Abstract
This study aimed to determine the connection between polymorphisms of kallikrein kinin system including KLK1 (rs5516), KNG1 (rs710446, rs2304456) and ACE (rs4291, rs4309, rs4343) and late-onset Alzheimer’s disease (LOAD). The research was conducted as a case-control study, comprising 201 AD patients in the AD group, and 257 healthy subjects as the control group. PCR amplification and matrix-assisted laser desorption-ionization time-of-flight mass spectrometry (MALDI-TOF MS) were used to detect the six polymorphisms (rs5516 in KLK1; rs710446, rs2304456 in KNG1; rs4291, rs4309, rs4343 in ACE) from both groups. No statistically significant difference was found between the genotype and allelotype distributions of rs5516, rs710446, rs2304456, rs4291 and rs4343 (P>0.05). The differences between the genotype and allelotype distributions of the rs4309 were statistically significant (P<0.05). Haplotype analysis confirmed the existence of three haplotypes (AG, AT, GT) composed of rs710446/rs2304456, and six haplotypes (ATA, ACA, TCA, TCG, TTA, TTG) composed of rs4291/rs4309/rs4343, among which the distribution of ATA, ACA, TCA between the two groups was statistically significant difference (P<0.05). Our study showed that the polymorphisms of rs5516, rs710446, rs2304456, rs4291 and rs4343 is not related to the incidence of LOAD. The polymorphisms of rs4309 may be related to LOAD, as well as ATA, ACA, and TCA haplotype composed of rs4291/rs4309/rs4343.
Keywords: Alzheimer’s disease, kallikrein-kinin system, gene, polymorphism
Introduction
Alzheimer’s disease (AD) is a degenerative disease of the central nervous system with symptoms of progressive cognitive decline and memory impairment. The etiology and pathogenesis of AD are not completely determined, and are possibly correlated with multiple genetic and environmental factors. KKS is a very important regulatory system, consisting of a series of vasoactive peptides and enzymes, and plays an important function in the blood and tissues. In recent years, KKS has been discovered having the function in AD, ischemia re-perfusion injury and neurological protection, providing a new way for the treatment of neurological diseases. Research shows that the high molecular weight kininogen lysate in the cerebrospinal fluid of AD patients can activate kinin system [1]; there are a variety of tissue types of bradykinin in senile plaques in the brain of AD patients, and its synthesis was increased significantly [2]. The above findings suggest an association between KKS system and pathogenesis of AD.
It has been pointed out that the polymorphism of KLK1 gene rs5516 and KNG1 gene rs2304456 is associated with hypertension [3,4], and KNG1 gene rs710446 is related to coagulation and venous thrombosis [5,6]. The 2010 European Federation of Neurological Societies noted that hypertension is an independent risk factor for AD [7], but also the cause of AD progression. Hypertension can make 2.3 times increased risk of AD [8]. Given that the role of vascular mechanisms in the pathogenesis of AD is increasingly being confirmed, presumably these polymorphisms may also be involved in the pathogenesis of AD.
ACE may also take part in the multi-factor process of cognitive disorders [9]: studies showed that ACE activity in the cerebrospinal fluids of the AD and mild cognitive impairment patients is significantly higher than that of the control group, and is also linearly correlated to the Mini-Mental State Examination score of the patients, suggesting a possible role of ACE in the development of AD [10]. In animal AD cases, this increase could be suppressed by applying ACEI perindopril 2 d (1 mg·kg-1), which is capable of penetrating the blood-brain barrier and alleviating the cognitive impairment suffered by the AD mice [11], Besides, ACEI can improve the cognitive function of AD patients [12]. All of these point at a possible link between ACE and the occurrence of AD.
In this study, to examine the genetic background and pathogenesis of the disease at the genetic level, matrix-assisted laser desorption-ionization time-of-flight mass spectrometry (MALDI-TOF MS) technology was used to detect the six polymorphisms (rs5516 in KLK1; rs710446, rs2304456 in KNG1; rs4291, rs4309, rs4343 in ACE). The distribution of each genotype and its allelotype was analyzed in AD patients and in a healthy population to determine the correlation between these polymorphisms and the risk of developing AD. The results provide clues for the genetic diagnosis and preemptive treatment of AD.
