Abstract
The incidence and mortality of liver cancer increased year by year. Our country presents high incidence of liver cancer. MicroRNAs have tissue sensitivity as tumor biomarkers that play a role by promoting tumor growth as oncogenes or inhibit malignant cell growth as tumor suppressor genes. Studies showed that miR-15b abnormal expression in the tumor and can be treated as one of the tumor molecular markers. However, miR-15b expression and role in the liver cancer cells have not been elucidated. This study intended to explore the mechanism of miR-15b effect on liver cancer occurrence and development. Liver cancer cell line HepG2 was transfected with miR-15b mimic or inhibitor. Real-time PCR was applied to detect miR-15b expression. MTT was used to test cell proliferation. Transwell assay was performed to determine cell invasive ability. Real-time PCR and Western blot were used to detect BCL2 expression. MiR-15b mimic transfection promoted miR-15b overexpression and inhibited HepG2 cell proliferation significantly (P < 0.05). MiR-15b overexpression downregulated BCL2 mRNA and protein expression obviously (P < 0.05). On the contrary, miR-15b inhibitor transfection markedly reduced miR-15b expression in liver cancer cells (P < 0.05), promoted tumor cell proliferation, and increased BCL2 mRNA and protein expression. MiR-15b expression changes did not affect cell invasion (P > 0.05). MiR-15b can inhibit HepG2 cell proliferation and down-regulate BCL2 mRNA and protein expression.
Keywords: Liver cancer, miR-15b, BCL2, proliferation
Introduction
Liver cancer is one of the most common malignant tumors accounts for the fifth. Its mortality rate is the second in malignant tumor only behind lung cancer [1,2]. Our country has a high incidence of liver cancer that accounts for 55% of the whole world [3]. There are numerous therapy methods for liver cancer, including surgery, radiotherapy, chemotherapy, immunotherapy, and interventional therapy, etc. however, their efficacies are poor. Easy to metastasis and high recurrence result in low survival and poor quality of life of liver cancer patients. Therefore, it is a problem urgently to be solved about liver cancer occurrence, development, diagnosis, and treatment [4,5]. Following the development of molecular biology, investigation on new molecular targets for liver cancer diagnosis and treatment become another hope [6,7].
Recent study found that miRNA is associated with tumor occurrence and development by playing the role of oncogenes or tumor suppressor genes, especially in liver cancer [8-10]. MiRNA can act on the downstream target genes expression and regulate protein translation. MicroRNA-15 (miR-15) family includes miR-15a, miR-15b, and miR-192, etc [11,12]. MiR-15a has been proved to play an important role in regulation of liver cancer [13,14]. As a member of family highly homologous with miR-15a, miR-15b expression and function in liver cancer has not been fully elucidated. This study intended to explore miR-15b expression in liver cancer and its effect on liver cancer cells.
Materials and methods
Main instruments and reagents
Liver cancer cell line HepG2 was purchased from ATCC (USA). DMEM and penicillin-streptomycin were bought from Hyclone (USA). DMSO and MTT were from Gibco. Enzyme-EDTA was got from Sigma (USA). PVDF membrane was from Pall Life Sciences. Western blot related reagents were purchased from Beyotime (Shanghai, China). ECL reagent was from Amersham Biosciences. BCL2 primary antibody and HRP-tagged IgG secondary antibody were bought from Cell Signaling (USA). RNA extraction kit, reverse transcription kit, and lipo2000 were from Invitrogen (USA). MiR-15b mimics and inhibitor were synthesized by Genepharma (Shanghai, China). Transwell chamber was purchased from Hyclone (USA). Microplate reader was from BD Company (USA). DNA amplifier was from PE Gene Amp PCR System 2400 (USA). Other reagents were got from Sangon (Shanghai, China).
Methods
HepG2 cell culture and grouping
HepG2 cells stored in liquid nitrogen were unfreezed in 37°C water bath and centrifuged at 1000 rpm for 3 min. Then the cells were resuspended in medium and cultured in incubator for 24-48 h at 37°C and 5% CO2. After that, HepG2 cells were seeded in dish at 1 × 107 cells/cm2 and maintained in high glucose DMEM medium containing 10% FBS, 100 U/ml penicillin, and 100 μg/ml streptomycin. The cells were passaged every 2-3 days. HepG2 cells at 2nd-8th generation in logarithmic phase were randomly divided into four groups including miR-15b mimics NC group, miR-15b inhibitor NC group, miR-15b mimics group, and miR-15b inhibitor group.
