Abstract
Background: MiR-30a-5p has been reported to play vital roles in the carcinogenesis and progression of various malignancies via different molecular mechanisms. However, the role and target genes of miR-30a-5p in hepatocellular carcinoma (HCC) remain still unclear. In silico analysis finds that there are complementary sequences between the 3’-untrasnlated region of astrocyte elevated gene 1 (AEG-1) and miR-30a-5p. Herein, we investigated the biological function of miR-30a-5p, as well as the potential molecular mechanism via targeting AEG-1 in HCC cells. Materials and methods: MiR-30a-5p inhibitor, miR-30a-5p mimic, AEG-1 siRNAs, as well as their negative controls were transfected into HCC cell lines HepG2, SMMC-7221, HepB3 and SNU449. Then, the in vitro influence and mechanism of miR-30a-5p on cell viability, proliferation, caspase-3/7 activity and apoptosis were studied, as assessed by different methods, including spectrophotometry, fluorimetry, fluorescence microscopy of Hoechst 33342/propidium iodide double chromatin staining, western blot and dual luciferase reporter assay, respectively. Results: MiR-30a-5p mimic markedly inhibited cell growth, also induced caspase-3/7 activity and apoptosis in all four HCC cell lines tested. The strongest effect was observed in HepG2 and SMMC-7721 cells. However, this effect was slightly weaker than that of AEG-1 siRNAs. Transfection of miR-30a-5p mimic led to a markedly reduced AEG-1 protein level and further dual luciferase reporter assay confirmed that AEG-1 was one of the target genes of miR-30a-5p in HCC cells. Conclusions: MiR-30a-5p may play an essential role in the cell growth and apoptosis of HCC cells, partially via targeting AEG-1.
Keywords: miR-30a-5p, AEG-1, hepatocellular carcinoma, cell growth, apoptosis, caspase
Introduction
Hepatocellular carcinoma (HCC) is a major cancer burden worldwide, with estimated 550,000 to 600,000 increased cases every year, which ranks the sixth most common cancer and the second highest cause of cancer-related death worldwide [1]. Due to the challenge of early diagnosis, frequency of metastasis and recurrence, and poor response to variant treatments, the clinical outcome of HCC remains still grim. In recent years, the incidence of HCC-related deaths of USA has been growing. Furthermore, the 5-year survival rate maintains the ratio at 12% [2,3]. HCC develops within a complex background. In particular, chronic infections of hepatitis B virus (HBV) and/or HCV can facilitate the oncogenesis and progression of HCC. Other etiological factors involve alcoholism, aflatoxin, cirrhosis, nonalcoholic steatohepatitis, hemochromatosis, Wilson disease, diabetes and hemophilia [4,5]. A multistep process has been confirmed in the development of HCC. Some alterations at molecular level have been clarified to contribute to hepatocarcinogenesis and poor prognosis of HCC [6]. Unfortunately, the precise molecular mechanisms remain incompletely understood [7].
MicroRNAs (miRNAs) are a class of short non-coding RNAs that can regulate genes expression through combining with specific sequences in the 3’-untranslated region (UTR), resulting in mRNA degradation, mRNA translational suppression or even gene activation [8]. A number of studies have showed that miRNAs possess crucial regulatory functions in growth, proliferation, differentiation, apoptosis and stress response of cells [9]. It is a common sense that the abnormal expression of miRNAs plays a critical role in the development of cancers [10,11]. Recently, growing evidences have revealed that the aberrant expressions of miRNAs are related to tumorigenesis and deterioration of HCC. Reports showed that some of these miRNAs were decreased or increased in the HCC, and other specific miRNAs even were associated with clinical features, such as the HBV and HCV infection, cirrhosis, portal vein tumor thrombus (PVTT) and prognosis [12-14].
The function of microRNA-30a (miR-30a-5p) dysregulation in carcinoma deterioration and its downstream signaling remained unclear. MiR-30a-5p has been reported to be relevant to the inhibition of the epithelial-to-mesenchymal transition (EMT) in the non-small cell lung cancer (NSCLC) [15]. Zeng et al. established combinatorial regulatory networks of HCC without and with metastasis, which include both of transcription factor (TF) and miRNA regulation, and this differential combinatorial regulatory network analysis related to venous metastasis of HCC indicated that miR-30a-5p could be a key miRNA regulator contributed to metastasis in the HCC [16]. The relationship between miR-30a-5p and development of HCC has been only studied by only one group by far. Liu et al. found that the downregulation of miR-30a-5p was observed in HCC tissues and cell lines, and knock-down of miR-30a-5p could facilitate tumor cells migration, invasion and EMT processes in HCC via targeting SNAI1 [17]. However, the role and mechanism of miR-30a-5p on HCC is complex and many other potential target genes could exist. In silico analysis points out that the 3’-UTR of astrocyte elevated gene 1 (AEG-1) shares complementary sequence with miR-30a-5p. Additionally, the role of miR-30a-5p on the cell growth and apoptosis has not been illustrated. Herein, the effect of miR-30a-5p on cell biological function in HCC cell lines was explored. Furthermore, the hypothesis that AEG-1 could be a new potential target molecule of miR-30a-5p in HCC was investigated.
Materials and methods
Cell transfection and RT-qPCR
HepG2 (American Type Culture Collection, ATCC), SMMC-7221 (Chinese Academy of Medical Sciences), HepB3 (ATCC) and SNU449 (ATCC) cell lines were cultured as previously described [18-21]. In vitro experiments were performed in triplicate. HCC cells were planted 2.5 × 103 cells per well in 96-well plates and incubated at 37°C for 24 h before each transfection. The transfection was respectively performed in cells with blank control, mock control, miRNA inhibitor negative control, miR-30a-5p inhibitor, miRNA negative mimic control, miR-30a-5p mimic, scrambled siRNAs and AEG-1 siRNAs (Ambion, Life Technologies Grand Island, NY, USA) at a concentration of 200 nmol/L up to 96 h with combiMAGnetofection (OZBIOSCIENCES, Marseille Cedex 9, France) according to instructions of manufacturer. Sequence of miR-30a-5p was UGUAAACAUCCUCGACUGGAAG. Previously, we have tested different siRNA candidates to knock-down AEG-1 efficiently and found a pool with 4 siRNAs produced the best efficiency (data not shown), including siRNA-1033: AACTACAACCGCATCATT, siRNA-1455: ATGATGAATGGTCTGGGTT, siRNA-1967: AAGTCAAATACCAAGCAAA and siRNA-3566: CTTATTAATGGACAGCTTT. The pool of scrambled siRNAs was also obtained (data not shown) as negative controls.
