Abstract
Objectives: Published data on the association between Interleukin-8-251A/T polymorphism and gastric cancer (GC) risk are inconclusive. Thus, we conducted a meta-analysis to evaluate the relationship between cyclin D1 G870A polymorphism and GC risk. Methods: We searched PubMed, EMBASE, Web of science and the Cochrane Library up to July 12, 2015 for relevant studies. Odds ratios (ORs) and 95% confidence intervals (CIs) were used to estimate the strength of associations. Results: Twenty-six studies published from 2004 to 2015, with a total of 5286 cases and 8000 controls, were included in this meta-analysis. The pooled results showed that there was significant association between Interleukin-8-251A/T polymorphism and GC risk in any genetic model. In the subgroup analysis by ethnicity, the effects remained in Asians. However, no genetic models reached statistical association in Europeans. The subgroup analysis stratified by Source of controls showed an increased breast cancer risk in hospital-based (HB) studies in any genetic model except recessive model. However, there was no association in any genetic model in population based (PB) studies. When stratifying by Genotyping method, we found statistical association in Non-RFLP (restriction fragment length polymorphism) in any genetic model except heterozygote comparison, the effect was remain in PCR-RFLP in dominant model and heterozygote comparison. Conclusions: This meta-analysis suggests that Interleukin-8-251A/T polymorphism is a risk factor for susceptibility to GC in overall population, especially in Asians, in hospital populations and in Non-RFLP. While, there was no association in Europeans and in general population. Further large scale multicenter epidemiological studies are warranted to confirm this finding.
Keywords: Interleukin-8-251A/T, polymorphism, gastric cancer, susceptibility, meta-analysis
Introduction
Gastric cancer (GC) is one of the most common malignant tumors and the third leading cause of cancer-related death in the word, the 5-year survival rate is low, especially for advanced GC [1,2]. In the majority of developing countries, the incidence of GC is constantly increasing, as well as mortality [3,4]. For most GCs are diagnosed to be advanced stages, early detection seems particularly important [5]. While, the determination of the association between Interleukin-8-251A/T polymorphism and GC risk provides us a promising approach to achieve this goal.
Interleukin-8 is an important member of the chemokine superfamily, which belongs to the CXC chemokine family. Under the stimulation of a variety of factors (such as lipopolysaccharide, IL.1, etc.), many cells can produce IL-8, such as monocytes, endothelial cells and tumor cells [6,7]. In recent years, the expression of IL-8 in tumor cells was significantly increased. It is found that IL-8 can induce the migration and proliferation of endothelial cells to mediate tumor angiogenesis, which can promote tumor [8-10]. Human IL-8 gene is located on fourth chromosome, composed of four exons, three introns, and a proximal promoter region. In the promoter region of IL-8, there is a genetic polymorphism in -251T, which has been reported to be closely associated with altered expression levels of IL-8 through gene transcription regulation [11].
Previous functional studies have reported the association between Interleukin-8-251A/T polymorphism and GC risk, but the results remain inconclusive [12-37]. To clarify the role of Interleukin-8-251A/T polymorphism in GC risk, four meta-analysis on the associations between Interleukin-8-251A/T polymorphism and cancers [38-41]. However, number of their studies included in their meta-analysis about GC is small, and GC is just a small part of their study. In the subgroup of their analyses the sample size is extremely small, and some just no subgroup. Therefore, we decided to carry out a meta-analysis on all eligible case-control studies to make a more precise estimation of the association. Furthermore, we conducted the subgroup analysis by stratification according to the ethnicity, source of controls and genotyping method.
Materials and methods
Literature searching strategy
We searched PubMed, EMBASE, Web of science, the Cochrane Library for relevant studies published before July 12, 2015. The following keywords were used: interleukin*/IL*, variant/genotype/polymorphism/SNP, Gastric/stomach/cardia, cancer/carcinom*/neoplasm*/tumor and the combined phrases for all genetic studies on the association between the Interleukin-8-251A/T polymorphism and GC risk. The reference lists of all articles were also manually screened for potential studies. Abstracts and citations were screened independently by two authors, all the agreed articles need a second screen for full-text reports. The searching was done without restriction on language.
Selection and exclusion criteria
Inclusion criteria: A study was included in this meta-analysis if it meets the following criteria: i) independent case-control studies for humans; ii) the study evaluated the association between Interleukin-8-251A/T polymorphism and GC risk; iii) has available genotype frequencies in cancer cases and control subjects for risk estimate. We excluded comments, editorials, systematic reviews or studies lacking sufficient data. If the publications were duplicated or shared in more than one study, the most recent publications were included. All identified studies were screened by two investigators independently. What’s more, there was no limitation for publication language.
Data extraction and synthesis
We used endnote bibliographic software to construct an electronic library of citations identified in the literature search. All the PubMed, EMBASE, Web of science and the Cochrane Library searches were performed using Endnote; duplicates were found automatically by endnote and deleted manually. All data extraction were checked and calculated twice according to the inclusion criteria listed above by two independent investigators. Data extracted from the included studies were as follows: First author, year of publication, country, ethnicity, Source of controls, Genotyping method, number of cases and controls and evidence of HWE in controls. A third reviewer would participate if some disagreements were emerged, and a final decision was made by the majority of the votes.
Statistical analysis
All statistical analyses were performed using STATA version 11.0 software (StataCorp LP, College Station, TX) and Review Manage version 5.2.0 (The Cochrane Collaboration, 2012). Hardy-Weinberg equilibrium (HWE) was assessed by χ2 test in the control group of each study [42]. The strength of associations between the Interleukin-8-251A/T polymorphism and GC risk were measured by odds ratio (ORs) with 95% confidence interval (CIs). Z test was used to assess the significance of the ORs, I2 and Q statistics was used to determine the statistical heterogeneity among studies. A random-effect model was used if P value of heterogeneity tests was no more than 0.1 (P≤0.1), otherwise, a fixed-effect model was selected [42,43]. Sensitivity analyses were performed to assess the stability of the results. We used Begg’s funnel plot and Egger’s test to evaluate the publication bias [44,45]. The strength of the association was estimated in the allele model, the dominant model, the recessive model, the homozygous genetic model, and the heterozygous genetic model, respectively. P<0.05 was considered statistically significant. We performed subgroup according to Ethnicity, Source of controls, Genotyping method.