Liposome transfection
MiR-15b mimics and miR-15b inhibitor were transfected to HepG2 cells, respectively. Their sequences were as follows: miR-15b mimics, 5’-AGGUGCAAUCGGUGUUCA-3’; miR-15b inhibitor, 5’-AUCGGGAGGUGCAUUCUA-3’; miR-15b mimics NC, 5’-AUUCAGGUCGGUGCAAUG-3’; miR-15b inhibitor, 5’-ACAGGUUUCGCAAGGUUG-3’. MiR-15b mimics and miR-15b inhibitor were mixed with 200 μl FBS-free medium for 15 min under room temperature. Then the mixed lipo2000 were incubated with miR-15b mimics or miR-15b inhibitor for 30 min under room temperature. After washed with PBS, the cells were treated with mixture maintained in FBS-free medium and incubated for 6 h. The cells were further cultured for 48 h after changing the medium.
Real-time PCR detection of miR-15b and BCL-2 expression
Total RNA was extracted from the cells with Trizol and reverse transcripted to DNA according to the manual. Primer sequences were designed by Primer 6.0 and synthesized by Invitrogen (Shanghai, China) (Table 1). Real-time PCR was applied to detect the target genes. The reaction consists one cycle of 55°C for 1 min, followed by 35 cycles of 92°C for 30 s, 58°C for 45 s, and 72°C for 35 s. GAPDH was used as reference, and the results were calculated 2-ΔCt method.
Table 1.
Primer sequence
| Gene | Forward 5’-3’ | Reverse 5’-3’ |
|---|---|---|
| GADPH | AGTACCAGTCTGTTGCTGG | TAATAGACCCGGATGTCTGGT |
| miR-15b | ATGAACTTTCTCTGTCTTGG | TCACCG CCTCGGCTTGTCACA |
| BCL-2 | CTCCCACAGACTCTGTAAG | GCATTACCTGGGGCTGTAATT |
MTT assay
HepG2 cells in logarithmic phase were seeded in 96-well plate at 5 × 103 cells concentration. The cells in each group were added with 20 μl sterile MTT every 24 h with triplicate. After another 4 h incubation, the supernatant was removed and 150 μl DMSO was added for 10 min. The plate was read at 570 nm on microplate reader to calculate proliferation rate.
Transwell chamber assay
HepG2 cells were cultured for 24 h at 48 h after transfection. 50 mg/L Matrigel at 1:5 dilution was used to coat Transwell chamber membrane. 50 μl BSA at 10 g/L in FBS-free medium was added to each well for 30 min at 37°C. Then the transwell chamber was put into 24-well plate, and 500 μl DMEM with 10% FBS was added to the lower chamber. 100 μl tumor cell suspension in FBS-free DMEM medium was added to the upper chamber and cultured for 48 h. Then the chamber was fixed with ice ethanol and stained with crystal violet for 30 min. The cell number was counted under the microscopy for 10 fields of view.
Western blot detection of BCL-2, BAX, caspase3 protein expression
The cells in each group were cracked on ice for 15-30 min and ultrasonicated for 4 × 5 s to extract protein. After centrifuged at 10000 g and 4°C for 15 min, the protein was moved to a new Ep tube and store at -20°C. The protein was separated by 10% SDS-PAGE electrophoresis and transferred to PVDF membrane. After blocked by 5% skim milk for 2 h, the membrane was incubated in BCL-2 primary antibody at 1:1000 and 4°C over night. Then the membrane was incubated with secondary antibody at 1:2000 for 30 min and washed by PBST. At last, the membrane was treated with chemiluminescent agent for 1 min and imaged on X-ray. Protein image processing system and Quantity one software were used for data analysis. All experiments were repeated for four times.
Statistical analysis
All the statistical analysis was performed on SPSS11.5 software. The data was presented as x̅ ± s. ANOVA, Dunnett’s test, and chi-square test was used for mean comparison. P < 0.05 was considered as statistically significant.