The transfection efficiency was monitored with RT-qPCR. The total RNA including miRNA was extracted from HCC cells in different groups. Combination of RUN6B and let-7a was applied as the housekeeping reference for the detection of miR-30a-5p expression. The primers for miR-30a-5p, RNU6B and let-7a were maintained in TaqMan MicroRNA Assays (4427975, Applied Biosystems, Life Technologies Grand Island, NY 14072 USA). Sequence of miRNA and references were: miR-30a-5p: UGUAAACAUCCUCGACUGGAAG; RNU6B: CGCAAGGAUGA CACGCAAAUUCGUGAAGCGUUCCAUAUUUUU; let-7a: UGAGGUAGUAGGUUGUAUAGUU. A total volume of 10 μl was used in the step of reverse transcription with TaqMan MicroRNA Reverse Transcription Kit (4366596, Applied Biosystems, Life Technologies Grand Island, NY 14072 USA) with the same reverse primers. Real-time qPCR to detect miRNA level was performed with Applied Biosystems PCR7900 as reported [18-21].
Cell biological function influenced by miR-30a-5p
The biological function of miR-30a-5p inhibitor and miR-30a-5p mimic, including cell growth, caspase-3/7 activity, cellular apoptosis and nuclear morphology was accessed as described previously, with fluorimetric resorufin viability assay, MTS tetrazolium assay, Apo-ONE Homogeneous Caspase-3/7 Assay and Hoechst 33342/propidium iodide (PI) double florescent chromatin staining, respectively [18-27]. Western blot was performed as we described previously [18-27]. Primary antibodies in the present study included: AEG-1 and β-actin (Sigma-Aldrich N.V.).
Verification of AEG-1 as a direct target gene of miR-30a-5p
AEG-1 has been confirmed to be a direct gene targeting miR-30a-5p in breast cancer and lung cancer [28,29] with dual luciferase reporter assay. However, the role and mechanism of one miRNA could be tumor-specific. Thus, we repeated the dual luciferase reporter assay on HCC cell lines HepG2, SMMC-7221, HepB3 and SUN449 as reported [29]. Briefly, the 3’-UTR of AEG-1 with two putative miR-30a-5p-binding sites was amplified and cloned into pmirGLO vector separately (Promega, Madison, WI, USA). Two miR-30a-5p complementary sites with the sequence UGUUUACA in AEG-1 3’-UTR were mutated singly via complementarity dislodging to miR-30a-5p [29]. Cells were prepared in 96-well plates and co-transfected with 100 ng of wide-type or mutated AEG-1 3’-UTR constructs, or 50 mM of negative control or miR-30a-5p mimics up to 96 h. Luciferase activity was determined with the dual luciferase reporter assay system (Promega).
Statistical analysis
Statistical calculations were performed with SPSS software (version20.0). Values were presented as means ± standard deviation (SD) from at least three single experiments. For analyzing significance between different groups, one-way analysis of variance (ANOVA) test was used. If the probability for ANOVA was statistically significant, the least significant difference (LSD) method of multiple comparisons between two groups would be applied. A value of P < 0.05 was considered to indicate statistical significance.
Results
Effect of miR-30a-5p on inhibition of cell growth in HCC cells
The influence of different agents on the levels of miR-30a-5p was first detected with real time RT-qPCR and the transfection efficiency was confirmed to be optimal (data not shown). The effect of miR-30a-5p on cell growth was identified with three independent assays, including fluorimetric resorufin viability assay, MTS tetrazolium assay and Hoechst 33342/PI double fluorescent chromatin staining, respectively. Fluorimetric resorufin viability assay showed that cell viability increased slightly in HepG2 and SNU449 cells 72 and 96 h post transfection with miR-30a-5p inhibitor, as compared to blank and negative controls. In another 2 cell lines (SMMC-7221 and HepB3), only at 96 h after transfection, cell viability increased, however, less than 10%. In contract, miR-30a-5p mimic caused a large reduction in proliferation at 72 and 96 h in all the 4 cell lines tested, although with a lesser degree than the effect of siRNA targeting AEG-1 (Figure 1). The cell growth suppressive effect showed a time dependent manner (Figure 1) and also a dose dependent manner (data not shown) in all cell lines. To further confirm the effect of miR-30a on cell growth of HCC cells, MTS tetrazolium assay (Figure 2) and Hoechst 33342/PI double fluorescent chromatin staining (data not shown) were assessed, which almost mirrored the effects from the fluorimetric resorufin viability assay. The more potent impact of miR-30a-5p on cell growth was observed in the cell lines of HepG2 and SMMC-7721, for instance, 40% cell growth inhibition was achieved in SMMC-7721 96 h post transfection of miR-30a-5p (Figure 2B).
Figure 1.
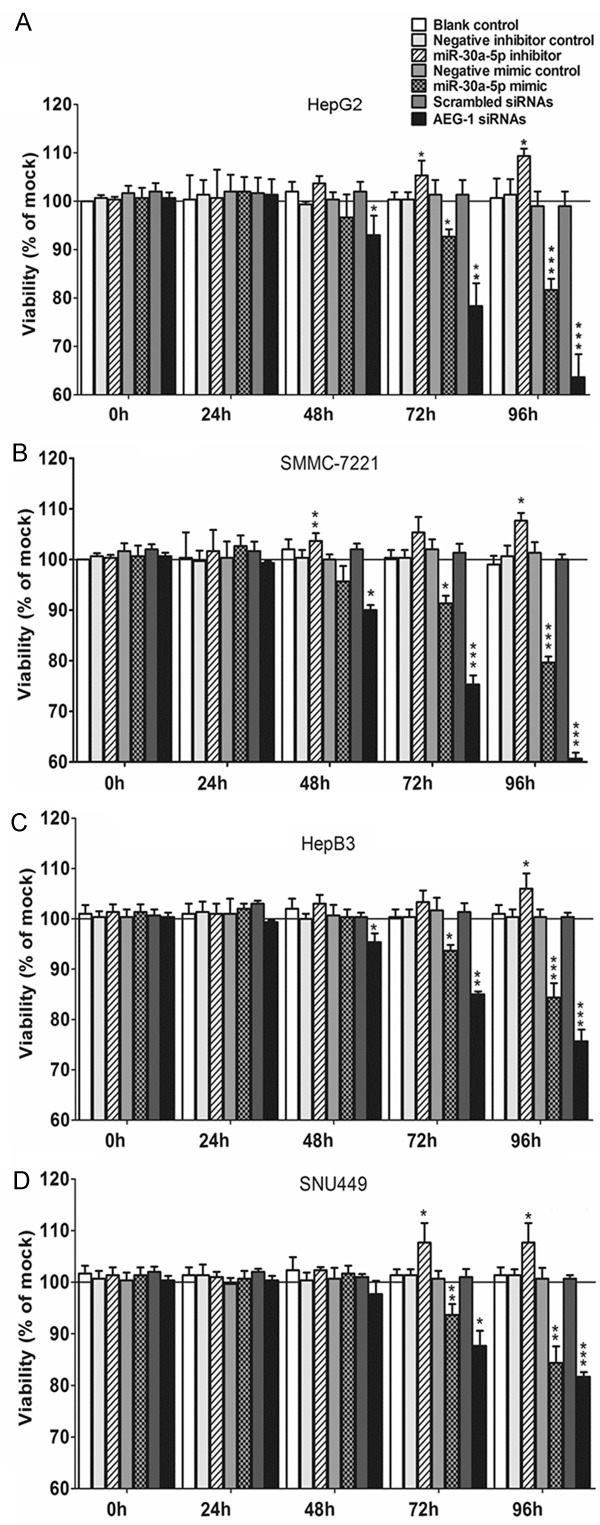
Time dependent effect of miR-30a-5p on cell viability in HCC cell lines. HepG2 (A), SMMC-7211 (B), HepB3 (C) and SNU449 (D) cells (2.5 × 103 cells per well in 96-well-plate) were cultured for 24 h and then transfected with miR-30a-5p inhibitor, miR-30a-5p mimic, AEG-1 siRNA and their negative controls (200 nM) up to another 96 h. Cell viability was monitored with fluorimetric detection of resorufin (CellTiter-Blue Cell Viability Assay). Columns were the average of three independent experiments. Bars represented standard deviation. *P < 0.05, ** P < 0.01 and ***P < 0.001, compared to blank and negative controls at the same time point.
