Abstract
Previous studies suggest that the RAD51 gene 135G/C polymorphism could be potentially associated with the risk of ovarian cancer. However, results from observational studies are conflicting rather than conclusive. We performed a meta-analysis of the literature aiming to clarify the relationship between the polymorphism of RAD51 gene 135G/C polymorphism and the risk of ovarian cancer. Summary odds ratios (ORs) and 95% confidence intervals (95% CIs) were calculated. We identified five eligible articles, 2336 ovarian cancer cases and 3548 controls. Meta-analysis results showed no significant association between 135G/C polymorphism in the RAD51 gene and ovarian cancer risk (GG vs CC: OR=0.42, 95% CI 0.16-1.06; GC vs CC: OR=0.37, 95% CI 0.12-1.16; Dominant model: OR=0.38, 95% CI 0.13-1.06; Recessive model: OR=1.20, 95% CI 0.91-1.58). No publication bias was found in the present study. This meta-analysis suggests that the RAD51 gene 135G/C polymorphism was not associated with risk of ovarian cancer. Further large and well-designed studies are needed to confirm this conclusion.
Keywords: Ovarian cancer, RAD51 gene, genetic variant, meta-analysis
Introduction
Ovarian cancer is the fifth leading cause of cancer deaths occurring in women and leading cause of mortality from gynecologic cancer [1]. Approximately 225,500 new cases and 140,200 deaths of ovarian cancer reported in the world annually [2,3]. After much investigation, the pathogenesis of ovarian cancer is not yet fully understood. Extensive epidemiology studies have shown that some factors may potentially result in ovarian cancer, such as age, childbearing status, infertility, dietary factors and gynaecological diseases (endometriosis, ovarian cysts, pelvic inflammatory disease) [4]. However, some women exposed to the same risk factors may not develop ovarian cancer while many cancer cases develop among individuals without those known risk factors. Recently, many studies show that genetic factors also play important role in the development of ovarian cancer [5,6]. This suggests that gene polymorphisms may explain individual differences in ovarian cancer risk.
DNA repair systems have been considered to maintain genomic integrity. Double-strand break repair is one of the key mechanisms of DNA repair, it can repair double-strand breaks through two major pathways: homologous recombination (HR) and non-homologous end-rejoining [7]. The RAD51 protein plays a irreplaceable role during HR repair via binding to DNA to promote ATP-dependent homologous pairing and strand transfer reactions [8]. The RAD51 gene is located at the human chromosome 15q15.1, and thought to participate in HR repair pathway. The RAD51 gene 135G/C polymorphism (rs1801320) is a G to C transversion at position 135 of the human RAD51 cDNA [9]. Previous meta-analyses demonstrated that RAD51 gene 135G/C polymorphism was associated with susceptibility to breast cancer and head-and-neck cancer [10,11].
In the past decade, a number of molecular epidemiological studies have been done to evaluate the association between RAD51 gene 135G/C polymorphism and ovarian cancer risk, but the results remained controversial. Meta-analysis is a powerful and rigorous method to resolve these problems. Therefore, we performed a meta-analysis to precisely characterize whether or not RAD51 gene 135G/C polymorphism is associated with human ovarian cancer risk.
Materials and methods
Publication search
A comprehensive computerized literature search was conducted in multiple online databases including PubMed, EMBASE and ISI Web of Science databases with the combination of the following search terms: “RAD51”, “135G/C”, “rs1801320”, and “ovarian cancer” to identify potentially relevant studies (last search update: April, 2015). Only those published studies in English language with full text articles were included in this meta-analysis. Additional studies were identified by a manual search of references of original studies or review articles on the association between RAD51 gene 135G/C polymorphism and ovarian cancer.
Inclusion, exclusion criteria and Data extraction
Studies in our meta-analysis had to meet all of the following criteria: 1) Case control studies; 2) Studies assessing the association of 135G/C polymorphism in the RAD51 gene with ovarian cancer risk; 3) Providing sufficient information for estimating odds ratio (OR) with its 95% confidence interval (95% CI); 4) Providing available data to acquire genotype frequency of 135G/C polymorphism. Any study with wrong data or inconsistent data was excluded. Two reviewers independently performed data extraction and then checked the results together. For conflicting evaluations, an agreement was reached following a discussion. The following information was extracted from the included studies: first author, year of publication, area, number of cases and controls, genotype frequencies in cases and controls, and evidence of Hardy-Weinberg equilibrium (HWE) in controls (Table 1).
