Abstract
MicroRNAs (miRNAs) are small noncoding RNA that have diverse functions in different biological process. The aim of this study was to evaluate the predictive ability of miR-29c, miR-124, miR-135a and miR-148a for lymph node metastasis (LNM) and tumor stage in gastric cancer. The expression of these miRNAs was detected and quantitated in gastric cancer tissues and in adjacent normal tissues from 60 patients by quantitative real-time reverse transcription-polymerase chain reaction. CT imaging and clinicopathologic characteristics of these patients were performed. The result of this study was that these miRNAs were down-regulated in gastric cancer tissues; The low expression of miR-124 and miR-135a in LNM group and tumor III-IV stages (P < 0.01) presented the potential correlation with LNM and tumor stage; The two miRNAs were highly correlated with r = 0.730. Receiver operating characteristic curve analysis showed that miR-124 had better predictive ability to identify LNM and tumor stage. It could discriminate non-LNM from LNM with 80.0% sensitivity and 80.0% specificity and discriminate tumor Ι-II stages from tumor III-IV stages with 71.9% sensitivity and 75.0% specificity at the best cut-off value of 0.0125. Compared with CT imaging, miR-124 had similar specificity (0.800 versus 0.900, P = 0.508) but higher sensitivity (0.800 versus 0.500, P = 0.022) for lymph node assessment; Combined of miR-124 and CT imaging, The sensitivity and specificity of assessing LNM were raised to 83.3% and 90.0% respectively. Taken together, miR-124 may be a predictor for LNM and tumor stage in gastric cancer.
Keywords: microRNAs, lymph node metastasis, tumor stage, CT imaging, gastric cancer
Introduction
Gastric cancer (GC) is one of the most common cancers, there are approximate 951,600 new GC cases and 723,100 deaths every year [1]. Although GC incidence and mortality rate have a steady decline and many achievements have been made in the field of GC therapy, we are still not satisfied with the prognosis of GC patients. Lymphatic metastasis is an important prognostic factor of GC patients [2,3]. However, there is no effective method to detect lymph node metastasis (LNM). Although computed Tomography (CT) imaging is commonly used to detect LNM in GC, the sensitivity of CT imaging is only 6.1% while metastasis lymph nodes are smaller than 5 mm [4]. Moreover, it can be influenced by reactively or benignly enlarged lymph nodes, radiologist’s ability and clinical experience. These lead to a wide variation for metastatic lymph node detection, the range of variation is about 38.5%-86.26% [5-8]. Therefore, we need to search some objective markers to identify LNM in GC.
MicroRNAs (miRNAs) are evolutionarily conserved, small, noncoding RNA molecules, about 22 nucleotides in length [9]. MiRNAs have diverse functions including the regulation of cellular differentiation, development, proliferation and apoptosis [10]. They can combine with 3’-untranslated-regions (3’-UTR) of target mRNAs to inhibit protein translation or induce mRNA degradation [11]. So, miRNAs are considered as “regulators” in numerous biological events including tumor suppressors and oncogenes. Although not each function or mechanism of miRNAs is fully understood, some studies suggest that abnormal expression of miRNAs is associated with a variety of tumors [12-15]. Therefore, it is possible that some miRNAs have a role in predicting LNM and tumor stage.
MiR-29c, miR-124, miR-135a and miR-148a belong to these kinds of tumor suppressors’ miRNAs, they have been found down-regulated in GC [16-20]. However, these studies didn’t illuminate the quantitative relationship between these miRNAs expression levels and LNM and tumor stage in GC. So our aim was to explore these miRNAs expression levels and their correlation with clinicopathological factors; Furthermore, we will link the expression levels of these miRNAs to LNM, tumor stage quantitatively, and comparison of miRNAs with CT imaging in assessing LNM of GC.
Material and methods
Tissue samples and clinical data
Sixty patients diagnosed with GC at the Affiliated Tumor Hospital of Guangxi Medical University between 2010 and 2012 were recruited in our study. These patients were treated by subtotal or total gastrectomy with lymphadenectomy. For accurate N staging, the number of cleaned lymph nodes was more than 15 in a patient. The clinical stage of postoperative patients was evaluated on the basis of the 7th edition of American Joint Committee on Cancer TNM staging system. None of these patients had undergone chemotherapy, radiotherapy or other treatment prior to surgery. Human tissues including sixty gastric cancer tissues and thirty matched adjacent normal tissues were immediately frozen in liquid nitrogen after surgical resection and stored at -80°C. The CT imaging and clinicopathologic characteristics of these patients were collected from electronic medical records. The clinicopathologic characteristics of these patients showed at Table 2. This study was approved by the Guangxi Medical University Institutional Review Board.
Table 2.
