Abstract
Background: KPNA2 has effects on carcinogenesis, cell differentiation and transcriptional regulation. KPNA2 has been linked to DNA damage repair by its role to import the DNA double strand break repair complex MRN into the nucleus. The aim of our study was to evaluate the prognostic value of KPNA2 expression in both cytoplasmic and nuclear location in patients with HNSCC treated with radio(chemo)therapy. Material and methods: 225 patients with HNSCC treated with neoadjuvant, definitive or adjuvant radio(chemo)therapy were included. Immunohistochemical staining was performed on tissue micro arrays to evaluate nuclear and cytoplasmic KPNA2 expression. Results: The median fraction of tumor cells with nuclear KPNA2 expression was 15%. 47% of tumor samples showed positive cytoplasmic staining. Patients with low nuclear as well as negative cytoplasmic expression tended to have an unfavorable prognosis. There was no correlation between nuclear and cytoplasmic KPNA2 expression. Low nuclear combined with negative cytoplasmic KPNA2 had a clearly unfavorable prognostic effect in local failure-free survival (P=0.014), metastasis-free survival (P=0.001) and no evidence of disease (P=0.008). A combination of low nuclear/negative cytoplasmic with high nuclear/high cytoplasmic KPNA2 expression was prognostically unfavorable with regard to tumor specific survival (P=0.021) and to a lower extent to overall survival (P=0.18). In multivariate analysis low nuclear/negative cytoplasmic versus any high KPNA2 (P=0.008) and T-category (P=0.002) proved as independent prognostic variables. Conclusion: The combination of nuclear and cytoplasmic KPNA2 expression is a potential excellent prognostic parameter in HNSCC treated with radio(chemo)therapy.
Keywords: Radiotherapy, head and neck cancer, HNSCC, KPNA2, prognostic marker, radiotherapy
Introduction
Locally advanced head and neck squamous cell carcinoma (HNSCC) is still a disease with poor prognosis. In recent years, several new biomarkers have been identified to improve treatment response and prognosis. Karyopherin α 2 (KPNA2) is a promising biomarker [1-3] which has been studied in a variety of cancers [4]. So far only one study dealt with the role of KPNA2 in HNSCC [5]. KPNA2 has been linked to DNA damage repair by its role in importing the DNA double strand break repair complex MRN into the nucleus. This complex consists of Mre11, Rad50 and Nbs1. Deficiency of these proteins leads to an increased radiosensitivity and cancer proneness in patients with genetic alterations like Nijmegen Breakage syndrome, ataxia-telangiectasia-like disorder [6] or Nijmegen breakage syndrome-like disorder [7]. So far, KPNA2 has not been investigated as prognostic marker in HNSCC patients receiving radiotherapy. The aim of our study was to evaluate the prognostic value of KPNA2 expression in HNSCC patients treated by neoadjuvant, definitive or adjuvant radio(chemo)therapy.
Material and methods
Human specimens
Head and neck squamous cell carcinoma (HNSCC) tissue samples from 225 patients were evaluated. Patients originated from five different cohorts. The five HNSCC cohorts were characterized as follows: (i) low risk, early disease, treated by surgery and adjuvant radiotherapy (RT) [8]; (ii) high risk, advanced disease, treated by definitive radiochemotherapy (RCT) [8]; (iii) metastatic disease, treated by surgery and adjuvant RT or RCT [9]; (iv) advanced disease without distant metastasis, treated by neoadjuvant RCT [10]; (v) tumors treated by surgery and adjuvant RCT (Table 1). Patient characteristics of four of the cohorts were published previously [8-10].
Table 1.
HNSCC patient’s characteristics
| Treatment | All (%) | (i) pre RT | (ii) pre | (iii) pre RCT | (iv) | (v) adjuvant | |
|---|---|---|---|---|---|---|---|
|
|
|
||||||
| Group | Tumor resection early disease (%) | RCT biopsy advanced disease (%) | Tumor resection metastatic disease (%) | Neoadjuvant biopsy metastatic disease (%) | RCT biopsy early disease (%) | ||
