are the critical components of ribosome. Up to 42 genes in this category were up-regulated. A similar pattern of expression was found for five genes related to aminoacyl-tRNA biosynthesis, which, respectively, involved in the synthesis of L-Aspartyl-tRNA (Asp), L-Glutamyl-tRNA (Glu), L-Glutaminyl-tRNA (Gln), L-Asparaginyl-tRNA (Asn), L-Prolyl-tRNA (Pro), and L-Phenylalanyl-tRNA (Phe). These observations implied that SWU1 also hijacked the bacterial protein expression system to produce virions in the later stage of infection.
Iron uptake system was disrupted by SWU1
Phage SWU1 infected M. smegmatis mc2155 down-regulated the genes actively involved in iron homeostasis (eccC3, mbtF, mbtE, mbtD, mbtB). MbtF, mbtE, mbtD, and mbtB are key genes for mycobactin biosynthesis, a class of siderophores for iron uptake in pathogenic and unpathogenic Mycobacterium (Snow, 1965). EccC3 is a crucial component of the ESX-3 secretion system important for iron concentration adaptation of Mycobacterium (Serafini et al., 2009, 2013). In the absence of Esx-3, Mycobacteria failed to use iron-bound mycobactin. The down-regulation of these genes might represent phage tactics to control the bacterial growth.
The infection of SWU1 activates the metabolism of Mycobacterium
M. tuberculosis dormancy can benefit the pathogen surviving the stress induced by the accumulation of nitrite under hypoxia within macrophage. The up-regulation of 57 genes and down-regulation of 63 genes were detected in M. tuberculosis treated with nitrate (Cunningham-Bussel et al., 2013). These nitrate-regulated genes might underlie the M. tuberculosis growth inhibition. Upon SWU1 infection 25 M. smegmatis mc2155 homologs were differentially expressed (Table S7). Interestingly, 21 genes (84%) showed distinct expression upon the two conditions. This is consistent with previous reports that phages resuscitate and promote growth of dormancy host (Pedulla et al., 2003). The 21 genes might play important roles in regulating the growth of Mycobacterium.
Verification of RNA-seq by real-time PCR analysis
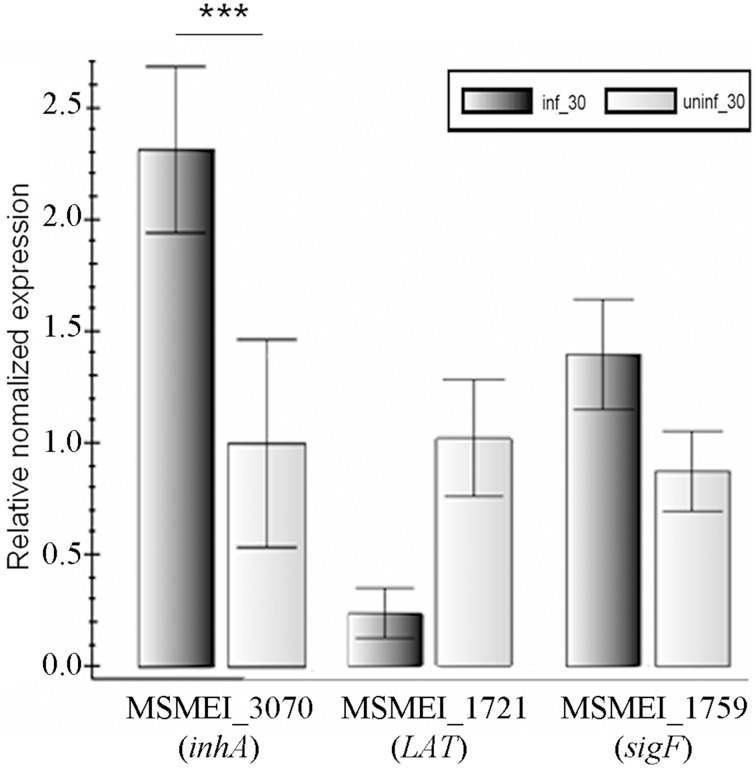
Real-time analyses were performed for selected genes to confirm the RNA-seq results. Three genes, namely MSMEI_3070 (inhA), MSMEI_1721 (L-lysine-epsilon aminotransferase gene, LAT), and MSMEI_1759 (sigF) were selected for real-time PCR verification. The three genes were chosen that offered a good representation of the difference in regulation on phage infection. They are involved in cell wall biosynthesis, persistence (metabolism), and signaling, respectively. These genes showed an increase of 5.87 times, decrease of 10.21 times, and increase of 4.62 times, respectively. The real-time PCR results are as following: up-regulated (2.31 times), down-regulated (4.34 times), and up-regulated (1.60 times), which (Figure 4) are in good agreement with the RNA-seq data.
