Abstract
Gentamicin (Gm) is an aminoglycoside commonly used to treat bacterial infections such as tularemia – the disease caused by Francisella tularensis. In addition to being pathogenic, F. tularensis is found in environmental niches such as soil where this bacterium likely encounters Gm producers (Micromonospora sp.). Here we show that F. tularensis exhibits increased resistance to Gm at ambient temperature (26°C) compared to mammalian body temperature (37°C). To evaluate whether F. tularensis was less permeable to Gm at 26°C, a fluorescent marker [Texas Red (Tr)] was conjugated with Gm, yielding Tr-Gm. Bacteria incubated at 26°C showed reduced fluorescence compared to those at 37°C when exposed to Tr-Gm suggesting that uptake of Gm was reduced at 26°C. Unconjugated Gm competitively inhibited uptake of Tr-Gm, demonstrating that this fluorescent compound was taken up similarly to unconjugated Gm. Lysates of F. tularensis bacteria incubated with Gm at 37°C inhibited the growth of Escherichia coli significantly more than lysates from bacteria incubated at 26°C, further indicating reduced uptake at this lower temperature. Other facultative pathogens (Listeria monocytogenes and Klebsiella pneumoniae) exhibited increased resistance to Gm at 26°C suggesting that the results generated using F. tularensis may be generalizable to diverse bacteria. Regulation of the uptake of antibiotics provides a mechanism by which facultative pathogens survive alongside antibiotic-producing microbes in nature.
Keywords: Francisella, gentamicin, antibiotic resistance, Listeria monocytogenes, Klebsiella, gentamicin uptake, temperature
Introduction
Gentamicin (Gm) is an aminoglycoside antibiotic used to treat bacterial infections, including tularemia, a disease caused by Francisella tularensis (Alford et al., 1972; Jackson and Lester, 1978; Mason et al., 1980; Hassoun et al., 2006). This antibiotic binds to the bacterial 30S ribosomal subunit, which interferes with nascent polypeptide elongation (Mingeot-Leclercq et al., 1999). This binding impairs the machinery that prevents misreading of mRNA as well as premature termination of translation (Mingeot-Leclercq et al., 1999). In addition to being used clinically, Gm is also used in the “Gm protection assay” – a common laboratory test designed to quantify the survival and growth of intracellular pathogens in host cells (Isberg and Falkow, 1985; Hilbi et al., 2001; Horzempa et al., 2008b, 2011; Wisner et al., 2012; Schmitt et al., 2013).
Due to the highly polar nature of aminoglycosides, these antibiotics pass through the gram-negative outer membrane by disrupting Mg2+ bridges that stabilize the lipopolysaccharide molecules (Hancock, 1981; Mingeot-Leclercq et al., 1999). Import of aminoglycosides such as Gm through the cytoplasmic membrane is mediated through an energy-dependent process powered by proton motive force (PMF; Hancock, 1981; Taber et al., 1987). The specific cytoplasmic membrane transport machinery for Gm uptake has not yet been elucidated. Bacterial resistance to aminoglycosides can be facilitated by diminished permeability (Davies, 1971), and therefore understanding the molecular mechanism mediating Gm transport, and the regulation of this process is important.
As the body of an animal that died from tularemia decomposes, the F. tularensis bacteria are likely to inhabit the soil that absorbs the decaying material. Although F. tularensis has been identified in soil environments (Barns et al., 2005; Berrada and Telford, 2010; Broman et al., 2011), little is known about interactions with the microbiota, or how F. tularensis survives the chemical and physical stresses associated with a terrestrial inhabitance. For instance, the aminoglycoside Gm is naturally produced by the soil bacterium, Micromonospora sp. (Luedemann and Brodsky, 1963). Presumably, F. tularensis and other pathogenic bacteria that transiently occupy a soil niche would exhibit a means to survive this and other antibiotics produced by competing microorganisms prior to the colonization of a subsequent host.
Here we show that F. tularensis is more resistant to Gm at 26°C (a lower environmental temperature) than at 37°C (mammalian body temperature). This resistance is mediated by diminished antibiotic uptake at the lower temperature. Other pathogenic bacteria (Listeria monocytogenes and Klebsiella pneumoniae) exhibited similar enhanced resistance at 26°C, suggesting that the temperature-dependent uptake of Gm is generalizable to other microbes. We propose that bacteria that experience both a pathogenic and a temporary terrestrial life cycle evolved to utilize low temperature as a cue to decrease aminoglycoside uptake, increasing survival during habitation with competing bacteria that produce aminoglycosides and other antibiotics. During infection, these pathogenic bacteria inhabit an environment free of antibiotic-producing competing microbes (until antibiotics were introduced for clinical use approximately 75 years ago).
Materials and Methods
Bacterial Strains and Media
The strains of bacteria used in this study are listed in Table 1. Francisella bacteria cultivated on chocolate II agar (BD) were used to inoculate trypticase soy broth (TSB, BD) supplemented with 0.1% cysteine HCl (TSBc). L. monocytogenes bacteria cultivated on chocolate II agar were used to inoculate TSB supplemented with 0.6% yeast extract. K. pneumoniae bacteria were grown in TSB and Escherichia coli bacteria were cultured in LB broth. Broth cultures were incubated at 37°C with agitation. Kanamycin (Km) was included in the media at 10 μg/ml when necessary. All work with F. tularensis Schu S4 was conducted at the University of Pittsburgh in a biosafety level 3 facility with approval from the Centers for Disease Control and Prevention Select Agent Program.
Table 1.
Bacterial strains used in this study.
| Bacterial Strain | Reference |
|---|---|
| Francisella tularensis subsp. holarctica live vaccine strain | Karen Elkins |
| F. tularensis subsp. tularensis Schu S4 (NR-643) | NIH BEI Resources Repositorya |
| F. tularensis LVS emrA1transposon mutant KmR | Ma et al., 2014 |
| F. tularensis LVS ΔpyrF | Horzempa et al., 2010 |
| Escherichia coli DH5α | Invitrogen |
| F. novicida U112 | Karen Elkins |
| Listeria monocytogenes EGD | Douglas Drevets |
| Klebsiella pneumoniae | WLU-MCCb |
aNational Institutes of Health Biodefense and Emerging Infections (NIH BEI) Research Resources Repository, National Institute of Allergy and Infectious Diseases.
bWLU-MCC, West Liberty University Microbiology Culture Collection, bacterial species routinely verified by standard metabolic and physiological tests.
