Abstract
MALAT1 is an important long noncoding RNA in tumor progression. Here we showed that the expression of MALAT1 was upregulated in non-small cell lung cancer cells (NSCLCs) or tissues as compared with the normal lung cell or tissues. Thus, the knockdown of MALAT1 led to decreased cell migration and invasion. Next we also found that CXCL5 as a downstream gene of MALAT1 regulated cell migration and invasion. However the regulation of MALAT1 expression was rarely known. Here we found that the treatment with SAM suppressed of MALAT1 expression. Finally, we showed that the methylated forms of MALAT1 promoter in lung cancer cells or tissues decreased compared with normal lung cells or tissues. These demonstrated that the expression of MALAT1 was dependent on the methylation. Overall, our findings illuminate the oncogenic function of MALAT1 which is regulated by DNA methylation that might provide potential clinical application in NSCLC.
Keywords: MALAT1, non-small cell lung cancer, CXCL5, methylation, cellular migration and invasion
Introduction
Mammalian genomes can transcribe into large numbers of non-coding RNAs including long non-coding RNA [1]. Recently, an increasing number of long non-coding RNAs have been identified [2,3]. There are growing evidence that many long ncRNAs have the potential function in diverse biological processes, such as invasiveness and metastasis [4,5].
Metastasis-associated lung adenocarcinoma transcript 1 (MALAT-1) is a long non-coding RNA (~8.7 kb in humans) that was originally found to be overexpressed in early-stage NSCLCs as a prognostic marker for metastasis [6-8]. MALAT1 can be processed into a stable nuclear-retained non-coding RNA and a small tRNA-like cytoplasmic RNA [9]. MALAT1 is up-regulated in many human carcinomas such as these cancers of the breast, pancreas, lung, colon, prostate, and liver [10,11]. MALAT1 has been also found to be involved in epigenetic process. MALAT1 localizes to hundreds of genomic sites in human cells and binds the active chromatin sites [12]. Importantly MALAT1 regulates the chromatin histone methylations status through binding to PRC2 complex to modulate target genes [13,14]. However, the epigenetic regulation of MALAT1 expression is less known.
In present study we focused on the role of long non-coding RNA MALAT1 in non-small cell lung cancer. We investigated the expression of MALAT1 and its function in lung cancer. We identified MALAT1 as an upregualted lncRNA promoted cell invasion by regulating expression of CXCL5 and further found the regulation of MALAT1 expression by promoter methylation.
Materials and methods
Cells and human tissue samples
NSCLC cells A549 were purchased from ATCC. The cells were cultured in RPMI-1640 (Gibco) supplemented with 10% bovine calf serum (BCS) (Gibco) at 37°C in a humidified atmosphere containing 5% CO2. Fifteen paired cancerous and adjacent normal lung tissues were obtained from NSCLC patients receiving surgical treatment at Department of Lung Cancer Surgery, Tianjin Medical University General Hospital. Informed consent was obtained from all patients with regards to surgical treatment and acquisition of tissue specimens. Histological typing of tumor tissue and adjacent normal tissue were pathologically confirmed in all cases. Acquisition of tissue specimens was approved by local Institutional Review Boards at the authors’ affiliated institutions. Human tissue acquisition and use in this study complied with the National Regulations on the Use of Clinical Samples in China.
Real-time RT-PCR
Total cellular RNA was isolated from tissues and cells using a Trizol reagent (Invitrogen, Carlsbad, CA, USA), and reversely transcribed using a reverse transcription-PCR System (Takara, Dalian, China). MALAT1 were analyzed by real-time RT-PCR. Quantitative real-time RT-PCR (qRT-pcr) was performed by the ∆∆Ct method using a SYBR® Green PCR Master Mix kit (Takara). In brief, 0.2 μg total RNA was mixed with the SYBR Green reaction mix and specific primers. Quantitative RT-PCR was run using an ABI PRISM 7900HT Sequence Detection System. Data were analyzed with StepOne Software V2.1 (Applied Biosystems, CA). Each PCR was performed in triplicate and for at least 3 times independently. GAPDH was used to normalize the expression of MALAT1.
