Abstract
Several studies have focused on the correlation between the programmed death-1 (PD-1) rs2227981 C > T polymorphism and the risk of cancer; however, the results of such studies remain conflicting. To address this gap, we performed this meta-analysis to identify the potential association. Search strategies were performed in PubMed and EMBASE using appropriate terms. In total, 2,977 cancer cases and 2,642 controls from seven publications were recruited in our study. According to the seven eligible publications, the odds ratios (ORs) and 95% confidence intervals (CIs) on the risk of cancer for the TT vs. CC and TT vs. CT+CC genotypes were 0.67 and 0.50-0.91 and 0.65 and 0.47-0.90, respectively. In a subgroup analysis by cancer type, PD-1 rs2227981 C > T polymorphism was associated with a significantly decreased risk of breast cancer (OR, 0.82; 95% CI, 0.71-0.95; P = 0.009 for T vs. C and OR, 0.76; 95% CI, 0.63-0.92; P = 0.005 for TT+CT vs. CC) and of other cancer (OR, 0.58; 95% CI, 0.36-0.92; P = 0.004 for TT vs. CT+CC). In a subgroup analysis by ethnicity, a significant decreased cancer risk was identified among Asians (OR, 0.74; 95% CI, 0.63-0.86; P < 0.001 for T vs. C and OR, 0.71; 95% CI, 0.59-0.87; P = 0.001 for TT+CT vs. CC) and among Caucasians (OR, 0.66; 95% CI, 0.44-0.99; P = 0.047 for TT vs. CT+CC). These findings highlight the fact that the T allele of PD-1 rs2227981 C > T polymorphism modestly decreases the susceptibility of cancer. Nevertheless, further large and well-designed studies are needed to enrich the evidence of this association.
Keywords: Polymorphism, programmed death-1, cancer risk
Introduction
It is estimated that about 14.1 million new cancer cases and 8.2 million cancer-associated deaths occurred in 2012 worldwide [1]. With new cases and mortality arising annually, cancer constitutes an enormous public health burden worldwide. These situations encourage researchers to explore the association of the latent environmental and genetic factors with the susceptibility of cancer. The aetiology of cancer is very elusive and has not been clarified thoroughly, although a number of investigations have focused on the function of the immune system [2,3]. Immune-related genetic mutations may also affect the risk of cancer [4,5].
Programmed death-1 (PD-1, also named CD279 or PDCD1), a co-inhibitory receptor that suppresses the activation of T lymphocytes and leads to peripheral immune tolerance, has been suggested to be involved in influencing the tumor cells to escape the host immune system after interaction with its two ligands, PD-1 ligand 1 (PD-L1) and PD-L2. PD-Ls are expressed in various malignancies [6-10]. In addition, up-regulation of PD-Ls in some cancers can contribute to tumor evasion and is associated with poor prognosis of malignancies [11-14]. Additionally, the function of regulatory T (Treg) cells also can be regulated through PD-1 pathway in cancer patients. Several recent investigations highlighted a correlation of PD-1 blockade with down-regulation of foxp3 expression by Treg cells in malignancy patients to correct immune escape [15-17].
The PD-1 gene lies in chromosome 2q37.3, encoding a 55 KDa type I transmembrane glycoprotein. Several researchers have reported single nucleotide polymorphisms (SNPs) within the PD-1 gene, such as rs36084323 A > G (PD-1.1), rs11568821 G > A (PD-1.3), rs2227981 C > T (PD-1.5), rs10204525 A > G (PD-1.6), rs7421861 T > C (PD-1.7), and rs2227982 C > T (PD-1.9) et al. Among these polymorphisms of the PD-1 gene, one of the most widely studied SNPs is rs2227981 C > T polymorphism with minor allele frequency (MAF) > 0.05. This very SNP is located in exon 5 and does not affect the final amino acid residue (a synonymous mutation; Ala to Ala). To date, a few studies have explored the correlation of PD-1 rs2227981 C > T polymorphism with caner susceptibility [18-24]; however, the results were conflicting. Thus, we performed this meta-analysis by pooling data from eligible case-control studies to further clarify the role of the PD-1 rs2227981 C > T polymorphism in cancer.