Time-Kill Assays
Stationary phase broth cultures were suspended in phosphate buffered saline (PBS) or TSBc as indicated to a concentration of 2 × 108 CFU/ml (F. tularensis) or 2 × 106 CFU/ml (L. monocytogenes or K. pneumoniae). This phase was selected to minimize the effect of temperature on growth rate and metabolism of the bacteria (Horzempa et al., 2008a). Bacteria were normalized using OD600 and treated with 0, 50, 100, or 500 μg/ml Gm sulfate and incubated at 26 or 37°C. Antibiotic concentrations capable of showing significant bacterial killing over 2 h were selected. PABN (25 μM Phenylalanine-Arginine Beta-Naphthylamide) or CCCP (25 μM cyanide-m-chlorophenylhydrazone) was added when indicated. At the times indicated, suspensions of bacteria were serially diluted in PBS and plated to determine CFU.
Antibiotic Disk Diffusion Assays
Stationary phase broth cultures were diluted to an optical density (A600) of 0.3. 100 μl of this suspension was spread plated onto solid medium (chocolate II agar for F. tularensis, LB agar for E. coli, trypticase soy agar for K. pneumoniae). Sterile Whatman filter disks infused with antibiotics were placed onto the surface of these plates that were subsequently incubated at 37°C overnight. The diameters of the zones of inhibition were measured using a metric ruler.
A variation of this disk diffusion assay was used to determine whether F. tularensis exhibited diminished uptake of Gm at 26°C. Here, F. tularensis LVS broth cultures were grown to stationary phase. Cultures were diluted to an optical density (A600) of 0.3 in TSBc and were incubated with 500 μg/ml Gm at 26 or 37°C. After 15 min, these bacteria were washed twice with PBS (each wash consisted of centrifuging the bacterial suspension at 16,300 × g for 3 min, and suspending the pellet in PBS). After the second wash, bacteria were centrifuged similarly and the pellet was suspended in 25 μl deionized water. This suspension was sonicated (3 s burst) to lyse cells. Lysates were added to a filter disk that was placed onto solid medium that had been spread plated with E. coli. These plates were incubated at 37°C overnight and subsequently, zones of inhibition were measured using a metric ruler.
Uptake of Gm Conjugated to Texas Red
Texas Red®-X, succinimidyl ester (Life Technologies) was conjugated to Gm as previously described (Sandoval et al., 1998; Li et al., 2011). Briefly, a 300:1 molar ratio (Gm sulfate: Texas Red succinimidyl ester) were incubated together in K2CO3 (100 mM, pH = 10; Li et al., 2011). These conditions minimize the addition of multiple fluorescent moieties to a single Gm molecule and also ensure a minimal amount of free, unbound Texas Red (Sandoval et al., 1998; Li et al., 2011). Unbound Texas Red was removed using PierceTM Dye Removal Columns (Thermo Scientific) according to protocol of the manufacturer. Conjugation and removal of unbound Texas Red were verified by agarose gel electrophoresis (data not shown).
Francisella tularensis LVS bacteria were suspended in TSBc and treated with Tr-Gm (at an amount equivalent to 128 μg/ml Gm) at either 26 or 37°C. To measure bacterial fluorescence at the times indicated, F. tularensis cells were centrifuged at 16,300 × g for 3 min, the pellets were washed twice with PBS, and after an additional centrifugation were suspended in PBS. The fluorescence was determined using an Eppendorf plate reader using a 535/595 filter.
When indicated, bacteria were incubated with both Tr-Gm (at an amount equivalent to 128 μg/ml Gm) and increasing concentrations of Gm (0, 50, 100, 150, or 200 μg/ml) for 15 min at 37°C. Bacteria were centrifuged, washed twice in PBS, and then fluorescence was determined using an Eppendorf plate reader with a 535/595 filter.
Statistical Analyses
Data were analyzed for significant differences using GraphPad Prism software. The statistical tests used are indicated in the figure legends.
Results
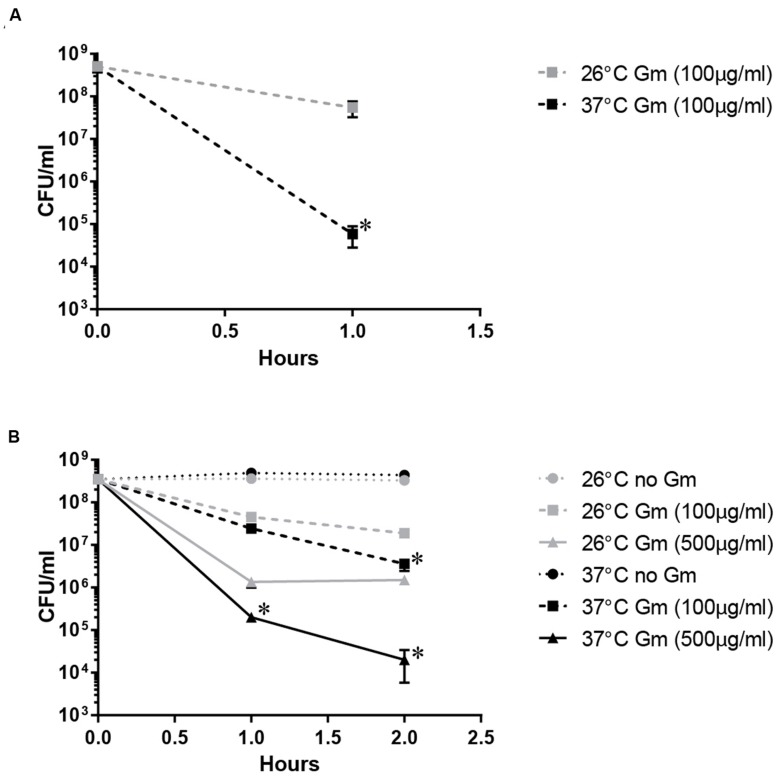
The Gm protection assay is used to quantify uptake and intracellular replication of pathogenic bacteria (Isberg and Falkow, 1985; Horzempa et al., 2011). This method involves the utilization of Gm, an aminoglycoside that permeates poorly into eukaryotic cells, to kill extracellular bacteria. Cultured host cells that had been incubated with pathogenic bacteria for a period of time are treated with Gm, washed, lysed, and intracellular CFU are determined by plating serial dilutions. When working out the conditions for this assay using F. tularensis, we observed that the incubation temperature substantially affected the ability of Gm to reduce bacterial viability (Figure 1). Here, bacteria were incubated with Gm suspended in PBS and incubated at 37°C (mammalian body temperature) or at 26°C (ambient temperature). Incubation of F. tularensis Schu S4 with Gm (100 μg/ml in PBS) at 37°C for 1 h resulted in a 10,000-fold reduction of CFU, whereas incubation with this same antibiotic at 26°C only reduced CFU by 10-fold (Figure 1A). This indicated that the F. tularensis Schu S4 bacteria were more resistant to Gm at ambient versus mammalian body temperature. Incubation of F. tularensis Schu S4 with 20 μg/ml Gm for 1 h at either temperature did not result in a significant reduction of viability (data not shown). In addition, the attenuated F. tularensis LVS strain exhibited a similar phenotype with increased Gm resistance at 26°C (Figure 1B). Notably, fewer F. tularensis LVS bacteria were killed by the 100 μg/ml Gm than Schu S4, indicating an intrinsic disparity in Gm susceptibility between these strains, which is consistent with other published reports (Kreizinger et al., 2013). However, at both 100 and 500 μg/ml Gm, F. tularensis LVS bacteria appeared to be more resistant at the lower temperature (26°C) than at mammalian body temperature (Figure 1B).