Table 1.
Characteristics of the included studies for meta-analysis
| Study included | Year | Area | Race | Cases/Controls | Genotypes for cases | Genotypes for controls | HWE test | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
|||||||||||
| GG | GC | CC | GG | GC | CC | ||||||
| Auranen | 2005 | Danish | Caucasian | 278/699 | 241 | 36 | 1 | 616 | 78 | 5 | 0.15 |
| Auranen | 2005 | UK | Caucasian | 729/847 | 642 | 84 | 3 | 745 | 100 | 2 | 0.48 |
| Auranen | 2005 | USA | Caucasian | 326/419 | 270 | 52 | 4 | 357 | 61 | 1 | 0.34 |
| Webb | 2005 | Australia | Mixed | 546/1126 | 457 | 85 | 4 | 971 | 145 | 10 | 0.08 |
| Jakubowska | 2007 | Poland | Caucasian | 127/127 | 104 | 23 | 0 | 89 | 37 | 1 | 0.17 |
| Romanowicz-Makowska | 2012 | Poland | Caucasian | 120/120 | 13 | 15 | 92 | 33 | 69 | 18 | 0.07 |
| Smolarz | 2013 | Poland | Caucasian | 210/210 | 43 | 45 | 122 | 63 | 99 | 48 | 0.45 |
Hardy-Weinberg Equilibrium.
Statistical analysis
The association between the RAD51 gene 135G/C polymorphism and ovarian cancer was compared using the OR corresponding to a 95% CI under a co-dominant model (GG vs CC, GC vs CC), a dominant model (GG+GC vs CC), and a recessive model (CC+GC vs GG). We tested whether genotype frequencies of controls were in HWE using the χ2 test. Between-study heterogeneities were estimated using I2 test. I2 values of 25, 50 and 75% were defined as low, moderate, and high estimates, respectively. If heterogeneity was found among the studies, the fixed effect model (the Mantel-Haenszel method) was utilized to calculate the pooled ORs. Otherwise, the random-effects model (the Der Simonian and Laird method) was used to estimate the pooled OR. Sensitivity analysis was performed through random effect model values compared to the fixed effect. Publication bias was investigated by Begg’s test (P<0.05 was considered statistically significant).Statistical analysis was performed using STATA version 12 (Stata Corporation, College Station, TX).
Results
Eligible studies
A database search yielded 143 publications, of which both of the reviewers considered 10 to be potentially eligible. We excluded 5 of the articles during the second phase of the inclusion process. The remaining 5 articles were included in the meta-analysis [12-16]. The flow chart of study selection is summarized in Figure 1. All the articles were written in English. The source of controls was mainly based on healthy populations. The genotype frequencies of the controls from six studies were in agreement with HWE. The study characteristics are presented in Table 1.
Figure 1.
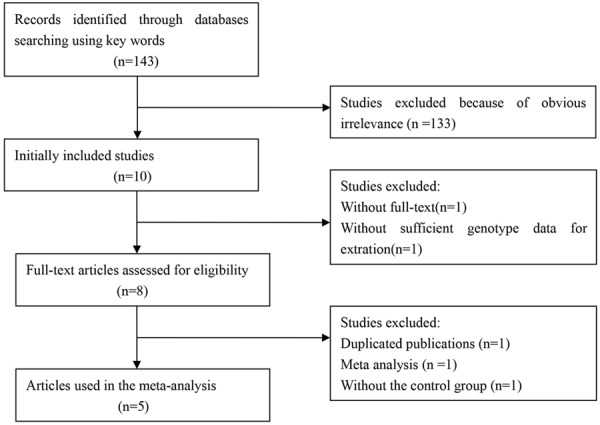
The flow chart of study selection.