Correlations between clinicopathological parameters and miRNAs expression in GC tissues
| Parameters | Patient | Median (25-75th) | |||||||
|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||
| miR-29c | P-value | miR-124 | P-value | miR-135a | P-value | miR-148a | P-value | ||
| Gender | |||||||||
| Female | 24 | 0.331 (0.143-1.000) | 0.497 | 0.023 (0.005-0.051) | 0.051 | 0.052 (0.017-0.227) | 0.325 | 0.529 (0.149-4.790) | 0.541 |
| Male | 36 | 0.351 (0.222-1.300) | 0.007 (0.003-0.025) | 0.031 (0.009-0.045) | 0.516 (0.164-1.454) | ||||
| Age | |||||||||
| < 60 | 32 | 0.232 (0.143-1.005) | 0.335 | 0.011 (0.001-0.038) | 0.059 | 0.028 (0.008-0.110) | 0.313 | 0.420 (0.143-2.023) | 0.750 |
| ≥ 60 | 28 | 0.416 (0.235-1.176) | 0.016 (0.007-0.029) | 0.036 (0.013-0.067) | 0.603 (0.166-1.174) | ||||
| Tumor location | |||||||||
| Upper third | 7 | 0.231 (0.079-0.237) | 0.136 | 0.004 (0.001-0.005) | 0.072 | 0.015 (0.009-0.031) | 0.234 | 0.230 (0.105-0.420) | 0.216 |
| Middle third | 16 | 0.240 (0.157-1.000) | 0.007 (0.004-0.029) | 0.038 (0.007-0.110) | 1.811 (0.132-6.421) | ||||
| Lower third | 37 | 0.467 (0.222-1.312) | 0.015 (0.007-0.037) | 0.036 (0.012-0.086) | 0.529 (0.164-1.369) | ||||
| Tumor size | |||||||||
| < 5 cm | 45 | 0.351 (0.222-1.120) | 0.602 | 0.013 (0.004-0.038) | 0.174 | 0.036 (0.012-0.110) | 0.286 | 0.529 (0.164-1.454) | 0.746 |
| ≥ 5 cm | 15 | 0.235 (0.150-1.007) | 0.008 (0.004-0.018) | 0.024 (0.003-0.045) | 0.529 (0.218-2.085) | ||||
| Differentiation | |||||||||
| Well and moderately | 16 | 0.240 (0.119-2.458) | 0.881 | 0.015 (0.002-0.177) | 0.607 | 0.035 (0.006-0.202) | 0.496 | 0.566 (0.135-14.873) | 0.249 |
| Poorly | 44 | 0.409 (0.132-0.997) | 0.006 (0.003-0.016) | 0.017 (0.008-0.043) | 0.388 (0.118-1.266) | ||||
| Depth of invasion | |||||||||
| T3 | 41 | 0.351 (0.143-1.028) | 0.505 | 0.006 (0.002-0.029) | 0.824 | 0.024 (0.006-0.047) | 0.704 | 0.420 (0.123-1.454) | 0.899 |
| T4 | 15 | 0.235 (0.111-0.674) | 0.008 (0.002-0.029) | 0.014 (0.003-0.059) | 0.252 (0.122-1.283) | ||||
| Lymph node involvement | |||||||||
| Negative | 30 | 0.384 (0.234-1.240) | 0.088 | 0.027 (0.013-0.055) | 0.000 | 0.079 (0.024-0.153) | 0.000 | 0.725 (0.420-4.790) | 0.007 |
| Positive | 30 | 0.257 (0.099-0.971) | 0.005 (0.002-0.012) | 0.014 (0.005-0.037) | 0.251 (0.109-1.232) | ||||
| Tumor stage | |||||||||
| Ι-II | 32 | 0.384 (0.232-1.512) | 0.049 | 0.020 (0.007-0.038) | 0.003 | 0.040 (0.015-0.153) | 0.005 | 0.603 (0.268-1.516) | 0.065 |
| III-IV | 28 | 0.257 (0.088-0.997) | 0.005 (0.002-0.017) | 0.015 (0.005-0.051) | 0.304 (0.107-1.450) | ||||
RNA extraction
Total RNA were isolated from frozen tissue samples using TRIzoI Reagent (Invitrogen, Carlsbad, USA), according to the manufacturer’s guide. The total RNA was dissolved in 80 μl of diethylpyrocarbonate (DEPC)-treated water and stored at -80°C. The concentration and purity of total RNA were quantified by Nanodrop 2000 (PeqLab Biotechnology, Erlangen, Germany). Only the RNA samples with ratio of 1.8 < A260/A280 < 2.1 was used for the next experiments.
Reverse transcription
RNA was reverse transcribed to synthesize cDNA using miScript II RT kit (Qiagen, Hilden, Germany) according to the instructions. In brief, the 20 ul reverse transcription reaction system contains the following reagents: 2 ug of total RNA, 4 ul of 5 × miScript HiSpec Buffer, 2 ul of 10 × Nucleics Mix, 2 ul of miScript Reverse Transcriptase Mix, and RNase-free water. The reaction conditions were: 60°C for 37 min, 95°C for 5 min. The cDNA was stored at -80°C after the reaction.