FIGURE 1.
Temperature-dependent Gm resistance of Francisella tularensis. F. tularensis Schu S4 (A) and LVS (B) showed increased resistance to gentamicin (Gm) at ambient temperature (26°C) compared to mammalian body temperature (37°C). Bacteria were cultivated in general growth media, suspended in PBS, and treated with Gm at the indicated temperature. Bacteria were diluted and plated for viable CFU at the indicated times. Plotted values represent mean CFU ± SD. Log-transformed data were analyzed using a two-way ANOVA and a Sidak’s multiple comparisons test. ∗P < 0.05 Schu S4 1 h 26°C vs. 37°C (A); LVS 1 h 26°C vs. 37°C Gm (500 μg/ml), 2 h 26°C vs. 37°C Gm (100 and 500 μg/ml; B).
Because the attenuated F. tularensis LVS also exhibited temperature-dependent Gm resistance, we utilized this strain to investigate the mechanism of this resistance throughout this study. Resistance to aminoglycosides can be manifested through drug inactivation, ribosomal alteration, antibiotic efflux, metabolic dormancy, or by diminished uptake (Mingeot-Leclercq et al., 1999; Allison et al., 2011). The genome of F. tularensis does not encode any proteins homologous to those known to inactivate aminoglycosides (data not shown). Moreover mutations that lead to modification of the ribosome structure are unlikely to cause this resistance at 26°C given the short duration of this assay and magnitude of surviving bacteria at 26°C compared to 37°C. Therefore, we first experimentally tested whether multi-drug efflux was enhanced at 26°C. EmrA1 comprises a component of a multidrug efflux pump of F. tularensis (Ma et al., 2014). This membrane fusion protein actively expels toxic materials, such as various types of antibiotics, from the bacterial cytosol (Ma et al., 2014). Similar to wild-type bacteria, the F. tularensis emrA1 mutant was more resistant to Gm at 26°C compared to 37°C (Figure 2). This suggests that enhanced EmrA1-mediated efflux is not responsible for the increased resistance to Gm at 26°C.
FIGURE 2.
Mutation of emrA1 gene in LVS does not change this bacterium’s resistance to Gm at 26°C. EmrA1 is a member of the membrane fusion protein family and is associated with the outer membrane TolC homolog SilC, to form a multidrug efflux pump. Increased Gm-resistance at 26°C is not mediated via efflux through the encoded efflux pump. Bacteria were suspended in PBS, and treated with Gm (100 μg/ml) for 2 h at the indicated temperature. Bacteria were diluted and plated for viable CFU at the times indicated. Plotted values represent mean CFU ± SD. Log-transformed data were analyzed using a two-way ANOVA and a Sidak’s multiple comparisons test. ∗P < 0.05 LVS 2 h 26 vs. 37°C Gm (100 μg/ml), emrA1 2 h 26°C vs. 37°C Gm (100 μg/ml). LVS vs. emrA1 not significant.
To further investigate whether enhanced Gm efflux mediated the increased resistance at 26°C, bacteria were treated with PABN, a non-specific inhibitor against RND-type multidrug efflux pumps (Renau et al., 1999; Lamers et al., 2013; Opperman and Nguyen, 2015). Together with the PABN, these bacteria were treated with Gm, incubated at 26 or 37°C, and were then plated to determine viable CFU (Figure 3A). Inclusion of PABN did not reduce resistance at 26°C, providing further evidence that enhanced efflux at this lower temperature does not mediate the observed resistance. F. tularensis LVS has been shown to utilize multidrug efflux to expel nalidixic acid (NA) and tetracycline (Tc) (Forslund et al., 2006; Gil et al., 2006; Bina et al., 2008; Platz et al., 2010). Antibiotic disk diffusion assays indicated the inclusion of PABN increased the susceptibility of F. tularensis LVS to both NA (Figure 3B) and Tc (Figure 3C) – findings consistent with the inhibition of efflux machinery. These data verify that PABN was likely inhibiting antibiotic efflux machinery during the assay depicted in Figure 3A. Altogether, Figures 2 and 3 indicate that the increased resistance to Gm at 26°C by F. tularensis is not likely mediated by enhanced efflux.
FIGURE 3.
Inhibition of multidrug efflux does not reduce the resistance of F. tularensis LVS to Gm at 26°C. Bacteria were cultivated in general growth media, suspended in PBS, and treated with Gm, or Gm + PABN (25 μM Phenylalanine-Arginine Beta-Naphthylamide; efflux pump inhibitor) at the indicated temperature (A). Bacteria were diluted and plated for viable CFU at the indicated times. Plotted values represent mean CFU ± SD. F. tularensis LVS bacteria were subjected to disk diffusion assays in the presence or absence of PABN to show that this compound was capable of inhibiting multi-drug efflux (B,C) as reported for other bacteria F. tularensis LVS is known to utilize multidrug efflux to expel nalidixic acid (NA) and tetracycline (Tc). Disks contained 30 μg of the indicated antibiotic. Bars represent the mean of the zones of inhibition ±SE. Media containing PABN significantly increased the zones of inhibition produced by NA (B; ∗P < 0.0001, unpaired t-test) and Tc (C; ∗P = 0.0271, unpaired t-test) likely due to reduced efflux activity.