Materials and methods
Subjects
Based on the probability of AD in the National Institute of Neurological and Communicative Disorders and Stroke and the Alzheimer’s disease and Related Disorders Association (NINCDS-ADRAD), we chose 201 patients confirmed by the neurology department of the 3rd Xiangya Hospital of Central South University between July 2009-July 2013 as the AD group. While those in the control group were healthy subjects receiving physical checks in the Health Management Center of the same hospital. Clinical data and peripheral blood samples were obtained from the members of both groups. The AD group comprised 201 members, including 90 men and 111 women, aged 76.79±5.65 years. AD group individuals had been affected with AD for 1-15 years, and had Mini-Mental State Examination scores of 15.36±3.48, Activities of Daily Living scores of 54.24±7.82. The 257 members of the control group included 121 men and 136 women, aged 75.88±6.50 years, of normal intellect. All subjects were Hunan Han Chinese, with no incidence of myocardial infarction, congestive heart failure, cerebral apoplexy, type 2 diabetes, atherosclerosis, or autoimmune disease. This study was approved by the Ethics Committee of the Third Xiangya Hospital, and informed consent was obtained by the subjects or their relatives.
Laboratory data collection
Fasting venous blood was separated into two 3-ml tubes. One was preserved by EDTA anticoagulant at -20°C for genomic DNA extraction, while the second was used to test for fasting blood glucose, TC, TG, LDL-c, and HDL-c.
DNA isolation and primer design
Genomic DNA was isolated using the Blood Gen Midi Kit (Kangwei Shiji, Beijing, China) according to the manufacturer’s instructions and stored at -80°C. PCR primers were designed using Assay Design 3.1 software (Sequenom Inc., San Diego, CA, USA), and synthesized by Beijing Gene-Cloud Biotechnology Co., Ltd.
SNP genotyping
The Sequenom MassARRAY system was used for SNP genotyping with a SpectroREADER matrix assisted laser desorption ionization time of flight mass spectrometer (Sequenom). PCR amplification reactions (5 μL) were performed by real-time quantitative PCR amplification (Eppendorf, Hamburg, Germany) in standard 384-well plates (ABI,) and included 10 ng genomic DNA, 0.5 μL 10 × PCR buffer, 0.4 μL MgCl2, 0.1 μL dNTP mix, 1 μL primer mix, 0.2 U HotStar Taq, and 1.8 μL water. PCR thermal cycling was carried out for 15 min at 94°C, followed by 45 cycles of 94°C for 20 s, 56°C for 30 s, 72°C for 1 min, and then 72°C for a final 3 min. Next, 0.5 U of shrimp alkaline phosphatase and 0.17 μL of SAP buffer were added to the PCR reaction products, incubated for 40 min at 37°C, then inactivated for 5 min at 85°C to remove free dNTP.
Single-base extension reactions were performed in a final volume of 9 μL including 7 μL of SAP+PCR reaction, 0.041 μL iPLEX enzyme (all iPLEX reagents from Sequenom), 0.94 µL iPLEX Extend Primer Mix, 0.2 μL iPLEX Buffer Plus, 0.2 μL of iPLEX Termination Mix, and 0.619 µL Nanopure™ water. The reactions consisted of 40 cycles of denaturation at 94°C and a final 3-min extension step at 72°C. The products were purified by Clean Resin (Sequenom) and were then spotted on a SpectroChip (Sequenom). Data were processed and analyzed by MassARRAY TYPER 4.0 software (Sequenom).
Statistical analysis
Statistical analysis was performed using the SPSS19.0 statistical package (IBM, Armonk, NJ, USA). SHEsis software (http://analysis.bio-x.cn/myAnalysis.php) was used for Hardy-Weinberg analysis, linkage disequilibrium, and haplotype analysis. Measurement data were compared using the Student’s t-test, while genotype and allele distributions between the two groups were compared with the χ2 test. All statistical tests were two-tailed and statistical significance was P<0.05.
Results
Comparison of clinical characteristics AD
Clinical characteristics of the study population are shown in Table 1. No significant differences were found between the AD and control groups in terms of sex, mean age, widowed status, body mass index (BMI), head trauma, hypertension, triglycerides (TG), low-density lipoprotein cholesterol (LDL-c), high-density lipoprotein cholesterol (HDL-c), or fasting blood glucose levels (P>0.05). However, the differences between the two groups with respect to level of education and total cholesterol (TC) were significant (P<0.05), indicating that a low level of education or high cholesterol are risk factors for AD.