Results
Characteristics of included studies
Detailed search procedures are summarized in Figure 1. A total of 2499 references were preliminarily identified at first based on our selection strategy. We also identified 1 paper through other source. After excluding duplicate articles, we reviewed titles and abstracts of all identified studies to exclude those that were clearly irrelevant. Next, the full texts of the remaining articles were examined according to the inclusion and exclusion criteria. Finally, 26 studies [12-37] on Interleukin-8-251A/T polymorphism and GC risk were finally identified in this meta-analysis, including 5286 cases and 8000 controls. The characteristics of the included studies are listed in Table 1. The 26 case-control studies were published between 2004 and 2015, among them, 9 studies were performed in European and 17 in Asians. All studies were case-controlled. 13 studies were hospital-based and 13 were population-based studies.
Figure 1.
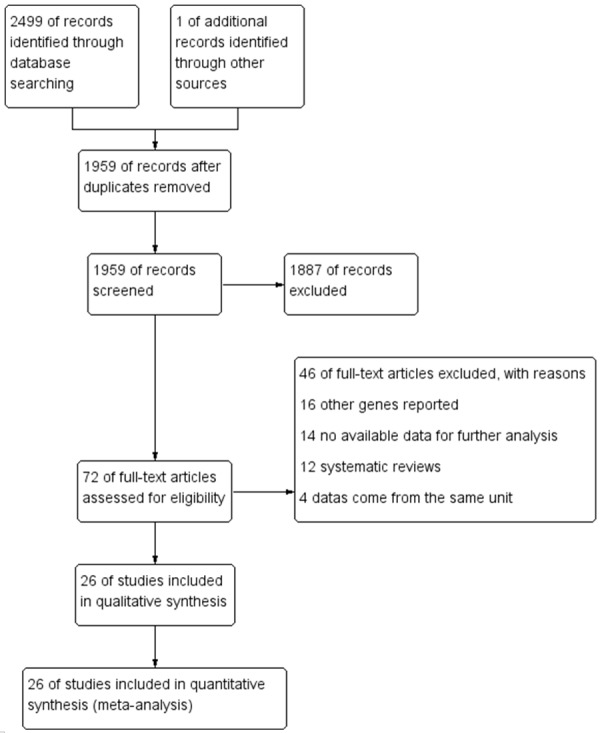
Flow chart of studies selection in this meta-analysis.
Table 1.
Characteristics of the studies included in the meta-analysis
| First author | Year | Country | Ethnicity | Source of controls | Genotyping method | Number (case/control) | HWE | Quality assessment score | Published language |
|---|---|---|---|---|---|---|---|---|---|
| Bo [37] | 2010 | China | Asian | HB | PCR-RFLP | 208/190 | 0.389403209 | 7 | English |
| Canedo [36] | 2008 | Portugal | European | PB | Taq Man-PCR | 333/693 | 0.459719109 | 8 | English |
| Crusius [35] | 2008 | Caucasia | European | PB | Real-time | 236/1139 | 0.705567725 | 8 | English |
| de Oliveira [34] | 2015 | Brazil | European | HB | PCR-RFLP | 207/240 | 0.059480986 | 7 | English |
| Felipe [33] | 2012 | Brazil | European | PB | PCR-RFLP | 104/196 | 0.065528611 | 8 | English |
| Garza-Gonzalez [32] | 2007 | Mexico | European | HB | ARMS-PCR | 78/189 | 0.538816094 | 7 | English |
| Kamali-Sarvestani [31] | 2006 | Iran | Asian | HB | ASO-PCR | 19/153 | 0.797575578 | 7 | English |
| Kamangar [30] | 2006 | Finland | European | PB | TaqMan-PCR | 112/207 | 0.054934096 | 8 | English |
| Kang [29] | 2009 | Korea | Asian | PB | PCR-RFLP | 334/322 | 0.22569954 | 8 | English |
| Ko [28] | 2009 | Korea | Asian | PB | Snapshot | 81/308 | 0.155354832 | 8 | English |
| Lee [27] | 2005 | Taiwan | Asian | HB | PCR-RFLP | 470/308 | 0.14303682 | 7 | English |
| Liu [12] | 2009 | China | Asian | HB | Taq Man-PCR | 138/137 | 0.145093518 | 7 | Chinese |
| Lu [26] | 2005 | China | Asian | PB | PCR-DHPLC | 250/300 | 0.515848398 | 8 | English |
| Ohyauchi [25] | 2005 | Japan | Asian | HB | Direct | 212/346 | 0.549317592 | 7 | English |
| Pan [24] | 2014 | China | Asian | HB | SBE | 308/308 | 0.715713139 | 7 | English |
| Qadri [23] | 2014 | India | Asian | PB | PCR-CTPP | 130/200 | 0.066109789 | 8 | English |
| Ramis [22] | 2015 | Brazil | European | PB | PCR-RFLP | 9/38 | 0.691258974 | 8 | English |
| Savage [21] | 2004 | China | Asian | PB | SBE | 88/429 | 0.884813104 | 8 | English |
| Savage [20] | 2006 | Poland | European | PB | Taqman or MGB Eclipse | 287/428 | 0.391465014 | 8 | English |
| Shirai [19] | 2006 | Japan | Asian | HB | PCR-RFLP | 181/468 | 0.830460367 | 7 | English |
| Song [18] | 2009 | China | Asian | HB | PCR-RFLP | 125/140 | 0.720157181 | 7 | English |
| Taguchi [17] | 2005 | Japan | Asian | HB | PCR-RFLP | 396/252 | 0.994013656 | 7 | English |
| Vinagre [16] | 2011 | Brazil | European | HB | PCR-RFLP | 102/103 | 0.150502334 | 7 | English |
| Ye [15] | 2009 | Korea | Asian | HB | PCR-RFLP | 153/206 | 0.552934095 | 7 | English |
| Zeng [14] | 2005 | China | Asian | PB | PCR-RDB | 206/196 | 0.02187681 | 8 | Chinese |
| Zhang [13] | 2010 | China | Asian | PB | PCR-RFLP | 519/504 | 0.75413968 | 8 | English |
HWE: Hardy-Weinberg equilibrium; PB: population based; HB: hospital-based; PCR: polymerase chain reaction; RFLP: restriction fragment length polymorphism; SBE, single base extension; PCR-RDB, polymerase chain reaction-reverse dot blot. DHPLC: PCR-based denaturing high-performance liquid chromatography; Direct: Direct sequence analysis of polymerase chain reaction; ASO: oligonucleotide allele specific polymerase chain reaction; MGB Eclipse: MGB Eclipse Assay polymerase chain reaction method; ARMS: Amplification refractory mutation system polymerase chain reaction; Snapshot: the Snapshot assay which provides detection of certain SNPs. CTPP: confronting two-pair primers. The quality of studies included in this meta-analysis was assessed using Newcastle-Ottawa scale, which graded the quality of a study from 0 to 10 points. Articles exceeding 6 points were considered as high quality.