Quantitative real-time PCR
The PCR was performed in 20 ul reaction system including 10 μl SYBR-Green qPCR Master Mix (Thermo Fisher Scientific, Waltham, USA), 1 μl specific forward primer for each miRNA (Invitrogen, Carlsbad, USA): hsa-miR-29c CTAGCACCATTTGAAATCGGTTA, hsa-miR-135a ACGGGGCGATATGGATTTTT, hsa-miR-148a ACGGGGATGGTCAGTGCACT, hsa-miR-124 TAAGGCACGCGGTGAATG, U6 CGCAAGGATGACACGCAAATTCGT, 1 μl miScript Universal primer (Qiagen), 0.5 ul cDNA product and 7.5 μL nuclease-freewater in triplicate. The reaction condition was as follows: incubated at 95°C for 7 min, followed by 43 cycles at 95°C for 10 sec, and 60°C for 30 sec in Mx3000P Real-Time Quantitative PCR system (Agilent Technologies, Santa Clara, USA). Melting curves were created for each real-time qPCR to verify the specificity of each PCR reaction. U6 snRNA was used as normalize the expression of target miRNA. The Ct value was based on the automatic threshold of MxPro-Mx3000P software setting. Delta Ct (ΔCt) represented the different expression of the normalizer and target miRNA: ΔCt = CtmiRNA-Ctnormalizer. The relative quantification of miRNA was calculated by the 2-ΔCt method.
Statistical analysis
The expression levels of normalized miRNAs were presented for the median and quartiles (25th-75th percentile) because they were not Gaussian distribution. Mann-Whitney U test or Kruskal-Wallis H test was used to analyze the differential expression of different groups. Spearman correlation was used to analyze the correlation of two miRNAs. Univariate and Multivariate logistic regression were used to further determine the correlations between miRNAs and LNM, tumor stage. Receiver-operating-characteristic (ROC) curve was used to identify which 2-ΔCt value of miRNAs could effectively discriminate LNM and tumor stage, the cut-off value was based on the ROC curve with Youden’s index (J, J = sensitivity + specificity - 1). The McNemar test was used to assess the difference between miRNA and CT imaging for predicting LNM. P < 0.05 was considered to have statistically significant difference. All statistical analyses were performed with IBM SPSS 19.0 software (IBM SPSS, Inc, Chicago).
Results
Expression of miRNAs in GC tissues
MiR-29c, miR-124, miR-135a and miR-148a had a statistical difference between the GC tissues and matched normal tissues (P < 0.01). The expression levels of miR-29c, miR-124, miR-135a and miR-148a were significantly decreased in GC tissues (Table 1). The results suggested that miR-29c, miR-124, miR-135a and miR-148a had a role as tumor suppressors in GC.
Table 1.
The relative expression of miRNAs (2-ΔCt) in cancer group and normal group
| MiRNAs | Median (25-75th) | P-value | |
|---|---|---|---|
|
| |||
| Cancer group | Normal group | ||
| MiR-29c | 0.3511 (0.1900-0.9966) | 1.5212 (0.7820-4.9933) | < 0.000 |
| MiR-124 | 0.0094 (0.0029-0.0280) | 0.0380 (0.0141-0.1092) | < 0.000 |
| MiR-135a | 0.0235 (0.0090-0.1002) | 0.0565 (0.0164-0.1030) | 0.034 |
| MiR-148a | 0.5088 (0.1638-1.4113) | 3.5988 (1.0065-7.5676) | < 0.000 |
Correlations between miRNAs expression and clinicopathological factors in GC specimens
To investigate the correlation between clinicopathological factors and miRNAs expression, the different groups were generated in Table 2. The results showed that miR-29c was different in the tumor stage group (P = 0.049); MiR-148a had a statistically difference in the lymph node involvement group (P = 0.007); Only miR-124 and miR-135a were different in lymph node involvement group and tumor stage group (P < 0.01). The results suggested that the low expression of miR-124 and miR-135a in LNM group and tumor III-IV stages presented the potential correlation with LNM and tumor stage.
For the two candidate miRNAs: miR-124 and miR-135a, univariate logistic regression and multivariate logistic regression were used to further analysis the correlations between miR-124/miR-135a and LNM, tumor stage. The result revealed that miR-124 and miR-135a were closely correlated with LNM and tumor stage in univariate logistic regression model (P < 0.05, Tables 3, 4). However, the correlations between the two miRNAs and LNM, tumor stage were attenuated in multivariate regression analysis, which only contained miR-124 and miR-135a as show in Tables 3 and 4. Furthermore, Spearman correlation analysis showed a highly linear correlation between miR-124 and miR-135a with r = 0.730 as show in Figure 1.
Table 3.
Univariate and multivariate logistic regression analysis of miR-124/miR-135a about LNM in gastric cancer
| Variables | Univariate logistic regression | Multivariate logistic regression | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
|
|
|
|||||||||
| Regression coefficient (β) | SE | Wald | P-value | Exp (β) | Regression coefficient (β) | SE | Wald | P-value | Exp (β) | |
| MiR-124 | -0.626 | 0.182 | 11.861 | 0.001 | 0.535 | -0.383 | 0.210 | 3.326 | 0.068 | 0.682 |
| MiR-135a | -0.744 | 0.205 | 13.157 | 0.000 | 0.475 | -0.512 | 0.236 | 4.707 | 0.030 | 0.599 |
LNM: lymph node metastasis.
Table 4.
Univariate and multivariate logistic regression analysis of miR-124/miR-135a about tumor stage in gastric cancer
| Variables | Univariate logistic regression model | Multivariate logistic regression model | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
|
|
|
|||||||||
| Regression coefficient (β) | SE | Wald | P-value | Exp (β) | Regression coefficient (β) | SE | Wald | P-value | Exp (β) | |
| MiR-124 | -0.376 | 0.136 | 7.701 | 0.006 | 0.686 | -0.205 | 0.173 | 1.397 | 0.237 | 0.815 |
| MiR-135a | -0.457 | 0.156 | 8.521 | 0.004 | 0.633 | -0.305 | 0.199 | 2.354 | 0.125 | 0.737 |
Figure 1.