Table 1.
Comparison of clinical characteristics in AD group and control group
| Index | AD group (n=201) | control group (n=257) | P |
|---|---|---|---|
| Sex (male/female) | 90/111 | 121/136 | 0.623 |
| Mean age (years) | 76.79±5.65 | 75.88±6.50 | 0.113 |
| Education (≤primary school/>primary school) | 117/84 | 120/137 | 0.014* |
| Widowed (yes/no) | 89/112 | 92/165 | 0.065 |
| head trauma (yes/no) | 8/193 | 11/246 | 0.873 |
| Hypertension (yes/no) | 62/139 | 65/192 | 0.188 |
| BMI (kg/m2) | 22.65±1.94 | 22.35±1.48 | 0.068 |
| TC (mmol/L) | 5.12±0.98 | 4.87±0.69 | 0.002* |
| TG (mmol/L) | 1.85±1.12 | 1.98±1.40 | 0.294 |
| LDL (mmol/L) | 2.60±1. 09 | 2.46±1.13 | 0.227 |
| HDL (mmol/L) | 1.32±0.36 | 1.33±0.77 | 0.722 |
| Fasting blood glucose (mmol/L) | 5.34±0.90 | 5.20±0.71 | 0.065 |
Compared with the control group, P<0.05.
Hardy-Weinberg analysis
Hardy-Weinberg analysis was conducted on the genotypes of rs5516, rs710446, rs2304456, rs4291, rs4309 and rs4291 in the AD and control groups, revealing Hardy-Weinberg equilibrium in both groups. Therefore, the selected samples are suitable to represent the population and for genetic analysis.
SNP distributions
The genotype and allele distributions of the SNPs in the AD and control groups are shown in Table 2. No significant differences were found in the genotype or allele of rs5516 distributions between the two groups (χ2=2.148, P=0.342; χ2=0.807, P=0.369, respectively). No significant differences were found in the genotype and allele distributions of rs710446 between the two groups (χ2=1.373, P=0.503; χ2=0.024, P=0.876, respectively). No significant differences were found in the genotype and allele distributions of rs2304456 between the two groups (χ2=1.286, P=0.526; χ2=1.043, P=0.307, respectively).
Table 2.
Distribution of SNPs in the AD group and the control group
| SNP ID | Allele1/2 | AD 1/2 | Control 1/2 | X2 | P | AD 11/12/22 | Control 11/12/22 | X2 | P |
|---|---|---|---|---|---|---|---|---|---|
| rs5516 | C/G | 310/92 | 409/105 | 0.807 | 0.369 | 123/64/14 | 162/85/10 | 2.148 | 0.342 |
| rs710446 | A/G | 271/131 | 344/170 | 0.024 | 0.876 | 88/95/18 | 117/110/30 | 1.373 | 0.503 |
| rs2304456 | G/T | 61/341 | 91/423 | 1.043 | 0.307 | 8/45/148 | 11/69/177 | 1.286 | 0.526 |
| rs4291 | A/T | 268/134 | 372/142 | 3.490 | 0.062 | 84/100/17 | 130/112/15 | 3.903 | 0.142 |
| rs4309 | C/T | 141/261 | 113/401 | 19.288 | <0.001 | 20/101/80 | 9/95/153 | 20.69 | <0.001 |
| rs4343 | A/G | 283/119 | 391/123 | 3.733 | 0.053 | 92/95/12 | 144/103/10 | 4.225 | 0.120 |
No significant differences were found in the genotype or allele of rs4291 distributions between the two groups (χ2=3.903, P=0.142; χ2=3.490, P=0.062, respectively). Similarly, no significant differences were found in the genotype and allele distributions of rs4343 between the two groups (χ2=4.225, P=0.120; χ2=3.733, P=0.053, respectively). The difference between the genotype and allele distributions of rs4309 in the AD group and the control group was statistically significant, with a noticeable increase in the C allele frequency being detected in the AD group (OR=1.917, 95% CI=1.431-2.568, P<0.05).
Mass array analysis of spectrometric peak charts
Spectrometric peak chart of rs5516 genotypes
The MassArray system detected CC, GC, and GG rs5516 genotypes in the AD and control groups as shown in the spectrometric peak chart (Figure 1).
Figure 1.