| All | 225 | 55 | 35 | 20 | 26 | 89 | |
| Gender | Male | 191 | 45 | 29 | 19 | 24 | 74 |
| Female | 34 | 10 | 6 | 1 | 2 | 15 | |
| Age | Median | 58.1 | 56.0 | 60.5 | 55.4 | 55.9 | 61.2 |
| T | T1 | 35 (15.4) | 12 (21.0) | 0 (0) | 5 (24.1) | 2 (8.6) | 16 (18.0) |
| T2 | 75 (33.4) | 27 (48.4) | 1 (3.8) | 6 (27.6) | 7 (25.7) | 35 (39.2) | |
| T3 | 58 (25.7) | 12 (22.6) | 17 (48.1) | 4 (20.7) | 4 (17.1) | 20 (22.5) | |
| T4 | 57 (25.5) | 4 (8.1) | 17 (48.1) | 6 (27.6) | 13 (48.6) | 18 (20.3) | |
| N | N0 | 81 (36) | 20 (37.1) | 3 (9.6) | 0 (0) | 5 (20.0) | 52 (58.5) |
| N1 | 33 (14.8) | 12 (22.6) | 3 (7.7) | 3 (13.8) | 1 (5.7) | 14 (15.8) | |
| N2 | 98 (43.7) | 22 (40.3) | 28 (78.8) | 16 (79.3) | 19 (71.4) | 14 (15.8) | |
| N3 | 12 (5.5) | 0 (0) | 1 (3.8) | 1 (6.9) | 1 (2.9) | 9 (10.1) | |
| M | M0 | 170 (75.7) | 55 (100) | 26 (75.0) | 4 (20.7) | 24 (91.4) | 61 (68.6) |
| M1 | 55 (24.4) | 0 (0) | 9 (25.0) | 16 (79.3) | 2 (8.6) | 28 (31.5) | |
| Grading | G1/2 | 145 (64.5) | 33 (59.7) | 25 (71.2) | 10 (51.7) | 20 (77.1) | 57 (64.2) |
| G3/4 | 80 (35.5) | 22 (40.3) | 10 (28.8) | 10 (48.3) | 6 (22.9) | 32 (36.0) | |
| UICC97 | 1 | 10 (4.5) | 5 (9.7) | 0 (0) | 0 (0) | 1 (2.9) | 4 (4.5) |
| 2 | 22 (9.7) | 12 (21.0) | 0 (0) | 1 (3.4) | 1 (5.7) | 8 (9.0) | |
| 3 | 39 (17.2) | 15 (27.4) | 5 (13.5) | 1 (6.9) | 1 (5.7) | 16 (18.0) | |
| 4 | 155 (68.7) | 23 (41.9) | 30 (86.5) | 18 (89.7) | 22 (85.7) | 61 (68.7) | |
The cancer tissues were derived from pretherapeutic biopsies or the tumor resection specimen before radiochemotherapy. All samples were processed into tissue microarrays (TMA) with at least two 2 mm diameter cores per tumor as described previously and reviewed by one pathologist (K. B.). Clinical data were obtained from the Erlangen Tumour Centre Database. Written informed consent was obtained from all patients. The study was approved by the Ethics Review Committee of the University Hospital Erlangen, Erlangen, Germany.
Antibodies and immunohistochemistry
Immunohistochemistry (IHC) was performed on formalin-fixed paraffin-embedded tissue on tissue microarray sections. After standard demasking, sections were incubated with the primary polyclonal goat anti-KPNA2 (SC-6917, Santa Cruz Biotechnology; dilution 1:200) antibody as previously described [1,2,11].
KPNA2 expression
KPNA2 expression was assessed by one pathologist (K. B.) blinded to the clinical data. For each sample, both nuclear and cytoplasmic staining was evaluated. For each sample, both the presence and absence of cytoplasmic staining and the percentage of positive stained nuclei were evaluated. The median value of 15% was defined as cutoff for low and high nuclear KPNA2 expression.
Statistical analysis
Statistical analyses were performed with the SPSS for Windows software (version 21.0 SPSS, IBM, Munich, Germany). No evidence of disease, local failure-free, metastasis-free, tumor-specific and overall survivals were calculated according to Kaplan Meier. The log rank test was applied to compare survival curves between subgroups of patients. The median was used as cut-off value. Univariate and multivariate regression analyses of overall survival were performed using Cox’s proportional hazards model. The proportional hazards assumption was tested through plotting log-minus-log curves. P-values < 0.05 were considered to be significant.
Results
Distinct differences among the KPNA2 stained tissues were observed in the cohort of 225 HNSCC patients treated with neoadjuvant, definitive of adjuvant radio(chemo)therapy. (Figure 1A-D). In addition to the nuclear staining (present in 220 of 225 cases) a cytoplasmic staining was prominent in 104 cases. This prompted us to evaluate the percentage of positive stained nuclei as well as the presence/absence of a cytoplasmic staining. Between the cohorts there were differences both in nuclear staining and cytoplasmic staining (Figure 1E, 1F).
Figure 1.