The observed increased resistance to Gm at 26°C was manifested in bacteria that were suspended in PBS resulting in starvation conditions. Because lack of nutrients can trigger tolerance to antibiotics (Nguyen et al., 2011), we hypothesized that the lower temperature enhanced the starvation response, increasing the number of bacteria surviving at 26°C. To test whether starvation conditions augmented resistance at 26°C, we incubated bacteria treated with Gm in TSBc – a rich medium routinely used for the cultivation of F. tularensis. Similarly to when bacteria were incubated in PBS, suspension in TSBc resulted in an increased resistance to Gm at 26°C compared to 37°C (Figure 4). This suggests that an enhanced starvation response at 26°C did not increase resistance to Gm.
FIGURE 4.
Incubation in a rich medium maintains the observed Gm resistance at 26°C. Bacteria were cultivated in general growth media, suspended in trypticase soy broth supplemented with 0.1% cysteine (TSBc), and treated with Gm at the indicated temperature. Bacteria were diluted and plated for viable CFU at the indicated times. Plotted values represent mean CFU ± SD. Log-transformed data were analyzed using a two-way ANOVA and a Sidak’s multiple comparisons test. ∗P < 0.05 LVS 26°C vs. 37°C Gm (100 μg/ml) at 1 and 2 h.
To further assess whether a reduced metabolic state mediated the observed resistance at ambient temperature, we treated the F. tularensis LVS ΔpyrF mutant with Gm, incubated at 26 or 37°C, and determined the number of viable CFU (Figure 5). This strain is incapable of synthesizing uracil, and therefore grows at a reduced rate (Horzempa et al., 2010) which is indicative of a diminished metabolism. At 26°C, the F. tularensis LVS ΔpyrF bacteria were similarly resistant to Gm as wild type bacteria (Figure 5) indicating that a reduced metabolism was not responsible for the resistance to Gm at 26°C. Notably, the F. tularensis LVS ΔpyrF mutant bacteria were more sensitive to Gm than wild type bacteria at 37°C, indicating that a diminished metabolic capability seemingly increases sensitivity to this antibiotic under the conditions tested. Altogether, the results from Figures 4 and 5 indicate that an enhanced starvation response at 26°C is not likely responsible for increasing F. tularensis resistance to Gm.
FIGURE 5.
Francisella tularensisΔpyrF is not more resistant to Gm than wild type LVS. Bacteria were cultivated in general growth media, suspended in TSBc, and treated with Gm at the indicated temperature. Bacteria were diluted and plated for viable CFU at the indicated times. Plotted values represent mean CFU ± SD. Log-transformed data were analyzed using a two-way ANOVA and a Sidak’s multiple comparisons test. LVS 26°C vs. ΔpyrF Gm (100 μg/ml) not significant. ∗P < 0.05 LVS 26°C vs. 37°C Gm (100 μg/ml) at 1 and 2 h; ∗P < 0.05 LVS 37°C vs. ΔpyrF Gm (100 μg/ml) at 1 and 2 h.
We next sought to test whether decreasing uptake of Gm at 26°C mediated the observed resistance. Gram-negative bacteria import Gm utilizing PMF (Taber et al., 1987). Utilization of protonophores inhibit PMF, and thusly reduce bacterial uptake of aminoglycosides (Taber et al., 1987). To determine whether F. tularensis takes up Gm similarly to other Gram-negative bacteria, the protonophore, CCCP was used. Here, bacteria were treated with CCCP in combination with Gm, incubated at 26 or 37°C, and viable CFU were determined at the times indicated (Figure 6). The bacteria treated with CCCP survived Gm treatment, regardless of the incubation temperature, suggesting that this protonophore likely prevented uptake of Gm. This indicated that PMF mediates the uptake of Gm in F. tularensis similarly to other bacteria.
FIGURE 6.
Francisella tularensis LVS takes up Gm using proton motive force (PMF) similarly to other Gram-negative bacteria. Bacteria were cultivated in general growth media, suspended in TSBc, and treated with Gm, or Gm + CCCP (25 μM cyanide-m-chlorophenylhydrazone; a PMF inhibitor) at the indicated temperature. Bacteria were diluted and plated for viable CFU at the indicated times. Plotted values represent mean CFU ± SD. Log-transformed data were analyzed using a two-way ANOVA and a Sidak’s multiple comparisons test. ∗P < 0.05 LVS 26°C vs. 37°C Gm (100 μg/ml) at 1 and 2 h. LVS 26°C vs. 37°C CCCP + Gm (100 μg/ml) not significant.
To determine whether F. tularensis took up less Gm at 26°C, we fluorescently labeled Gm with Texas Red (Tr-Gm) and subsequently analyzed cellular fluorescence similarly to previous investigations (Sandoval et al., 1998; Steyger et al., 2003; Allison et al., 2011). F. tularensis LVS bacteria were treated with Tr-Gm, incubated at 26 or 37°C, and at the indicated times, were washed three times with PBS, and analyzed for red fluorescence using a plate reader (Figure 7A). At each time point, bacteria incubated at 37°C were more fluorescent than those incubated at 26°C (Figure 7A). This suggested that at the lower temperature, bacteria took up less Tr-Gm. Inclusion of unconjugated Gm competitively inhibited uptake of Tr-Gm (Figure 7B). This indicated that the fluorescently labeled form of this antibiotic was taken up similarly to the unmodified Gm. These data indicate that the increased resistance of F. tularensis to Gm at 26°C (Figures 1 and 4) was likely due to diminished uptake of this antibiotic.
FIGURE 7.