Meta-analysis results
Table 2 shows the interleukin-8-251A/T polymorphisms genotype distribution and allele frequency in cases and controls. The main results of this meta-analysis were listed in Table 3. There were 26 studies with 5286 cases and 8000 controls for Interleukin-8-251A/T polymorphism. As shown in Table 3, The pooled results showed that there was significant association between Interleukin-8-251A/T polymorphism and GC risk in any genetic model: Allele model (A vs. T: OR=1.16, 95% CI=1.05-1.27, P=0.002), dominant model (AA + AT vs. TT: OR=1.22, 95% CI=1.07-1.39, P=0.003) recessive model (AA vs. AT + TT: OR=1.20, 95% CI=1.01-1.41, P=0.03) homozygous genetic model (AA vs. TT: OR=1.31, 95% CI=1.08-1.59, P=0.006) heterozygote comparison (AT vs. TT: OR=1.19, 95% CI=1.04-1.35, P=0.01). In the subgroup analysis by ethnicity, the effects remained in Asians (A vs. T: OR=1.23, 95% CI=1.10-1.37, P=0.0002; AA + AT vs. TT: OR=1.30, 95% CI=1.13-1.49, P=0.0002; AA vs. AT + TT: OR=1.33, 95% CI=1.07-1.64, P=0.009; AA vs. TT: OR=1.49, 95% CI=1.18-1.87, P=0.0009; AT vs. TT: OR=1.25, 95% CI=1.09-1.43, P=0.001). However, no genetic models reached statistical association in Europeans (Table 3).
Table 2.
Interleukin-8-251A/T polymorphisms genotype distribution and allele frequency in cases and controls
| First author | Genotype (N) | Allele frequency (N) | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
| ||||||||||||
| Case | Control | Case | Control | |||||||||
|
| ||||||||||||
| Total | AA | AT | TT | Total | AA | AT | TT | A | T | A | T | |
| Bo | 208 | 36 | 108 | 64 | 190 | 26 | 96 | 68 | 180 | 236 | 148 | 232 |
| Canedo | 333 | 53 | 169 | 111 | 693 | 137 | 353 | 203 | 275 | 391 | 627 | 759 |
| Crusius | 236 | 48 | 113 | 75 | 1139 | 250 | 574 | 315 | 209 | 263 | 1074 | 1204 |
| de Oliveira | 207 | 47 | 98 | 62 | 240 | 45 | 134 | 61 | 192 | 222 | 224 | 256 |
| Felipe | 104 | 15 | 58 | 31 | 196 | 52 | 85 | 59 | 88 | 120 | 189 | 203 |
| Garza-Gonzalez | 78 | 16 | 47 | 15 | 189 | 33 | 87 | 69 | 79 | 77 | 153 | 225 |
| Kamali-Sarvestani | 19 | 9 | 6 | 4 | 153 | 22 | 74 | 57 | 24 | 14 | 118 | 188 |
| Kamangar | 112 | 14 | 56 | 42 | 207 | 24 | 111 | 72 | 84 | 140 | 159 | 255 |
| Kang | 334 | 49 | 159 | 126 | 322 | 27 | 148 | 147 | 257 | 411 | 202 | 442 |
| Ko | 81 | 12 | 35 | 34 | 308 | 27 | 146 | 135 | 59 | 103 | 200 | 416 |
| Lee | 470 | 59 | 213 | 198 | 308 | 62 | 138 | 108 | 331 | 609 | 262 | 354 |
| Liu | 138 | 23 | 89 | 26 | 137 | 15 | 72 | 50 | 135 | 141 | 102 | 172 |
| Lu | 250 | 54 | 102 | 94 | 300 | 37 | 144 | 119 | 210 | 290 | 218 | 382 |
| Ohyauchi | 212 | 13 | 106 | 93 | 346 | 20 | 118 | 208 | 132 | 292 | 158 | 534 |
| Pan | 308 | 48 | 168 | 92 | 308 | 59 | 148 | 101 | 264 | 352 | 266 | 350 |
| Qadri | 130 | 12 | 68 | 50 | 200 | 12 | 94 | 94 | 92 | 168 | 118 | 282 |
| Ramis | 9 | 4 | 1 | 4 | 38 | 7 | 20 | 11 | 9 | 9 | 34 | 42 |
| Savage | 88 | 23 | 39 | 26 | 429 | 75 | 207 | 147 | 85 | 91 | 357 | 501 |
| Savage | 287 | 76 | 140 | 71 | 428 | 117 | 205 | 106 | 292 | 282 | 439 | 417 |
| Shirai | 181 | 20 | 78 | 83 | 468 | 49 | 208 | 211 | 118 | 244 | 306 | 630 |
| Song | 125 | 20 | 72 | 33 | 140 | 23 | 70 | 47 | 112 | 138 | 116 | 164 |
| Taguchi | 396 | 44 | 191 | 161 | 252 | 22 | 105 | 125 | 279 | 513 | 149 | 355 |
| Vinagre | 102 | 25 | 56 | 21 | 103 | 19 | 42 | 42 | 106 | 98 | 80 | 126 |
| Ye | 153 | 17 | 82 | 54 | 206 | 23 | 86 | 97 | 116 | 190 | 132 | 280 |
| Zeng | 206 | 59 | 110 | 37 | 196 | 39 | 114 | 43 | 228 | 184 | 192 | 200 |
| Zhang | 519 | 128 | 261 | 130 | 504 | 93 | 251 | 160 | 517 | 521 | 437 | 571 |
Table 3.