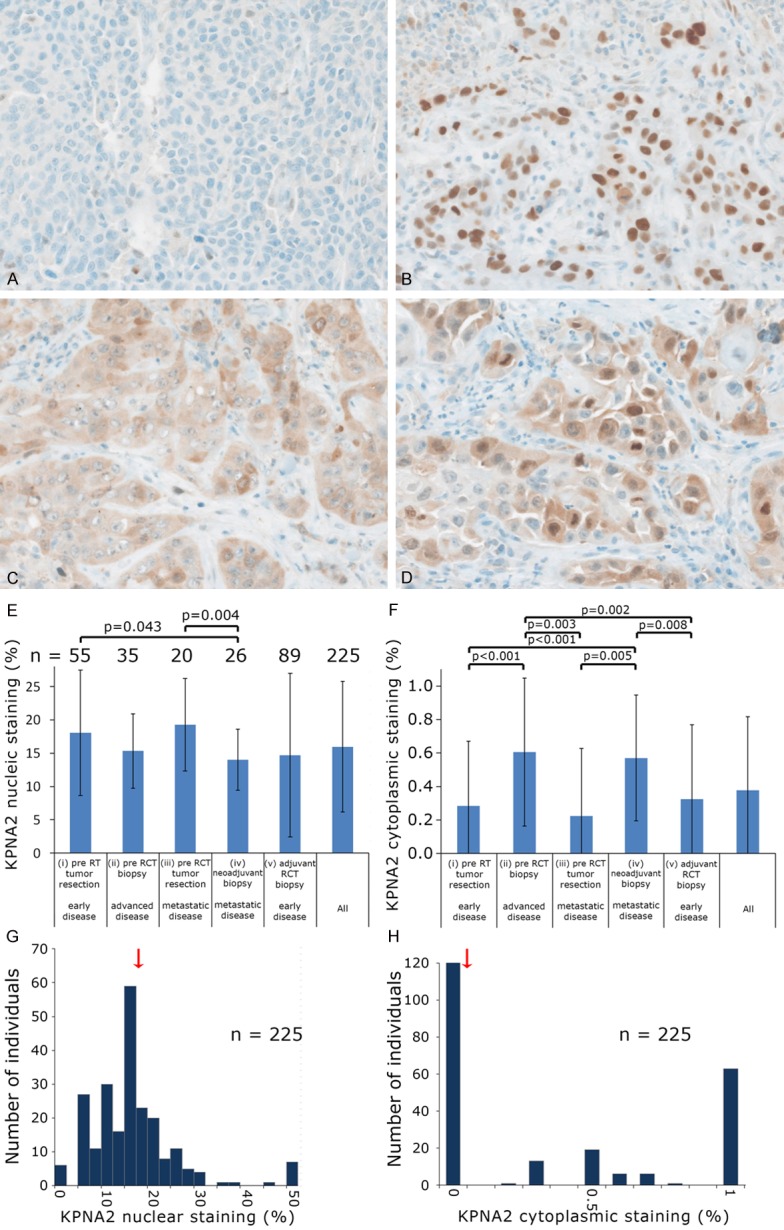
Representative images of staining patterns for KPNA2: (A) Nucleus low/cytoplasm negative; (B) Nucleus high/cytoplasm negative; (C) Nucleus low/cytoplasm positive; (D) Nucleus high/cytoplasm positive. Frequency of (E) nuclear and (F) cytoplasmic KPNA2 staining in different HNSCC cohorts. Distributions of (G) nuclear and (H) cytoplasmic KPNA2 staining. Arrow indicates the median, which was used as cut-off value.
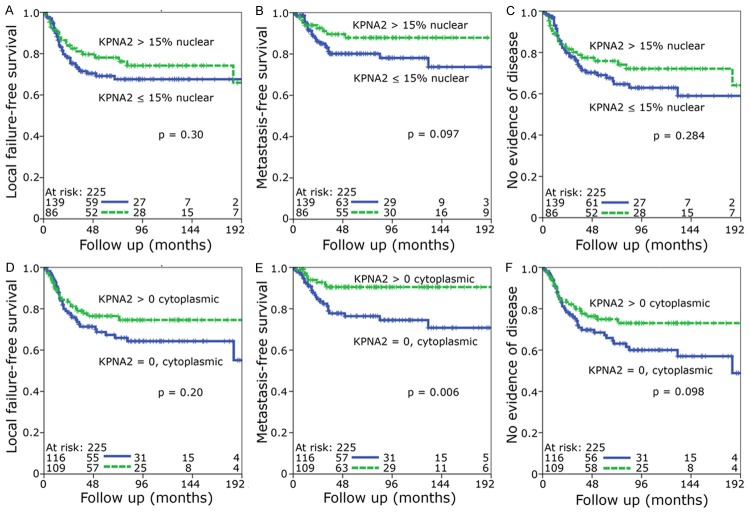
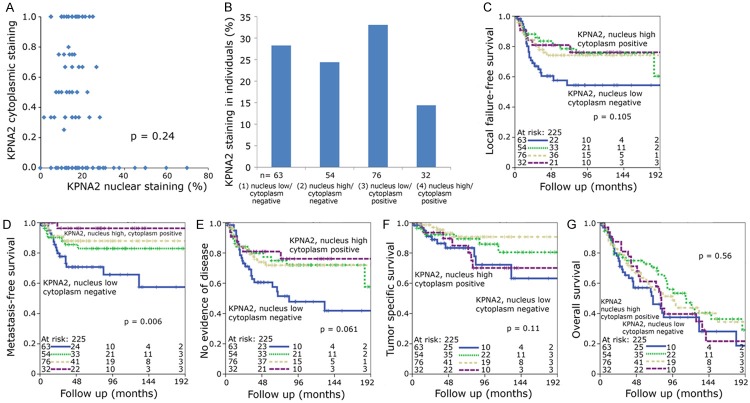
Some of the patient cohorts were quite small, so all patients were combined into one group. Most tumor samples showed a nuclear staining graded from 0% to 100% with a median value of 15% (Figure 1G) and a negative cytoplasmic staining (Figure 1H). The median value of 15% was defined as cutoff for low and high nuclear KPNA2 expression. Kaplan Meier analyses tended towards an improved local failure-free survival, metastasis-free survival and no evidence of disease for both high nuclear (defined as nuclear staining > 15%) and positive cytoplasmic KPNA2 (Figure 2A-F) with only the metastasis-free survival reaching a significant value in case of cytoplasmic staining (P=0.006) (Figure 2E). A scatter diagram analysis did not show a significant correlation between nuclear and cytoplasmic staining (P=0.24) (Figure 3A). This prompted us to define four different groups i.e. group 1 nucleus low/cytoplasm negative, group 2 nucleus high/cytoplasm negative, group 3 nucleus low/cytoplasm positive and group 4 nucleus high/cytoplasm positive for further Kaplan Meier analyses (Figure 3B).