Materials and methods
Search strategy
PubMed and EMBASE databases were used to search the potential papers which were published before March 4, 2015 without any language restriction. The following terms were used: ‘polymorphism’ or ‘variant’ or ‘SNP’ and ‘programmed death-1’, ‘PD-1’ or ‘PDCD1’ and ‘cancer’ or ‘carcinoma’ or ‘malignance’.
Inclusion criteria and exclusion criteria
The major criteria were used to include eligible studies: (a) case-control studies; (b) studies that provided sufficient data to calculate crude odds ratios (ORs) and 95% confidence intervals (CIs) and (c) those assessing the correlation between the PD-1 rs2227981 C > T polymorphism and the risk of cancer. The major excluded criteria were: (1) case reports, system reviews, editorials, letters, and comments; (2) not case-control study and (3) duplicated publications.
Data extraction
Two reviewers (W. Tang and Y. Wang) screened and extracted data independently. If there were any discrepancies, differences were adjudicated through discussions between all reviewers. The following information was extracted from every study: PD-1 rs2227981 C > T polymorphism information, first author’s surname, year of publication, country, ethnicity and sample size.
Statistical analysis
Hardy-Weinberg equilibrium (HWE) in the controls for individual study was assessed using an internet-based HWE programme (http://ihg.gsf.de/cgi-bin/hw/hwa1.pl) and violation of HWE was defined by P < 0.05. Correlation between PD-1 rs2227981 C > T polymorphism and cancer susceptibility was assessed using crude ORs together with corresponding 95% CIs. The Q-statistic and I2 statistical tests were harnessed to measure the heterogeneity among studies. If the value of I2 > 50% or P < 0.10 suggests substantial heterogeneity, random-effects models using DerSimonian-Laird method were used [25], otherwise, fixed-effects models using Mantel-Haenszel methods was performed [26]. The Begg’s funnel plot [27] and Egger’s linear regression [28] were used to assessed the potential publication bias. One-way sensitivity analysis was harnessed to assess the stability of our results. All analyses were conducted by the Stata 12.0 statistical software (Stata Corp LP, College Station, TX, USA). A P < 0.05 (two-sided) was defined as a statistically significant difference.
Results
Study characteristics
According to the search keywords and subject terms from the databases of PubMed and EMBASE, one hundred and eighty-eight potential correlated publications were enrolled. Based on the included criteria, seven studies were identified [18-24] (Figure 1). Among them, two case-control studies deviated from HWE [18,19]. Five case-control studies focused on Caucasians [19,20,22-24], two focused on Asias [18,21]. Of these articles, two investigated breast cancer [20,21], the others investigated lung cancer [18], colorectal cancer [22], gastric cancer [23], gestational trophoblastic neoplasm [19] and cervical cancer [24]. Characteristics from each included study were listed in Table 1. The genotype numbers and P value of HWE for the eligible studies were summarized in Table 2.
Figure 1.
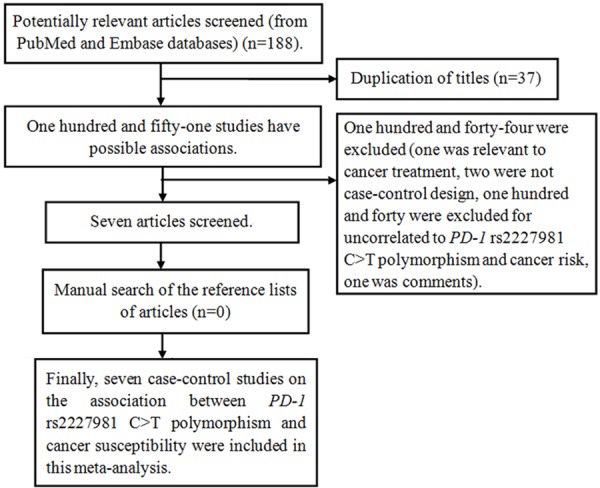
Flow diagram of articles selection process for metaanalysis.