Meta-analysis results
| Subgroup | OR | 95% CI | P value | Heterogeneity | Effects model | ||
|---|---|---|---|---|---|---|---|
|
| |||||||
| I2 | P value | ||||||
| Allele model A vs. T | |||||||
| Overall | 1.16 | 1.05-1.27 | 0.002 | 66% | P<0.00001 | R | |
| Ethnicity | European | 1.01 | 0.87-1.17 | 0.90 | 54% | 0.03 | R |
| Asian | 1.23 | 1.10-1.37 | 0.0002 | 61% | 0.0005 | R | |
| Source of controls | PB | 1.10 | 0.98-1.24 | 0.09 | 58% | 0.004 | R |
| HB | 1.23 | 1.05-1.43 | 0.01 | 72% | P<0.0001 | R | |
| Genotyping method | PCR-RFLP | 1.13 | 0.98-1.30 | 0.10 | 68% | 0.0003 | R |
| Non-RFLP | 1.18 | 1.04-1.35 | 0.01 | 66% | 0.0002 | R | |
| Dominant model AA + AT vs. TT | |||||||
| Overall | 1.22 | 1.07-1.39 | 0.003 | 61% | P<0.0001 | R | |
| Ethnicity | European | 1.05 | 0.81-1.35 | 0.71 | 64% | 0.005 | R |
| Asian | 1.30 | 1.13-1.49 | 0.0002 | 49% | 0.01 | R | |
| Source of controls | PB | 1.09 | 0.98-1.21 | 0.13 | 27% | 0.17 | F |
| HB | 1.40 | 1.12-1.76 | 0.004 | 73% | P<0.0001 | R | |
| Genotyping method | PCR-RFLP | 1.20 | 1.00-1.46 | 0.06 | 62% | 0.002 | R |
| Non-RFLP | 1.23 | 1.02-1.49 | 0.03 | 63% | 0.0007 | R | |
| Recessive model AA vs. AT + TT | |||||||
| Overall | 1.20 | 1.01-1.41 | 0.03 | 60% | P<0.0001 | R | |
| Ethnicity | European | 0.94 | 0.80-1.10 | 0.43 | 38% | 0.12 | F |
| Asian | 1.33 | 1.07-1.64 | 0.009 | 61% | 0.0005 | R | |
| Source of controls | PB | 1.25 | 0.99-1.58 | 0.06 | 65% | 0.0005 | R |
| HB | 1.14 | 0.90-1.44 | 0.29 | 54% | 0.01 | R | |
| Genotyping method | PCR-RFLP | 1.12 | 0.87-1.45 | 0.39 | 63% | 0.002 | R |
| Non-RFLP | 1.27 | 1.01-1.58 | 0.04 | 60% | 0.002 | R | |
| Homozygous genetic model AA vs. TT | |||||||
| Overall | 1.31 | 1.08-1.59 | 0.006 | 63% | P<0.00001 | R | |
| Ethnicity | European | 1.01 | 0.76-1.34 | 0.97 | 50% | 0.04 | R |
| Asian | 1.49 | 1.18-1.87 | 0.0009 | 60% | 0.0008 | R | |
| Source of controls | PB | 1.27 | 0.98-1.64 | 0.07 | 63% | 0.001 | R |
| HB | 1.38 | 1.02-1.88 | 0.04 | 66% | 0.0005 | R | |
| Genotyping method | PCR-RFLP | 1.24 | 0.91-1.67 | 0.17 | 67% | 0.0005 | R |
| Non-RFLP | 1.38 | 1.06-1.80 | 0.02 | 62% | 0.001 | R | |
| Heterozygote comparison AT vs. TT | |||||||
| Overall | 1.19 | 1.04-1.35 | 0.01 | 57% | 0.0002 | R | |
| Ethnicity | European | 1.07 | 0.81-1.42 | 0.63 | 67% | 0.002 | R |
| Asian | 1.25 | 1.09-1.43 | 0.001 | 41% | 0.04 | R | |
| Source of controls | PB | 1.04 | 0.93-1.16 | 0.54 | 5% | 0.39 | F |
| HB | 1.39 | 1.11-1.74 | 0.004 | 68% | 0.0002 | R | |
| Genotyping method | PCR-RFLP | 1.20 | 1.00-1.45 | 0.05 | 55% | 0.01 | R |
| Non-RFLP | 1.18 | 0.97-1.42 | 0.09 | 60% | 0.002 | R | |
F-fixed effects model; R-random effects model.
The subgroup analysis stratified by Source of controls showed an increased breast cancer risk in hospital-based (HB) studies in any genetic model except recessive model (A vs. T: OR=1.23, 95% CI=1.05-1.43, P=0.01; AA + AT vs. TT: OR=1.40, 95% CI=1.12-1.76, P=0.004; AA vs. TT: OR=1.38, 95% CI=1.02-1.88, P=0.04; AT vs. TT: OR=1.39, 95% CI=1.11-1.74, P=0.004). However, there was no association in any genetic model in population based (PB) studies (Table 3).