Figure 2.
Kaplan-Meier analysis of low versus high nuclear KPNA2 expression (A-C) and negative versus positive cytoplasmic KPNA2 expression (D-F). The median was used as cut-off value. Statistical significance for the (A, D) local failure-free survival, (B-E) metastasis-free survival and (C-F) no evidence of disease were determined by the log rank test.
Figure 3.
(A) Scatter diagram comparing individuals’ KPNA2 nuclear and cytoplasmic staining. (B) Percentage of individuals with low or high nuclear KPNA2 combined with negative or positive cytoplasmic KPNA2. Kaplan-Meier graphs of (C) local failure-free survival, (D) metastasis-free survival, (E) no evidence of disease, (F) tumor specific and (G) overall survival analyzed by the log rank test. Individuals were grouped into high nuclear and positive cytoplasmic KPNA2 (long dashed line), low nuclear and positive cytoplasmic KPNA2 (short dashed line), high nuclear and negative cytoplasmic KPNA2 (dotted line) and low nuclear and negative cytoplasmic KPNA2 (solid line).
The nucleus low/cytoplasm negative group 1 was associated with an adverse local failure-free, metastasis-free survival and no evidence of disease compared to the other three groups. In case of metastasis-free survival (P=0.006) a clear difference was observed (Figure 3C-E). After dichotomization into only two different groups with only nucleus low and cytoplasm negative vs. any high KPNA2 the first group was significantly associated with a poor outcome with regard to local failure-free, metastasis-free survival and no evidence of disease (Figure 4A-C). In tumor-specific survival and overall survival-analysis the nucleus low/cytoplasm negative and nucleus high/cytoplasm positive groups tended to have an inferior prognosis (Figure 3F, 3G) which could be demonstrated again even better after dichotomization into two groups: A combination of low nuclear/negative cytoplasmic with high nuclear/high cytoplasmic KPNA2 expression was prognostically unfavorable with regard to tumor specific survival (P=0.021) and to a lower extent to overall survival (P=0.18) (Figure 4D, 4E). Multivariate analysis was performed including gender, age, TNM-category, stage, grading and KPNA2 as covariates. Only KPNA2 and T-category were independent significant variables with impact on metastasis-free survival (Table 2) (P=0.008) and no evidence of disease (Table 3) (P=0.039).
Figure 4.
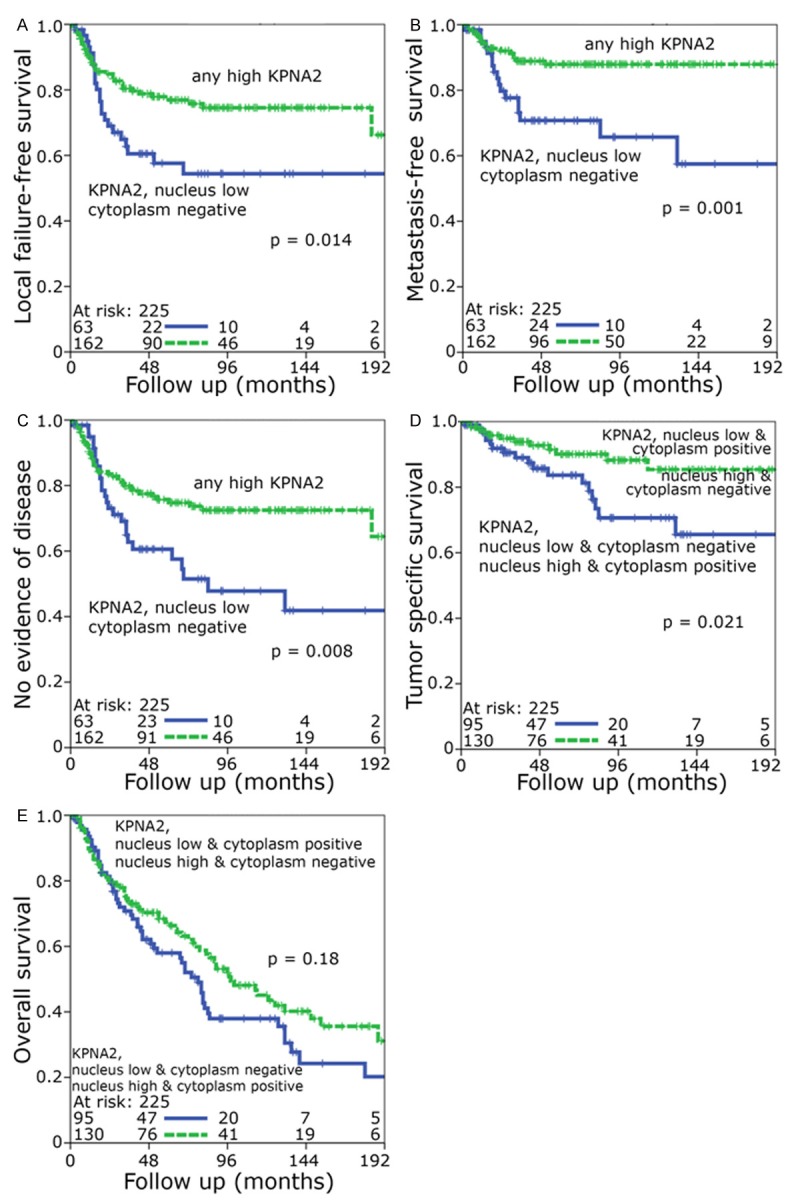
Kaplan-Meier analysis of (A) local failure-free survival, (B) metastasis-free survival and (C) no evidence of disease. Individuals with any high nuclear or positive cytoplasmic KPNA2 staining were compared to low nuclear staining combined with negative cytoplasmic staining. Kaplan-Meier analysis of (D) tumor specific survival and (E) overall survival. Individuals with low nuclear and positive cytoplasmic as well as high nuclear and negative cytoplasmic staining were compared to low nuclear and negative cytoplasmic as well as high nuclear and positive cytoplasmic staining.