Figure 4.
Verification of RNA-seq results by real-time PCR. Numbers means the numbering of gene in M. smegmatis mc2155. The data were averaged from three independent experiments ± s.d. Significant differences (***p < 0.001).
The shared differentially regulated host genes between SWU1 and other phages
To determine whether there is unifying theme among bacteria–phage interaction, we compared the subset of processes that responded to SWU1 infection with those responding to other phages infection. Genetic response to phage infection has been studied in Lactococcus lactis (c2 infection; Fallico et al., 2011), E. coli (PRD1 infection) (Poranen et al., 2006), and Pseudomonas aeruginosa (PRR1 infection) (Ravantti et al., 2008). We found that some processes are overlap in different phage–host interaction. L. lactis, E. coli, and M. smegmatis mc2155 all regulated stress response genes. Although those stress genes are different, the stress signal transduction is necessary for host response to phage infection. Ribosomal protein encoding genes were significantly up-regulated in P. aeruginosa and M. smegmatis mc2155, which are essential for the production of phage proteins. L. lactis and M. smegmatis mc2155 increases expression of genes involved in cell wall. That suggested that host repaired cell wall to defense phage infection. In addition, Salmonella typhimurium (PRD1 infection; (Daugelavicius et al., 1997)), L. lactis and M. smegmatis mc2155 changed cell energetics in response to phage. Manipulating the cell energetics in the bacterial cytosol might represent a phage reproduction strategy. However, the uniqueness of SWU1 might be the control of iron uptake system and the activation the metabolism of dormancy host.
Discussion
In this paper, we used RNA-seq and functional assay to characterize the reciprocal reprogramming between SWU1 and Mycobacterium. Phage SWU1 early transcripts, the key effector proteins which shut off the expression of some host genes, were analyzed. Phage early proteins have been reported to be involved in bacteriophage–host interactions within the infected cell (Rybniker et al., 2008; Roucourt and Lavigne, 2009). The search of candidate effectors altering the host metabolism among the early transcripts of SWU1 will pinpoint the key players in such interaction and inspire future antibiotics or drug target discovery.
To characterize the landscape of host response to phage infection and find the possible key targets phage hijacked during infection, we performed data mining. Significant changes occurred during phage early infection were highlighted. The major targets phage SWU1 hijacked are signal transduction, cell energetics and ions fluxes, cell wall biosynthesis, DNA, RNA, and protein synthesis system, iron uptake system, and some metabolic processes.
During SWU1 infection, M. smegmatis mc2155 down-regulates some genes about signal transduction. The overexpression of PknA and PknB can hinder the growth of mycobacteria and altered cell morphology (Kang et al., 2005). Phosphorylated substrates of PknA and PknB are involved in diverse cellular events such as the synthesis of polar cell wall, cell shape maintenance, cell division, and peptidoglycan synthesis (Kang et al., 2005; Molle and Kremer, 2010; Prisic et al., 2010). The PknH plays an important role in the LAM/LM (lipoarabinomannan/lipomannan) balance (Molle and Kremer, 2010). PknH can phosphorylate and activate EmbR, the transcriptional repressor of the embCAB operon (Sharma et al., 2006). This was consistent with our data which the embCAB operon is down-regulated. Decrease of PknF can lead to aberrant septum formation and higher uptake of D-glucose (Deol et al., 2005). SigA is a main sigma factor mediating the enhanced growth of M. tuberculosis in vivo (Wu et al., 2004). The high intracellular level of SigF might result from: the expression of sigF or the gene of anti-sigma F factor antagonist. SigF is one of the stress response σ factors. M. smegmatis sigF can be significant induced under a variety of stress conditions (Singh and Singh, 2008). SigF is also required for the biosynthesis of cell wall in M. smegmatis. WhiA could regulate several genes involved in cell division, cell development, and chromosome segregation in Streptomyces venezuelae and B. subtilis (Bush et al., 2013). FadR-like regulators are involved in amino acid metabolism and some branch point in various metabolic pathways (Vindal et al., 2007). Lrp/AsnC family regulators regulate multiple cellular metabolism globally, such as amino acid metabolism, pili synthesis, DNA metabolism during DNA repair, and recombination (Deng et al., 2011). MerR family regulator is known to be involved in the regulation of gene expression in response to resistance and detoxification (Ramos et al., 2005). The RpiR family of transcriptional regulators can affect virulence determinant synthesis and PPP activity (Zhu et al., 2011). In addition, dosR a gene encoding response regulator of a two-component system with two sensor kinases—DosS and DosT, was also up-regulated. The two-component system can respond to nitric oxide, hypoxia, and carbon monoxide stress (Braunstein et al., 2003). IpsA, a LacI-type regulator, which control the biogenesis of cell wall in Mycobacteria (Baumgart et al., 2013), was elevated too.