Figure 2.
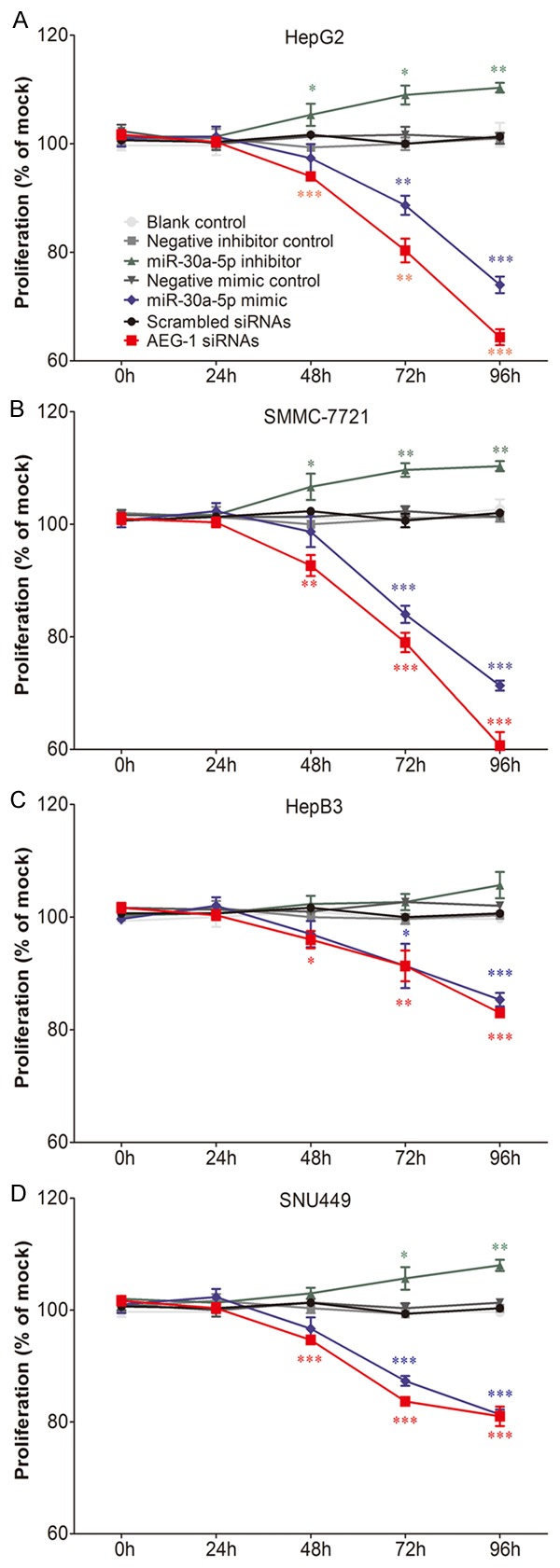
Cell proliferation in HCC cell lines influenced by miR-30a-5p. HepG2 (A), SMMC-7211 (B), HepB3 (C) and SNU449 (D) cells (2.5 × 103 cells per well in 96-well-plate) were cultured for 24 h and transfected with miR-30a-5p inhibitor, miR-30a-5p mimic, AEG-1 siRNA and their negative controls (200 nM) for another 96 h. Cell proliferation was assessed per day with MTS assay (CellTiter96 Aqueous One Solution Cell Proliferation Assay). Points were the average of three independent experiments. Bars represented standard deviation. *P < 0.05, **P < 0.01 and ***P < 0.001, compared to blank and negative controls at the same time point.
Apoptosis induction and activation of caspase-3/7 activity of miR-30a-5p in HCC cells
To further evaluate the effect of miR-30a-5p on apoptosis and activated caspase activity of HCC cells, the CellTiter-Blue assay was multiplexed with a fluorescent caspase-3/7 assay. With miR-30a-5p inhibitor, no variation of caspase-3/7 activity between different groups was observed. However, with miR-30a-5p mimic, caspase-3/7 activity was evidently enhanced in all 4 HCC cell lines tested with a time (Figure 3) and dose dependent manner (data not shown). Analogous to the result of cell growth, the effect of miR-30a-5p on caspase activity was much slighter than that of siRNA targeting AEG-1. The time and dose dependent effect on apoptosis was supported by Hoechst 33342 and PI double fluorescent staining microscopically (Figures 4, 5). Again, stronger effect was observed in the cells of HepG2 and SMMC-7221.
Figure 3.