Meta-analysis
Figure 2 and Table 2 listed the main results of the meta-analysis. Overall, no significant association of the RAD51 gene 135G/C polymorphism with ovarian cancer risk was found (GG vs CC: OR=0.42, 95% CI 0.16-1.06; GC vs CC: OR=0.37, 95% CI 0.12-1.16; Dominant model: OR=0.38, 95% CI 0.13-1.06; Recessive model: OR=1.20, 95% CI 0.91-1.58). Sensitivity analyses were conducted by altering the statistic models. No material alteration was detected, indicating that our results were statistically robust. The Begg’s funnel plot was performed to assess the publication bias in the reports included for meta-analysis. The shapes of the funnel plots in all genetic models did not reveal any evidence of obvious asymmetry (Figure 3; Table 2).
Figure 2.
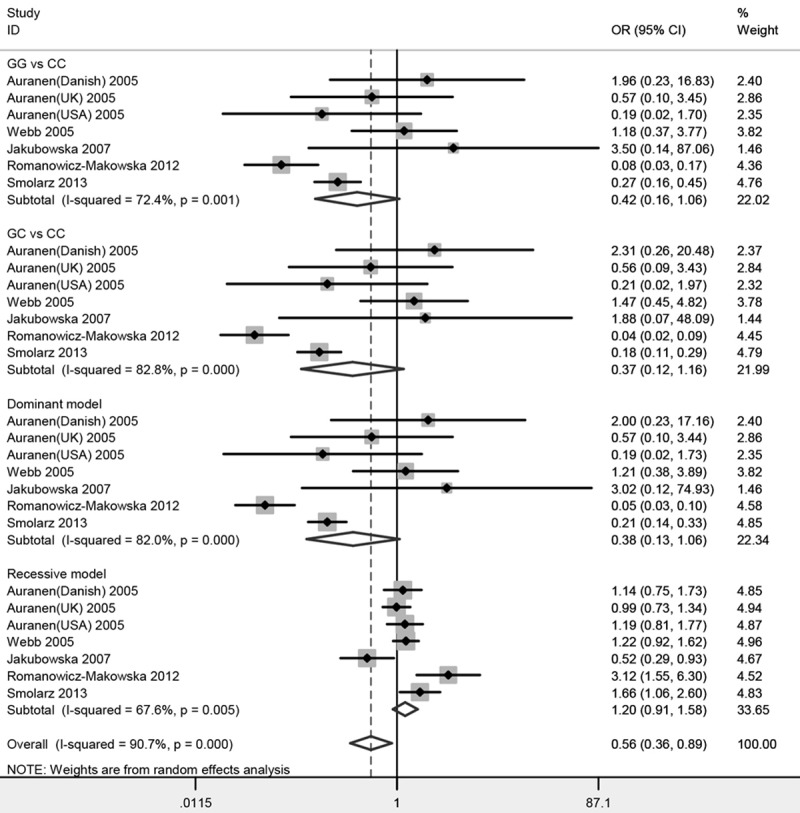
Forest plots of the association between RAD51 gene 135G/C polymorphism and ovarian cancer risk.
Table 2.
Summary ORs and 95% CI of RAD51 gene 135G/C polymorphism and ovarian cancer risk
| Genetic model | Sample size | Type of model | Test of heterogeneity | Test of association | Test of publication bias | ||||
|---|---|---|---|---|---|---|---|---|---|
|
|
|
||||||||
| Case | Control | I2 | P | OR | 95% CI | z | P | ||
| GG vs CC | 2336 | 3548 | Random | 72.4% | 0.00 | 0.42 | 0.16-1.06 | 0.00 | 1.00 |
| GC vs CC | Random | 82.8% | 0.00 | 0.37 | 0.12-1.16 | 0.00 | 1.00 | ||
| Dominant model | Random | 82.0% | 0.00 | 0.38 | 0.13-1.06 | 0.00 | 1.00 | ||
| Recessive model | Random | 67.6% | 0.00 | 1.20 | 0.91-1.58 | 0.00 | 1.00 | ||
Figure 3.