Francisella tularensis LVS takes up less Gm conjugated to Texas Red (Tr-Gm) at 26°C. F. tularensis LVS showed an increase in Tr-Gm fluorescence at mammalian body temperature (37°C) compared to ambient temperature (26°) (A). Bacteria were suspended in TSBc and treated with Tr-Gm (at an amount equivalent to 128 μg/ml Gm) at the indicated temperature. Bacteria were centrifuged, the pellets were washed twice and then suspended in PBS at the indicated times. The fluorescence was determined using an Eppendorf plate reader with a 535/595 filter. Plotted values represent mean relative fluorescence units ±SE. Data were analyzed using a two-way ANOVA and a Sidak’s multiple comparisons test. ∗P < 0.05 37°C vs. 26°C at 2 h; 37°C at 2 h vs. 37°C 15 min or 1 h. 26°C 2 h vs. 26°C 15 min or 26°C 1 h, not significant. To show that Tr-Gm was taken up through the Gm transporter (B), bacteria were incubated with Tr-Gm and increasing concentrations of Gm for 15 min at 37°C. Bacteria were centrifuged, washed in PBS, and then fluorescence was determined using an Eppendorf plate reader with a 535/595 filter. Plotted values represent mean relative fluorescence units ±SE. Data were analyzed using a one-way ANOVA and a Dunnetts’s multiple comparisons test. ∗P = 0.0150, 0 μg/ml vs. all other conditions.
To confirm that F. tularensis took up less Gm at 26°C compared to 37°C, F. tularensis LVS bacteria were incubated with Gm at 26 or 37°C, and after a short period of time (15 min), these cells were washed twice with PBS, suspended in deionized water, and then lysed. Lysates were added to filter disks that were placed onto LB agar plates that had been lawn-streaked with E. coli. After incubation, zones of inhibition were measured. Disks infused with lysates of LVS bacteria that had been treated with Gm and incubated at 37°C were significantly larger than those produced by lysates of antibiotic-treated bacteria incubated at 26°C (Figure 8). Notably, lysates from bacteria not treated with antibiotic did not produce zones of inhibition (data not shown). This experiment confirms that the increased resistance to Gm at 26°C is due to diminished uptake by F. tularensis LVS.
FIGURE 8.
Growth of Escherichia coli is inhibited more by F. tularensis LVS lysates from bacteria incubated with Gm at 37°C. F. tularensis were suspended in TSBc and incubated with 500 μg/ml Gm at 26°C or 37°C. Subsequently, these bacteria were washed twice with PBS, then suspended in deionized water and sonicated for 3 s to lyse. Lysates were added to a filter disk that was placed onto a lawn-streaked agar plate of E. coli. These plates were incubated at 37°C overnight. E. coli grew up to the filter disk on plates containing F. tularensis LVS lysates from bacteria that were incubated in TSBc at 26°C and 37°C without Gm (not shown). Bars represent the mean of the zones of inhibition ±SE. p = 0.0114, unpaired t-test.
Determining whether other bacteria possess increased Gm resistance at lower ambient temperature could provide further insight into the mechanism and the prevalence of this phenomenon. We therefore tested the impact of temperature on the Gm resistance of F. novicida, a bacterium related to F. tularensis that has been isolated from saltwater and rarely from human infections (Whitehouse et al., 2012; Kingry and Petersen, 2014). F. novicida bacteria were exposed to Gm followed by incubation at 26 or 37°C. Unlike F. tularensis, F. novicida showed a slight, but significantly increased resistance to Gm at 37°C 1 h after exposure (Figure 9A). However, after 2 h, bacteria incubated at either temperature were equally susceptible (Figure 9A). These data suggest that although genetically similar, F. tularensis and F. novicida regulate the uptake of Gm differently. We subsequently tested whether diverse bacteria exhibited increased Gm resistance at lower ambient temperatures similar to F. tularensis. L. monocytogenes is an intracellular pathogen that has been isolated from environmental sources. However, this bacterium is gram-positive (Firmicutes) indicating a great deal of taxonomic separation from F. tularensis. Similarly to F. tularensis, K. pneumoniae is a gram-negative γ-proteobacterium. However, K. pneumoniae is an opportunistic pathogen that is not associated with an intracellular lifestyle. Both L. monocytogenes (Figure 9B) and K. pneumoniae (Figure 9C) exhibited increased resistance to Gm at 26°C suggesting that diverse bacteria have developed a temperature-dependent resistance to this antibiotic. To confirm these findings, we conducted antibiotic disk diffusion assays in which plates that had been lawn-streaked with bacteria were incubated at 37 or 26°C. Incubation at the lower temperature yielded inconsistent and sparse growth of F. tularensis LVS, therefore negating the possibility of using this confirmatory test (data not shown). However, antibiotic disk diffusion assays for F. novicida, L. monocytogenes, and K. pneumoniae mirrored the time kill assays at 1 h post-exposure to Gm (Figures 9D–F). In other words, F. novicida was more resistant to Gm at 37°C, while L. monocytogenes and K. pneumonaie bacteria were more resistant at 26°C.
FIGURE 9.
Francisella novicida, Listeria monocytogenes and Klebsiella pneumoniae exhibit temperature-dependent Gm resistance. F. novicida (A), L. monocytogenes (B), or K. pneumoniae (C) bacteria were cultivated in general growth media overnight, suspended in PBS, and treated with Gm at the indicated temperature. Bacteria were diluted and plated for viable CFU at the indicated times. Plotted values represent mean CFU ± SD. Log-transformed data were analyzed using a two-way ANOVA and a Sidak’s multiple comparisons test. ∗P < 0.05 F. novicida 26°C vs. 37°C Gm (100 μg/ml) at 1 h (A); L. monocytogenes 1 h 26°C vs. 37°C Gm (100 and 500 μg/ml), 2 h 26°C vs. 37°C Gm (100 and 500 μg/ml); K. pneumoniae 26°C vs. 37°C Gm (50 μg/ml) at 1 and 2 h. Antibiotic disk diffusion assays were carried out in which F. novicida (D), L. monocytogenes (E), or K. pneumoniae (F) bacteria were lawn streaked onto solid media, and incubated with filter disks containing the indicated amount of Gm. These plates were incubated at the temperature indicated, and zones of inhibition were measured. Bars represent the mean of the zones of inhibition ±SD. Disk diffusion data were analyzed using an unpaired t-test. ∗P = 0.0009 (D); ∗P = 0.0026 (E); ∗P = 0.0050 (F).