Table 1.
Characteristics of the individual studies included in the meta-analysis
| Study | Year | Country | Ethnicity | Cancer type | Case/control | Genotype method |
|---|---|---|---|---|---|---|
| Yin et al. [18] | 2014 | China | Asians | Lung cancer | 324/330 | PCR-RFLP |
| Savabkar et al. [23] | 2013 | Iran | Caucasians | Gastric cancer | 122/166 | PCR-RFLP |
| Mojtahedi et al. [22] | 2012 | Iran | Caucasians | Colorectal cancer | 200/200 | PCR-RFLP |
| Haghshenas et al. [20]% | 2011 | Iran | Caucasians | Breast cancer | 443/328 | PCR-RFLP |
| Hua et al. [21] | 2011 | China | Asians | Breast cancer | 490/512 | PCR-RFLP |
| Ivansson et al. [24] | 2010 | Sweden | Caucasians | Cervical cancer | 1306/811 | TaqMan |
| Dehaghani et al. [19] | 2009 | Iran | Caucasians | Gestational trophoblastic neoplasm | 92/295 | PCR-RFLP |
PCR-RFLP: polymerase chain reaction-restriction fragment length polymorphism.
Table 2.
Distribution of PD-1 rs2227981 C > T polymorphism genotype and allele
| Study | Year | Case | Control | Case | Control | HWE | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
| ||||||||||||
| CC | CT | TT | CC | CT | TT | T | C | T | C | |||
| Yin et al. [18] | 2014 | 198 | 106 | 20 | 181 | 105 | 44 | 146 | 502 | 193 | 467 | No |
| Savabkar et al. [23] | 2013 | 50 | 66 | 6 | 89 | 70 | 7 | 78 | 166 | 84 | 248 | Yes |
| Mojtahedi et al. [22] | 2012 | 59 | 109 | 32 | 75 | 89 | 36 | 173 | 227 | 161 | 239 | Yes |
| Haghshenas et al. [20] | 2011 | 194 | 191 | 50 | 137 | 145 | 46 | 291 | 579 | 237 | 419 | Yes |
| Hua et al. [21] | 2011 | 295 | 169 | 22 | 244 | 210 | 24 | 213 | 759 | 258 | 698 | Yes |
| Ivansson et al. [24] | 2010 | 471 | 603 | 226 | 257 | 375 | 178 | 1055 | 1545 | 731 | 889 | Yes |
| Dehaghani et al. [19] | 2009 | 42 | 37 | 13 | 118 | 56 | 121 | 63 | 121 | 298 | 292 | No |
Quantitative synthesis
A total of 2,977 cancer cases and 2,642 controls from seven eligible investigations were enrolled. Our findings highlighted the statistical evidence of association between PD-1 rs2227981 C > T variants and a decreased risk of malignance in two genetic models: TT vs. CC (OR, 0.67; 95% CI, 0.50-0.91; P = 0.011) and TT vs. CT+CC (OR, 0.65; 95% CI, 0.47-0.90; P = 0.009) (Table 3 and Figure 2). In a subgroup analysis by cancer type, PD-1 rs2227981 C > T polymorphism was associated with a significantly decreased risk of breast cancer (OR, 0.82; 95% CI, 0.71-0.95; P = 0.009 for T vs. C and OR, 0.76; 95% CI, 0.63-0.92; P = 0.005 for TT+CT vs. CC) and of other cancer (OR, 0.58; 95% CI, 0.36-0.92; P = 0.004 for TT vs. CT+CC). In a subgroup analysis by ethnicity, a significant decreased cancer risk was identified among Asians (OR, 0.74; 95% CI, 0.63-0.86; P < 0.001 for T vs. C and OR, 0.71; 95% CI, 0.59-0.87; P = 0.001 for TT+CT vs. CC) and among Caucasians (OR, 0.66; 95% CI, 0.44-0.99; P = 0.047 for TT vs. CT+CC).
Table 3.