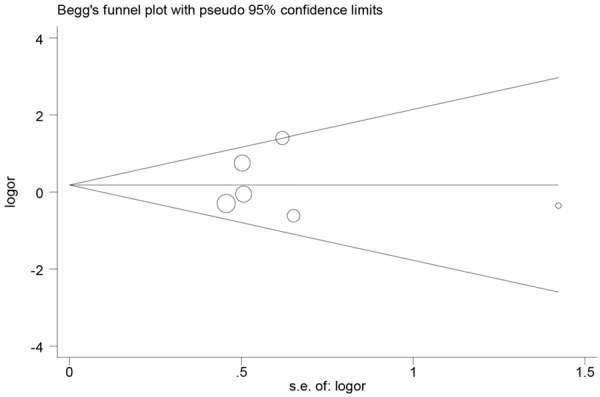
Begg’s funnel plot for publication bias test (GG vs CC).
Discussion
Previous studies have already found that DNA repair system is crucial for genomic integrity by countering threats posed by DNA lesions. Deficiency in the DNA repair pathways might make these lesions unrepaired or repaired incorrectly, eventually leading to genome instability or mutations which may lead to an increased susceptibility to cancer [17]. RAD51, a kind of ubiquitous strand exchange protein, is known to be a core component involved in DNA double-strand break repair in HR repair pathway [18]. The RAD51 gene 135G/C polymorphism may be related with RAD51 protein over-expression and DNA repair increase [19]. Recently, a variety of studies have focused on the association between the 135G/C polymorphism in the RAD51 gene and ovarian cancer. However, the observed associations of these studies were inconclusive. The most likely reason for the inconsistencies among these studies is that they are single case-control studies with small sample sizes. Therefore, we conducted a meta-analysis to summarize the effects of Rad51 gene 135G/C polymorphism on risk of ovarian cancer.
To the best of our knowledge, this study for the first time investigated this issue in a meta-analyzing way. Our meta-analysis quantitatively assessed the association between 135G/C polymorphism and ovarian cancer risk. It is worthwhile to mention that we performed an efficient searching strategy based on computer and manual searches, which allowed us to include studies as many as possible. The current meta-analysis included 2336 ovarian cancer patients and 3548 controls and explored the association between the 135G/C polymorphism of the Rad51 gene and ovarian cancer risk. The results of the present meta-analysis revealed that the Rad51 gene 135G/C polymorphism is not associated with increased or decreased risk of ovarian cancer. Previous meta-analyses were carried out to assess the effect of Rad51 gene G135C polymorphism on the risk of cancer, and the RAD51 gene 135G/C polymorphism was associated with the risk of breast cancer and head-and-neck cancer [10,11]. Inconsistent results might be due to a different role of Rad51 gene G135C polymorphism in different cell types or tissues. And another explanation for the different findings may be based on gene-gene and gene-environment interactions. Further large and well-designed studies are needed to confirm this conclusion.
Some limitations of our meta-analysis should be addressed. First of all, only published studies and papers written in English were searched in this meta-analysis, some unpublished studies or studies written in other language that might also meet the inclusion criteria were overlooked. Second, OR value was obtained without correction. More accurate OR should be corrected by age, menopause status, ethnicity and other exposure factors that are potentially associated with ovarian cancer risk. Third, the existence of gene-gene and gene-environment interactions may affect the accuracy of our results. Lacking of the original data limited our further evaluation of potential interactions among gene-gene and gene-environment.
To sum up, our meta-analysis suggests that the G135C polymorphism in the Rad51 gene may be not associated with ovarian cancer risk. Based on the limitations mentioned earlier, high-quality studies are needed to confirm our findings.
Disclosure of conflict of interest
None.