Spectrometric genotype-feature peak charts of rs5516 (CC, CG, GG).
Spectrometric peak chart of rs710446 genotypes
The MassArray system detected AA, AG, and GG rs710446 genotypes in the AD and control groups as shown in the spectrometric peak chart (Figure 2).
Figure 2.

Spectrometric genotype-feature peak charts of rs710446 (AA, AG, GG).
Spectrometric peak chart of rs2304456 genotypes
The MassArray system detected GG, GT, and TT rs2304456 genotypes in the AD and control groups as shown in the spectrometric peak chart (Figure 3).
Figure 3.

Spectrometric genotype-feature peak charts of rs2304456 (GG, GT, TT).
Spectrometric peak chart of rs4291 genotypes
The MassArray system detected AA, AT, and TT rs4291 genotypes in the AD and control groups as shown in the spectrometric peak chart (Figure 4).
Figure 4.

Spectrometric genotype-feature peak charts of rs4291 (AT, AT, TT).
Spectrometric peak chart of rs4309 genotypes
The MassArray system detected GG, GC, and CC rs4309 genotypes in the AD and control groups as shown in the spectrometric peak chart (Figure 5).
Figure 5.
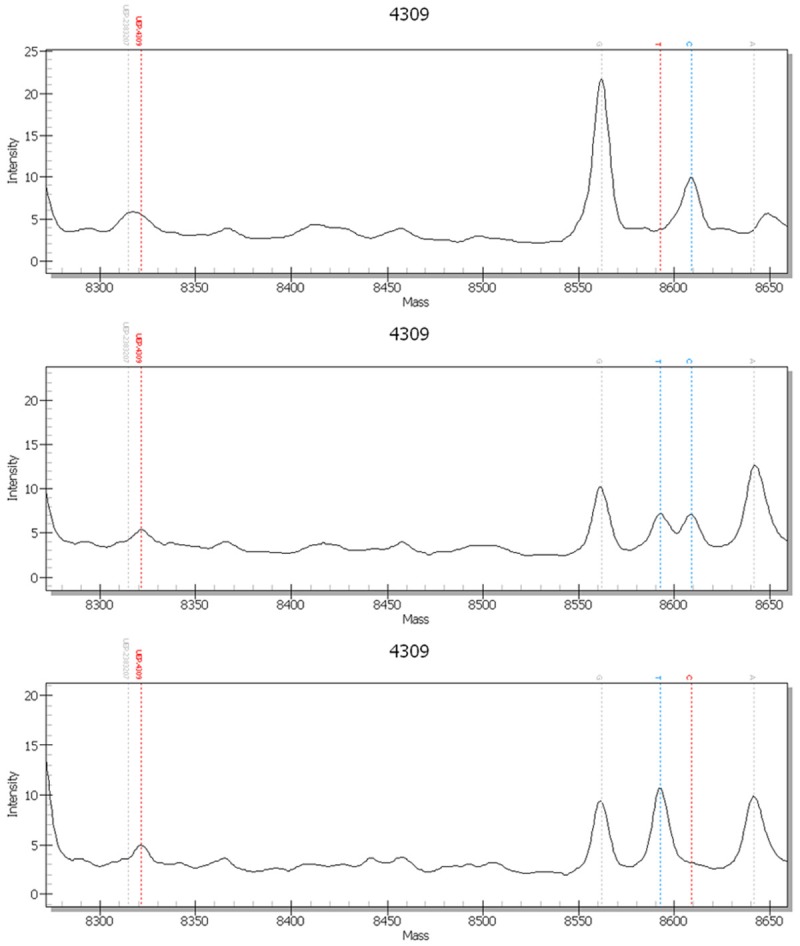
Spectrometric genotype-feature peak charts of rs4309 (CC, TC, TT).
Spectrometric peak chart of rs4343 genotypes
The MassArray system detected AA, AG, and GG rs4343 genotypes in the AD and control groups as shown in the spectrometric peak chart (Figure 6).
Figure 6.

Spectrometric genotype-feature peak charts of rs4343 (AA, AG, GG).