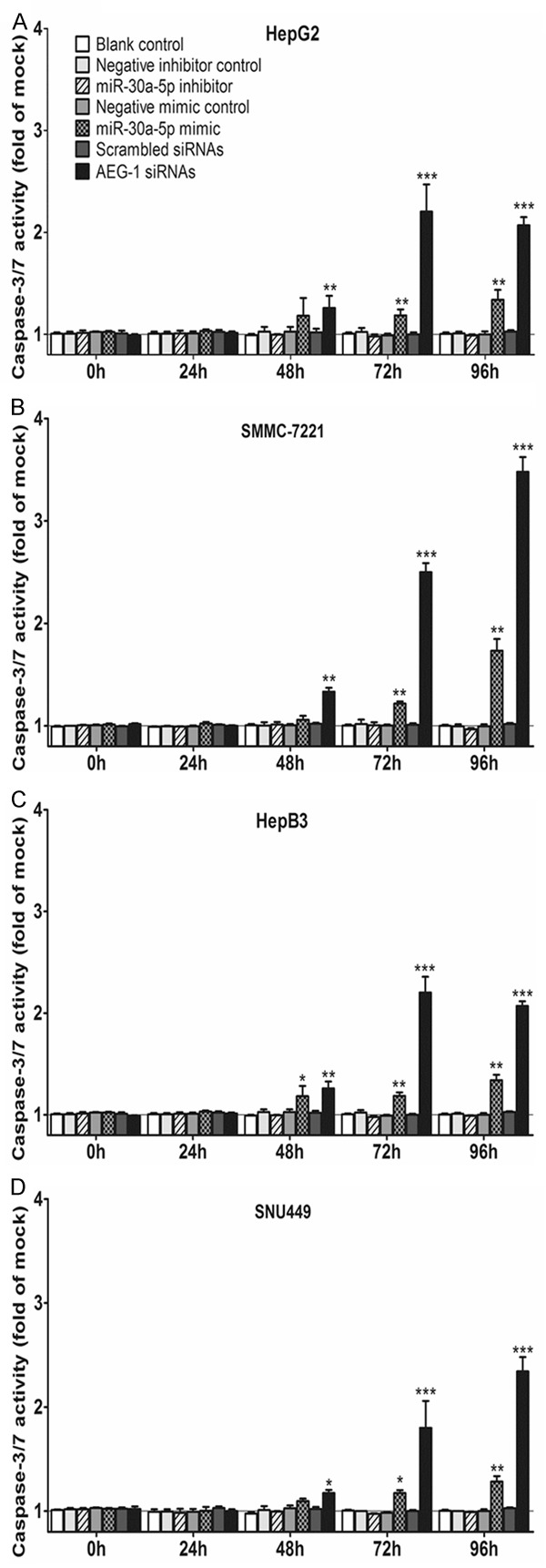
Effect of miR-30a-5p on caspase-3/7 activity in HCC cell lines. HepG2 (A), SMMC-7211 (B), HepB3 (C) and SNU449 (D) cells (2.5 × 103 cells per well in 96-well-plate) were treated as aforementioned in Figure 1. After the cell viability was monitored with fluorimetric detection of resorufin, caspase-3/7 activity (Apo-ONE Homogeneous Caspase-3/7 Assay) was detected with the same samples. Columns were the average of three independent experiments. Bars represented standard deviation. *P < 0.05, **P < 0.01 and ***P < 0.001, compared to blank and negative controls at the same time point.
Figure 4.
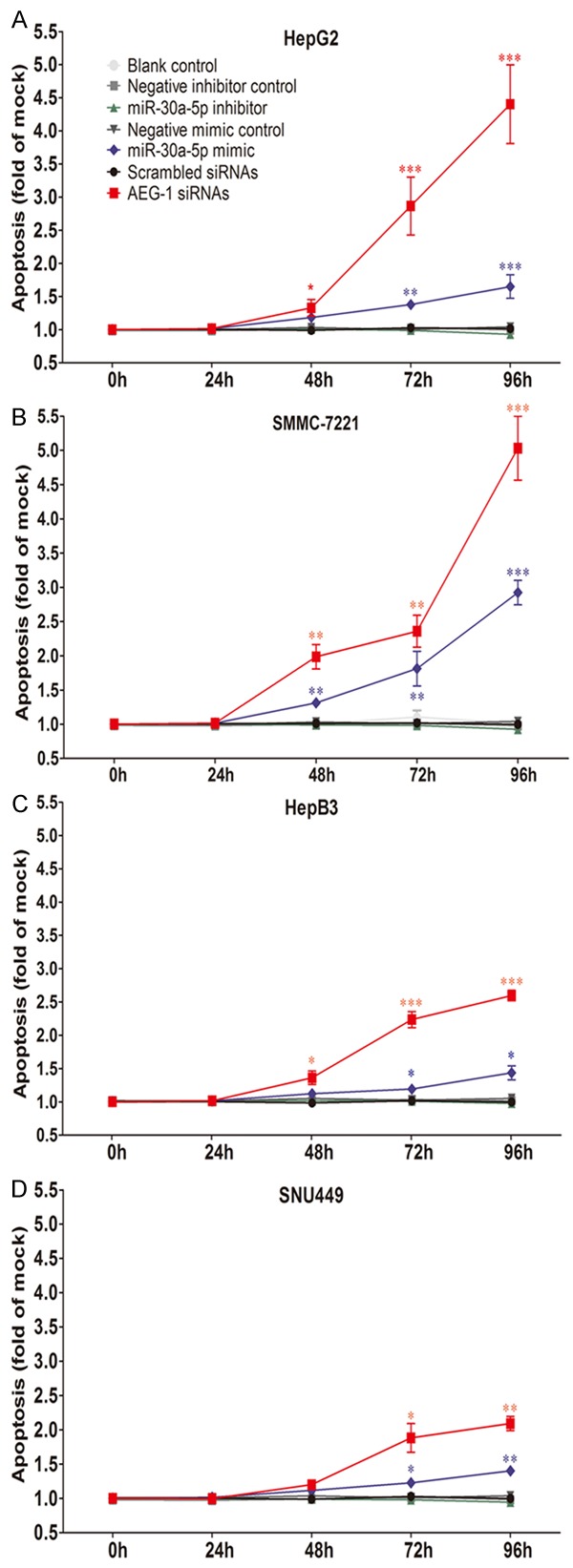
Apoptosis induced by miR-30a-5p in HCC cell lines. HepG2 (A), SMMC-7211 (B), HepB3 (C) and SNU449 (D) cells (2.5 × 103 cells per well in 96-well-plate) were cultured for 24 h and transfected with miR-30a-5p inhibitor, miR-30a-5p mimic, AEG-1 siRNA and their negative controls (200 nM) for another 96 h. Cellular apoptosis was assessed with Hoechst 33342/propidium iodide (PI) double fluorescent chromatin staining. Points were the average of three independent experiments. Bars represented standard deviation. *P < 0.05, **P < 0.01 and ***P < 0.001, compared to blank and negative controls at the same time point.
Figure 5.