When stratifying by Genotyping method, we found statistical association in Non-RFLP (restriction fragment length polymorphism) in any genetic model except heterozygote comparison (A vs. T: OR=1.18, 95% CI=1.04-1.35, P=0.01; AA + AT vs. TT: OR=1.23, 95% CI=1.02-1.49, P=0.03; AA vs. AT + TT: OR=1.27, 95% CI=1.01-1.58, P=0.04; AA vs. TT: OR=1.38, 95% CI=1.06-1.80, P=0.02), the effect was remain in PCR-RFLP in dominant model and heterozygote comparison (AA + AT vs. TT: OR=1.20, 95% CI=1.00-1.46, P=0.06; AT vs. TT: OR=1.20, 95% CI=1.00-1.45, P=0.05).
Sensitivity analyses
As shown in Table 1, all the studies conformed to he balance of Hardy-Weinberg equilibrium (HWE) in controls except Zeng’s (P<0.05), however, after performing the sensitivity analyses, The overall results did not show quantitative changes when excluding any study, suggesting the stability and reliability of this meta-analysis.
Detection for heterogeneity
Statistically significant heterogeneity was observed between trials of the following analyses using Q statistic: allele model (A vs. T: P<0.00001, I2=66%), the dominant model (AA + AT vs. TT: P<0.0001, I2=61%), the recessive model (AA vs. AT + TT: P<0.0001, I2=60%), the homozygous genetic model (AA vs. TT: P<0.00001, I2=63%), and the heterozygous genetic model (AT vs. TT: P=0.0002, I2=57%), and the random-effects model was performed in these studies.
Publication bias
Begg’s funnel plot and Egger’s test were employed to assess the publication bias. As shown in Figure 2, the funnel plots failed to reveal any obvious asymmetry in all genotypes in overall population. Neither Begg’s test nor Egger’s test showed statistical evidence for publication bias in our meta-analysis (P>0.05).
Figure 2.

Funnel plot assessing evidence of publication bias from 26 studies (A vs. T). Abbreviations: SE, standard error; OR, odds ratio; A vs. G, Allele model.
Discussion
A large amount of evidence suggests that genetics is important in determining the risk of cancer. Related research is to search for the susceptibility genes associated with cancer [46]. It is believed that single nucleotide polymorphism is the most common source of human genetic variation, which may contribute to the susceptibility of individuals to cancer [38-41,47]. In recent years, genetic susceptibility to cancer has caused people’s great interest, and the study on the genetic polymorphism of the tumor is increasing.
Recently, a growing number of epidemiological studies have been performed to assess the association of Interleukin-8-251A/T polymorphisms with GC risk [12-37]. However, the results are conflicting. Thus, we conducted a comprehensive meta-analysis involving published data, to assess the strength of association between the polymorphisms and GC risk.
In this present meta-analysis, 26 studies with 5286 cases and 8000 controls were included. And we explored the association between the potentially functional polymorphisms of Interleukin-8-251A/T and GC risk. In the overall population, the pooled results showed that there was significant association between Interleukin-8-251A/T polymorphism and GC risk in any genetic model: Allele model (A vs. T: OR=1.16, 95% CI=1.05-1.27, P=0.002), dominant model (AA + AT vs. TT: OR=1.22, 95% CI=1.07-1.39, P=0.003) recessive model (AA vs. AT + TT: OR=1.20, 95% CI=1.01-1.41, P=0.03) homozygous genetic model (AA vs. TT: OR=1.31, 95% CI=1.08-1.59, P=0.006) heterozygote comparison (AT vs. TT: OR=1.19, 95% CI=1.04-1.35, P=0.01). In a previous meta-analysis by Wang et al. [39], they failed to find association between Interleukin-8-251A/T polymorphism and gastric cancer susceptibility. This contradiction may result from different sample size and ethnic groups.
In the subgroup analysis by ethnicity, the effects remained in Asians (A vs. T: OR=1.23, 95% CI=1.10-1.37, P=0.0002; AA + AT vs. TT: OR=1.30, 95% CI=1.13-1.49, P=0.0002; AA vs. AT + TT: OR=1.33, 95% CI=1.07-1.64, P=0.009; AA vs. TT: OR=1.49, 95% CI=1.18-1.87, P=0.0009; AT vs. TT: OR=1.25, 95% CI=1.09-1.43, P=0.001). However, no genetic models reached statistical association in Europeans (Table 3). It was partially in line with the results of Cheng (2013)’s [41], Xue (2012)’s [38] and Wang (2012)’s [40] finding. However, there were only 18 studies respectively in their meta-analysis, while 26 studies were involved in our meta-analysis.
The subgroup analysis stratified by Source of controls showed an increased breast cancer risk in hospital-based (HB) studies in any genetic model except recessive model (A vs. T: OR=1.23, 95% CI=1.05-1.43, P=0.01; AA + AT vs. TT: OR=1.40, 95% CI=1.12-1.76, P=0.004; AA vs. TT: OR=1.38, 95% CI=1.02-1.88, P=0.04; AT vs. TT: OR=1.39, 95% CI=1.11-1.74, P=0.004). However, there was no association in any genetic model in population based (PB) studies (Table 3). When stratifying by Genotyping method, we found statistical association in Non-RFLP (restriction fragment length polymorphism) in any genetic model except heterozygote comparison (A vs. T: OR=1.18, 95% CI=1.04-1.35, P=0.01; AA + AT vs. TT: OR=1.23, 95% CI=1.02-1.49, P=0.03; AA vs. AT + TT: OR=1.27, 95% CI=1.01-1.58, P=0.04; AA vs. TT: OR=1.38, 95% CI=1.06-1.80, P=0.02), the effect was remain in PCR-RFLP in dominant model and heterozygote comparison (AA + AT vs. TT: OR=1.20, 95% CI=1.00-1.46, P=0.06; AT vs. TT: OR=1.20, 95% CI=1.00-1.45, P=0.05). Above findings was reported first by the paper.