Meta-analysis of the PD-1 rs2227981 C > T polymorphism and cancer risk
| No. of study | T vs. C | TT vs. CC | TT+CT vs. CC | TT vs. CT+CC | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
|
|
|
||||||||||
| OR (95% CI) | P | P (Q-test) | OR (95% CI) | P | P (Q-test) | OR (95% CI) | P | P (Q-test) | OR (95% CI) | P | P (Q-test) | ||
| Total | 7 | 0.84 (0.71-1.00) | 0.050 | 0.001 | 0.67 (0.50-0.91) | 0.011 | 0.032 | 0.91 (0.74-1.12) | 0.381 | 0.007 | 0.65 (0.47-0.90) | 0.009 | 0.008 |
| Ethnicity | |||||||||||||
| Asians | 2 | 0.74 (0.63-0.86) | < 0.001 | 0.647 | 0.56 (0.31-1.00) | 0.051 | 0.154 | 0.71 (0.59-0.87) | 0.001 | 0.510 | 0.61 (0.30-1.27) | 0.188 | 0.073 |
| Caucasians | 5 | 0.90 (0.71-1.14) | 0.367 | 0.001 | 0.73 (0.49-1.07) | 0.101 | 0.032 | 1.03 (0.79-1.34) | 0.843 | 0.015 | 0.66 (0.44-0.99) | 0.047 | 0.009 |
| Cancer type | |||||||||||||
| Breast cancer | 2 | 0.82 (0.71-0.95) | 0.009 | 0.301 | 0.76 (0.53-1.10) | 0.147 | 0.975 | 0.76 (0.63-0.92) | 0.005 | 0.159 | 0.83 (0.59-1.17) | 0.292 | 0.750 |
| Other cancer | 5 | 0.85 (0.66-1.11) | 0.238 | < 0.001 | 0.64 (0.41-1.01) | 0.056 | 0.010 | 1.00 (0.75-1.33) | 0.994 | 0.009 | 0.58 (0.36-0.92) | 0.022 | 0.003 |
| HWE | |||||||||||||
| Yes | 5 | 0.93 (0.79-1.10) | 0.400 | 0.019 | 0.77 (0.63-0.93) | 0.006 | 0.446 | 0.98 (0.74-1.29) | 0.871 | 0.002 | 0.79 (0.66-0.94) | 0.007 | 0.905 |
| No | 2 | 0.61 (0.45-0.84) | 0.002 | 0.138 | 0.36 (0.24-0.56) | < 0.001 | 0.476 | 0.78 (0.60-1.01) | 0.060 | 0.927 | 0.32 (0.21-0.49) | < 0.001 | 0.165 |
HWE: Hardy-Weinberg equilibrium. Bold values are statistically significant (P < 0.05).
Figure 2.
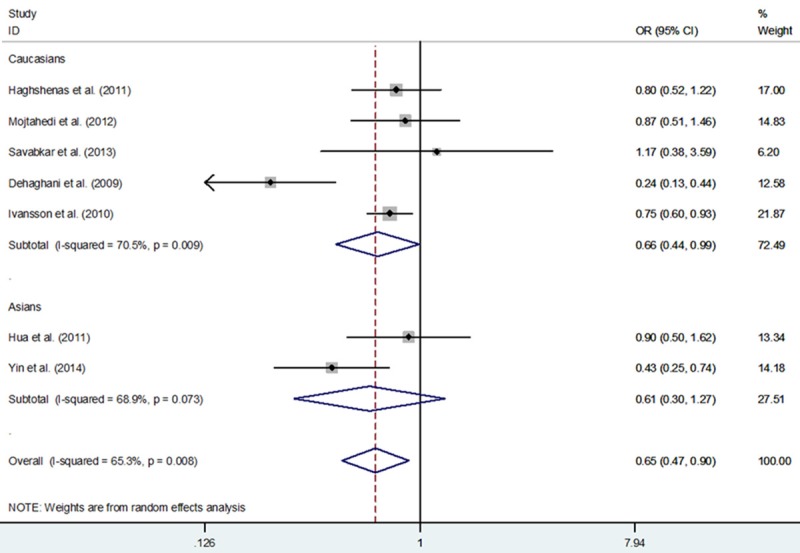
Meta-analysis with a fixed-effects model for the association between PD-1 rs2227981 C > T polymorphism and cancer risk (TT vs. CT+CC genetic model).