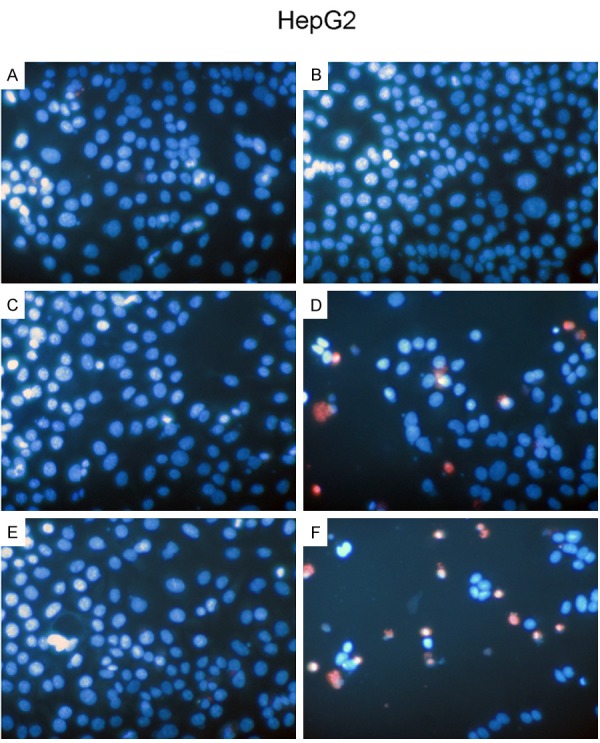
HCC HepG2 cells morphology influenced by miR-30a-5p. HepG2 cells (2.5 × 103 cells per well in 96-well-plate) were cultured for 24 h and transfected with miR-30a-5p inhibitor, miR-30a-5p mimic, AEG-1 siRNA and their negative controls (200 nM) for another 96 h. Change of morphology was evaluated with Hoechst 33342/propidium iodide (PI) double fluorescent chromatin staining. A: Negative inhibitor control; B: miR-30a-5p inhibitor; C: Negative mimic control; D: miR-30a-5p mimic; E: Scrambled siRNAs; F: AEG-1 siRNAs.
AEG-1 as a direct target gene of miR-30a-5p in HCC
Within silico prediction, AEG-1 was identified to gain the potential to act as one of the target genes of miR-30a-5p and it was also of specific interest since our previous studies indicated its vital role in the carcinogenesis and deterioration of tumors [30]. Also, in breast cancer and lung cancer, the targeting relationship between AEG-1 and miR-30a-5p has been validated. Thus, the hypothesis of AEG-1 being also a target gene of miR-30a-5p in HCC cells was verified with 4 cell lines: HepG2, SMMC-7221, HepB3 and SNU449. As reported previously, the miRNA: mRNA alignment analysis showed that the 3’-UTR of AEG-1 contains two putative binding sites for miR-30a-5p located at 287–295 nt and 1569–1577 nt [29] (data not shown). To validate direct targeting of AEG-1 by miR-30a-5p, each of the putative binding sites of miR-30a-5p was cloned individually into the pmiRGLO vector. Co-transfection of HepG2 cells with miR-30a-5p mimic and the pmiRGLO-wt1 and wt2 3’-UTR vector showed a remarkably reduced luciferase activity than the negative controls (Figure 6A), indicating both of the putative binding sites were functional. In line with the results, the equivalent effect was observed in other 3 cell lines: SMMC-7221, HepB3 and SNU449 (data not shown). Consistent with the dual luciferase assay, an obvious reduction of AEG-1 protein expression in HepG2 cells was observed (Figure 6B). Other cell lines yielded the similar outcomes (data not shown).
Figure 6.
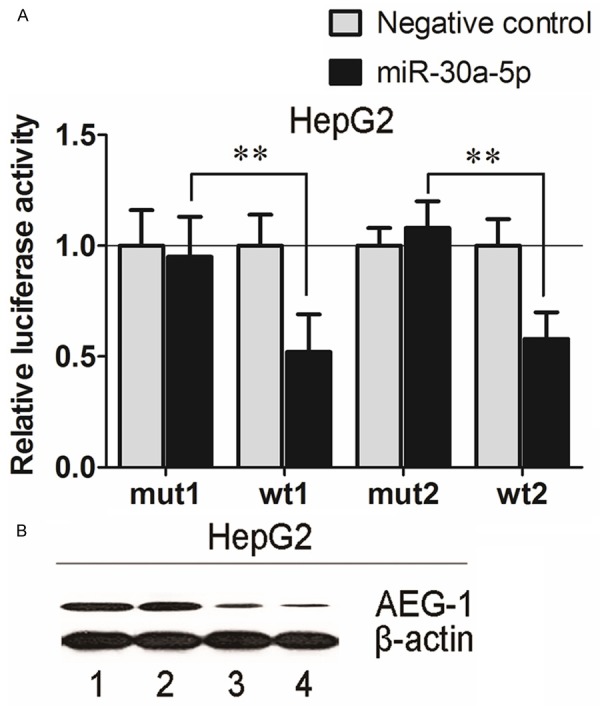
AEG-1 as a direct target gene for miR-30a-5p in HCC HepG2 cells. A: Firefly luciferase activity was normalized to the internal Renilla luciferase activity. Relative luciferase activity of HepG2 cells post transfection with the wt or mut 3’-UTR of AEG-1 genes. Luciferase reporter assay revealed miR-30a-5p suppressed AEG-1 3’-UTR luciferase activity of both wt constructs, but not mut constructs. wt: wide type; mut: mutated. Columns were the average of three independent experiments. Bars represented standard deviation. *P < 0.05, **P < 0.01 and ***P < 0.001. B: MiR-30a-5p mimic, AEG-1 siRNA and their negative controls (200 nM) were transfected into HepG2 cells for 96 h and western blot was performed to detect the protein level of AEG-1, a potential target gene of miR-30a-5p. (1): Negative mimic control; (2): Scrambled siRNAs; (3): miR-30a-5p mimic; (4): AEG-1 siRNAs.
Discussion
In the present study, RT-qPCR was performed to confirm the transfection efficiency of miR-30a-5p inhibitor and miR-30a-5p mimic in HCC HepG2, SMMC-7221, HepB3 and SNU499 cells. Further in vitro experiments confirmed that miR-30a-5p is closely associated with the malignant biological phenotypes of HCC cells, such as cell viability, proliferation, activated caspase-3/7 activity and apoptosis. The results suggested that miR-30a-5p has the capacity to depress the cell growth of HCC cells and induce the apoptosis, partially via targeting AEG-1.