Construction and analysis of KNG1 and ACE SNPs haplotypes
The SHEsis software was used to perform linkage disequilibrium analysis and haplotype analysis on KNG1 and ACE. For KNG1, linkage disequilibrium analysis suggesting D’=0.86 (Figure 7). The haplotypes identified were AG, AT and GT. No significant differences were found in the distributions between the two groups (Table 3). For ACE, with the former suggesting D’ values among any two of the three loci as follows: D’ (rs4291, rs4309)=0.70, D’ (rs4291, rs4343)=0.87, D’ (rs4309, rs4343)=0.66 (Figure 7). The haplotypes identified were ATA, ACA, TCA, TCG, TTA and TTG, among which the ATA haplotype may be negatively correlated with risk of AD (OR=0.554, 95% CI=0.420-0.730, P<0.05). The ACA and TCA haplotypes may be positively correlated with the risk of AD (ACA:OR=5.231, 95% CI=2.447-11.183, P<0.05; TCA: OR=2.429, 95% CI=1.126-5.240, P<0.05) (Table 3).
Figure 7.
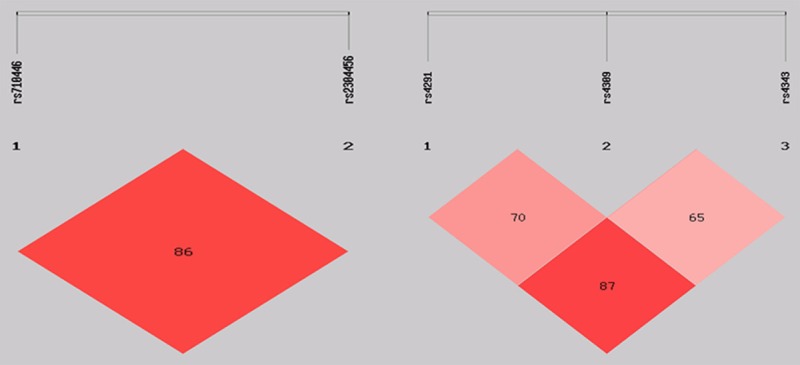
Linkage disequilibrium of KNG1 and ACE SNPs. The number in each box stands for the pairwise LD as assessed by the D’ value.
Table 3.
Estimated haplotypes in the case-control samples in KNG1and ACE
| Haplotype | Freq (Case) | Freq (Control) | X2 | p | OR 95% Cl | |
|---|---|---|---|---|---|---|
| KNG1* | AG | 0.130 | 0.177 | 3.291 | 0.070 | 0.711 [0.492~1.029] |
| AT | 0.544 | 0.492 | 3.688 | 0.055 | 1.294 [0.994~1.685] | |
| GT | 0.304 | 0.331 | 0.407 | 0.523 | 0.912 [0.688~1.209] | |
| ACE** | ATA | 0.545 | 0.698 | 17.651 | <0.001 | 0.554 [0.420~0.730] |
| ACA | 0.080 | 0.017 | 22.210 | <0.001 | 5.231 [2.447~11.183] | |
| TCA | 0.046 | 0.020 | 5.433 | 0.020 | 2.429 [1.126~5.240] | |
| TCG | 0.203 | 0.178 | 1.439 | 0.230 | 1.226 [0.879~1.711] | |
| TTA | 0.032 | 0.025 | 0.572 | 0.449 | 1.352 [0.617~2.962] | |
| TTG | 0.052 | 0.053 | 0.002 | 0.961 | 1.014 [0.565~1.821] |
The KNG1 haplotype composed of the rs710446-rs2304456;
The ACE haplotype composed of rs4291-rs4309-rs4343.
Discussion and conclusion
Senile plaques caused by excessive accumulation of Ab, along with neurofibrillary tangles and extensive neural attrition, establish the pathological basis of AD symptoms, such as progressive memory impairment. However, in recent years, the role of vascular factors in the development of AD has been revealed [13-15]. Improved cerebral circulation can result in reduced risk of AD and improved prognosis [16,17]. In this study, PCR amplification and MALDI-TOF MS detection were conducted on polymorphisms rs5516, rs710446, rs234456, rs4291, rs4309 and rs4343 from 201 AD patients and 257 control individuals to determine their possible correlation with AD.
No statistically significance difference was found between the genotype and allele distributions of rs5516, rs710446, rs2304456, rs4291 and rs4343 in our study. We consider that the rs5516, rs710446, rs2304456, rs4291 and rs4343 polymorphisms are not associated with AD pathogenesis in a Hunan Han Chinese population.