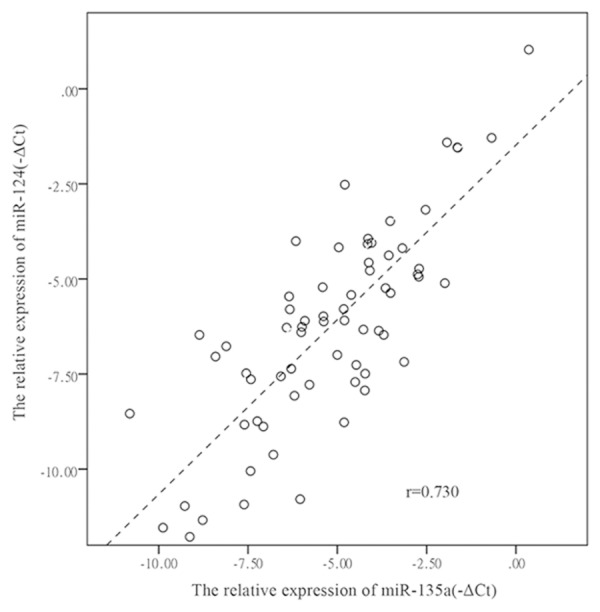
Relative expression levels of miR-124 and miR-135a are presented as -ΔCt in 60 GC tissues and 30 controls, the spearman correlation analysis indicates highly correlation between the two miRNAs with r = 0.730 as shown in the scatter plot.
The predictive value of miR-124/miR-135a for LNM and tumor stage in GC
To further evaluate the ability of miR-124/miR-135a for identifying LNM and tumor stage, receiver operating characteristics (ROC) curve analysis was generated. As shown in Figures 2 and 3. MiR-124 could discriminate non-LNM from LNM with 80.0% sensitivity and 80.0% specificity and discriminate tumor Ι-II stages from tumor III-IV stages with 71.9% sensitivity and 75.0% specificity at the best cut-off value of 0.0125; MiR-135a could discriminate non-LNM from LNM with 50.0% sensitivity and 100.0% specificity at the best cut-off value of 0.0901 and discriminate tumor Ι-II stages from tumor III-IV stages with 37.5% sensitivity and 100.0% specificity at the best cut-off value of 0.1122. The combination of the two miRNAs generated the sensitivity and specificity, which were not significantly difference from that of miRNA-124. These results suggested that miR-124 had a better predictive ability to identify LNM and tumor stage in GC.
Figure 2.
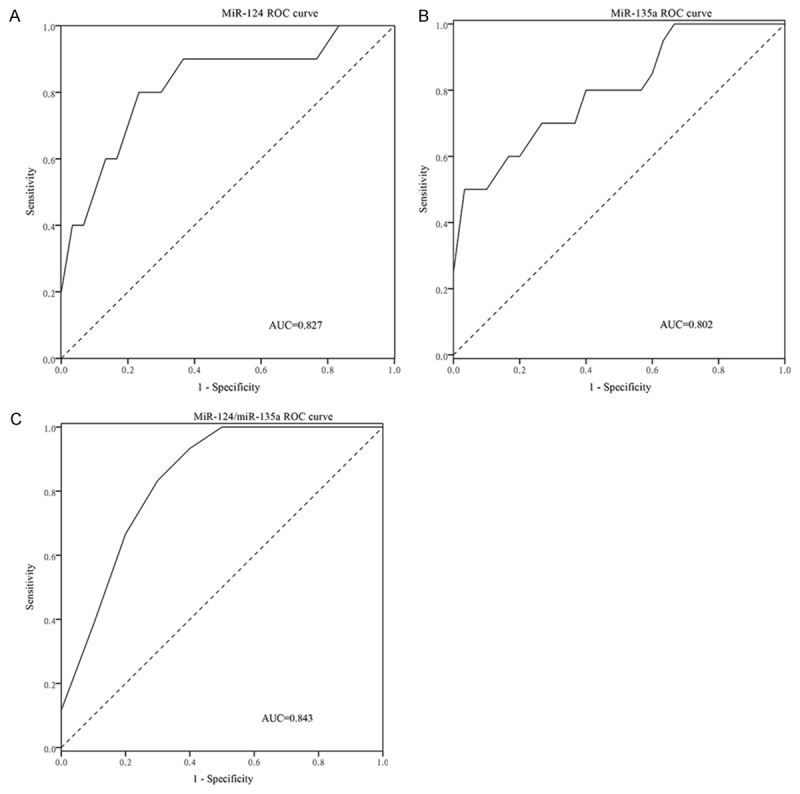
The GC patients with non-lymph node metastasis were distinguished from the GC patients with lymph node metastasis by the two miRNAs. MiR-124 shows 0.827 AUC (A), the best cut-off value is 0.0125 with 80.0% sensitivity and 80.0% specificity; MiR-135a shows 0.802 AUC (B), the best cut-off value is 0.0901 with 50.0% sensitivity and 100.0% specificity. Combination of the two miRNAs produced 0.843AUC (C), the best cut-off value is 0.5910 with 80.0% sensitivity and 80.0% specificity.