There are some limitations in this meta-analysis. Firstly, this meta-analysis was based on pooled data. We could not assess the risk of cancer according to stratification of age, Sex and H. pylori infection, smoking, alcohol consumption, environment factors, and other risk factors. Secondly, we just included the published studies in the meta-analysis. It is possible that we missed some related unpublished studies that might meet the inclusion criteria. Moreover, small study effect, in which effects reported in small studies are larger, could not be avoided in that some studies were of a relative small size. Further large scale multicenter studies are warranted to further validate on Interleukin-8-251A/T polymorphisms and GC risk.
Conclusions
In conclusion, this meta-analysis suggests that Interleukin-8-251A/T polymorphism is a risk factor for susceptibility to GC in overall population, especially in Asians, in hospital populations and in Non-RFLP. While, there was no association in Europeans and in general population. Further large scale multicenter epidemiological studies are warranted to confirm this finding.
Disclosure of conflict of interest
None.
References
- 1.Nagini S. Carcinoma of the stomach: A review of epidemiology, pathogenesis, molecular genetics and chemoprevention. World J Gastrointest Oncol. 2012;4:156–169. doi: 10.4251/wjgo.v4.i7.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 3.Nicolas C, Sylvain M, Come L, Jean F, Anne-Marie B, Valerie J. Trends in gastric cancer incidence: a period and birth cohort analysis in a well-defined French population. Gastric Cancer. 2015 doi: 10.1007/s10120-015-0509-9. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 4.Morais S, Ferro A, Bastos A, Castro C, Lunet N, Peleteiro B. Trends in gastric cancer mortality and in the prevalence of Helicobacter pylori infection in Portugal. Eur J Cancer Prev. 2015 doi: 10.1097/CEJ.0000000000000183. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 5.Kim HS, Lee H, Jeung HC, Noh SH, Chung HC, Roh JK, Nam CM, Rha SY. Advanced detection of recent changing trends in gastric cancer survival: up-to-date comparison by period analysis. Jpn J Clin Oncol. 2011;41:1344–1350. doi: 10.1093/jjco/hyr153. [DOI] [PubMed] [Google Scholar]
- 6.Shimizu Y, Kondo S, Shirai A, Furukawa M, Yoshizaki T. A single nucleotide polymorphism in the matrix metalloproteinase-1 and interleukin-8 gene promoter predicts poor prognosis in tongue cancer. Auris Nasus Larynx. 2008;35:381–389. doi: 10.1016/j.anl.2007.12.002. [DOI] [PubMed] [Google Scholar]
- 7.Xue H, Liu J, Lin B, Wang Z, Sun J, Huang G. A meta-analysis of interleukin-8 -251 promoter polymorphism associated with gastric cancer risk. PLoS One. 2012;7:e28083. doi: 10.1371/journal.pone.0028083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Choe JY, Park KY, Lee SJ, Park SH, Kim SK. Rebamipide inhibits tumor necrosis factor-alpha-induced interleukin-8 expression by suppressing the NF-kappaB signal pathway in human umbilical vein endothelial cells. Inflamm Res. 2010;59:1019–1026. doi: 10.1007/s00011-010-0221-5. [DOI] [PubMed] [Google Scholar]
- 9.Jain M, LoGerfo FW, Guthrie P, Pradhan L. Effect of hyperglycemia and neuropeptides on interleukin-8 expression and angiogenesis in dermal microvascular endothelial cells. J Vasc Surg. 2011;53:1654–1660. e1652. doi: 10.1016/j.jvs.2011.02.019. [DOI] [PubMed] [Google Scholar]
- 10.Xie K. Interleukin-8 and human cancer biology. Cytokine Growth Factor Rev. 2001;12:375–391. doi: 10.1016/s1359-6101(01)00016-8. [DOI] [PubMed] [Google Scholar]
- 11.Simeonova PP, Luster MI. Asbestos induction of nuclear transcription factors and interleukin 8 gene regulation. Am J Respir Cell Mol Biol. 1996;15:787–795. doi: 10.1165/ajrcmb.15.6.8969274. [DOI] [PubMed] [Google Scholar]
- 12.Liu J, Xing C, Sun L, Gong Y, Bai X, Zhang Y, Chen W, Yuan Y. The relationship of interleukin-8-251 with gastric cancer and precancerous lesion. Chin J Dig Endoscopy. 2009;26:310–312. [Google Scholar]
- 13.Zhang L, Du C, Guo X, Yuan L, Niu W, Yu W, Er L, Wang S. Interleukin-8-251A/T polymorphism and Helicobacter pylori infection influence risk for the development of gastric cardiac adenocarcinoma in a high-incidence area of China. Mol Biol Rep. 2010;37:3983–3989. doi: 10.1007/s11033-010-0057-7. [DOI] [PubMed] [Google Scholar]
- 14.Zeng Z, Zhou S, Liao S, Chen B, Li C, Chen F, Hu P. Correlation of Polymorphism of Interleukin 8 Gene-251 Locus and Gastric Cancer in High and Low Prevalence Regions in China. Journal of Sun Yat-sen University Med Sci. 2005;26:537–540. [Google Scholar]
- 15.Ye BD, Kim SG, Park JH, Kim JS, Jung HC, Song IS. The Interleukin-8-251 A Allele is Associated With Increased Risk of Noncardia Gastric Adenocarcinoma in Helicobacter pyloriinfected Koreans. J Clin Gastroenterol. 2009;43:233–239. doi: 10.1097/MCG.0b013e3181646701. [DOI] [PubMed] [Google Scholar]