References
- 1.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2012. CA Cancer J Clin. 2012;62:10–29. doi: 10.3322/caac.20138. [DOI] [PubMed] [Google Scholar]
- 2.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 3.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. CA Cancer J Clin. 2013;63:11–30. doi: 10.3322/caac.21166. [DOI] [PubMed] [Google Scholar]
- 4.Risch HA, Howe GR. Pelvic inflammatory disease and the risk of epithelial ovarian cancer. Cancer Epidemiol Biomarkers Prev. 1995;4:447–51. [PubMed] [Google Scholar]
- 5.Bolton KL, Tyrer J, Song H, Ramus SJ, Notaridou M, Jones C, Sher T, Gentry-Maharaj A, Wozniak E, Tsai YY, Weidhaas J, Paik D, Van Den Berg DJ, Stram DO, Pearce CL, Wu AH, Brewster W, Anton-Culver H, Ziogas A, Narod SA, Levine DA, Kaye SB, Brown R, Paul J, Flanagan J, Sieh W, McGuire V, Whittemore AS, Campbell I, Gore ME, Lissowska J, Yang HP, Medrek K, Gronwald J, Lubinski J, Jakubowska A, Le ND, Cook LS, Kelemen LE, Brook-Wilson A, Massuger LF, Kiemeney LA, Aben KK, van Altena AM, Houlston R, Tomlinson I, Palmieri RT, Moorman PG, Schildkraut J, Iversen ES, Phelan C, Vierkant RA, Cunningham JM, Goode EL, Fridley BL, Kruger-Kjaer S, Blaeker J, Hogdall E, Hogdall C, Gross J, Karlan BY, Ness RB, Edwards RP, Odunsi K, Moyisch KB, Baker JA, Modugno F, Heikkinenen T, Butzow R, Nevanlinna H, Leminen A, Bogdanova N, Antonenkova N, Doerk T, Hillemanns P, Dürst M, Runnebaum I, Thompson PJ, Carney ME, Goodman MT, Lurie G, Wang-Gohrke S, Hein R, Chang-Claude J, Rossing MA, Cushing-Haugen KL, Doherty J, Chen C, Rafnar T, Besenbacher S, Sulem P, Stefansson K, Birrer MJ, Terry KL, Hernandez D, Cramer DW, Vergote I, Amant F, Lambrechts D, Despierre E, Fasching PA, Beckmann MW, Thiel FC, Ekici AB, Chen X Australian Ovarian Cancer Study Group; Australian Cancer Study (Ovarian Cancer); Ovarian Cancer Association Consortium. Johnatty SE, Webb PM, Beesley J, Chanock S, Garcia-Closas M, Sellers T, Easton DF, Berchuck A, Chenevix-Trench G, Pharoah PD, Gayther SA. Common variants at 19p13 are associated with susceptibility to ovarian cancer. Nat Genet. 2010;42:880–4. doi: 10.1038/ng.666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Song H, Ramus SJ, Tyrer J, Bolton KL, Gentry-Maharaj A, Wozniak E, Anton-Culver H, Chang-Claude J, Cramer DW, DiCioccio R, Dörk T, Goode EL, Goodman MT, Schildkraut JM, Sellers T, Baglietto L, Beckmann MW, Beesley J, Blaakaer J, Carney ME, Chanock S, Chen Z, Cunningham JM, Dicks E, Doherty JA, Dürst M, Ekici AB, Fenstermacher D, Fridley BL, Giles G, Gore ME, De Vivo I, Hillemanns P, Hogdall C, Hogdall E, Iversen ES, Jacobs IJ, Jakubowska A, Li D, Lissowska J, Lubiński J, Lurie G, McGuire V, McLaughlin J, Medrek K, Moorman PG, Moysich K, Narod S, Phelan C, Pye C, Risch H, Runnebaum IB, Severi G, Southey M, Stram DO, Thiel FC, Terry KL, Tsai YY, Tworoger SS, Van Den Berg DJ, Vierkant RA, Wang-Gohrke S, Webb PM, Wilkens LR, Wu AH, Yang H, Brewster W, Ziogas A Australian Cancer (Ovarian) Study; Australian Ovarian Cancer Study Group; Ovarian Cancer Association Consortium. Houlston R, Tomlinson I, Whittemore AS, Rossing MA, Ponder BA, Pearce CL, Ness RB, Menon U, Kjaer SK, Gronwald J, Garcia-Closas M, Fasching PA, Easton DF, Chenevix-Trench G, Berchuck A, Pharoah PD, Gayther SA. A genome-wide association study identifies a new ovarian cancer susceptibility locus on 9p22.2. Nat Genet. 2009;41:996–1000. doi: 10.1038/ng.424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Agarwal S, Tafel AA, Kanaar R. DNA doublestrand break repair and chromosome translocations. DNA Repair. 2006;5:1075–81. doi: 10.1016/j.dnarep.2006.05.029. [DOI] [PubMed] [Google Scholar]