Table 2.
Univariate and multivariate analysis of metastasis-free survival according to Cox’s proportional hazards model
| HNSCC tumors | Univariate analysis | Multivariate analysis | ||||
|---|---|---|---|---|---|---|
|
|
||||||
| Variable | Hazard ratio | 95% C.I. | P | Hazard ratio | 95% C.I. | P |
| Gender (male [n=191] v. female [n=34]) | 1.971 | 0.834-4.658 | 0.122 | 1.992 | 0.855-4.645 | 0.110 |
| Age. years (younger 58 years [n=114] v. older 58 years [n=111]) | 1.538 | 0.709-3.336 | 0.276 | 1.569 | 0.744-3.309 | 0.237 |
| T category (T1/T2 [n=110] v. T3/T4 [n=115]) | 3.106 | 1.314-7.341 | 0.010 | 3.491 | 1.565-7.787 | 0.002 |
| N category (N0 [n=81] v. N+ [n=144]) | 0.696 | 0.236-2.052 | 0.512 | --- | --- | --- |
| Stage (UICC I-III [n=70] v. UICC IV [n=155]) | 1.039 | 0.309-3.495 | 0.951 | --- | --- | --- |
| Grad (1 + 2 [n=145] v. 3 + 4 [n=80]) | 0.876 | 0.415-1.852 | 0.730 | --- | --- | --- |
| KPNA2 (low/negative [n=139] v. any high [n=86]) | 3.629 | 1.104-11.927 | 0.034 | 2.821 | 1.303-6.106 | 0.008 |
| KPNA2 (< 15% [n=115] v. ≥ 15% [n=110]) | 2.084 | 0.684-6.35 | 0.196 | 1.832 | 0.844-3.976 | 0.126 |
| KPNA2 (no cytoplasmic staining [n=120] v. positive [n=105]) | 1.249 | 0.363-4.298 | 0.725 | --- | --- | --- |
Table 3.
Univariate and multivariate analysis of no evidence of disease according to Cox’s proportional hazards model
| HNSCC tumor | Univariate analysis | Multivariate analysis | ||||
|---|---|---|---|---|---|---|
|
|
||||||
| Variable | Hazard ratio | 95% C.I. | P | Hazard ratio | 95% C.I. | P |
| Gender (male [n=191] v. female [n=34]) | 1.255 | 0.627-2.51 | 0.521 | --- | --- | --- |
| Age, years (younger 58 years [n=114] v. older 58 years [n=111]) | 0.842 | 0.493-1.438 | 0.529 | --- | --- | --- |
| T category (T1/T2 [n=110] v. T3/T4 [n=115]) | 1.710 | 0.967-3.023 | 0.065 | 1.580 | 0.941-2.652 | 0.083 |
| N category (N0 [n=81] v. N+ [n=144]) | 0.753 | 0.3-1.891 | 0.546 | 0.756 | 0.449-1.273 | 0.292 |
| Stage (UICC I [n=70] v. UICC II and higher [n=155]) | 1.013 | 0.37-2.776 | 0.980 | --- | --- | --- |
| Grad (1 + 2 [n=145] v. 3 + 4 [n=80]) | 0.866 | 0.501-1.497 | 0.606 | --- | --- | --- |
| KPNA2 (low/low [n=139] v. any high [n=86]) | 0.634 | 0.266-1.511 | 0.304 | 0.577 | 0.342-0.974 | 0.039 |
| KPNA2 (< 15% [n=115] v. ≥ 15% [n=110]) | 0.902 | 0.413-1.974 | 0.797 | --- | --- | --- |
| KPNA2 (no cytoplasmic staining [n=120] v. positive [n=105]) | 0.886 | 0.452-1.738 | 0.725 | --- | --- | --- |
Discussion
To our knowledge, this is the first study investigating the impact of KPNA2 in HNSCC patients undergoing radiotherapy. We found a limited prognostic value of both nuclear and cytoplasmic KPNA2 expression in HNSCC. Cytoplasmic expression of KPNA2 has not been investigated so far. In all our settings a high expression of both nuclear and cytoplasmic KPNA2 for itself tended to be related to an improved prognosis (Figure 2). This is, however, in contrast to several previous studies in a variety of malignancies, where a high nuclear expression was associated with an adverse outcome. The KPNA2 nuclear import pathway was reported to be frequently activated in various cancers and to be associated with tumourigenesis and cancer progression by transporting the cancer progression-related gene product NBS1 into the nucleus [12,13].