Phages or DNA-containing infectious particles could induce the leakage of K+ from the cytosol, a much more complex system than non-specific channel (Boulanger and Letellier, 1992; Daugelavicius et al., 1997). The infection of SWU1 also can up-regulate genes involved in K+ ions fluxes. The K+ efflux was reported to be associated with an influx of H+ and Na+ or Li+ which compete for entry through the channel. These ion fluxes might underlie the cell depolarization. This selective regulation might underlie a role of host cell energetics in bacteriophage DNA entry. Phage SWU1 infection up-regulated M. smegmatis mc2155 genes actively involved in the synthesis of F-type ATPase. F-type ATPase can drive an influx of protons (H+) through the cell membrane to generate ATP (Fallico et al., 2011). This is consistent with the up-regulation of the biosynthetic genes AsnB. AsnB is a synthetase of asparagines, whose metabolic product is ammonia (Song et al., 2011), a pH buffer. The F-type ATPase-mediated import of protons would result in lower intracellular pH. To maintain the pH homeostasis, M. smegmatis mc2155 up-regulated the biosynthesis of asparagines, which is the ammonia source for mycobacteria at lower pH in vitro (Song et al., 2011). Transient depolarization and repolarization of host membrane was a common effect induced by phage infection (Labedan and Letellier, 1981; Kalasauskaite et al., 1983; Fallico et al., 2011). This might result from the influx or efflux of H+ or K+. Based on our transcriptome data, we proposed following model. Upon the adsorption of SWU1 to the cell wall of host, the pahge DNA traverses the membranes, an ATP-driven process (Daugelavicius et al., 1997; González-Huici et al., 2004). On this occasion, F-type ATPase is synthesized and H+ influxes the cell. The balanced proton motive force is thereby disrupted and the membrane depolarizes. The infected bacteria respond to the signal of depolarization by a repolarization strategy, that opens the potassium channel (Fallico et al., 2011). Subsequently, to resist the influx of H+, the infected bacteria induce the biosynthetic genes of asparagines, which leads to ammonia production and is proposed to maintain pH homeostasis. However, evidences are needed to support this largely educated guesswork.
The data of RNA-seq showed that SWU1 disrupted iron uptake system of M. smegmatis. However, the experiment (Figure 3B) did not support this hypothesis. We think the major reason is that bacteria have just begun to respond to SWU1 infection from transcription level rather than from translation level after SWU1 infected 30 min. So RNA-seq data revealed that M. smegmatis down-regulated the genes involved in iron uptake system. The experiment of ferri concentration measurement showed that the reduction of ferri concentration was not statistically significant.
The infection of SWU1 can up-regulate genes involved in cell wall biosynthesis. As we know, the unique Mycobacterium cell wall with three layers represents the first barrier the phage must overcome for successful infection (Brennan, 2003). The cell wall core is the mycolyl arabinogalactan–peptidoglycan (mAGP) complex consisting of peptidoglycan (PG), which is close to the membrane, arabinogalactan (AG) and mycolate. The upper segment of mAGP consists of free lipids, of which cord factor (trehalose 6-dimycolate, TDM) is the most abundant glycolipid and is long held as a virulence factor in the cell wall. The third layer is scattered components, such as, some cell-wall proteins, the phosphatidylinositol mannosides (PIMs), the phthiocerol-containing lipids, lipomannan (LM), and lipoarabinomannan (LAM). Phages must penetrate the host cell wall to deliver their genetic materials. This might mirror that the host is desperate to repair and reinforce the cell wall disrupted by phage to stop the efflux of cytoplasm. Alternatively, the synthesis of the host cell wall might represent a mechanism of superinfection immunity.