The study of Helbecque et al. [18] suggested that allele T of rs4291 is a protective factor of AD for people aged over 73 years; however, the correlation between rs4291 and AD has not been substantiated in our research, which is a conclusion shared by Belbin [19] and Meng et al. [20]. The distribution pattern of the allele frequencies of the healthy individuals in the control group showed the frequency of allele T of rs4291 as 27.6%, which is not significantly different from the 33% result reported by Meng et al. [20], yet is significantly different from the 45.9% result from Helbecque et al. [18], the 37% result from Belbin et al. [19] and Marguerite et al. [21] and the 37% result from Miners et al. [22], implying a noticeable ethnic or regional difference in the allele frequency of the genetic locus, which may provide part of the explanation for the discrepancy in the experimental results.
Currently, no study concerning a link between rs4309 and AD has been published, which makes this research the first to uncover such a correlation. The frequency of rs4309 C was 35.1% for the AD group, which was significantly higher than the 22.0% for the control group, suggesting a significant positive correlation between AD and allele C, with an OR=1.917, meaning the likelihood of AD morbidity for those who have allele C is 1.917 times greater than those who do not.
No agreement was reached by previous studies on the rs4343 polymorphism of ACE and AD: Belbin et al. [19] suggested no correlation between the polymorphism and the morbidity of AD, yet Ning et al. [23] confirmed rs4343 A as a risk factor for AD. Helbecque et al. [18], after looking into different age groups, identified rs4343 A as a protective factor against AD for people aged over 73 years. Another study tagged rs4343 G as a risk factor for AD [20]. The results of this study indicated rs4343 polymorphism is not associated with AD. The distribution pattern of allele frequencies of the control group showed the frequency of allele G of rs4291 as 23.9%, with no significant difference from the 18% result from the research of Meng et al. [20]; however, it is significantly different from the 48.1% result from Helbecque et al. [18], the 49% result from Belbin et al. [19], the 44% result from Marguerite et al. [21], and the 59% result from Miners et al. [22], implying a noticeable ethnic or regional difference in allele frequency of the genetic locus, which may explain the discrepancy in the experimental results.
The haplotypes constructed by rs710446/rs2304456 was not found to be associated with AD in this study. Correlations between ACE haplotypes and AD have already been reported. Helbecque et al. [18] suggested ATI as a protective factor against AD in haplotypes constructed by rs4343/rs4291/rs1799752 for people aged over 73 years. Meng et al. [20] indicated that the GA haplotype, composed of rs4343/rs4351, was a risk factor for AD. A case-control study performed on 144 LOAD patients and 476 healthy control individuals in Shanghai suggested the existence of five different haplotypes (CAI, CGD, TAI, TGD, and TGI) composed of rs1800764/rs4343/rs1799752, among which CGD served as a protective factor against AD, and CAI, TGD, and TGI were risk factors for the disease [23]. The results of the present research reveal a possible link between haplotypes ATA, ACA, and TCA (composed of rs4291/rs4309/rs4343) and the morbidity of AD. However, the hypothesis of the increased AD risk caused by ACA and TCA should be considered carefully, because the experiment did not assess an adequate number of subjects carrying such haplotypes. Even so, the results still indicate likely approaches for further studies.
In conclusion, we report the possible association of the ACE SNP rs4309 and haplotypes ATA, ACA, and TCA (composed of rs4291/rs4309/rs4343) with AD in a Hunan Han Chinese population. However, our study has several limitations. First, because it is a case-control study, all participants were Hunan Han Chinese. Because the types and frequencies of polymorphisms reflect different geographical and racial backgrounds, this could affect the analysis. Second, our study was conducted at a single institute so the number of patients was small. Therefore, further replication studies will be necessary to evaluate the association between the polymorphisms and haplotypes and AD risk. This will provide a theoretical basis for clarifying the role of kallikrein-kinin system in AD pathogenesis, and will also contribute to the discovery of AD-susceptible populations for which early intervention and treatment will be valuable.
Acknowledgements
The study was supported by the Science and Technology Projects from Science and Technology Agency, Hunan Province, China (2014SK3090). The study was supported by the New Xiangya Talent Project of the Third xiangya hospital of Central South University.
Disclosure of conflict of interest
None.