Results
miR-15b expression in HepG2 cells
Real time PCR was used to test miR-15b mimics and inhibitor effect on miR-15b expression in HepG2 cells. As shown in Figure 1, miR-15b transfection promoted miR-15b expression in HepG2 cells obviously compared with control (P < 0.05). On the other side, miR-15b inhibitor transfection suppressed miR-15b level significantly (P < 0.05).
Figure 1.

miR-15b expression in HepG2 cells. *P < 0.05, compared with mimics NC group; #P < 0.05, compared with inhibitor NC group.
miR-15b impact on HepG2 cell proliferation
MTT assay was performed to detect miR-15b impact on HepG2 cell proliferation. It was found that miR-15b mimics transfection upregulated miR-15b and inhibited HepG2 cell proliferation markedly (P < 0.05). On the contrary, miR-15b inhibitor transfection reduced miR-15b expression and promoted HepG2 cell proliferation (P < 0.05) (Figure 2). The results suggested that miR-15b overexpression in HepG2 was in favor of liver cancer cell proliferation.
Figure 2.

miR-15b impact on HepG2 cell proliferation. *P < 0.05, compared with mimics NC group; #P < 0.05, compared with inhibitor NC group.
miR-15b effect on HepG2 cell invasion
Transwell chamber assay was applied to determine miR-15b effect on HepG2 cell invasive ability. The results showed that miR-15b mimics transfection upregulated miR-15b but did not affect HepG2 cell invasive ability (P > 0.05). Similarly, miR-15b inhibitor transfection decreased miR-15b expression and also did not change HepG2 cell invasive ability (P > 0.05) (Figures 3 and 4). It indicated that miR-15b level changes do not affect tumor cell invasive ability.
Figure 3.
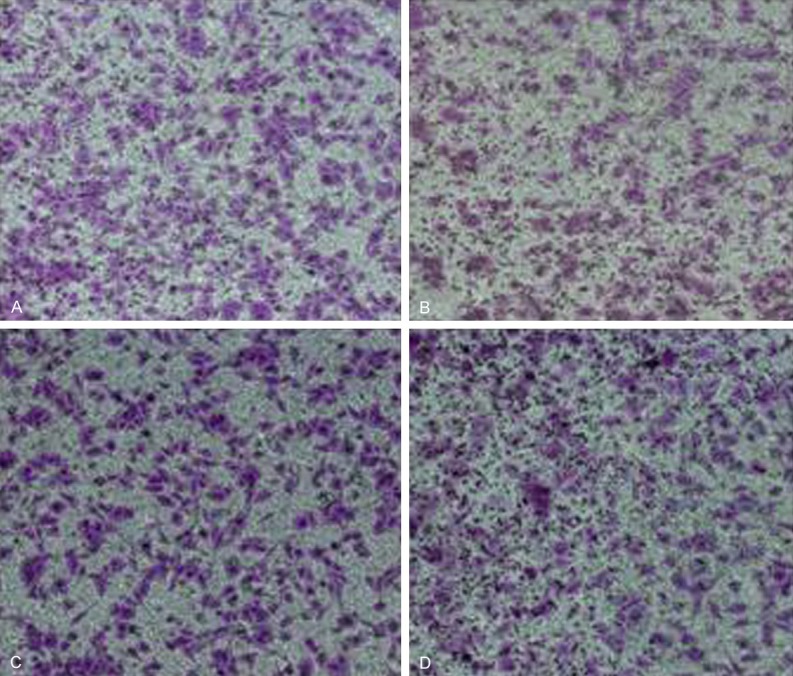
miR-15b effect on HepG2 cell invasion. A. mimics NC group; B. miR-15b mimics group; C. Inhibitor NC group; D. miR-15b inhibitor group.
Figure 4.