KPNA2 was reported to be elevated in multiple forms of cancer [14]. Rachidi et al. found elevated levels of KPNA2 in oral and laryngeal squamous cell carcinomas [5] compared to a control group. Ma and Zhao [14] found elevated expression of KPNA2 in esophageal squamous cell carcinoma (ESCC) and also found elevated concentrations of KPNA2 in serum from ESCC patients compared to control groups. It was reported to be an independent prognostic marker of poor survival e.g. in endometrial cancer [15], ovarian carcinoma [16], prostate cancer [17], gastric cancer [12] and bladder cancer [18]. However, only a few of these studies were performed in cohorts including a radio(chemo)therapeutic regime, e.g. two studies focusing on astrocytomas [19,20]. To our knowledge malignant epithelial tumors undergoing RCT has not been investigated so far.
It remains unclear whether the type of treatment (surgery, radiotherapy or radiochemotherapy) or the stage of disease may correlate with the prognostic influence of KPNA2-expression. Although there are not yet any studies focusing on KPNA2 in carcinoma patients receiving RCT, some linked molecules have already been studied in RCT-treated cohorts [21]. KPNA2 plays a key role as a so called importin transporting the MRN complex, which consists of Mre11, Rad50 and Nbs1, into the nucleus. Therefore, similar effects on tumor prognosis might be expected with regard to these linked proteins. Interestingly a significant association of low tumor Mre11 expression with worse cancer-specific survival in a radiotherapy-treated cohort was described [21]. Our at first glance contradictory observation of an association of adverse outcome with low KPNA2 expression may be explained analogously to the previously described adverse effect of Mre11 expression: Reduced KPNA2 may result in reduced nuclear availability of the double strand break repair complex MRN which may potentially cause a failure of DNA damage signaling cascade and consequently less activation of the downstream apoptotic cell death pathway. Due to the lack of cell death pathway activation, the tumor may continue to proliferate, resulting in radioresistance.
Alteration of KPNA2-mediated tumor-suppressor functions of the NBS1-complex by decreased KPNA2-expression could explain a worse response to radio(chemo)therapy. In fact, KPNA2 has been demonstrated to interact with a number of proteins including those with tumor-suppressive as well as oncogenic properties [22-25]. Consequently the multifunctional role of KPNA2 should be kept in mind. Of course further studies are required to gain more insights into the function and interaction of importin-proteins and their related molecules in DNA repair both in normal and cancer cells. Beyond doubt the influence of radiation on this system is of great interest.
The aim of this study was to evaluate the prognostic influence of KPNA2 expression in R(C)T-treated HNSCC of different stages surveying relapses, appearance of metastasis and overall-survival. The combination of low nuclear and low cytoplasmic expression was significantly associated with an adverse prognosis in our RCT-treated cohorts with regard to local failure-free, metastasis-free survival and no evidence of disease. Interestingly, in the overall survival and tumor specific-survival analysis both the low nuclear/low cytoplasmic and the high nuclear/high cytoplasmic groups tended to have an adverse prognosis. This leads to the possible assumption that rather the alteration per se irrespective of the direction of alteration may be relevant in tumor progression. It has to be discussed if possibly any aberration in KPNA2 expression concerning both the cytoplasm and the nucleus may contribute to radioresistance by dysregulation of MRN complex transport and of other cargo proteins.
These results demonstrate that the relation of the cytoplasmic and nuclear sublocations of proteins might play a so far underestimated role. However, the expression of KPNA2 in cancer tissue appears to be predominantly nuclear [4], which is in line with our finding that most tumor samples showed a negative cytoplasmic staining.
As KPNA2-expression may be a general prognostic marker in HNSCC rather than being predictive of RT treatment response alone, further studies dealing with a cohort treated by surgery only in comparison with a RT-cohort would be of interest. Promising results were already published concerning the Mre11-protein [21] where significant differences between a RT-cohort and a cystectomy cohort in bladder cancer were demonstrated.
In multivariate analysis including well accepted prognostic clinicopathologic parameters only KPNA2 and T-category were independent significant predictive variables concerning metastasis-free survival and no evidence of disease. Further studies will be needed to estimate whether KPNA2 expression is useful as a supplement to conventional clinicopathologic risk factors. Future work should also focus on functional studies in order to gain more insights into the unique function of KPNA2 and to evaluate its potential as a therapeutic target in cancer treatment with multimodal treatment schemes. In summary the combination of nuclear and cytoplasmic KPNA2 expression is a potential excellent prognostic parameter in HNSCC treated with radio(chemo)therapy.