- 8.Baumann P, West SC. Role of the human RAD51 protein in homologous recombination and double-stranded-break repair. Trends Biochem Sci. 1998;23:247–51. doi: 10.1016/s0968-0004(98)01232-8. [DOI] [PubMed] [Google Scholar]
- 9.Cheng D, Shi H, Zhang K, Yi L, Zhen G. RAD51 Gene 135G/C polymorphism and the risk of four types of common cancers: a meta-analysis. Diagn Pathol. 2014;9:18. doi: 10.1186/1746-1596-9-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kayani MA, Khan S, Baig RM, Mahjabeen I. Association of RAD51 135G/C, 172G/T and XRCC3 Thr241Met gene polymorphisms with increased risk of head and neck cancer. Asian Pac J Cancer Prev. 2014;15:10457–62. doi: 10.7314/apjcp.2014.15.23.10457. [DOI] [PubMed] [Google Scholar]
- 11.Zhou GW, Hu J, Peng XD, Li Q. RAD51 135G>C polymorphism and breast cancer risk: a metaanalysis. Breast Cancer Res Treat. 2011;125:529–35. doi: 10.1007/s10549-010-1031-8. [DOI] [PubMed] [Google Scholar]
- 12.Auranen A, Song H, Waterfall C, Dicioccio RA, Kuschel B, Kjaer SK, Hogdall E, Hogdall C, Stratton J, Whittemore AS, Easton DF, Ponder BA, Novik KL, Dunning AM, Gayther S, Pharoah PD. Polymorphisms in DNA repair genes and epithelial ovarian cancer risk. Int J Cancer. 2005;117:611–8. doi: 10.1002/ijc.21047. [DOI] [PubMed] [Google Scholar]
- 13.Webb PM, Hopper JL, Newman B, Chen X, Kelemen L, Giles GG, Southey MC, Chenevix-Trench G, Spurdle AB. Double-strand break repair gene polymorphisms and risk of breast or ovarian cancer. Cancer Epidemiol Biomarkers Prev. 2005;14:319–23. doi: 10.1158/1055-9965.EPI-04-0335. [DOI] [PubMed] [Google Scholar]
- 14.Jakubowska A, Gronwald J, Menkiszak J, Górski B, Huzarski T, Byrski T, Edler L, Lubiñski J, Scott RJ, Hamann U. The RAD51 135 G>C polymorphism modifies breast cancer and ovarian cancer risk in Polish BRCA1 mutation carriers. Cancer Epidemiol Biomarkers Prev. 2007;16:270–5. doi: 10.1158/1055-9965.EPI-06-0562. [DOI] [PubMed] [Google Scholar]
- 15.Romanowicz-Makowska H, Smolarz B, Samulak D, Michalska M, Lewy J, Burzyński M, Kokołaszwili G. A single nucleotide polymorphism in the 5’ untranslated region of RAD51 and ovarian cancer risk in Polish women. Eur J Gynaecol Oncol. 2012;33:406–10. [PubMed] [Google Scholar]
- 16.Smolarz B, Makowska M, Samulak D, Michalska MM, Mojs E, Romanowicz H, Wilczak M. Association between polymorphisms of the DNA repair gene RAD51 and ovarian cancer. Pol J Pathol. 2013;64:290–5. doi: 10.5114/pjp.2013.39338. [DOI] [PubMed] [Google Scholar]
- 17.Wood RD, Mitchell M, Lindahl T. Human DNA repair genes, 2005. Mutat Res. 2005;577:275–83. doi: 10.1016/j.mrfmmm.2005.03.007. [DOI] [PubMed] [Google Scholar]
- 18.Karpenshif Y, Bernstein KA. From yeast to mammals: recent advances in genetic control of homologous recombination. DNA Repair (Amst) 2012;11:781–8. doi: 10.1016/j.dnarep.2012.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Richardson C, Stark JM, Ommundsen M, Jasin M. Rad51 overexpression promotes alternative double-strand break repair pathways and genome instability. Oncogene. 2004;23:546–553. doi: 10.1038/sj.onc.1207098. [DOI] [PubMed] [Google Scholar]