Genes predicted to be related to DNA, RNA and protein synthesis system were up-regulated. These data suggested that SWU1 control host bacteria to synthetise DNA, RNA, and protein of phage. Those synthesis systems of M. smegmatis mc2155 had been hijacked by SWU1. After virus infection, transfer RNA pool of cell will change (Clarkson and Runner, 1971; Kunisawa, 1992). The change is an important guarantee for the reproduction of virus (Wang et al., 2001). We speculated that the up-regulation of those genes related to tRNA synthesis was the survival mechanism of SWU1 for creating favorable environment.
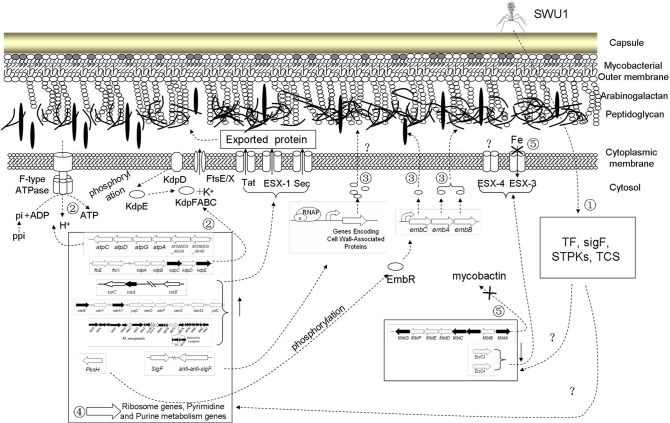
Based on the transcription profiles of both mycobacteriophage SWU1 and Mycobacterium during the infection, a usurp strategy employed by the phage and countermeasures mobilized by the host are evident. Phage early transcripts may be the key effector proteins to shut off the expression of some host genes. The major targets phage SWU1 hijacked are DNA, RNA, and protein synthesis system, iron uptake system, and some metabolic processes (Figure 5). Our results also suggested that host regulated some signal transduction system, changed host cell energetics, and inducted the synthetic gene of cell wall to defend the attack of SWU1 (Figure 5). The overall response of M. smegmatis mc2155 to phage infection involves a complex integrated regulation.
Figure 5.
Putative working model of M. smegmatis mc2155 response to phage SWU1 infection. Numbers stand for host response to SWU1 infection, which is described in summary. Hollow-out wide arrows stand for those regulated genes. Dotted lines stand for the process of regulatory network. Forks represent that the process is inhibited.
Author contributions
XF participated in the design of the study, did experiments, analyzed data, and wrote the paper. XD analyzed data and wrote the paper. YT did experiments. QH, MZ, and HW analyzed data. LZ and RY participated in the design of the study and helped to modify the manuscript. JX designed the research and wrote the paper. All authors read and approved the final manuscript.
Funding
This work was supported by National Natural Science Foundation [grant numbers 81511120001, 81371851, 81071316, 81271882, 81301394], New Century Excellent Talents in Universities [grant number NCET-11-0703], National Megaprojects for Key Infectious Diseases [grant number 2008ZX10003-006], Excellent Ph.D. thesis fellowship of Southwest University [grant numbers kb2010017, ky2011003], the Fundamental Research Funds for the Central Universities [grant numbers XDJK2016D025, XDJK2011D006, XDJK2012D011, XDJK2012D007, XDJK2013D003, XDJK2014D040], Graduate research and innovation project of graduate in Chongqing [grant number CYS14044], The Chongqing Municipal Committee of Education for postgraduates excellence program [grant number YJG123104], The undergraduates teaching reform program [grant number 2013JY201], National Institute of General Medical Sciences of the National Institutes of Health under Award Number R01GM107597, and Shandong Excellent Young Scientist Award Fund [grant number BS2014YY031].