Acknowledgements
We thank Christa Winkelmann and Rudi Jung for excellent technical assistance. We thank the Tumor Centre at the Friedrich-Alexander University Erlangen-Nürnberg, Erlangen, Germany for providing us with patient data. The present work was performed in fulfillment of the requirements for obtaining the degree “Dr. med”.
Disclosure of conflict of interest
None.
References
- 1.Dahl E, Kristiansen G, Gottlob K, Klaman I, Ebner E, Hinzmann B, Hermann K, Pilarsky C, Durst M, Klinkhammer-Schalke M, Blaszyk H, Knuechel R, Hartmann A, Rosenthal A, Wild PJ. Molecular profiling of laser-microdissected matched tumor and normal breast tissue identifies karyopherin alpha2 as a potential novel prognostic marker in breast cancer. Clin Cancer Res. 2006;12:3950–3960. doi: 10.1158/1078-0432.CCR-05-2090. [DOI] [PubMed] [Google Scholar]
- 2.Dankof A, Fritzsche FR, Dahl E, Pahl S, Wild P, Dietel M, Hartmann A, Kristiansen G. KPNA2 protein expression in invasive breast carcinoma and matched peritumoral ductal carcinoma in situ. Virchows Arch. 2007;451:877–881. doi: 10.1007/s00428-007-0513-5. [DOI] [PubMed] [Google Scholar]
- 3.Gluz O, Wild P, Meiler R, Diallo-Danebrock R, Ting E, Mohrmann S, Schuett G, Dahl E, Fuchs T, Herr A, Gaumann A, Frick M, Poremba C, Nitz UA, Hartmann A. Nuclear karyopherin alpha2 expression predicts poor survival in patients with advanced breast cancer irrespective of treatment intensity. Int J Cancer. 2008;123:1433–1438. doi: 10.1002/ijc.23628. [DOI] [PubMed] [Google Scholar]
- 4.Christiansen A, Dyrskjot L. The functional role of the novel biomarker karyopherin alpha 2 (KPNA2) in cancer. Cancer Lett. 2013;331:18–23. doi: 10.1016/j.canlet.2012.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rachidi SM, Qin T, Sun S, Zheng WJ, Li Z. Molecular profiling of multiple human cancers defines an inflammatory cancer-associated molecular pattern and uncovers KPNA2 as a uniform poor prognostic cancer marker. PLoS One. 2013;8:e57911. doi: 10.1371/journal.pone.0057911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Regal JA, Festerling TA, Buis JM, Ferguson DO. Disease-associated MRE11 mutants impact ATM/ATR DNA damage signaling by distinct mechanisms. Hum Mol Genet. 2013;22:5146–5159. doi: 10.1093/hmg/ddt368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Waltes R, Kalb R, Gatei M, Kijas AW, Stumm M, Sobeck A, Wieland B, Varon R, Lerenthal Y, Lavin MF, Schindler D, Dork T. Human RAD50 deficiency in a Nijmegen breakage syndrome-like disorder. Am J Hum Genet. 2009;84:605–616. doi: 10.1016/j.ajhg.2009.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Distel LV, Fickenscher R, Dietel K, Hung A, Iro H, Zenk J, Nkenke E, Buttner M, Niedobitek G, Grabenbauer GG. Tumour infiltrating lymphocytes in squamous cell carcinoma of the oro- and hypopharynx: prognostic impact may depend on type of treatment and stage of disease. Oral Oncol. 2009;45:e167–174. doi: 10.1016/j.oraloncology.2009.05.640. [DOI] [PubMed] [Google Scholar]
- 9.Pretscher D, Distel LV, Grabenbauer GG, Wittlinger M, Buettner M, Niedobitek G. Distribution of immune cells in head and neck cancer: CD8+ T-cells and CD20+ B-cells in metastatic lymph nodes are associated with favourable outcome in patients with oro- and hypopharyngeal carcinoma. BMC Cancer. 2009;9:292. doi: 10.1186/1471-2407-9-292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tabachnyk M, Distel LV, Buttner M, Grabenbauer GG, Nkenke E, Fietkau R, Lubgan D. Radiochemotherapy induces a favourable tumour infiltrating inflammatory cell profile in head and neck cancer. Oral Oncol. 2012;48:594–601. doi: 10.1016/j.oraloncology.2012.01.024. [DOI] [PubMed] [Google Scholar]
- 11.Tehrany N, Kitz J, Rave-Frank M, Lorenzen S, Li L, Kuffer S, Hess CF, Burfeind P, Reichardt HM, Canis M, Beissbarth T, Wolff HA. Highgrade acute organ toxicity and p16 expression as positive prognostic factors in primary radio(chemo)therapy for patients with head and neck squamous cell carcinoma. Strahlenther Onkol. 2015;191:566–572. doi: 10.1007/s00066-014-0801-3. [DOI] [PubMed] [Google Scholar]