Publication bias
Begg’s funnel plots and Egger’s linear regression tests were harnessed to assess the publication bias (Figure 3). Significant publication bias was found in some genetic models (T vs. C: Begg’s test P = 0.764, Egger’s test P = 0.828; TT vs. CC: Begg’s test P = 1.000, Egger’s test P = 0.969; TT+CT vs. CC: Begg’s test P = 0.133, Egger’s test P = 0.148; TT vs. CT+CC: Begg’s test P = 0.548, Egger’s test P = 0.624).
Figure 3.
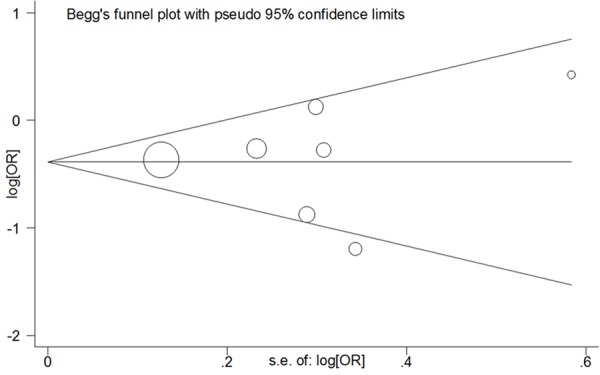
Begg’s funnel plot of meta-analysis of the association between PD-1 rs2227981 C > T polymorphism and the risk of cancer (TT vs. CC genetic model).
Sensitivity analyses
Sensitivity analysis was conducted to assess the influence of anyone study on the pooled ORs and CIs by omitting an individual study in turn. Our findings showed that these results were robust and reliable (Figure 4) (data not shown).
Figure 4.
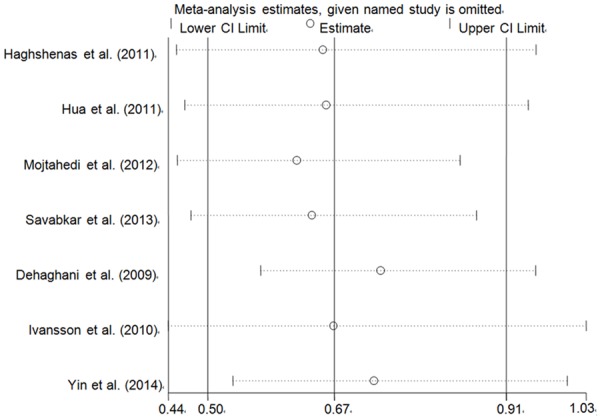
Sensitivity analysis of the influence of TT vs. CC compare genetic model (random-effects estimates for PD-1 rs2227981 C > T polymorphism).
Heterogeneity
As shown in Table 3, heterogeneity across the studies was significant in the current study. Thus, we assessed the sources of heterogeneity by race, the origin of cancer cells and HWE (Table 3). The findings showed that Caucasians and other cancer may contribute to the major sources of heterogeneity.
Discussion
In total, 2,977 cancer cases and 2,642 controls from seven eligible publications were recruited to investigate the correlation between the PD-1 rs2227981 C > T polymorphism and the risk of cancer.
According to the results, the PD-1 rs2227981 C > T polymorphism was suggested to be associated with a significantly decreased risk of cancer. The TT homozygote carriers suggested lower cancer incident susceptibility in comparison with the CC and CC+CT genotype carriers. The crude ORs and 95% CIs were 0.67 and 0.50-0.91, and 0.65 and 0.47-0.90, respectively. Although no statistical correlation between the PD-1 rs2227981 C > T polymorphism and the risk of cancer was identified, when the genetic comparisons were carried out between the TT+CT and CC homozygotes or between the T and C alleles, there remained a latent effect from the TT+CT genotypes or the T allele on the susceptibility of cancer. The crude ORs and 95% CIs were 0.91 and 0.74-1.12, respectively, for the TT+CT versus the CC homozygotes and 0.84 and 0.71-1.00, respectively, for the T versus C alleles.