- 16.Vinagre RM, Corvelo TC, Arnaud VC, Leite AC, Barile KA, Martins LC. Determination of strains of Helicobacter pylori and of polymorphism in the interleukin-8 gene in patients with stomach cancer. Arq Gastroenterol. 2011;48:46–51. doi: 10.1590/s0004-28032011000100010. [DOI] [PubMed] [Google Scholar]
- 17.Taguchi A, Ohmiya N, Shirai K, Mabuchi N, Itoh A, Hirooka Y, Niwa Y, Goto H. Interleukin-8 promoter polymorphism increases the risk of atrophic gastritis and gastric cancer in Japan. Cancer Epidemiol Biomarkers Prev. 2005;14:2487–2493. doi: 10.1158/1055-9965.EPI-05-0326. [DOI] [PubMed] [Google Scholar]
- 18.Song B, Zhang D, Wang S, Zheng H, Wang X. Association of Interleukin-8 with Cachexia from Patients with Low-Third Gastric Cancer. Comp Funct Genomics. 2009:212345. doi: 10.1155/2009/212345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shirai K, Ohmiya N, Taguchi A, Mabuchi N, Yatsuya H, Itoh A, Hirooka Y, Niwa Y, Mori N, Goto H. Interieukin-8 gene polymorphism associated with susceptibility to non-cardia gastric carcinoma with microsatellite instability. J Gastroenterol Hepatol. 2006;21:1129–1135. doi: 10.1111/j.1440-1746.2006.04443.x. [DOI] [PubMed] [Google Scholar]
- 20.Savage SA, Hou LF, Lissowska J, Chow WH, Zatonski W, Chanock SJ, Yeager M. Interleukin-8 polymorphisms are not associated with gastric cancer risk in a Polish population. Cancer Epidemiol Biomarkers Prev. 2006;15:589–591. doi: 10.1158/1055-9965.EPI-05-0887. [DOI] [PubMed] [Google Scholar]
- 21.Savage SA, Abnet CC, Mark SD, Qiao YL, Dong ZW, Dawsey SM, Taylor PR, Chanock SJ. Variants of the IL8 and IL8RB genes and risk for gastric cardia adenocarcinoma and esophageal squamous cell carcinoma. Cancer Epidemiol Biomarkers Prev. 2004;13:2251–2257. [PubMed] [Google Scholar]
- 22.Ramis IB, Vianna JS, Goncalves CV, von Groll A, Dellagostin OA, da Silva PE. Polymorphisms of the IL-6, IL-8 and IL-10 genes and the risk of gastric pathology in patients infected with Helicobacter pylori. J Microbiol Immunol Infect. 2015 doi: 10.1016/j.jmii.2015.03.002. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 23.Qadri Q, Rasool R, Afroze D, Naqash S, Gulzar GM, Yousuf A, Siddiqi MA, Shah ZA. Study of TLR4 and IL-8 Gene Polymorphisms in H-pylori-Induced Inflammation in Gastric Cancer in an Ethnic Kashmiri Population. Immunol Invest. 2014;43:324–336. doi: 10.3109/08820139.2013.854378. [DOI] [PubMed] [Google Scholar]
- 24.Pan XF, Wen Y, Loh M, Wen YY, Yang SJ, Zhao ZM, Tian Z, Huang H, Lan H, Chen F, Soong R, Yang CX. Interleukin-4 and -8 gene polymorphisms and risk of gastric cancer in a population in Southwestern China. Asian Pac J Cancer Prev. 2014;15:2951–2957. doi: 10.7314/apjcp.2014.15.7.2951. [DOI] [PubMed] [Google Scholar]
- 25.Ohyauchi M, Imatani A, Yonechi M, Asano N, Miura A, Iijima K, Koike T, Sekine H, Ohara S, Shimosegawa T. The polymorphism interleukin 8-251 A/T influences the susceptibility of Helicobacter pylori related gastric diseases in the Japanese population. Gut. 2005;54:330–335. doi: 10.1136/gut.2003.033050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lu WL, Pan KF, Zhang L, Lin DX, Miao XP, You WC. Genetic polymorphisms of interleukin (IL)-1B, IL-1RN, IL-8, IL-10 and tumor necrosis factor alpha and risk of gastric cancer in a Chinese population. Carcinogenesis. 2005;26:631–636. doi: 10.1093/carcin/bgh349. [DOI] [PubMed] [Google Scholar]
- 27.Lee WP, Tai DI, Lan KH, Li AF, Hsu HC, Lin EJ, Lin YP, Sheu ML, Li CP, Chang FY, Chao Y, Yen SH, Lee SD. The -251T allele of the interleukin-8 promoter is associated with increased risk of gastric carcinoma featuring diffusetype histopathology in Chinese population. Clin Cancer Res. 2005;11:6431–6441. doi: 10.1158/1078-0432.CCR-05-0942. [DOI] [PubMed] [Google Scholar]
- 28.Ko KP, Park SK, Cho LY, Gwack J, Yang JJ, Shin A, Kim CS, Kim Y, Kang D, Chang SH, Shi HR, Yoo KY. Soybean Product Intake Modifies the Association between Interleukin-10 Genetic Polymorphisms and Gastric Cancer Risk. J Nutr. 2009;139:1008–1012. doi: 10.3945/jn.108.101865. [DOI] [PubMed] [Google Scholar]
- 29.Kang JM, Kim N, Lee DH, Park JH, Lee MK, Kim JS, Jung HC, Song IS. The Effects of Genetic Polymorphisms of IL-6, IL-8, and IL-10 on Helicobacter pylori-induced Gastroduodenal Diseases in Korea. J Clin Gastroenterol. 2009;43:420–428. doi: 10.1097/MCG.0b013e318178d1d3. [DOI] [PubMed] [Google Scholar]
- 30.Kamangar F, Abnet CC, Hutchinson AA, Newschaffer CJ, Helzlsouer K, Shugart YY, Pietinen P, Dawsey SM, Albanes D, Virtamo J, Taylor PR. Polymorphisms in inflammation-related genes and risk of gastric cancer (Finland) Cancer Causes Control. 2006;17:117–125. doi: 10.1007/s10552-005-0439-7. [DOI] [PubMed] [Google Scholar]