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2016.00041
References
- Barrangou R., Fremaux C., Deveau H., Richards M., Boyaval P., Moineau S., et al. (2007). CRISPR provides acquired resistance against viruses in prokaryotes. Science 315, 1709–1712. 10.1126/science.1138140 [DOI] [PubMed] [Google Scholar]
- Baumgart M., Luder K., Grover S., Gätgens C., Besra G. S., Frunzke J. (2013). IpsA, a novel LacI-type regulator, is required for inositol-derived lipid formation in Corynebacteria and Mycobacteria. BMC Biol. 11:122. 10.1186/1741-7007-11-122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belanger A. E., Besra G. S., Ford M. E., Mikusová K., Belisle J. T., Brennan P. J., et al. (1996). The embAB genes of Mycobacterium avium encode an arabinosyl transferase involved in cell wall arabinan biosynthesis that is the target for the antimycobacterial drug ethambutol. Proc. Natl. Acad. Sci. U.S.A. 93, 11919–11924. 10.1073/pnas.93.21.11919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulanger P., Letellier L. (1992). Ion channels are likely to be involved in the two steps of phage T5 DNA penetration into Escherichia coli cells. J. Biol. Chem. 267, 3168–3172. [PubMed] [Google Scholar]
- Braunstein M., Espinosa B. J., Chan J., Belisle J. T., and R., Jacobs W. (2003). SecA2 functions in the secretion of superoxide dismutase A and in the virulence of Mycobacterium tuberculosis. Mol. Microbiol. 48, 453–464. 10.1046/j.1365-2958.2003.03438.x [DOI] [PubMed] [Google Scholar]
- Brennan P. (2003). Structure, function, and biogenesis of the cell wall of Mycobacterium tuberculosis. Tuberculosis 83, 91–97. 10.1016/S1472-9792(02)00089-6 [DOI] [PubMed] [Google Scholar]
- Bush M. J., Bibb M. J., Chandra G., Findlay K. C., Buttner M. J. (2013). Genes required for aerial growth, cell division, and chromosome segregation are targets of WhiA before sporulation in Streptomyces venezuelae. MBio 4:e00684-13. 10.1128/mBio.00684-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen T., Zhao Q., Li W., Xie J. (2013). Mycobacterium tuberculosis PE_PGRS17 promotes the death of host cell and cytokines secretion via Erk kinase accompanying with enhanced survival of recombinant Mycobacterium smegmatis. J. Interferon Cytokine Res. 33, 452–458. 10.1089/jir.2012.0083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarkson S. G., Runner M. N. (1971). Transfer RNA changes in HeLa cells after vaccinia virus infection. Biochim. Biophys. Acta 238, 498–502. 10.1016/0005-2787(71)90629-0 [DOI] [PubMed] [Google Scholar]
- Crick D. C., Mahapatra S., Brennan P. J. (2001). Biosynthesis of the arabinogalactan-peptidoglycan complex of Mycobacterium tuberculosis. Glycobiology 11, 107R–118R. 10.1093/glycob/11.9.107R [DOI] [PubMed] [Google Scholar]
- Cunningham-Bussel A., Zhang T., Nathan C. F. (2013). Nitrite produced by Mycobacterium tuberculosis in human macrophages in physiologic oxygen impacts bacterial ATP consumption and gene expression. Proc. Natl. Acad. Sci. U.S.A. 110, E4256–E4265. 10.1073/pnas.1316894110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daugelavicius R., Bamford J., Bamford D. H. (1997). Changes in host cell energetics in response to bacteriophage PRD1 DNA entry. J. Bacteriol. 179, 5203–5210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng W., Wang H., Xie J. (2011). Regulatory and pathogenesis roles of Mycobacterium Lrp/AsnC family transcriptional factors. J. Cell. Biochem. 112, 2655–2662. 10.1002/jcb.23193 [DOI] [PubMed] [Google Scholar]