MiR-30a-5p has been implicated in several human malignancies, underscoring its potential roles in diagnostics, progression prediction, as well as molecular target for novel therapeutics for different tumors [31-38]. Previously, our group detected significantly lower level of miR-30a-5p in lung cancer, as compared to non-cancerous lung tissues [39]. MiR-30a-5p level was also negatively associated to tumor size, lymphatic metastasis, clinical TNM stage, pathological grading, histological classification and overall survival, indicating miR-30a-5p may be regarded as a tumor suppressor in lung cancer [39]. When concerning the role of miR-30a-5p in HCC, only Liu et al. [17] found that miR-30 family members were significantly downregulated in 63 cases HCC tissues and also several HCC cell lines, especially miR-30a-5p, which was consistent with our previous finding that miR-30a-5p was downregulated in HCC, as compared to non-cancerous liver tissues or cells (data on file). Furthermore, Liu et al. [17] reported that miR-30a-5p downregulation was significantly related with worse disease-free survival (DFS) of HCC patients. With gain- and loss-of-function studies, silencing of miR-30a-5p facilitated tumor cell migration, invasion and epithelial-mesenchymal transition (EMT). This could partly explain the mechanism of miR-30a-5p on the tumorigenesis and poor prognosis of HCC. Besides that, miR-30a-5p has been identified as a key miRNA regulator contributed to HCC metastasis via a “differential combinatorial regulatory network analysis” [16]. However, no report has been available investigating the effect of miR-30a-5p on the cell growth and apoptosis, which are also important cancer biological phenotypes, in HCC cells. In the current study, the miR-30a-5p mimic could decelerate the cell growth in all the cell lines tested (HepG2, SMMC-7721, HepB3 and SNU449), with three independent assays. In addition, miR-30a-5p mimic enhanced the caspase-3/7 activity and induced apoptosis in HCC cell lines, with both dose and time-dependent manners. The in vitro effect of miR-30a-5p in the current study, together with literatures [17], gives an account of the vital role of miR-30a-5p in the carcinogenesis and deterioration of HCC via influencing the cell growth, apoptosis, motility and metastasis.
MiRNAs majorly regulate the expression of downstream genes to play a critical role in the physiological and pathological processes. So the understanding of the target genes of miR-30a-5p probably indicates its potential mechanisms in the HCC. The possible target genes have been validated in several other malignancies. Autophagy related gene (ATG) was reported to be a target gene of miR-30a-5p in the chronic myelogenous leukemia cells [40]. In the research of breast cancer, Ouzounova et al. found that miR-30 family can modulate redundantly the expression of proliferation and apoptosis -related genes. It was an interesting finding that not only anti-apoptotic protein AVEN was the most significantly downregulated genes post miR-30a-5p overexpression, but also AVEN can partially revert the effect of miR-30a-5p overexpression [41]. Chen et al. [42] found that miR-30a-5p inhibits cell migration and invasion by decreasing the expression of vimentin expression in breast cancer. In HCC, SNAI1 was identified as a direct target of miR-30a, and miR-30a was also demonstrated as a novel regulator of EMT by targeting SNAI1, showing its potential therapeutic value for decreasing invasion and metastasis of HCC [17]. Via in silico prediction, AEG-1 had the potential to be a target gene of miR-30a-5p, which has been confirmed in breast cancer and lung cancer [28,29]. AEG-1 is a single-pass transmembrane protein and it is composed of 582 amino acids, which locate at chromosome 8q22.5 [43]. Studies uncovered that AEG-1 played a vital role in several crucial aspects of HCC progression, including growth, transformation, cell survival, invasion, metastasis and chemoresistance. Both of AEG-1 mRNA and protein expression were higher in most of the HCC cancerous tissues than their paired adjacent non-tumor tissues. Additionally, AEG-1 overexpression activates the Wnt/β-catenin, mitogen-activated protein kinase (MAPK), nuclear factor (NF)-κB, and PI3K/Akt signaling pathways, and accelerates its downstream gene expression to promote malignant potential. More importantly, transgenic mice with hepatocyte-specific expression of AEG-1 (Alb/AEG-1) and AEG-1-knockout mouse both explored new aspects of the functions of AEG-1 in an in vivo context [44-46]. In the current study, we primarily revealed that AEG-1 could be also one of miR-30a-5p target genes in HCC with both dual luciferase reporter assay and western blot.
Conclusions
In the current study, we demonstrate the role of miR-30a-5p in cell growth and apoptosis in HCC HepG2, SMMC-7221, HepB3 and SNU449 cells. MiR-30a-5p can reduce cell growth and induce cell apoptosis, via directly targeting AEG-1 in HCC cells. MiR-30a-5p may be a potential noteworthy target for the molecular therapy of HCC.
Acknowledgements
The study was supported partly by the Funds of Guangxi Zhuang Autonomous Region University Student Innovative Plan (No. 201510598016), Natural Science Foundation of Guangxi, China (2015GXNSFBA139157), Guangxi Provincial Health Bureau Scientific Research Project (Z2014054), Youth Science Foundation of Guangxi Medical University (GXMUYSF201311), Guangxi University Science and Technology Research Projects (LX2014075), and National Natural Science Foundation of China (NSFC 81360327, 81560489). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Disclosure of conflict of interest
None.