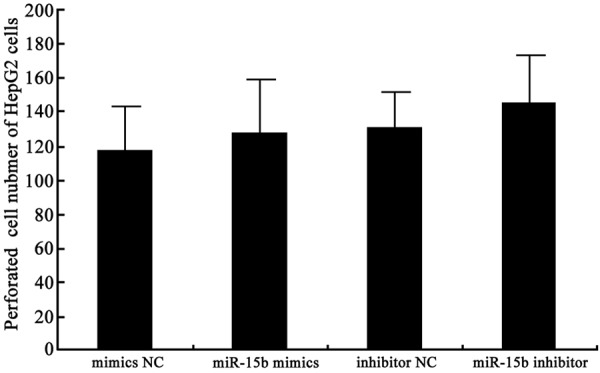
miR-15b effect on HepG2 cell invasion analysis.
miR-15b impact on target gene BCL2 mRNA and protein expression
Real time PCR and Western blot were used to detect miR-15b impact on its target gene BCL-2 mRNA and protein expression. miR-15b mimics transfection down-regulated BCL2 mRNA and protein significantly compared with control (P < 0.05), while miR-15b inhibitor increased BCL2 mRNA and protein levels obviously (P < 0.05) (Figures 5 and 6). It revealed that miR-15b overexpression in HepG2 cells help suppress liver cancer cell proliferation by regulating its target gene BCL2 mRNA and protein expression.
Figure 5.

miR-15b impact on target gene BCL2 mRNA expression.
Figure 6.
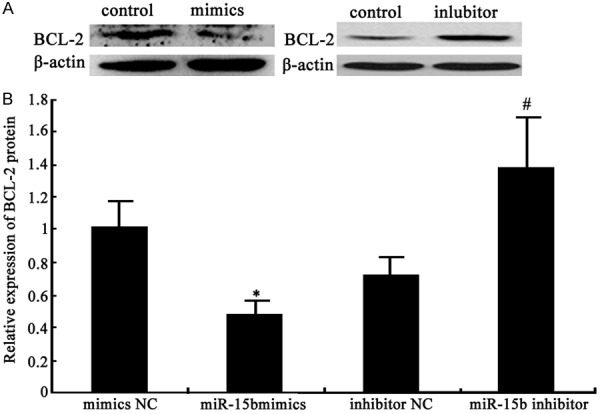
miR-15b impact on target gene BCL2 protein expression. A. miR-15b impact on BCL2 protein expression; B. miR-15b impact on BCL2 protein expression analysis *P < 0.05, compared with mimics NC group; #P < 0.05, compared with inhibitor NC group.
Discussion
The incidence of liver cancer increased day by day following the speeding up of the pace of life and the change of life style. Liver cancer’s occurrence and development is a multiple factors and multiple gene regulation process [2]. Finding the molecular pathogenesis of HCC, therefore, has important clinical significance for liver cancer early diagnosis, prevention and prognosis. Apoptosis plays a quite important role in regulating liver cancer cell occurrence and development, thus causing increasing attention [15]. The antiapoptotic mechanism of liver cancer cells is closely related to the Bcl-2 overexpression [16]. Apoptosis is a regulation to body self-stabilization that can inhibit tumor growth. Bcl-2 overexpression can let damaged cell escape from apoptosis and progress to liver cancer under related genes effect. Bcl-2 overexpression can prevent liver cancer cell apoptosis. It was thought that Bcl-2 overexpression can inhibit nucleus transfer and reduce intracellular calcium concentration, leading to antioxidation damage and imbalance of proliferation and apoptosis. It further resulted in abnormal cell increase and prolonged cell survival time, thus breaking the cell dynamic balance, promoting deterioration, and forming liver cancer [17,18].
Numerous evidences revealed that miRNA abnormal expression is involved in various biological processes, including animal and plant growth and development, cell proliferation and apoptosis, cell growth and differentiation, and cell cycle, etc [19]. MiRNAs can regulate cell death, proliferation and differentiation, development and metabolism through regulating some signal molecules, such as cell growth factor, transcription factors, and cell death genes [20]. MiR-15b widely distributed in multiple tissues and organs without specific tissue specificity, including the heart, lung, liver, kidney, and spleen. Its expression level fluctuated with body development, growth, and proliferation [21]. Our study confirmed that miR-15b transfection can promote its expression in HepG2 cells and inhibit HepG2 cell proliferation, while miR-15b inhibitor transfection resulted in the opposite results. However, miR-15b presented no impact on liver cancer cell invasive ability, suggesting that miR-15b plays its role mainly through regulating tumor cell proliferation instead of invasion.