The findings of this pooled study were supported by some investigations. In a previous study in China conducted by Hua et al [21], compared with the C allele, the T allele was a protective factor of breast cancer. Our findings were also supported by a Sweden study [24]. In this study (1, 306 cervical cancer cases and 811 controls), the TT homozygote reduced the risk of cervical cancer significantly. The ORs and 95% CIs were 0.69 and 0.54-0.89, respectively, for TT vs. CC and 0.75 and 0.60-0.93, respectively, for TT vs. CC+CT. The majority of our findings demonstrated the protective effect from the T allele on the susceptibility of cancer. In the future, further investigations based on a larger population and detailed gene-environment data are needed to be undertaken to confirm or refute our findings.
In the current study, two studies deviated from the HWE in controls, which showed the presence of population stratification and/or genotyping errors [18,19]. When we omitted these two studies, the correlation between PD-1 rs2227981 C > T polymorphism and cancer risk was also significant with respect to the two genetic models (OR, 0.77; 95% CI, 0.63-0.93; P = 0.006 for TT vs. CC and OR, 0.79; 95% CI, 0.66-0.94; P = 0.007 for TT vs. CT+CC; Table 3), attesting the robustness of our findings.
Several limitations of our study should be addressed when interpreting these findings. Due to the limited number of publications recruited in this pooled study, these findings should be interpreted with very caution. In addition, there were only two case-control studies conducted in Asians, which may generate a fluctuated assessment or restrict the statistical power to detect a real influence. Moreover, in this meta-analysis, large heterogeneities across the studies included in the current analysis should also be taken into consideration. Finally, in this study, we only focused on PD-1 rs2227981 C > T polymorphism, and did not ponder other PD-1 polymorphisms or risk genes. However, our study also had several merits. First, to date, this is the first meta-analysis detecting the association of PD-1 rs2227981 C > T polymorphism with the risk of cancer. The results demonstrated that this polymorphism was associated with the decreased risk of cancer. Second, although the large heterogeneities were identified in our study, the results of sensitivity analysis attesting the robustness of our findings.
In conclusion, our findings suggest that the T allele modestly decrease the susceptibility of cancer. Nevertheless, for practical reasons, further evidence from epidemiological studies across different populations incorporating with the functional assessments is required in order to confirm or refute the findings of this study.
Acknowledgements
This study was supported in part by National Natural Science Foundation of China (81472332, 81341006), Fujian Province Natural Science Foundation (2013J01126, 2013J05116), Fujian Medical University professor fund (JS12008), The Fund of Union Hospital (2015TC-1-048 and 2015TC-2-004), Fujian Province Science and Technology Programmed Fund (2012Y0030), Fujian Medical Innovation Fund (2014-CX-15) and Jiangsu University Clinical Medicine Science and Technology Development Fund (JLY20140012).
Disclosure of conflict of interest
None.