Figure 3.
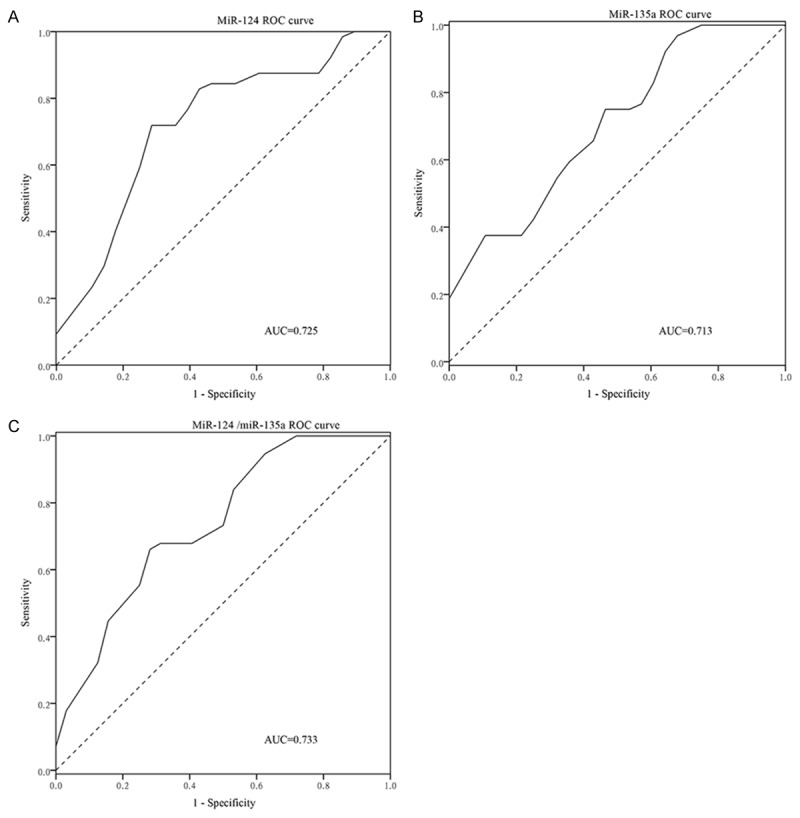
The GC patients with tumor Ι-II stages were distinguished from the GC patients with tumor III-IV stage by the two miRNAs. MiR-124 shows 0.725 AUC (A), the best cut-off value is 0.0125 with 71.9% sensitivity and 75.0% specificity; MiR-135a shows 0.713 AUC (B), the best cut-off value is 0.1122 with 37.5% sensitivity and 100.0% specificity. Combination of the two miRNAs produced 0.733 AUC (C), the best cut-off value is 0.5566 with 67.9% sensitivity and 71.9% specificity.
The comparison of miR-124 with CT imaging in predicting lymph node metastasis
To compare miR-124 with CT imaging in lymph node metastasis assessment, 2 × 2 × 2 chi-square test was used, as showed in Table 5. Compared with CT imaging, miR-124 had similar specificity (0.800 versus 0.900, P= 0.508) but higher sensitivity (0.800 versus 0.500, P = 0.022) for lymph node assessment; At last, combination of miR-124 and CT imaging produced 0.893AUC (95% CI: 0.811-0.976), the sensitivity and specificity of diagnosing LNM were 83.3% and 90% respectively (Figure 4).
Table 5.
The comparison between miRNA-124 and CT in predicting lymph node status of gastric cancer
| Pathologic LNM | Pathologic non-LNM | |||||||
|
|
|
|||||||
| MiRNA-124 | MiRNA-124 | |||||||
|
|
|
|||||||
| LNM | Non-LNM | Total | LNM | Non-LNM | Total | |||
|
|
|
|||||||
| CT | LNM | 13 | 2 | 15 | 0 | 3 | 3 | |
| Non-LNM | 11 | 4 | 15 | 6 | 21 | 27 | ||
| Total | 24 | 6 | 30 | 6 | 24 | 30 | ||
|
| ||||||||
| Groups | Sen | Spe | α | β | J | CR | (+)LR | (-)LR |
|
| ||||||||
| MiRNA-124 | 0.800 | 0.800 | 0.200 | 0.200 | 0.600 | 80.0% | 4.000 | 0.250 |
| CT | 0.500 | 0.900 | 0.100 | 0.500 | 0.400 | 70.0% | 5.000 | 0.556 |
| P-value | 0.022* | 0.508 | - | - | - | - | - | - |
CT identified the lymph node metastasis when the short-axis diameter of perigastric lymph nodes greater than 6 mm and larger than 8 mm for the extraperigastric lymph nodes, especially enhancement on contrast-enhanced CT and lymph nodes with arounded shape; LNM = lymph node metastasis; α = false positive rate; β = false negative rate; CR = concordance rate; J = Youden index; (+)LR = positive likelihood ratio; (-)LR = negative likelihood ratio; Sen = sensitivity; Spe = specificity.
P < 0.05.
Figure 4.