MiRNAs usually can regulate multiple downstream target genes expression in mRNA or protein level, so as to activate different signaling pathways that affect tumor cell biology behavior [22]. BCL2 is one of the target genes of MiR15 family. In chronic lymphocytic leukemia, miR-15a expression is negatively correlated with BCL2 expression. They both can induce cell apoptosis through inhibiting BCL2 to activate the intrinsic apoptotic pathway [23]. Nevertheless, whether miR-15b can regulate liver cancer cell proliferation through BCL2 has not been confirmed. Our research proved that miR-15b overexpression can decline BCL2 mRNA and protein expression obviously. On the contrary, miR-15b inhibitor transfection can significantly decrease miR-15b expression in liver cancer cells, thus promoting BCL2 mRNA and protein levels. It confirmed that miR-15b can regulate its target gene BCL2 expression in liver cancer occurrence and development.
To sum up, miR-15b can downregulate BCL2 protein expression, promote tumor cell apoptosis, and inhibit HepG2 cell proliferation. It provides new theoretical basis for liver cancer mechanism investigation and molecular target selection.
Disclosure of conflict of interest
None.
References
- 1.Takada Y, Tohyama T, Watanabe J. Biological markers of hepatocellular carcinoma for use as selection criteria in liver transplantation. J Hepatobiliary Pancreat Sci. 2015;22:279–286. doi: 10.1002/jhbp.195. [DOI] [PubMed] [Google Scholar]
- 2.Ling D, Xia H, Park W, Hackett MJ, Song C, Na K, Hui KM, Hyeon T. pH-sensitive nanoformulated triptolide as a targeted therapeutic strategy for hepatocellular carcinoma. ACS Nano. 2014;8:8027–8039. doi: 10.1021/nn502074x. [DOI] [PubMed] [Google Scholar]
- 3.Alsaied OA, Sangwan V, Banerjee S, Krosch TC, Chugh R, Saluja A, Vickers SM, Jensen EH. Sorafenib and triptolide as combination therapy for hepatocellular carcinoma. Surgery. 2014;156:270–279. doi: 10.1016/j.surg.2014.04.055. [DOI] [PubMed] [Google Scholar]
- 4.Chan EW, Cheng SC, Sin FW, Xie Y. Triptolide induced cytotoxic effects on human promyelocytic leukemia, T cell lymphoma and human hepatocellular carcinoma cell lines. Toxicol Lett. 2001;122:81–87. doi: 10.1016/s0378-4274(01)00353-8. [DOI] [PubMed] [Google Scholar]
- 5.Huang W, He T, Chai C, Yang Y, Zheng Y, Zhou P, Qiao X, Zhang B, Liu Z, Wang J, Shi C, Lei L, Gao K, Li H, Zhong S, Yao L, Huang ME, Lei M. Triptolide inhibits the proliferation of prostate cancer cells and down-regulates SUMOspecific protease 1 expression. PLoS One. 2012;7:e37693. doi: 10.1371/journal.pone.0037693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cao L, Chang Y, Lin W, Zhou J, Tan H, Yuan Y, Zeng W. Long-term survival after resection of hepatocelluar carcinoma: a potential risk associated with the choice of postoperative analgesia. Anesth Analg. 2014;118:1309–1316. doi: 10.1213/ANE.0000000000000207. [DOI] [PubMed] [Google Scholar]
- 7.Orang AV, Barzegari A. MicroRNAs in colorectal cancer: from diagnosis to targeted therapy. Asian Pac J Cancer Prev. 2014;15:6989–6999. doi: 10.7314/apjcp.2014.15.17.6989. [DOI] [PubMed] [Google Scholar]
- 8.Gandhi NS, Tekade RK, Chougule MB. Nanocarrier mediated delivery of siRNA/miRNA in combination with chemotherapeutic agents for cancer therapy: current progress and advances. J Control Release. 2014;194:238–256. doi: 10.1016/j.jconrel.2014.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gallach S, Calabuig-Farinas S, Jantus-Lewintre E, Camps C. MicroRNAs: promising new antiangiogenic targets in cancer. Biomed Res Int. 2014;2014:878450. doi: 10.1155/2014/878450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li E, Ji P, Ouyang N, Zhang Y, Wang XY, Rubin DC, Davidson NO, Bergamaschi R, Shroyer KR, Burke S, Zhu W, Williams JL. Differential expression of miRNAs in colon cancer between African and Caucasian Americans: implications for cancer racial health disparities. Int J Oncol. 