- Deol P., Vohra R., Saini A. K., Singh A., Chandra H., Chopra P., et al. (2005). Role of Mycobacterium tuberculosis Ser/Thr kinase PknF: implications in glucose transport and cell division. J. Bacteriol. 187, 3415–3420. 10.1128/JB.187.10.3415-3420.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fallico V., Ross R. P., Fitzgerald G. F., McAuliffe O. (2011). Genetic response to bacteriophage infection in Lactococcus lactis reveals a four-strand approach involving induction of membrane stress proteins, D-alanylation of the cell wall, maintenance of proton motive force, and energy conservation. J. Virol. 85, 12032–12042. 10.1128/JVI.00275-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan X., Teng T., Wang H., Xie J. (2012). Biology of a novel mycobacteriophage, SWU1, isolated from Chinese soil as revealed by genomic characteristics. J. Virol. 86, 10230–10231. 10.1128/JVI.01568-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan X., Yan J., Xie L., Zeng L., Young R. F., Xie J. (2015). Genomic and proteomic features of mycobacteriophage SWU1 isolated from China soil. Gene 561, 45–53. 10.1016/j.gene.2015.02.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garber M., Grabherr M. G., Guttman M., Trapnell C. (2011). Computational methods for transcriptome annotation and quantification using RNA-seq. Nat. Methods 8, 469–477. 10.1038/nmeth.1613 [DOI] [PubMed] [Google Scholar]
- García P., Martinez B., Obeso J., Rodriguez A. (2008). Bacteriophages and their application in food safety. Lett. Appl. Microbiol. 47, 479–485. 10.1111/j.1472-765X.2008.02458.x [DOI] [PubMed] [Google Scholar]
- González-Huici V., Salas M., Hermoso J. M. (2004). The push–pull mechanism of bacteriophage Ø29 DNA injection. Mol. Microbiol. 52, 529–540. 10.1111/j.1365-2958.2004.03993.x [DOI] [PubMed] [Google Scholar]
- Hatfull G. F., Pedulla M. L., Jacobs-Sera D., Cichon P. M., Foley A., Ford M. E., et al. (2006). Exploring the mycobacteriophage metaproteome: phage genomics as an educational platform. PLoS Genet. 2:e92. 10.1371/journal.pgen.0020092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jankute M., Grover S., Rana A. K., Besra G. S. (2012). Arabinogalactan and lipoarabinomannan biosynthesis: structure, biogenesis and their potential as drug targets. Future Microbiol. 7, 129–147. 10.2217/fmb.11.123 [DOI] [PubMed] [Google Scholar]
- Kalasauskaite E. V., Kadisaite D. L., Daugelavicius R. J., Grinius L. L., Jasaitis A. A. (1983). Studies on energy supply for genetic processes. Requirement for membrane potential in Escherichia coli infection by phage T4. Eur. J. Biochem. 130, 123–130. 10.1111/j.1432-1033.1983.tb07126.x [DOI] [PubMed] [Google Scholar]
- Kanehisa M., Goto S. (2000). KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 28, 27–30. 10.1093/nar/28.1.27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang C.-M., Abbott D. W., Park S. T., Dascher C. C., Cantley L. C., Husson R. N. (2005). The Mycobacterium tuberculosis serine/threonine kinases PknA and PknB: substrate identification and regulation of cell shape. Genes Dev. 19, 1692–1704. 10.1101/gad.1311105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunisawa T. (1992). Synonymous codon preferences in bacteriophage T4: a distinctive use of transfer RNAs from T4 and from its host Escherichia coli. J. Theor. Biol. 159, 287–298. 10.1016/S0022-5193(05)80725-8 [DOI] [PubMed] [Google Scholar]
- Labedan B., Letellier L. (1981). Membrane potential changes during the first steps of coliphage infection. Proc. Natl. Acad. Sci. U.S.A. 78, 215–219. 10.1073/pnas.78.1.215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laermann V., Cudic E., Kipschull K., Zimmann P., Altendorf K. (2013). The sensor kinase KdpD of Escherichia coli senses external K+. Mol. Microbiol. 88, 1194–1204. 10.1111/mmi.12251 [DOI] [PubMed] [Google Scholar]
- Liao G., Liu Q., Xie J. (2013). Transcriptional analysis of the effect of exogenous decanoic acid stress on Streptomyces roseosporus. Microb. Cell Fact. 12:19. 10.1186/1475-2859-12-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Dehbi M., Moeck G., Arhin F., Bauda P., Bergeron D., et al. (2004). Antimicrobial drug discovery through bacteriophage genomics. Nat. Biotechnol. 22, 185–191. 10.1038/nbt932 [DOI] [PubMed] [Google Scholar]
- Molle V., Kremer L. (2010). Division and cell envelope regulation by Ser/Thr phosphorylation: Mycobacterium shows the way. Mol. Microbiol. 75, 1064–1077. 10.1111/j.1365-2958.2009.07041.x [DOI] [PubMed] [Google Scholar]
- Monk A., Rees C., Barrow P., Hagens S., Harper D. (2010). Bacteriophage applications: where are we now? Lett. Appl. Microbiol. 51, 363–369. 10.1111/j.1472-765X.2010.02916.x [DOI] [PubMed] [Google Scholar]