References
- 1.Bergamaschini L, Donarini C, Gobbo G, Parnetti L, Gallai V. Activation of complement and contact system in Alzheimer’s disease. Mech Ageing Dev. 2001;122:1971–83. doi: 10.1016/s0047-6374(01)00311-6. [DOI] [PubMed] [Google Scholar]
- 2.Menendez-Gonzalez M, Castro-Santos P, Suarez A, Calatayud MT, Perez-Pinera P, Martinez M, Ribacoba R, Gutierrez C. Value of measuring plasmatic levels of neurosin in the diagnosis of Alzheimer’s disease. J Alzheimers Dis. 2008;14:59–67. doi: 10.3233/jad-2008-14106. [DOI] [PubMed] [Google Scholar]
- 3.Jiang S, Hsu YH, Venners SA, Zhang Y, Xing H, Wang X, Xu X. Effects of protein coding polymorphisms in the kallikrein 1 gene on baseline blood pressure and antihypertensive response to irbesartan in Chinese hypertensive patients. J Hum Hypertens. 2011;25:327–33. doi: 10.1038/jhh.2010.70. [DOI] [PubMed] [Google Scholar]
- 4.Zhao W, Wang Y, Wang L, Lu X, Yang W, Huang J, Chen S, Gu D. Gender-specific association between the kininogen 1 gene variants and essential hypertension in Chinese Han population. J Hypertens. 2009;27:484–90. doi: 10.1097/hjh.0b013e32831e19f9. [DOI] [PubMed] [Google Scholar]
- 5.Sabater-Lleal M, Martinez-Perez A, Buil A, Folkersen L, Souto JC, Bruzelius M, Borrell M, Odeberg J, Silveira A, Eriksson P, Almasy L, Hamsten A, Soria JM. A genome-wide association study identifies KNG1 as a genetic determinant of plasma factor XI Level and activated partial thromboplastin time. Arterioscler Thromb Vasc Biol. 2012;32:2008–16. doi: 10.1161/ATVBAHA.112.248492. [DOI] [PubMed] [Google Scholar]
- 6.Morange PE, Oudot-Mellakh T, Cohen W, Germain M, Saut N, Antoni G, Alessi MC, Bertrand M, Dupuy AM, Letenneur L, Lathrop M, Lopez LM, Lambert JC, Emmerich J, Amouyel P, Trégouët DA. KNG1 Ile581Thr and susceptibility to venous thrombosis. Blood. 2011;117:3692–4. doi: 10.1182/blood-2010-11-319053. [DOI] [PubMed] [Google Scholar]
- 7.Hort J, O’Brien JT, Gainotti G, Pirttila T, Popescu BO, Rektorova I, Sorbi S, Scheltens P EFNS Scientist Panel on Dementia. EFNS guidelines for the diagnosis and management of Alzheimer’s disease. Eur J Neurol. 2010;17:1236–48. doi: 10.1111/j.1468-1331.2010.03040.x. [DOI] [PubMed] [Google Scholar]
- 8.Schiffrin EL. Blood pressure lowering in PROGRESS (Perindopril Protection Against Recurrent Stroke Study) and white matter hyperintensities: should this progress matter to patients? Circulation. 2005;112:1525–6. doi: 10.1161/CIRCULATIONAHA.105.566489. [DOI] [PubMed] [Google Scholar]
- 9.Yang L, Zhou HH, Ye YF, Fan XW, Wang YJ, Meng Y. Association of PS1 1/2, ACE I/D, and LRP C/T polymorphisms with disease in the Chinese population: a meta-analysis of casecontrol studies. Genet Mol Res. 2015;14:1017–24. doi: 10.4238/2015.February.6.5. [DOI] [PubMed] [Google Scholar]
- 10.Jochemsen HM, van der Flier WM, Ashby EL, Teunissen CE, Jones RE, Wattjes MP, Scheltens P, Geerlings MI, Kehoe PG, Muller M. Angiotensin-converting enzyme in cerebrospinal fluid and risk of brain atrophy. J Alzheimers Dis. 2015;44:153–62. doi: 10.3233/JAD-131496. [DOI] [PubMed] [Google Scholar]
- 11.Dong YF, Kataoka K, Tokutomi Y, Nako H, Nakamura T, Toyama K, Sueta D, Koibuchi N, Yamamoto E, Ogawa H, Kim-Mitsuyama S. Perindopril, a centrally active angiotensin-converting enzyme inhibitor, prevents cognitive impairment in mouse models of Alzheimer’s disease. FASEB J. 2011;25:2911–20. doi: 10.1096/fj.11-182873. [DOI] [PubMed] [Google Scholar]