References
- 1.van Meer S, de Man RA, Siersema PD, van Erpecum KJ. Surveillance for hepatocellular carcinoma in chronic liver disease: evidence and controversies. World J Gastroenterol. 2013;19:6744–6756. doi: 10.3748/wjg.v19.i40.6744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Saraswat VA, Pandey G, Shetty S. Treatment algorithms for managing hepatocellular carcinoma. J Clin Exp Hepatol. 2014;4(Suppl 3):S80–89. doi: 10.1016/j.jceh.2014.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Han MS, Moon KS, Lee KH, Cho SB, Lim SH, Jang WY, Jung TY, Kim IY, Jung S. Brain metastasis from hepatocellular carcinoma: the role of surgery as a prognostic factor. BMC Cancer. 2013;13:567. doi: 10.1186/1471-2407-13-567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chittmittrapap S, Chieochansin T, Chaiteerakij R, Treeprasertsuk S, Klaikaew N, Tangkijvanich P, Komolmit P, Poovorawan Y. Prevalence of aflatoxin induced p53 mutation at codon 249 (R249s) in hepatocellular carcinoma patients with and without hepatitis B surface antigen (HBsAg) Asian Pac J Cancer Prev. 2013;14:7675–7679. doi: 10.7314/apjcp.2013.14.12.7675. [DOI] [PubMed] [Google Scholar]
- 5.Yang L, Ji J, Chen Z, Wang H, Li J. Transcriptome profiling of malignant transformed rat hepatic stem-like cells by aflatoxin B1. Neoplasma. 2014;61:193–204. doi: 10.4149/neo_2014_025. [DOI] [PubMed] [Google Scholar]
- 6.Luo Y, Ren F, Liu Y, Shi Z, Tan Z, Xiong H, Dang Y, Chen G. Clinicopathological and prognostic significance of high Ki-67 labeling index in hepatocellular carcinoma patients: a meta-analysis. Int J Clin Exp Med. 2015;8:10235–10247. [PMC free article] [PubMed] [Google Scholar]
- 7.Tarocchi M, Polvani S, Marroncini G, Galli A. Molecular mechanism of hepatitis B virus-induced hepatocarcinogenesis. World J Gastroenterol. 2014;20:11630–11640. doi: 10.3748/wjg.v20.i33.11630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Moura J, Borsheim E, Carvalho E. The Role of MicroRNAs in Diabetic Complications-Special Emphasis on Wound Healing. Genes. 2014;5:926–956. doi: 10.3390/genes5040926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang J, Raimondo M, Guha S, Chen J, Diao L, Dong X, Wallace MB, Killary AM, Frazier ML, Woodward TA, Wang J, Sen S. Circulating microRNAs in Pancreatic Juice as Candidate Biomarkers of Pancreatic Cancer. J Cancer. 2014;5:696–705. doi: 10.7150/jca.10094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Aghanoori MR, Mirzaei B, Tavallaei M. MiRNA molecular profiles in human medical conditions: connecting lung cancer and lung development phenomena. Asian Pac J Cancer Prev. 2014;15:9557–9565. doi: 10.7314/apjcp.2014.15.22.9557. [DOI] [PubMed] [Google Scholar]
- 11.Jin D, Fang Y, Li Z, Chen Z, Xiang J. Epithelialmesenchymal transitionassociated microRNAs in colorectal cancer and drug-targeted therapies (Review) Oncol Rep. 2015;33:515–525. doi: 10.3892/or.2014.3638. [DOI] [PubMed] [Google Scholar]
- 12.Wang W, Lin H, Zhou L, Zhu Q, Gao S, Xie H, Liu Z, Xu Z, Wei J, Huang X, Zheng S. MicroRNA-30a-3p inhibits tumor proliferation, invasiveness and metastasis and is downregulated in hepatocellular carcinoma. Eur J Surg Oncol. 2014;40:1586–1594. doi: 10.1016/j.ejso.2013.11.008. [DOI] [PubMed] [Google Scholar]
- 13.Fan HX, Tang H. Complex interactions between microRNAs and hepatitis B/C viruses. World J Gastroenterol. 2014;20:13477–13492. doi: 10.3748/wjg.v20.i37.13477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Qi W, Liang W, Jiang H, Miuyee Waye M. The function of miRNA in hepatic cancer stem cell. Biomed Res Int. 2013;2013:358902. doi: 10.1155/2013/358902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kumarswamy R, Mudduluru G, Ceppi P, Muppala S, Kozlowski M, Niklinski J, Papotti M, Allgayer H. MicroRNA-30a inhibits epithelial-tomesenchymal transition by targeting Snai1 and is downregulated in non-small cell lung cancer. Int J Cancer. 2012;130:2044–2053. doi: 10.1002/ijc.26218. [DOI] [PubMed] [Google Scholar]
- 16.Zeng L, Yu J, Huang T, Jia H, Dong Q, He F, Yuan W, Qin L, Li Y, Xie L. Differential combinatorial regulatory network analysis related to venous metastasis of hepatocellular carcinoma. BMC Genomics. 2012;13(Suppl 8):S14. doi: 10.1186/1471-2164-13-S8-S14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu Z, Tu K, Liu Q. Effects of microRNA-30a on migration, invasion and prognosis of hepatocellular carcinoma. FEBS Lett. 2014;588:3089–3097. doi: 10.1016/j.febslet.2014.06.037. [DOI] [PubMed] [Google Scholar]
- 18.Dang Y, Luo D, Rong M, Chen G. Underexpression of miR-34a in hepatocellular carcinoma and its contribution towards enhancement of proliferating inhibitory effects of agents targeting c-MET. PLoS One. 2013;8:e61054. doi: 10.1371/journal.pone.0061054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rong M, Chen G, Dang Y. Increased miR-221 expression in hepatocellular carcinoma tissues and its role in enhancing cell growth and inhibiting apoptosis in vitro. BMC Cancer. 2013;13:21. doi: 10.1186/1471-2407-13-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang S, He R, Rong M, Dang Y, Chen G. Synergistic effect of MiR-146a mimic and cetuximab on hepatocellular carcinoma cells. Biomed Res Int. 2014;2014:384121. doi: 10.1155/2014/384121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dang YW, Zeng J, He RQ, Rong MH, Luo DZ, Chen G. Effects of miR-152 on cell growth inhibition, motility suppression and apoptosis induction in hepatocellular carcinoma cells. Asian Pac J Cancer Prev. 2014;15:4969–4976. doi: 10.7314/apjcp.2014.15.12.4969. [DOI] [PubMed] [Google Scholar]
- 22.Chen G, Kronenberger P, Teugels E, De Greve J. Influence of RT-qPCR primer position on EGFR interference efficacy in lung cancer cells. Biol Proced Online. 2011;13:1. doi: 10.1186/1480-9222-13-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen G, Kronenberger P, Teugels E, Umelo IA, De Greve J. Targeting the epidermal growth factor receptor in non-small cell lung cancer cells: the effect of combining RNA interference with tyrosine kinase inhibitors or cetuximab. BMC Medicine. 2012;10:28. doi: 10.1186/1741-7015-10-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen G, Kronenberger P, Teugels E, Umelo IA, De Greve J. Effect of siRNAs targeting the EGFR T790M mutation in a non-small cell lung cancer cell line resistant to EGFR tyrosine kinase inhibitors and combination with various agents. Biochem Biophys Res Commun. 2013;431:623–629. doi: 10.1016/j.bbrc.2012.12.070. [DOI] [PubMed] [Google Scholar]