- Munshi T., Gupta A., Evangelopoulos D., Guzman J. D., Gibbons S., Keep N. H., et al. (2013). Characterisation of ATP-dependent Mur ligases involved in the biogenesis of cell wall peptidoglycan in Mycobacterium tuberculosis. PLoS ONE 8:e60143. 10.1371/journal.pone.0060143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedulla M. L., Ford M. E., Houtz J. M., Karthikeyan T., Wadsworth C., Lewis J. A., et al. (2003). Origins of highly mosaic mycobacteriophage genomes. Cell 113, 171–182. 10.1016/S0092-8674(03)00233-2 [DOI] [PubMed] [Google Scholar]
- Pope W. H., Bowman C. A., Russell D. A., Jacobs-Sera D., Asai D. J., Cresawn S. G., et al. (2015). Whole genome comparison of a large collection of mycobacteriophages reveals a continuum of phage genetic diversity. Elife 4:e06416. 10.7554/eLife.06416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poranen M. M., Ravantti J. J., Grahn A. M., Gupta R., Auvinen P., Bamford D. H. (2006). Global changes in cellular gene expression during bacteriophage PRD1 infection. J. Virol. 80, 8081–8088. 10.1128/JVI.00065-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prisic S., Dankwa S., Schwartz D., Chou M. F., Locasale J. W., Kang C.-M., et al. (2010). Extensive phosphorylation with overlapping specificity by Mycobacterium tuberculosis serine/threonine protein kinases. Proc. Natl. Acad. Sci. U.S.A. 107, 7521–7526. 10.1073/pnas.0913482107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos J. L., Martínez-Bueno M., Molina-Henares A. J., Terán W., Watanabe K., Zhang X., et al. (2005). The TetR family of transcriptional repressors. Microbiol. Mol. Biol. Rev. 69, 326–356. 10.1128/MMBR.69.2.326-356.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravantti J. J., Ruokoranta T. M., Alapuranen A. M., Bamford D. H. (2008). Global transcriptional responses of Pseudomonas aeruginosa to phage PRR1 infection. J. Virol. 82, 2324–2329. 10.1128/JVI.01930-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson M. D., McCarthy D. J., Smyth G. K. (2010). edgeR: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140. 10.1093/bioinformatics/btp616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothenbücher M. C., Facey S. J., Kiefer D., Kossmann M., Kuhn A. (2006). The cytoplasmic C-terminal domain of the Escherichia coli KdpD protein functions as a K+ sensor. J. Bacteriol. 188, 1950–1958. 10.1128/JB.188.5.1950-1958.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roucourt B., Lavigne R. (2009). The role of interactions between phage and bacterial proteins within the infected cell: a diverse and puzzling interactome. Environ. Microbiol. 11, 2789–2805. 10.1111/j.1462-2920.2009.02029.x [DOI] [PubMed] [Google Scholar]
- Rybniker J., Plum G., Robinson N., Small P. L., Hartmann P. (2008). Identification of three cytotoxic early proteins of mycobacteriophage L5 leading to growth inhibition in Mycobacterium smegmatis. Microbiology 154, 2304–2314. 10.1099/mic.0.2008/017004-0 [DOI] [PubMed] [Google Scholar]
- Samaddar S., Grewal R. K., Sinha S., Ghosh S., Roy S., Gupta S. K. D. (2016). Dynamics of mycobacteriophage-mycobacterial host interaction: evidence for secondary mechanisms for host lethality. Appl. Environ. Microbiol. 82, 124–133. 10.1128/AEM.02700-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serafini A., Boldrin F., Palù G., Manganelli R. (2009). Characterization of a Mycobacterium tuberculosis ESX-3 conditional mutant: essentiality and rescue by iron and zinc. J. Bacteriol. 191, 6340–6344. 10.1128/JB.00756-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serafini A., Pisu D., Palù G., Rodriguez G. M., Manganelli R. (2013). The ESX-3 secretion system is necessary for iron and zinc homeostasis in Mycobacterium tuberculosis. PLoS ONE 8:e78351. 10.1371/journal.pone.0078351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma K., Gupta M., Pathak M., Gupta N., Koul A., Sarangi S., et al. (2006). Transcriptional control of the mycobacterial embCAB operon by PknH through a regulatory protein, EmbR, in vivo. J. Bacteriol. 188, 2936–2944. 10.1128/JB.188.8.2936-2944.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh A. K., Singh B. N. (2008). Conservation of sigma F in mycobacteria and its expression in Mycobacterium smegmatis. Curr. Microbiol. 56, 574–580. 10.1007/s00284-008-9126-8 [DOI] [PubMed] [Google Scholar]