References
- 1.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 2.Vendrame E, Martinez-Maza O. Assessment of pre-diagnosis biomarkers of immune activation and inflammation: Insights on the etiology of lymphoma. J Proteome Res. 2011;10:113–119. doi: 10.1021/pr100729z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.de Visser KE, Eichten A, Coussens LM. Paradoxical roles of the immune system during cancer development. Nat Rev Cancer. 2006;6:24–37. doi: 10.1038/nrc1782. [DOI] [PubMed] [Google Scholar]
- 4.Clifford RJ, Zhang J, Meerzaman DM, Lyu MS, Hu Y, Cultraro CM, Finney RP, Kelley JM, Efroni S, Greenblum SI, Nguyen CV, Rowe WL, Sharma S, Wu G, Yan C, Zhang H, Chung YH, Kim JA, Park NH, Song IH, Buetow KH. Genetic variations at loci involved in the immune response are risk factors for hepatocellular carcinoma. Hepatology. 2010;52:2034–2043. doi: 10.1002/hep.23943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brenner AV, Neta G, Sturgis EM, Pfeiffer RM, Hutchinson A, Yeager M, Xu L, Zhou C, Wheeler W, Tucker MA, Chanock SJ, Sigurdson AJ. Common single nucleotide polymorphisms in genes related to immune function and risk of papillary thyroid cancer. PLoS One. 2013;8:e57243. doi: 10.1371/journal.pone.0057243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zou W, Chen L. Inhibitory b7-family molecules in the tumour microenvironment. Nat Rev Immunol. 2008;8:467–477. doi: 10.1038/nri2326. [DOI] [PubMed] [Google Scholar]
- 7.Gao Q, Wang XY, Qiu SJ, Yamato I, Sho M, Nakajima Y, Zhou J, Li BZ, Shi YH, Xiao YS, Xu Y, Fan J. Overexpression of pd-l1 significantly associates with tumor aggressiveness and postoperative recurrence in human hepatocellular carcinoma. Clin Cancer Res. 2009;15:971–979. doi: 10.1158/1078-0432.CCR-08-1608. [DOI] [PubMed] [Google Scholar]
- 8.Karim R, Jordanova ES, Piersma SJ, Kenter GG, Chen L, Boer JM, Melief CJ, van der Burg SH. Tumor-expressed b7-h1 and b7-dc in relation to pd-1+ t-cell infiltration and survival of patients with cervical carcinoma. Clin Cancer Res. 2009;15:6341–6347. doi: 10.1158/1078-0432.CCR-09-1652. [DOI] [PubMed] [Google Scholar]
- 9.Hua D, Sun J, Mao Y, Chen LJ, Wu YY, Zhang XG. B7-h1 expression is associated with expansion of regulatory t cells in colorectal carcinoma. World J Gastroenterol. 2012;18:971–978. doi: 10.3748/wjg.v18.i9.971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ohigashi Y, Sho M, Yamada Y, Tsurui Y, Hamada K, Ikeda N, Mizuno T, Yoriki R, Kashizuka H, Yane K, Tsushima F, Otsuki N, Yagita H, Azuma M, Nakajima Y. Clinical significance of programmed death-1 ligand-1 and programmed death-1 ligand-2 expression in human esophageal cancer. Clin Cancer Res. 2005;11:2947–2953. doi: 10.1158/1078-0432.CCR-04-1469. [DOI] [PubMed] [Google Scholar]
- 11.Hamanishi J, Mandai M, Iwasaki M, Okazaki T, Tanaka Y, Yamaguchi K, Higuchi T, Yagi H, Takakura K, Minato N, Honjo T, Fujii S. Programmed cell death 1 ligand 1 and tumor-infiltrating cd8+t lymphocytes are prognostic factors of human ovarian cancer. Proc Natl Acad Sci U S A. 2007;104:3360–3365. doi: 10.1073/pnas.0611533104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen YB, Mu CY, Huang JA. Clinical significance of programmed death-1 ligand-1 expression in patients with non-small cell lung cancer: A 5-year-follow-up study. Tumori. 2012;98:751–755. doi: 10.1177/030089161209800612. [DOI] [PubMed] [Google Scholar]