- 25.Chen G, Kronenberger P, Umelo IA, Teugels E, De Grève J. Quantification of epidermal growth factor receptor T790M mutant transcripts in lung cancer cells by real-time reverse transcriptase-quantitative polymerase chain reaction. Anal Biochem. 2010;398:266–268. doi: 10.1016/j.ab.2009.11.034. [DOI] [PubMed] [Google Scholar]
- 26.Chen G, Noor A, Kronenberger P, Teugels E, Umelo IA, De Greve J. Synergistic effect of afatinib with su11274 in non-small cell lung cancer cells resistant to gefitinib or erlotinib. PLoS One. 2013;8:e59708. doi: 10.1371/journal.pone.0059708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen G, Umelo IA, Lv S, Teugels E, Fostier K, Kronenberger P, Dewaele A, Sadones J, Geers C, De Greve J. miR-146a inhibits cell growth, cell migration and induces apoptosis in nonsmall cell lung cancer cells. PLoS One. 2013;8:e60317. doi: 10.1371/journal.pone.0060317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu K, Guo L, Guo Y, Zhou B, Li T, Yang H, Yin R, Xi T. AEG-1 3’-untranslated region functions as a ceRNA in inducing epithelial-mesenchymal transition of human non-small cell lung cancer by regulating miR-30a activity. Eur J Cell Biol. 2015;94:22–31. doi: 10.1016/j.ejcb.2014.10.006. [DOI] [PubMed] [Google Scholar]
- 29.Zhang N, Wang X, Huo Q, Sun M, Cai C, Liu Z, Hu G, Yang Q. MicroRNA-30a suppresses breast tumor growth and metastasis by targeting metadherin. Oncogene. 2014;33:3119–3128. doi: 10.1038/onc.2013.286. [DOI] [PubMed] [Google Scholar]
- 30.Ren F, Ding H, Huang S, Wang H, Wu M, Luo D, Dang Y, Yang L, Chen G. Expression and clinicopathological significance of miR-193a-3p and its potential target astrocyte elevated gene-1 in non-small lung cancer tissues. Cancer Cell Int. 2015;15:80. doi: 10.1186/s12935-015-0227-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zheng B, Zhu H, Gu D, Pan X, Qian L, Xue B, Yang D, Zhou J, Shan Y. MiRNA-30a-mediated autophagy inhibition sensitizes renal cell carcinoma cells to sorafenib. Biochem Biophys Res Commun. 2015;459:234–239. doi: 10.1016/j.bbrc.2015.02.084. [DOI] [PubMed] [Google Scholar]
- 32.Lu N, Lin T, Wang L, Qi M, Liu Z, Dong H, Zhang X, Zhai C, Wang Y, Liu L, Xiang L, Qi L, Han B, Li J. Association of SOX4 regulated by tumor suppressor miR-30a with poor prognosis in lowgrade chondrosarcoma. Tumour Biol. 2015;36:3843–3852. doi: 10.1007/s13277-014-3026-2. [DOI] [PubMed] [Google Scholar]
- 33.Katz B, Reis ST, Viana NI, Morais DR, Moura CM, Dip N, Silva IA, Iscaife A, Srougi M, Leite KR. Comprehensive study of gene and microRNA expression related to epithelial-mesenchymal transition in prostate cancer. PLoS One. 2014;9:e113700. doi: 10.1371/journal.pone.0113700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huang Q, Jiang Z, Meng T, Yin H, Wang J, Wan W, Cheng M, Yan W, Liu T, Song D, Chen H, Wu Z, Xu W, Li Z, Zhou W, Xiao J. MiR-30a inhibits osteolysis by targeting RunX2 in giant cell tumor of bone. Biochem Biophys Res Commun. 2014;453:160–165. doi: 10.1016/j.bbrc.2014.09.076. [DOI] [PubMed] [Google Scholar]
- 35.Perez-Rivas LG, Jerez JM, Carmona R, de Luque V, Vicioso L, Claros MG, Viguera E, Pajares B, Sanchez A, Ribelles N, Alba E, Lozano J. A microRNA signature associated with early recurrence in breast cancer. PLoS One. 2014;9:e91884. doi: 10.1371/journal.pone.0091884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fu J, Xu X, Kang L, Zhou L, Wang S, Lu J, Cheng L, Fan Z, Yuan B, Tian P, Zheng X, Yu C, Ye Q, Lv Z. miR-30a suppresses breast cancer cell proliferation and migration by targeting Eya2. Biochem Biophys Res Commun. 2014;445:314–319. doi: 10.1016/j.bbrc.2014.01.174. [DOI] [PubMed] [Google Scholar]
- 37.Zhang HH, Zhang ZY, Che CL, Mei YF, Shi YZ. Array analysis for potential biomarker of gemcitabine identification in non-small cell lung cancer cell lines. Int J Clin Exp Pathol. 2013;6:1734–1746. [PMC free article] [PubMed] [Google Scholar]
- 38.Jia Z, Wang K, Wang G, Zhang A, Pu P. MiR-30a-5p antisense oligonucleotide suppresses glioma cell growth by targeting SEPT7. PLoS One. 2013;8:e55008. doi: 10.1371/journal.pone.0055008. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 39.Tang R, Liang L, Luo D, Feng Z, Huang Q, He R, Gan T, Yang L, Chen G. Downregulation of MiR-30a is Associated with Poor Prognosis in Lung Cancer. Med Sci Monit. 2015;21:2514–2520. doi: 10.12659/MSM.894372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yu Y, Cao LZ, Yang LC, Kang R, Lotze M, Tang DL. microRNA 30A promotes autophagy in response to cancer therapy. Autophagy. 2012;8:853–855. doi: 10.4161/auto.20053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ouzounova M, Vuong T, Ancey PB, Ferrand M, Durand G, Kelm FLC, Croce C, Matar C, Herceg Z, Hernandez-Vargas H. MiRcroRNA miR-30 family regulates non-attachment growth of breast cancer cells. BMC Genomics. 2013;14:139–153. doi: 10.1186/1471-2164-14-139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cheng CW, Wang HW, Chang CW, Chu HW, Chen CY, Yu JC, Chao JI, Liu HF, Ding SL, Shen CY. MicroRNA-30a inhibits cell migration and invasion by downregulating vimentin expression and is a potential prognostic marker in breast cancer. Breast Cancer Res Treat. 2012;134:1081–1093. doi: 10.1007/s10549-012-2034-4. [DOI] [PubMed] [Google Scholar]
- 43.Hu G, Wei Y, Kang Y. The multifaceted role of MTDH/AEG-1 in cancer progression. Clin Cancer Res. 2009;15:5615–5620. doi: 10.1158/1078-0432.CCR-09-0049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li WF, Ou Q, Dai H, Liu CA. Lentiviral-Mediated Short Hairpin RNA Knockdown of MTDHInhibits Cell Growth and Induces Apoptosis by Regulatingthe PTEN/AKT Pathway in Hepatocellular Carcinoma. Int J Mol Sci. 2015;16:19419–19432. doi: 10.3390/ijms160819419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhu HD, Liao JZ, He XX, Li PY. The emerging role of astrocyte-elevated gene-1 in hepatocellular carcinoma (Review) Oncol Rep. 2015;34:539–546. doi: 10.3892/or.2015.4024. [DOI] [PubMed] [Google Scholar]
- 46.Jung HI, Ahn T, Bae SH, Chung JC, Kim H, Chin S, Jeong D, Cho HD, Lee MS, Kim HC, Kim CH, Baek MJ. Astrocyte elevated gene-1 overexpression in hepatocellular carcinoma: an independent prognostic factor. Ann Surg Treat Res. 2015;88:77–85. doi: 10.4174/astr.2015.88.2.77. [DOI] [PMC free article] [PubMed] [Google Scholar]