- Snow G. (1965). Isolation and structure of mycobactin T, a growth factor from Mycobacterium tuberculosis. Biochem. J. 97, 166–175. 10.1042/bj0970166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song H., Huff J., Janik K., Walter K., Keller C., Ehlers S., et al. (2011). Expression of the ompATb operon accelerates ammonia secretion and adaptation of Mycobacterium tuberculosis to acidic environments. Mol. Microbiol. 80, 900–918. 10.1111/j.1365-2958.2011.07619.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steyn A. J., Joseph J., Bloom B. R. (2003). Interaction of the sensor module of Mycobacterium tuberculosis H37Rv KdpD with members of the Lpr family. Mol. Microbiol. 47, 1075–1089. 10.1046/j.1365-2958.2003.03356.x [DOI] [PubMed] [Google Scholar]
- Sun L., Young L. N., Zhang X., Boudko S. P., Fokine A., Zbornik E., et al. (2014). Icosahedral bacteriophage [Phi] X174 forms a tail for DNA transport during infection. Nature 505, 432–435. 10.1038/nature12816 [DOI] [PubMed] [Google Scholar]
- Takayama K., Wang C., Besra G. S. (2005). Pathway to synthesis and processing of mycolic acids in Mycobacterium tuberculosis. Clin. Microbiol. Rev. 18, 81–101. 10.1128/CMR.18.1.81-101.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ukai H., Matsuzawa H., Ito K., Yamada M., Nishimura A. (1998). ftsE (Ts) Affects translocation of K+-pump proteins into the cytoplasmic membrane of Escherichia coli. J. Bacteriol. 180, 3663–3670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vindal V., Suma K., Ranjan A. (2007). GntR family of regulators in Mycobacterium smegmatis: a sequence and structure based characterization. BMC Genomics 8:289. 10.1186/1471-2164-8-289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S., Xu H., Lu R., Meng X., Wang W., Wang Z., et al. (2001). Analysis of codon usage of vaccinia virus genome. Chin. J. Virol. 18, 227–234. 10.3321/j.issn:1000-8721.2002.03.007 [DOI] [Google Scholar]
- Whitman W. B., Coleman D. C., Wiebe W. J. (1998). Prokaryotes: the unseen majority. Proc. Natl. Acad. Sci. U.S.A. 95, 6578–6583. 10.1073/pnas.95.12.6578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams E. P., Lee J.-H., Bishai W. R., Colantuoni C., Karakousis P. C. (2007). Mycobacterium tuberculosis SigF regulates genes encoding cell wall-associated proteins and directly regulates the transcriptional regulatory gene phoY1. J. Bacteriol. 189, 4234–4242. 10.1128/JB.00201-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S., Howard S. T., Lakey D. L., Kipnis A., Samten B., Safi H., et al. (2004). The principal sigma factor sigA mediates enhanced growth of Mycobacterium tuberculosis in vivo. Mol. Microbiol. 51, 1551–1562. 10.1111/j.1365-2958.2003.03922.x [DOI] [PubMed] [Google Scholar]
- Xiang X., Li T., Wang J., Li J., Chen Y., Zhou F., et al. (2011). Geochemical characteristics of the overlying bedrock and soil, and its impact on hydro-chemistry of the drip waters in the Furong Cave, Chongqing. Carsolog. Sin. 30, 193–199. 10.3969/j.issn.1001-4810.2011.02.012 [DOI] [Google Scholar]
- Xie C., Mao X., Huang J., Ding Y., Wu J., Dong S., et al. (2011). KOBAS 2.0: a web server for annotation and identification of enriched pathways and diseases. Nucleic Acids Res. 39, W316–W322. 10.1093/nar/gkr483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young R. (2013). Phage lysis: do we have the hole story yet? Curr. Opin. Microbiol. 16, 790–797. 10.1016/j.mib.2013.08.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y., Nandakumar R., Sadykov M. R., Madayiputhiya N., Luong T. T., Gaupp R., et al. (2011). RpiR homologues may link Staphylococcus aureus RNAIII synthesis and pentose phosphate pathway regulation. J. Bacteriol. 193, 6187–6196. 10.1128/JB.05930-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.