- 12.Altan B, Yokobori T, Mochiki E, Ohno T, Ogata K, Ogawa A, Yanai M, Kobayashi T, Luvsandagva B, Asao T, Kuwano H. Nuclear karyopherinalpha2 expression in primary lesions and metastatic lymph nodes was associated with poor prognosis and progression in gastric cancer. Carcinogenesis. 2013;34:2314–2321. doi: 10.1093/carcin/bgt214. [DOI] [PubMed] [Google Scholar]
- 13.Nadler SG, Tritschler D, Haffar OK, Blake J, Bruce AG, Cleaveland JS. Differential expression and sequence-specific interaction of karyopherin alpha with nuclear localization sequences. J Biol Chem. 1997;272:4310–4315. doi: 10.1074/jbc.272.7.4310. [DOI] [PubMed] [Google Scholar]
- 14.Ma S, Zhao X. KPNA2 is a promising biomarker candidate for esophageal squamous cell carcinoma and correlates with cell proliferation. Oncol Rep. 2014;32:1631–1637. doi: 10.3892/or.2014.3381. [DOI] [PubMed] [Google Scholar]
- 15.Ikenberg K, Valtcheva N, Brandt S, Zhong Q, Wong CE, Noske A, Rechsteiner M, Rueschoff JH, Caduff R, Dellas A, Obermann E, Fink D, Fuchs T, Krek W, Moch H, Frew IJ, Wild PJ. KPNA2 is overexpressed in human and mouse endometrial cancers and promotes cellular proliferation. J Pathol. 2014;234:239–252. doi: 10.1002/path.4390. [DOI] [PubMed] [Google Scholar]
- 16.Huang L, Wang HY, Li JD, Wang JH, Zhou Y, Luo RZ, Yun JP, Zhang Y, Jia WH, Zheng M. KPNA2 promotes cell proliferation and tumorigenicity in epithelial ovarian carcinoma through upregulation of c-Myc and downregulation of FOXO3a. Cell Death Dis. 2013;4:e745. doi: 10.1038/cddis.2013.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grupp K, Habermann M, Sirma H, Simon R, Steurer S, Hube-Magg C, Prien K, Burkhardt L, Jedrzejewska K, Salomon G, Heinzer H, Wilczak W, Kluth M, Izbicki JR, Sauter G, Minner S, Schlomm T, Tsourlakis MC. High nuclear karyopherin alpha 2 expression is a strong and independent predictor of biochemical recurrence in prostate cancer patients treated by radical prostatectomy. Mod Pathol. 2014;27:96–106. doi: 10.1038/modpathol.2013.127. [DOI] [PubMed] [Google Scholar]
- 18.Jensen JB, Munksgaard PP, Sorensen CM, Fristrup N, Birkenkamp-Demtroder K, Ulhoi BP, Jensen KM, Orntoft TF, Dyrskjot L. High expression of karyopherin-alpha2 defines poor prognosis in non-muscle-invasive bladder cancer and in patients with invasive bladder cancer undergoing radical cystectomy. Eur Urol. 2011;59:841–848. doi: 10.1016/j.eururo.2011.01.048. [DOI] [PubMed] [Google Scholar]
- 19.Gousias K, Becker AJ, Simon M, Niehusmann P. Nuclear karyopherin a2: a novel biomarker for infiltrative astrocytomas. J Neurooncol. 2012;109:545–553. doi: 10.1007/s11060-012-0924-2. [DOI] [PubMed] [Google Scholar]
- 20.Gousias K, Niehusmann P, Gielen GH, Simon M. Karyopherin a2 and chromosome region maintenance protein 1 expression in meningiomas: novel biomarkers for recurrence and malignant progression. J Neurooncol. 2014;118:289–296. doi: 10.1007/s11060-014-1423-4. [DOI] [PubMed] [Google Scholar]
- 21.Choudhury A, Nelson LD, Teo MT, Chilka S, Bhattarai S, Johnston CF, Elliott F, Lowery J, Taylor CF, Churchman M, Bentley J, Knowles MA, Harnden P, Bristow RG, Bishop DT, Kiltie AE. MRE11 expression is predictive of cause-specific survival following radical radiotherapy for muscle-invasive bladder cancer. Cancer Res. 2010;70:7017–7026. doi: 10.1158/0008-5472.CAN-10-1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Marchenko ND, Hanel W, Li D, Becker K, Reich N, Moll UM. Stress-mediated nuclear stabilization of p53 is regulated by ubiquitination and importin-alpha3 binding. Cell Death Differ. 2010;17:255–267. doi: 10.1038/cdd.2009.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Narod SA, Foulkes WD. BRCA1 and BRCA2: 1994 and beyond. Nat Rev Cancer. 2004;4:665–676. doi: 10.1038/nrc1431. [DOI] [PubMed] [Google Scholar]
- 24.Wang CI, Chien KY, Wang CL, Liu HP, Cheng CC, Chang YS, Yu JS, Yu CJ. Quantitative proteomics reveals regulation of karyopherin subunit alpha-2 (KPNA2) and its potential novel cargo proteins in nonsmall cell lung cancer. Mol Cell Proteomics. 2012;11:1105–1122. doi: 10.1074/mcp.M111.016592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zannini L, Lecis D, Lisanti S, Benetti R, Buscemi G, Schneider C, Delia D. Karyopherinalpha2 protein interacts with Chk2 and contributes to its nuclear import. J Biol Chem. 2003;278:42346–42351. doi: 10.1074/jbc.M303304200. [DOI] [PubMed] [Google Scholar]