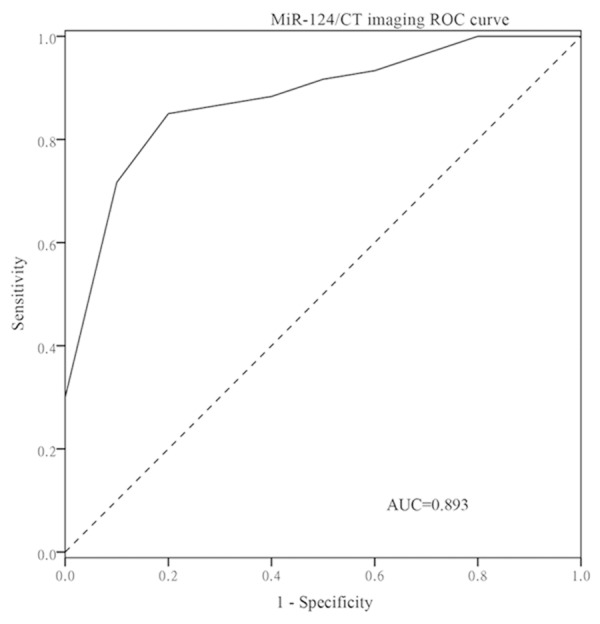
The GC patients without lymph node metastasis were distinguished from the GC patients with lymph node metastasis by the combination of miR-124 and CT imaging. Combination of miR-124 and CT imaging produced 0.893 AUC with 83.3% sensitivity and 90.0% specificity.
Discussion
Regional metastasis, especially through lymphatic system, is an important prognostic factor in GC patients [2,3]. However, there was no effective method to detect lymph node metastasis in GC. Several researches had explored the use of miRNAs expression in plasma samples or gastric tissues to enhance the prediction or diagnosis ability of LNM in GC [21-23]. However, they didn’t compare with CT imaging and focus on the diagnosis ability of quantifying miRNAs for tumor stage in GC. Moreover, not each function or mechanism of miRNAs that have been found is fully understood. Therefore, we explored the role of miR-29c, miR-124, miR-135a and miR-148a in GC.
We validated that miR-29c, miR-124, miR-135a and miR-148a were down-regulated in GC tissues, the low expression of miR-124 and miR-135a had a potential correlation with LNM and tumor stage (P < 0.01). These results are consistent with the previous studies [16,18-20,24].
Due to lack of evidence about the diagnostic ability of miR-124/135a for LNM and tumor stage in GC patients, we performed ROC curve analysis. MiR-124 could discriminate non-LNM from LNM with 80.0% sensitivity and 80.0% specificity and discriminate tumor Ι-II stages from tumor III-IV stages with 71.9% sensitivity and 75.0% specificity at the best cut-off value of 0.0125. MiR-135a could discriminate non-LNM from LNM with 50.0% sensitivity and 100.0% specificity at the best cut-off value of 0.0901. Shin et al found that miR-135a could predict LNM in early gastric cancer, the sensitivity and specificity of miR-135a were 75.0% and 73.0% [18]. This result was different from ours. The reason may be that not only the early gastric cancer patients but also the advanced gastric cancer patients are included in our study; Another reason may be that the selected cut-off value is different, while we selected the cut-off value as 0.0358, the sensitivity and specificity of miR-135a were 70.0%, 76.7%, which was similar to the previous study. The AUC of the two combined miRNAs showed that there was no significantly difference of single miRNA in predicting LNM and tumor stage. It might suggest that the single miRNA is enough for the identification, or this could be explained by the correlation of the two miRNAs (r = 0.730). Because the two miRNAs are highly correlated, this may lead to the presence of multicolinearity in multivariate logistic regression model. Therefore, compared with univariate logistic regression model, the correlation between the two miRNAs and LNM, tumor stage were attenuated and the estimation value of the predictive ability of individual miRNA will become inaccurate in multivariate logistic regression model (Tables 3, 4). Taken together, miR-124 had a better sensitivity and specificity in predicting LNM and tumor stage of GC.
CT imaging examination is a routine test for assessing tumor staging. However, it still remains difficult for assessing LNM. Due to that these factors can affect the diagnosis of CT imaging. Firstly, the sensitivity of CT imaging is only 6.1% while metastasis lymph nodes are smaller than 5 mm [4]; Secondly, the enlarged lymph nodes may be inflammatory hyperplasia or benign; Thirdly, the ability and clinical experience of radiologist will influence the result of CT imaging. Therefore, the range of variation for diagnosing LNM was 38.5%-86.26% [5-8]. MiR-124 had similar specificity (0.800 versus 0.900, P = 0.508) but higher sensitivity (0.800 versus 0.500, P = 0.022) for lymph node assessment; Moreover, due to the technologic advancement, the highly sensitivity and specificity of quantitative real-time reverse transcription-polymerase chain reaction also adapted to the detection of miRNAs [25,26]. This will be beneficial to clinical application of miR-124.
For molecular mechanism, miR-124 and miR-135a had a close correlation with tumor development. Some studies revealed that miR-124 and miR-135a were correlated with epithelial-mesenchymal transition (EMT) [18,27,28] and lymphangiogenesis [17]; They had a common target gene of ROCK1 [18,29]. ROCK1 had a variety of functions including cancer cell motility, invasion, and metastasis [30]. To a certain degree, these may explain why the two miRNAs were highly correlation (r = 0.730). Consequently, they are closely related to progress of carcinoma, especially lymphatic metastasis.