- 31.Kamali-Sarvestani E, Bazargani A, Masoudian M, Lankarani K, Taghavi AR, Saberifiroozi M. Association of H pylori cagA and vacA genotypes and IL-8 gene polymorphisms with clinical outcome of infection in Iranian patients with gastrointestinal diseases. World J Gastroenterol. 2006;12:5205–5210. doi: 10.3748/wjg.v12.i32.5205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Garza-Gonzalez E, Bosques-Padilla FJ, Mendoza-Ibarra SI, Flores-Gutierrez JP, Maldonado-Garza HJ, Perez-Perez GI. Assessment of the toll-like receptor 4 Asp299Gly, Thr399Ile and interleukin-8-251 polymorphisms in the risk for the development of distal gastric cancer. BMC Cancer. 2007;7:70. doi: 10.1186/1471-2407-7-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Felipe AV, Silva TD, Pimenta CA, Kassab P, Forones NM. Interleukin-8 gene polymorphism and susceptibility to gastric cancer in a brazilian population. Biol Res. 2012;45:369–374. doi: 10.4067/S0716-97602012000400007. [DOI] [PubMed] [Google Scholar]
- 34.de Oliveira JG, Rossi AF, Nizato DM, Cadamuro AC, Jorge YC, Valsechi MC, Venâncio LP, Rahal P, Pavarino ÉC, Goloni-Bertollo EM, Silva AE. Influence of functional polymorphisms in TNF-(alpha), IL-8, and IL-10 cytokine genes on mRNA expression levels and risk of gastric cancer. Tumor Biol. 2015 doi: 10.1007/s13277-015-3593-x. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 35.Crusius JBA, Canzian F, Capella G, Pena AS, Pera G, Sala N, Agudo A, Rico F, Del Giudice G, Palli D, Plebani M, Boeing H, Bueno-de-Mesquita HB, Carneiro F, Pala V, Save VE, Vineis P, Tumino R, Panico S, Berglund G, Manjer J, Stenling R, Hallmans G, Martinez C, Dorronsoro M, Barricarte A, Navarro C, Quiros JR, Allen N, Key TJ, Binghan S, Caldas C, Linseisen J, Kaaks R, Overvad K, Tjonneland A, Buechner FC, Peeters PHM, Numans ME, Clavel-Chapelon F, Trichopoulou A, Lund E, Jenab M, Rinaldi S, Ferrari P, Riboli E, Gonzalez CA. Cytokine gene polymorphisms and the risk of adenocarcinoma of the stomach in the European prospective investigation into cancer and nutrition (EPIC-EURGAST) Ann Oncol. 2008;19:1894–1902. doi: 10.1093/annonc/mdn400. [DOI] [PubMed] [Google Scholar]
- 36.Canedo P, Castanheira-Vale AJ, Lunet N, Pereira F, Figueiredo C, Gioia-Patricola L, Canzian F, Moreira H, Suriano G, Barros H, Carneiro F, Seruca R, Machado JC. The interieukin-8-251*T/*A polymorphism is not associated with risk for gastric carcinoma development in a Portuguese population. Eur J Cancer Prev. 2008;17:28–32. doi: 10.1097/CEJ.0b013e32809b4d0f. [DOI] [PubMed] [Google Scholar]
- 37.Bo S, Dianliang Z, Hongmei Z, Xinxiang W, Yanbing Z, Xiaobo L. Association of interleukin-8 gene polymorphism with cachexia from patients with gastric cancer. J Interferon Cytokine Res. 2010;30:9–14. doi: 10.1089/jir.2009.0007. [DOI] [PubMed] [Google Scholar]
- 38.Xue H, Liu J, Lin B, Wang Z, Sun J, Huang G. A Meta-Analysis of Interleukin-8-251 Promoter Polymorphism Associated with Gastric Cancer Risk. PLoS One. 2012;7:e28083. doi: 10.1371/journal.pone.0028083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang Z, Liu Y, Yang L, Yin S, Zang R, Yang G. The polymorphism interleukin-8-251A/T is associated with a significantly increased risk of cancers from a meta-analysis. Tumour Biol. 2014;35:7115–7123. doi: 10.1007/s13277-014-1881-5. [DOI] [PubMed] [Google Scholar]
- 40.Wang N, Zhou R, Wang C, Guo X, Chen Z, Yang S, Li Y. 2251 T/A polymorphism of the interleukin-8 gene and cancer risk: A HuGE review and meta-analysis based on 42 case-control studies. Mol Biol Rep. 2012;39:2831–2841. doi: 10.1007/s11033-011-1042-5. [DOI] [PubMed] [Google Scholar]
- 41.Cheng D, Hao Y, Zhou W, Ma Y. Positive association between Interleukin-8 -251A > T polymorphism and susceptibility to gastric carcinogenesis: a meta-analysis (Provisional abstract) Cancer Cell Int. 2013;13:100. doi: 10.1186/1475-2867-13-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mantel N, Haenszel W. Statistical aspects of the analysis of data from retrospective studies of disease. J Natl Cancer Inst. 1959;22:719–748. [PubMed] [Google Scholar]
- 43.DerSimonian R, Laird N. Meta-analysis in clinical trials. Control Clin Trials. 1986;7:177–188. doi: 10.1016/0197-2456(86)90046-2. [DOI] [PubMed] [Google Scholar]
- 44.Begg CB, Mazumdar M. Operating characteristics of a rank correlation test for publication bias. Biometrics. 1994;50:1088–1101. [PubMed] [Google Scholar]
- 45.Egger M, Davey Smith G, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ. 1997;315:629–634. doi: 10.1136/bmj.315.7109.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Risch N, Merikangas K. The future of genetic studies of complex human diseases. Science. 1996;273:1516–1517. doi: 10.1126/science.273.5281.1516. [DOI] [PubMed] [Google Scholar]
- 47.Eftang LL, Esbensen Y, Tannaes TM, Bukholm IRK, Bukholm G. Interleukin-8 is the single most up-regulated gene in whole genome profiling of H. pylori exposed gastric epithelial cells. BMC Microbiol. 2012;12:9. doi: 10.1186/1471-2180-12-9. [DOI] [PMC free article] [PubMed] [Google Scholar]