- 13.Zheng Z, Bu Z, Liu X, Zhang L, Li Z, Wu A, Wu X, Cheng X, Xing X, Du H, Wang X, Hu Y, Ji J. Level of circulating pd-l1 expression in patients with advanced gastric cancer and its clinical implications. Chin J Cancer Res. 2014;26:104–111. doi: 10.3978/j.issn.1000-9604.2014.02.08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zeng Z, Shi F, Zhou L, Zhang MN, Chen Y, Chang XJ, Lu YY, Bai WL, Qu JH, Wang CP, Wang H, Lou M, Wang FS, Lv JY, Yang YP. Upregulation of circulating pd-l1/pd-1 is associated with poor post-cryoablation prognosis in patients with hbv-related hepatocellular carcinoma. PLoS One. 2011;6:e23621. doi: 10.1371/journal.pone.0023621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang W, Lau R, Yu D, Zhu W, Korman A, Weber J. Pd1 blockade reverses the suppression of melanoma antigen-specific ctl by cd4+ cd25(hi) regulatory t cells. Int Immunol. 2009;21:1065–1077. doi: 10.1093/intimm/dxp072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guo Z, Wang X, Cheng D, Xia Z, Luan M, Zhang S. Pd-1 blockade and ox40 triggering synergistically protects against tumor growth in a murine model of ovarian cancer. PLoS One. 2014;9:e89350. doi: 10.1371/journal.pone.0089350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Duraiswamy J, Kaluza KM, Freeman GJ, Coukos G. Dual blockade of pd-1 and ctla-4 combined with tumor vaccine effectively restores t-cell rejection function in tumors. Cancer Res. 2013;73:3591–3603. doi: 10.1158/0008-5472.CAN-12-4100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yin L, Guo H, Zhao L, Wang J. The programmed death-1 gene polymorphism (pd-1.5 c/t) is associated with non-small cell lung cancer risk in a chinese han population. Int J Clin Exp Med. 2014;7:5832–5836. [PMC free article] [PubMed] [Google Scholar]
- 19.Dehaghani AS, Kashef MA, Ghaemenia M, Sarraf Z, Khaghanzadeh N, Fattahi MJ, Ghaderi A. Pdcd1, ctla-4 and p53 gene polymorphism and susceptibility to gestational trophoblastic diseases. J Reproduct Med. 2009;54:25–31. [PubMed] [Google Scholar]
- 20.Haghshenas MR, Naeimi S, Talei A, Ghaderi A, Erfani N. Program death 1 (pd1) haplotyping in patients with breast carcinoma. Mol Biol Rep. 2011;38:4205–4210. doi: 10.1007/s11033-010-0542-z. [DOI] [PubMed] [Google Scholar]
- 21.Hua Z, Li D, Xiang G, Xu F, Jie G, Fu Z, Jie Z, Da P, Li D. Pd-1 polymorphisms are associated with sporadic breast cancer in chinese han population of northeast china. Breast Cancer Res Treat. 2011;129:195–201. doi: 10.1007/s10549-011-1440-3. [DOI] [PubMed] [Google Scholar]
- 22.Mojtahedi Z, Mohmedi M, Rahimifar S, Erfani N, Hosseini SV, Ghaderi A. Programmed death-1 gene polymorphism (pd-1.5 c/t) is associated with colon cancer. Gene. 2012;508:229–232. doi: 10.1016/j.gene.2012.07.059. [DOI] [PubMed] [Google Scholar]
- 23.Savabkar S, Azimzadeh P, Chaleshi V, Nazemalhosseini Mojarad E, Aghdaei HA. Programmed death-1 gene polymorphism (pd-1.5 c/t) is associated with gastric cancer. Gastroenterol Hepatol Bed Bench. 2013;6:178–182. [PMC free article] [PubMed] [Google Scholar]
- 24.Ivansson EL, Juko-Pecirep I, Gyllensten UB. Interaction of immunological genes on chromosome 2q33 and ifng in susceptibility to cervical cancer. Gynecol Oncol. 2010;116:544–548. doi: 10.1016/j.ygyno.2009.10.084. [DOI] [PubMed] [Google Scholar]
- 25.DerSimonian R, Laird N. Meta-analysis in clinical trials. Control Clin Trials. 1986;7:177–188. doi: 10.1016/0197-2456(86)90046-2. [DOI] [PubMed] [Google Scholar]
- 26.Mantel N, Haenszel W. Statistical aspects of the analysis of data from retrospective studies of disease. J Natl Cancer Inst. 1959;22:719–748. [PubMed] [Google Scholar]
- 27.Begg CB, Mazumdar M. Operating characteristics of a rank correlation test for publication bias. Biometrics. 1994;50:1088–1101. [PubMed] [Google Scholar]
- 28.Egger M, Davey Smith G, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ. 1997;315:629–634. doi: 10.1136/bmj.315.7109.629. [DOI] [PMC free article] [PubMed] [Google Scholar]