Although the results are interesting, there are some limitations in our study. Firstly, the sample size of this study is relatively small, the larger sample size or a prospective study is necessary; Secondly, our study didn’t evaluate the different economic cost of testing miR-124 and CT imaging in clinical practice. In addition, serum miR-124 maybe more convenient for monitoring the course of the disease as a minimal invasive way. We will consider these problems in the following research.
In conclusion, our data confirm that miR-124 has a potential role in predicting LNM and tumor stage of GC. It can be used as the basis of further research, preferably in prospective studies and larger sample size.
Acknowledgements
The study was supported by the scientific research fund of Guangxi Zhuang Autonomous Region Education Department (no. 302304).
Disclosure of conflict of interest
None.
References
- 1.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 2.Lee SR, Kim HO, Son BH, Shin JH, Yoo CH. Prognostic significance of the metastatic lymph node ratio in patients with gastric cancer. World J Surg. 2012;36:1096–1101. doi: 10.1007/s00268-012-1520-5. [DOI] [PubMed] [Google Scholar]
- 3.Saito H, Fukumoto Y, Osaki T, Fukuda K, Tatebe S, Tsujitani S, Ikeguchi M. Prognostic significance of level and number of lymph node metastases in patients with gastric cancer. Ann Surg Oncol. 2007;14:1688–1693. doi: 10.1245/s10434-006-9314-3. [DOI] [PubMed] [Google Scholar]
- 4.Fukuya T, Honda H, Hayashi T, Kaneko K, Tateshi Y, Ro T, Maehara Y, Tanaka M, Tsuneyoshi M, Masuda K. Lymph-node metastases: efficacy for detection with helical CT in patients with gastric cancer. Radiology. 1995;197:705–711. doi: 10.1148/radiology.197.3.7480743. [DOI] [PubMed] [Google Scholar]
- 5.Fairweather M, Jajoo K, Sainani N, Bertagnolli MM, Wang J. Accuracy of EUS and CT imaging in preoperative gastric cancer staging. J Surg Oncol. 2015;111:1016–1020. doi: 10.1002/jso.23919. [DOI] [PubMed] [Google Scholar]
- 6.Kim HJ, Kim AY, Oh ST, Kim JS, Kim KW, Kim PN, Lee MG, Ha HK. Gastric cancer staging at multi-detector row CT gastrography: comparison of transverse and volumetric CT scanning. Radiology. 2005;236:879–885. doi: 10.1148/radiol.2363041101. [DOI] [PubMed] [Google Scholar]
- 7.Hasegawa S, Yoshikawa T, Shirai J, Fujikawa H, Cho H, Doiuchi T, Yoshida T, Sato T, Oshima T, Yukawa N, Rino Y, Masuda M, Tsuburaya A. A prospective validation study to diagnose serosal invasion and nodal metastases of gastric cancer by multidetector-row CT. Ann Surg Oncol. 2013;20:2016–2022. doi: 10.1245/s10434-012-2817-1. [DOI] [PubMed] [Google Scholar]
- 8.Yan C, Zhu ZG, Yan M, Zhang H, Pan ZL, Chen J, Xiang M, Chen MM, Liu BY, Yin HR, Lin YZ. Value of multidetector-row computed tomography in the preoperative T and N staging of gastric carcinoma: a large-scale Chinese study. J Surg Oncol. 2009;100:205–214. doi: 10.1002/jso.21316. [DOI] [PubMed] [Google Scholar]
- 9.Lagos-Quintana M, Rauhut R, Lendeckel W, Tuschl T. Identification of novel genes coding for small expressed RNAs. Science. 2001;294:853–858. doi: 10.1126/science.1064921. [DOI] [PubMed] [Google Scholar]
- 10.Cho WC. OncomiRs: the discovery and progress of microRNAs in cancers. Mol Cancer. 2007;6:60. doi: 10.1186/1476-4598-6-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet. 2004;5:522–531. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- 12.Kong W, He L, Richards EJ, Challa S, Xu CX, Permuth-Wey J, Lancaster JM, Coppola D, Sellers TA, Djeu JY, Cheng JQ. Upregulation of miRNA-155 promotes tumour angiogenesis by targeting VHL and is associated with poor prognosis and triple-negative breast cancer. Oncogene. 2014;33:679–689. doi: 10.1038/onc.2012.636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kitamura K, Seike M, Okano T, Matsuda K, Miyanaga A, Mizutani H, Noro R, Minegishi Y, Kubota K, Gemma A. MiR-134/487b/655 cluster regulates TGF-beta-induced epithelialmesenchymal transition and drug resistance to gefitinib by targeting MAGI2 in lung adenocarcinoma cells. Mol Cancer Ther. 2014;13:444–453. doi: 10.1158/1535-7163.MCT-13-0448. [DOI] [PubMed] [Google Scholar]
- 14.Kojima S, Enokida H, Yoshino H, Itesako T, Chiyomaru T, Kinoshita T, Fuse M, Nishikawa R, Goto Y, Naya Y, Nakagawa M, Seki N. The tumor-suppressive microRNA-143/145 cluster inhibits cell migration and invasion by targeting GOLM1 in prostate cancer. J Hum Genet. 2014;59:78–87. doi: 10.1038/jhg.2013.121. [DOI] [PubMed] [Google Scholar]