Discussion
Data presented here indicated that F. tularensis and other bacteria that exist as facultative pathogens exhibit increased resistance to Gm at a lower temperature associated with environmental niches, versus mammalian body temperature. Evidence indicated that this increased resistance was due to diminished permeability to Gm at the lower temperature. This work describes a mechanism in which an important pathogen interacts with its non-host environment. F. tularensis has been identified in various environmental niches, including the soil (Barns et al., 2005; Berrada and Telford, 2010; Broman et al., 2011) – an environment that also harbors antibiotic producers. One such microbe, Micromonospora, produces Gm (Luedemann and Brodsky, 1963; Jao and Jackson, 1964). We propose here that F. tularensis and other pathogenic microbes have evolved general mechanisms to resist antibiotics during their existence outside of hosts. And, as demonstrated by using K. pneumoniae and L. monocytogenes, these bacteria use temperature as a cue to decrease permeability to antibiotics such as aminoglycosides. Previously published data indicated that transcription of a F. tularensis β-lactamase gene is induced at 26°C vs. 37°C (Horzempa et al., 2008a). We have since confirmed that F. tularensis β-lactamase activity is more robust at 26°C (data not shown) indicating that lower environmental temperature induces general resistance against many types of antibiotics. Largely throughout the course of evolution (prior to the clinical use of antibiotics in the 1940s), these mechanisms were not necessary during mammalian infection as niches occupied here are devoid of antibiotic-producing microbes.
The concentration of Gm used in many of the studies represented in this manuscript are relatively high compared to clinical use and soil concentrations which reach ∼mg/kg quantities (Kümmerer, 2008). However, Micromonospora sp. bacteria are capable of producing concentrations of Gm over 7 g/l in culture (Meenavilli et al., 2008) indicating that in soil microenvironments adjacent to these antibiotic-producing microbes, quantities could reach or even exceed levels similar to those used in the studies outlined here.
This study models interactions that F. tularensis would have with its environment during and following the decomposition of an animal that perished as a result of tularemia. In this soil environment (Barns et al., 2005; Berrada and Telford, 2010; Broman et al., 2011), F. tularensis is at some point likely to encounter antibiotic-producing microbes. As this study presented evidence that F. tularensis bacteria take up less aminoglycosides at lower temperatures, future investigations should focus on whether lower temperatures cue this bacterium to restrict uptake of other antibiotics associated with soil inhabitation, such as Tc. Little is known of the duration F. tularensis exists outside of a host cell or organism, or whether F. tularensis grows free from a host. Certainly, F. tularensis is capable of being cultivated in the laboratory in culture media devoid of host cells. However, due to the fastidious nature of this organism, it is unlikely capable of replication in the absence of essential host factors. Therefore, the increased resistance to Gm at lower environmental temperatures demonstrated here may provide an advantage until F. tularensis is phagocytosed or engulfed by a protozoan or arthropod host (Abd et al., 2003; Mahajan et al., 2011; Ozanic et al., 2015) – a niche inhabited with a lower abundance of antibiotic-producing microbes.
Diminished uptake of Gm at 26°C correlated with the increased resistance of F. tularensis LVS at this lower temperature, associating these two phenomena. Because a genetically similar bacterium, F. novicida did not exhibit the same pattern of resistance, increased membrane fluidity at the higher temperature was not likely responsible for the disparity in Gm uptake. An alternative explanation would be differential activity of a Gm transporter. Identification of the protein responsible for aminoglycoside uptake in bacteria has been elusive. Perhaps this is because many different proteins are capable of non-specifically importing this category of antibiotics. A current focus of our laboratory is to identify F. tularensis proteins responsible for the import of Gm.
Lipopolysaccharide (LPS) structure substantially affects the permeability of bacteria to polar molecules (Nikaido, 2003). Temperature greatly influences the structure of the lipid A moiety of the Francisella LPS molecule (Shaffer et al., 2007; Li et al., 2012). Therefore, it is possible that the addition of the 3-OH C16 acyl group at the lower environmental temperature could be responsible for the decreased permeability to Gm. Investigations to test this possibility are ongoing. However, L. monocytogenes (a gram-positive bacterium) also exhibited increased resistance to Gm at 26°C. This indicates that LPS modification in response to temperature cues could not be the mechanism utilized under all circumstances. Further testing will focus on determining the extent of resistance mechanisms utilized to resist aminoglycosides in response to lower environmental temperatures.
Although data presented here indicated that the temperature-dependent Gm resistance of F. tularensis is mediated by uptake modulation, we cannot completely rule out the possibility that this resistance is, at least in part, due to enhanced efflux of aminoglycosides. In Pseudomonas aeruginosa, the MexXY multidrug efflux system is one of the primary determinants of aminoglycoside resistance (Morita et al., 2012). Although PABN is a non-specific inhibitor of RND-type efflux pumps, this compound antagonized the activity of aminoglycosides in a MexXY-dependent manner in P. aeruginosa (Mao et al., 2001). If F. tularensis possesses a homolog of the MexXY multidrug efflux system or a functional equivalent, this machinery may contribute to the temperature-dependent resistance observed here.
An unanswered question raised by the data presented here is: why have F. tularensis LVS and other pathogenic bacteria evolved to import more Gm at mammalian body temperature? We speculate that this is a consequence of an overall upregulation of uptake machinery that occurs in response to host temperature (Horzempa et al., 2008a). We hypothesize that the Gm enters the cell non-specifically through nutrient uptake transporters during this general upregulation. This hypothesis is currently being tested in our laboratory.
This work presented a novel observation – that F. tularensis and other facultative pathogenic bacteria are more resistant to Gm at lower ambient temperatures (26°C) compared to mammalian body temperature (37°C). We show that this resistance was mediated by diminished antibiotic uptake at the lower temperature. We hypothesize that bacteria experiencing both a pathogenic and a temporary terrestrial life cycle may have evolved to utilize low temperature as a cue to decrease aminoglycoside uptake, increasing survival during habitation with competing bacteria that produce aminoglycosides.
Author Contributions
JoH conceived and designed the experiments. KL, JeH, SK, DS, TrG, DMS, JB, TaG, BK, AF, JI, CB, and JoH constructed the mutants and performed the experiments. JoH, KL, DMS, and CB analyzed the data. JoH wrote the paper. JoH, KL, DMS, CB, and TaG edited the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors thank Peter Steyger for advice, sharing protocols, and corresponding with regard to the synthesis and purification of Tr-Gm.
Footnotes
Funding. This work was supported by NIH Grant 5P20RR016477 to the West Virginia IDeA Network for Biomedical Research Excellence, a grant from WV-INBRE [P20GM103434], a grant from NIH-NIAID [5K22AI087703], and West Liberty University faculty development funding.