- 12.Hebert PL, McBean AM, O’Connor H, Frank B, Good C, Maciejewski ML. Time until incident dementia among Medicare beneficiaries using centrally acting or non-centrally acting ACE inhibitors. Pharmacoepidemiol Drug Saf. 2013;22:641–8. doi: 10.1002/pds.3449. [DOI] [PubMed] [Google Scholar]
- 13.Levin OS, Trusova NA. [Vascular risk factors for Alzheimer’s disease] . Zh Nevrol Psikhiatr Im S S Korsakova. 2013;113:3–12. [PubMed] [Google Scholar]
- 14.Bidzan M, Bidzan L, Pachalska M. Neuropsychiatric symptoms in patients with Alzheimer’s disease with a vascular component. Ann Agric Environ Med. 2014;21:412–5. doi: 10.5604/1232-1966.1108615. [DOI] [PubMed] [Google Scholar]
- 15.Barker R, Ashby EL, Wellington D, Barrow VM, Palmer JC, Kehoe PG, Esiri MM, Love S. Pathophysiology of white matter perfusion in Alzheimer’s disease and vascular dementia. Brain. 2014;137:1524–32. doi: 10.1093/brain/awu040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lobanova I, Qureshi AI. The association between cardiovascular risk factors and progressive hippocampus volume loss in persons with Alzheimer’s disease. J Vasc Interv Neurol. 2014;7:52–5. [PMC free article] [PubMed] [Google Scholar]
- 17.de Bruijn RF, Ikram MA. Cardiovascular risk factors and future risk of Alzheimer’s disease. BMC Med. 2014;12:130. doi: 10.1186/s12916-014-0130-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Helbecque N, Codron V, Cottel D, Amouyel P. An age effect on the association of common variants of ACE with Alzheimer’s disease. Neurosci Lett. 2009;461:181–4. doi: 10.1016/j.neulet.2009.06.006. [DOI] [PubMed] [Google Scholar]
- 19.Belbin O, Brown K, Shi H, Medway C, Abraham R, Passmore P, Mann D, Smith AD, Holmes C, McGuinness B, Craig D, Warden D, Heun R, Kölsch H, Love S, Kalsheker N, Williams J, Owen MJ, Carrasquillo M, Younkin S, Morgan K, Kehoe PG. A multi-center study of ACE and the risk of late-onset Alzheimer’s disease. J Alzheimers Dis. 2011;24:587–97. doi: 10.3233/JAD-2011-101914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Meng Y, Baldwin CT, Bowirrat A, Waraska K, Inzelberg R, Friedland RP, Farrer LA. Association of polymorphisms in the Angiotensin-converting enzyme gene with Alzheimer disease in an IsraeliArabcommunity. Am J Hum Genet. 2006;78:871–7. doi: 10.1086/503687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Irvin MR, Lynch AI, Kabagambe EK, Tiwari HK, Barzilay JI, Eckfeldt JH, Boerwinkle E, Davis BR, Ford CE, Arnett DK. Pharmacogenetic association of hypertension candidate genes with fasting glucose in the GenHAT Study. J Hypertens. 2010;28:2076–83. doi: 10.1097/HJH.0b013e32833c7a4d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Miners S, Ashby E, Baig S, Harrison R, Tayler H, Speedy E, Prince JA, Love S, Kehoe PG. Angiotensin-converting enzyme levels and activity in Alzheimer’s disease: differences in brain and CSF ACE and association with ACE1 genotypes. Am J Transl Res. 2009;1:163–77. [PMC free article] [PubMed] [Google Scholar]
- 23.Ning M, Yang Y, Zhang Z, Chen Z, Zhao T, Zhang D, Zhou D, Xu J, Liu Z, Wang Y, Liu Y, Zhao X, Li W, Li S, He L. Amyloid-β-Related Genes SORL1 and ACE are Genetically Associated With Risk for Late-onset Alzheimer Disease in the Chinese Population. Alzheimer Dis Assoc Disord. 2010;24:390–6. doi: 10.1097/WAD.0b013e3181e6a575. [DOI] [PubMed] [Google Scholar]