- 15.Xia JT, Chen LZ, Jian WH, Wang KB, Yang YZ, He WL, He YL, Chen D, Li W. MicroRNA-362 induces cell proliferation and apoptosis resistance in gastric cancer by activation of NF-kappaB signaling. J Transl Med. 2014;12:33. doi: 10.1186/1479-5876-12-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li H, Xie S, Liu M, Chen Z, Liu X, Wang L, Li D, Zhou Y. The clinical significance of downregulation of mir-124-3p, mir-146a-5p, mir-155-5p and mir-335-5p in gastric cancer tumorigenesis. Int J Oncol. 2014;45:197–208. doi: 10.3892/ijo.2014.2415. [DOI] [PubMed] [Google Scholar]
- 17.Yang B, Jing C, Wang J, Guo X, Chen Y, Xu R, Peng L, Liu J, Li L. Identification of microRNAs associated with lymphangiogenesis in human gastric cancer. Clin Transl Oncol. 2014;16:374–379. doi: 10.1007/s12094-013-1081-6. [DOI] [PubMed] [Google Scholar]
- 18.Shin JY, Kim YI, Cho SJ, Lee MK, Kook MC, Lee JH, Lee SS, Ashktorab H, Smoot DT, Ryu KW, Kim YW, Choi IJ. MicroRNA 135a suppresses lymph node metastasis through down-regulation of ROCK1 in early gastric cancer. PLoS One. 2014;9:e85205. doi: 10.1371/journal.pone.0085205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang SH, Li X, Zhou LS, Cao ZW, Shi C, Zhou CZ, Wen YG, Shen Y, Li JK. microRNA-148a suppresses human gastric cancer cell metastasis by reversing epithelial-to-mesenchymal transition. Tumour Biol. 2013;34:3705–3712. doi: 10.1007/s13277-013-0954-1. [DOI] [PubMed] [Google Scholar]
- 20.Gong J, Li J, Wang Y, Liu C, Jia H, Jiang C, Wang Y, Luo M, Zhao H, Dong L, Song W, Wang F, Wang W, Zhang J, Yu J. Characterization of microRNA-29 family expression and investigation of their mechanistic roles in gastric cancer. Carcinogenesis. 2014;35:497–506. doi: 10.1093/carcin/bgt337. [DOI] [PubMed] [Google Scholar]
- 21.Wu WY, Xue XY, Chen ZJ, Han SL, Huang YP, Zhang LF, Zhu GB, Shen X. Potentially predictive microRNAs of gastric cancer with metastasis to lymph node. World J Gastroenterol. 2011;17:3645–3651. doi: 10.3748/wjg.v17.i31.3645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim SY, Jeon TY, Choi CI, Kim DH, Kim DH, Kim GH, Ryu DY, Lee BE, Kim HH. Validation of circulating miRNA biomarkers for predicting lymph node metastasis in gastric cancer. J Mol Diagn. 2013;15:661–669. doi: 10.1016/j.jmoldx.2013.04.004. [DOI] [PubMed] [Google Scholar]
- 23.Xu Y, Sun J, Xu J, Li Q, Guo Y, Zhang Q. miR-21 Is a Promising Novel Biomarker for Lymph Node Metastasis in Patients with Gastric Cancer. Gastroenterol Res Pract. 2012;2012:640168. doi: 10.1155/2012/640168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dong LL, Chen LM, Wang WM, Zhang LM. Decreased expression of microRNA-124 is an independent unfavorable prognostic factor for patients with breast cancer. Diagn Pathol. 2015;10:45. doi: 10.1186/s13000-015-0257-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schmittgen TD, Lee EJ, Jiang J, Sarkar A, Yang L, Elton TS, Chen C. Real-time PCR quantification of precursor and mature microRNA. Methods. 2008;44:31–38. doi: 10.1016/j.ymeth.2007.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zollner H, Hahn SA, Maghnouj A. Quantitative RT-PCR specific for precursor and mature miRNAs. Methods Mol Biol. 2014;1095:121–134. doi: 10.1007/978-1-62703-703-7_10. [DOI] [PubMed] [Google Scholar]
- 27.Matsuoka T, Yashiro M. Rho/ROCK signaling in motility and metastasis of gastric cancer. World J Gastroenterol. 2014;20:13756–13766. doi: 10.3748/wjg.v20.i38.13756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hu CB, Li QL, Hu JF, Zhang Q, Xie JP, Deng L. miR-124 inhibits growth and invasion of gastric cancer by targeting ROCK1. Asian Pac J Cancer Prev. 2014;15:6543–6546. doi: 10.7314/apjcp.2014.15.16.6543. [DOI] [PubMed] [Google Scholar]
- 29.Zhang Y, Zheng L, Huang J, Gao F, Lin X, He L, Li D, Li Z, Ding Y, Chen L. MiR-124 Radiosensitizes human colorectal cancer cells by targeting PRRX1. PLoS One. 2014;9:e93917. doi: 10.1371/journal.pone.0093917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Qin W, Pan Y, Zheng X, Li D, Bu J, Xu C, Tang J, Cui R, Lin P, Yu X. MicroRNA-124 regulates TGF-alpha-induced epithelial-mesenchymal transition in human prostate cancer cells. Int J Oncol. 2014;45:1225–1231. doi: 10.3892/ijo.2014.2506. [DOI] [PubMed] [Google Scholar]