2014;45:587–594. doi: 10.3892/ijo.2014.2469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mao C, Liu H, Chen P, Ye J, Teng L, Jia Z, Cao J. Cell-specific expression of artificial microRNAs targeting essential genes exhibit potent antitumor effect on hepatocellular carcinoma cells. Oncotarget. 2015;6:5707–5719. doi: 10.18632/oncotarget.3302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim SH, Park YN, Lim JH, Choi GH, Choi JS, Kim KS. Characteristics of combined hepatocelluar-cholangiocarcinoma and comparison with intrahepatic cholangiocarcinoma. Eur J Surg Oncol. 2014;40:976–981. doi: 10.1016/j.ejso.2014.04.016. [DOI] [PubMed] [Google Scholar]
- 13.Huang YH, Lin KH, Chen HC, Chang ML, Hsu CW, Lai MW, Chen TC, Lee WC, Tseng YH, Yeh CT. Identification of postoperative prognostic microRNA predictors in hepatocellular carcinoma. PLoS One. 2012;7:e37188. doi: 10.1371/journal.pone.0037188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu Y, Zhao JJ, Wang CM, Li MY, Han P, Wang L, Cheng YQ, Zoulim F, Ma X, Xu DP. Altered expression profiles of microRNAs in a stable hepatitis B virus-expressing cell line. Chin Med J (Engl) 2009;122:10–14. doi: 10.3901/jme.2009.11.010. [DOI] [PubMed] [Google Scholar]
- 15.Fu Q, Huang X, Shu B, Xue M, Zhang P, Wang T, Liu L, Jiang Z, Zhang L. Inhibition of mitochondrial respiratory chain is involved in triptolideinduced liver injury. Fitoterapia. 2011;82:1241–1248. doi: 10.1016/j.fitote.2011.08.019. [DOI] [PubMed] [Google Scholar]
- 16.Ke Z, Li J, Zhe J, Liang LB, Yan L, Yi M. Flow cytometric analysis of asialoglycoprotein receptor expression predicts hepatic functional reserve after hepatectomy. J Coll Physicians Surg Pak. 2014;24:820–824. [PubMed] [Google Scholar]
- 17.Song S, Luo M, Song Y, Liu T, Zhang H, Xie Z. Prognostic role of SIRT1 in hepatocellular carcinoma. J Coll Physicians Surg Pak. 2014;24:849–854. [PubMed] [Google Scholar]
- 18.Rich G, Williams D, Roche E. Gastrointestinal: hepatocellular carcinoma in an esophageal varix. J Gastroenterol Hepatol. 2014;29:1950. doi: 10.1111/jgh.12806. [DOI] [PubMed] [Google Scholar]
- 19.Quann K, Jing Y, Rigoutsos I. Posttranscriptional regulation of BRCA1 through its coding sequence by the miR-15/107 group of miRNAs. Front Genet. 2015;6:242. doi: 10.3389/fgene.2015.00242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ren F, Ding H, Huang S, Wang H, Wu M, Luo D, Dang Y, Yang L, Chen G. Expression and clinicopathological significance of miR-193a-3p and its potential target astrocyte elevated gene-1 in non-small lung cancer tissues. Cancer Cell Int. 2015;15:80. doi: 10.1186/s12935-015-0227-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sodhi KK, Bahl C, Singh N, Behera D, Sharma S. Functional genetic variants in pre-miR-146a and 196a2 genes are associated with risk of lung cancer in North Indians. Future Oncol. 2015;11:2159–2173. doi: 10.2217/fon.15.143. [DOI] [PubMed] [Google Scholar]
- 22.Li Y, Liu S, Zhang F, Jiang P, Wu X, Liang Y. Expression of the microRNAs hsa-miR-15a and hsa-miR-16-1 in lens epithelial cells of patients with age-related cataract. Int J Clin Exp Med. 2015;8:2405–2410. [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang DS, Li YY, Chen XJ, Li YJ, Liu ZY, Xie WJ, Sun ZL. BCL2 promotor methylation and miR-15a/16-1 upregulation is associated with sanguinarine-induced apoptotic death in rat HSC-T6 cells. J Pharmacol Sci. 2015;127:135–144. doi: 10.1016/j.jphs.2014.11.012. [DOI] [PubMed] [Google Scholar]