References
- Abd H., Johansson T., Golovliov I., Sandstrom G., Forsman M. (2003). Survival and growth of Francisella tularensis in Acanthamoeba castellanii. Appl. Environ. Microbiol. 69 600–606. 10.1128/AEM.69.1.600-606.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alford R. H., John J. T., Bryant R. E. (1972). Tularemia treated successfully with gentamicin. Am. Rev. Respir. Dis. 106 265–268. 10.1164/arrd.1972.106.2.265 [DOI] [PubMed] [Google Scholar]
- Allison K. R., Brynildsen M. P., Collins J. J. (2011). Metabolite-enabled eradication of bacterial persisters by aminoglycosides. Nature 473 216–220. 10.1038/nature10069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barns S. M., Grow C. C., Okinaka R. T., Keim P., Kuske C. R. (2005). Detection of diverse new Francisella-like bacteria in environmental samples. Appl. Environ. Microbiol. 71 5494–5500. 10.1128/AEM.71.9.5494-5500.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berrada Z. L., Telford S. R., III. (2010). Diversity of Francisella species in environmental samples from Martha’s Vineyard, Massachusetts. Microb. Ecol. 59 277–283. 10.1007/s00248-009-9568-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bina X. R., Lavine C. L., Miller M. A., Bina J. E. (2008). The AcrAB RND efflux system from the live vaccine strain of Francisella tularensis is a multiple drug efflux system that is required for virulence in mice. FEMS Microbiol. Lett. 279 226–233. 10.1111/j.1574-6968.2007.01033.x [DOI] [PubMed] [Google Scholar]
- Broman T., Thelaus J., Andersson A.-C., Bäckman S., Wikström P., Larsson E., et al. (2011). Molecular detection of persistent Francisella tularensis subspecies holarctica in natural waters. Int. J. Microbiol. 2011:10 10.1155/2011/851946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies J. (1971). Bacterial resistance to aminoglycoside antibiotics. J. Infect. Dis. 124(Suppl.), S7–S10. 10.1093/infdis/124.Supplement_1.S7 [DOI] [PubMed] [Google Scholar]
- Forslund A. L., Kuoppa K., Svensson K., Salomonsson E., Johansson A., Bystrom M., et al. (2006). Direct pilin repeat-mediated deletion of a type IV pilin gene results in major virulence attenuation of Francisella tularensis. Mol. Microbiol. 59 1818–1830. 10.1111/j.1365-2958.2006.05061.x [DOI] [PubMed] [Google Scholar]
- Gil H., Platz G. J., Forestal C. A., Monfett M., Bakshi C. S., Sellati T. J., et al. (2006). Deletion of TolC orthologs in Francisella tularensis identifies roles in multidrug resistance and virulence. Proc. Natl. Acad. Sci. U.S.A. 103 12897–12902. 10.1073/pnas.0602582103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hancock R. E. (1981). Aminoglycoside uptake and mode of action–with special reference to streptomycin and gentamicin. I. Antagonists and mutants. J. Antimicrob. Chemother. 8 249–276. 10.1093/jac/8.4.249 [DOI] [PubMed] [Google Scholar]
- Hassoun A., Spera R., Dunkel J. (2006). Tularemia and once-daily gentamicin. Antimicrob. Agents Chemother. 50:824 10.1128/AAC.50.2.824.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilbi H., Segal G., Shuman H. A. (2001). Icm/dot-dependent upregulation of phagocytosis by Legionella pneumophila. Mol. Microbiol. 42 603–617. 10.1046/j.1365-2958.2001.02645.x [DOI] [PubMed] [Google Scholar]
- Horzempa J., Carlson P. E., Jr., O’Dee D. M., Shanks R. M., Nau G. J. (2008a). Global transcriptional response to mammalian temperature provides new insight into Francisella tularensis pathogenesis. BMC Microbiol. 8:172 10.1186/1471-2180-8-172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horzempa J., Tarwacki D. M., Carlson P. E., Jr., Robinson C. M., Nau G. J. (2008b). Characterization and application of a glucose-repressible promoter in Francisella tularensis. Appl. Environ. Microbiol. 74 2161–2170. 10.1128/AEM.02360-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horzempa J., O’Dee D. M., Shanks R. M., Nau G. J. (2010). Francisella tularensis DeltapyrF mutants show that replication in nonmacrophages is sufficient for pathogenesis in vivo. Infect. Immun. 78 2607–2619. 10.1128/IAI.00134-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horzempa J., O’Dee D. M., Stolz D. B., Franks J. M., Clay D., Nau G. J. (2011). Invasion of erythrocytes by Francisella tularensis. J. Infect. Dis. 204 51–59. 10.1093/infdis/jir221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isberg R. R., Falkow S. (1985). A single genetic locus encoded by Yersinia pseudotuberculosis permits invasion of cultured animal cells by Escherichia coli K-12. Nature 317 262–264. 10.1038/317262a0 [DOI] [PubMed] [Google Scholar]
- Jackson R. T., Lester J. P. (1978). Case report. Tularemia presenting as unresponsive pneumonia: diagnosis and therapy with gentamicin. J. Tenn. Med. Assoc. 71 189–191. [PubMed] [Google Scholar]
- Jao R. L., Jackson G. G. (1964). Gentamicin sulfate, new antibiotic against gram-negative bacilli. laboratory, pharmacological, and clinical evaluation. JAMA 189 817–822. 10.1001/jama.1964.03070110019004 [DOI] [PubMed] [Google Scholar]
- Kingry L. C., Petersen J. M. (2014). Comparative review of Francisella tularensis and Francisella novicida. Front. Cell. Infect. Microbiol. 4:35 10.3389/fcimb.2014.00035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreizinger Z., Makrai L., Helyes G., Magyar T., Erdelyi K., Gyuranecz M. (2013). Antimicrobial susceptibility of Francisella tularensis subsp. holarctica strains from Hungary, Central Europe. J. Antimicrob Chemother. 68 370–373. 10.1093/jac/dks399 [DOI] [PubMed] [Google Scholar]
- Kümmerer K. (2008). Pharmaceuticals in the Environment: Sources, Fate, Effects, and Risks. Berlin: Springer. [Google Scholar]
- Lamers R. P., Cavallari J. F., Burrows L. L. (2013). The efflux inhibitor phenylalanine-arginine beta-naphthylamide (PAbetaN) permeabilizes the outer membrane of gram-negative bacteria. PLoS ONE 8:e60666 10.1371/journal.pone.0060666PONE-D-12-38648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Wang Q., Steyger P. S. (2011). Acoustic trauma increases cochlear and hair cell uptake of gentamicin. PLoS ONE 6:e19130 10.1371/journal.pone.0019130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Powell D. A., Shaffer S. A., Rasko D. A., Pelletier M. R., Leszyk J. D., et al. (2012). LPS remodeling is an evolved survival strategy for bacteria. Proc. Natl. Acad. Sci. U.S.A. 109 8716–8721. 10.1073/pnas.1202908109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luedemann G. M., Brodsky B. C. (1963). Taxonomy of gentamicin-producing micromonospora. Antimicrob. Agents Chemother. (Bethesda) 161 116–124. [PubMed] [Google Scholar]
- Ma Z., Banik S., Rane H., Mora V. T., Rabadi S. M., Doyle C. R., et al. (2014). EmrA1 membrane fusion protein of Francisella tularensis LVS is required for resistance to oxidative stress, intramacrophage survival and virulence in mice. Mol. Microbiol. 91 976–995. 10.1111/mmi.12509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahajan U. V., Gravgaard J., Turnbull M., Jacobs D. B., McNealy T. L. (2011). Larval exposure to Francisella tularensis LVS affects fitness of the mosquito Culex quinquefasciatus. FEMS Microbiol. Ecol. 78 520–530. 10.1111/j.1574-6941.2011.01182.x [DOI] [PubMed] [Google Scholar]
- Mao W., Warren M. S., Lee A., Mistry A., Lomovskaya O. (2001). MexXY-OprM efflux pump is required for antagonism of aminoglycosides by divalent cations in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 45 2001–2007. 10.1128/AAC.45.7.2001-2007.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason W. L., Eigelsbach H. T., Little S. F., Bates J. H. (1980). Treatment of tularemia, including pulmonary tularemia, with gentamicin. Am. Rev. Respir. Dis. 121 39–45. [DOI] [PubMed] [Google Scholar]
- Meenavilli H., Potumarthi R., Jetty A. (2008). Gentamicin production by Micromonospora echinospora (Me- 22) in stirred tank reactor: effect of various parameters. J. Basic Microbiol. 48 53–58. 10.1002/jobm.200700116 [DOI] [PubMed] [Google Scholar]
- Mingeot-Leclercq M. P., Glupczynski Y., Tulkens P. M. (1999). Aminoglycosides: activity and resistance. Antimicrob. Agents Chemother. 43 727–737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita Y., Tomida J., Kawamura Y. (2012). MexXY multidrug efflux system of Pseudomonas aeruginosa. Front. Microbiol. 3:408 10.3389/fmicb.2012.00408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen D., Joshi-Datar A., Lepine F., Bauerle E., Olakanmi O., Beer K., et al. (2011). Active starvation responses mediate antibiotic tolerance in biofilms and nutrient-limited bacteria. Science 334 982–986. 10.1126/science.1211037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikaido H. (2003). Molecular basis of bacterial outer membrane permeability revisited. Microbiol. Mol. Biol. Rev. 67 593–656. 10.1128/MMBR.67.4.593-656.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opperman T. J., Nguyen S. T. (2015). Recent advances toward a molecular mechanism of efflux pump inhibition. Front. Microbiol. 6:421 10.3389/fmicb.2015.00421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozanic M., Marecic V., Abu Kwaik Y., Santic M. (2015). The divergent intracellular lifestyle of Francisella tularensis in evolutionarily distinct host cells. PLoS Pathog. 11:e1005208 10.1371/journal.ppat.1005208PPATHOGENS-D-15-02017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platz G. J., Bublitz D. C., Mena P., Benach J. L., Furie M. B., Thanassi D. G. (2010). A tolC mutant of Francisella tularensis is hypercytotoxic compared to the wild type and elicits increased proinflammatory responses from host cells. Infect. Immun. 78 1022–1031. 10.1128/IAI.00992-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renau T. E., Leger R., Flamme E. M., Sangalang J., She M. W., Yen R., et al. (1999). Inhibitors of efflux pumps in Pseudomonas aeruginosa potentiate the activity of the fluoroquinolone antibacterial levofloxacin. J. Med. Chem. 42 4928–4931. 10.1021/jm9904598 [DOI] [PubMed] [Google Scholar]
- Sandoval R., Leiser J., Molitoris B. A. (1998). Aminoglycoside antibiotics traffic to the Golgi complex in LLC-PK1 cells. J. Am. Soc. Nephrol. 9 167–174. [DOI] [PubMed] [Google Scholar]
- Schmitt D. M., O’Dee D. M., Cowan B. N., Birch J. W., Mazzella L. K., Nau G. J., et al. (2013). The use of resazurin as a novel antimicrobial agent against Francisella tularensis. Front. Cell. Infect. Microbiol. 3:93 10.3389/fcimb.2013.00093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaffer S. A., Harvey M. D., Goodlett D. R., Ernst R. K. (2007). Structural heterogeneity and environmentally regulated remodeling of Francisella tularensis subspecies novicida lipid A characterized by tandem mass spectrometry. J. Am. Soc. Mass Spectrom. 18 1080–1092. 10.1016/j.jasms.2007.03.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steyger P. S., Peters S. L., Rehling J., Hordichok A., Dai C. F. (2003). Uptake of gentamicin by bullfrog saccular hair cells in vitro. J. Assoc. Res. Otolaryngol. 4 565–578. 10.1007/s10162-003-4002-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taber H. W., Mueller J. P., Miller P. F., Arrow A. S. (1987). Bacterial uptake of aminoglycoside antibiotics. Microbiol. Rev. 51 439–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitehouse C. A., Kesterson K. E., Duncan D. D., Eshoo M. W., Wolcott M. (2012). Identification and characterization of Francisella species from natural warm springs in Utah, USA. Lett. Appl. Microbiol. 54 313–324. 10.1111/j.1472-765X.2012.03214.x [DOI] [PubMed] [Google Scholar]
- Wisner A. L., Potter A. A., Koster W. (2012). Effect of the Salmonella pathogenicity island 2 type III secretion system on Salmonella survival in activated chicken macrophage-like HD11 cells. PLoS ONE 6:e29787 10.1371/journal.pone.0029787PONE-D-11-18196 [DOI] [PMC free article] [PubMed] [Google Scholar]