Small interfering RNA and transfection
Small interfering RNAs (siRNAs) targeting MALAT1 or the corresponding negative control were designed and synthesized by Guangzhou RiboBio (Guangzhou, China). For transfection, tumor cells were seeded in 24-well plates and transfected with 2 μl (20 μM) siRNA using a Lipofectamine™ 2000 reagent (Invitrogen). Each experiment was done in triplicate and at least three times independently.
Wound healing and invasion assays
For the healing assay, cells were grown in 10% RPMI-1640 in 60-mm plates in mono-layer to sub-confluence, and were wounded by scraping off the cells. The distance of cell migration was monitored at 24 h (relative to the edge immediately after the scrapping) and imaged under a microscope. The relative migrating distance of cells was measured. For the invasion assay, cells at 104/well were seeded in the upper chamber of the transwell insert coated with matrigel in 200 μL FBS-free RPMI-1640 and the lower wells were filled with 500 μL 10% FBS RPMI-1640 for inducing cell migration. Following incubation for 24 h, the cells on the filter surface were fixed with 4% formaldehyde, stained with 0.5% crystal violet, and examined under a microscope. Cells in at least six random microscopic fields (200×) were counted.
Methylation specific PCR (MSP)
DNA was extracted by using phenol-chloroform and treated with bisulfite using the EzDNA Methylation kit (Zymoresearch, Orange, CA) according to the manufacturer’s instruction. Bisulfite-treated DNA was then used for methylation specific PCR (MSP) reaction. PCR was performed at 95°C for 1 min followed by a total of 35 cycles of 95°C for 30 s, 60°C for 30 s and 68°C for 1 min 30 s with a final extension at 68°C for 10 min. The PCR products were analyzed on a 2% agarose gel.
Statistical analysis
Data are expressed as mean ± standard deviation, and analyzed with ANOVA, followed by Student’s t-test for pair-wise comparisons using the GraphPad Prism software version 4.0 (GraphPad Software, Inc., San Diego, CA). A value of P<0.05 was considered as statistical significant.
Results
MALAT1 expression was upregulated in lung cancer.
To identify the expression in lung cancer, the real-time PCR was performed. Firstly, we analyzed the expression of MAlALT1 in human normal lung cell (NC) and in lung cancer cells and observed that the expression of MALAT1 was upregulated in lung cancer cells as compared with NC (Figure 1A). Then the expression of MALAT1 in lung cancer and adjacent normal tissues were assessed. As shown in Figure 1B the expression of MALAT1 in lung cancer tissues was remarkably increased than the adjacent normal tissues. These data suggested that MALAT1 expression was upregulated in malignant lung cancer cell lines and tissues.
Figure 1.
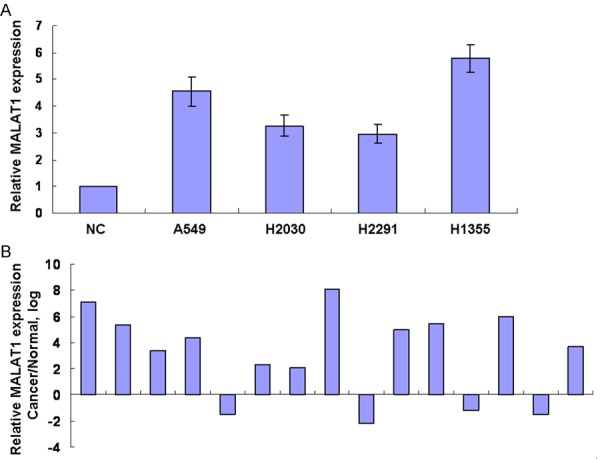
MALAT1 expression is upregulated in lung cancer. A. RNA was extracted from human normal lung cell (NC) and in lung cancer cells. MALAT1 expression was assessed by real time PCR. The expression was normalized to GAPDH. B. MALAT1 expression was analyzed in lung cancer and adjacent normal tissues by real time PCR and normalized to GAPDH.
Knockdown of MALAT1 decreases cell invasion
To explore the potential functional role of MALAT1 in lung cancer, we suppressed the expression of MALAT1 by RNA interference in lung cancer cells. A549 cells were transfected with siRNA targeting for MALAT1 (siM) or control siRNA (siCon). MALAT1 expression was decreased after 24 h of transfection in cells transfected with siRNA targeting for MALAT1 (Figure 2A). Downregulation of MALAT1 expression inhibited the migration of lung cells by wound healing analysis (Figure 2B). Next, we assessed the effect of MALAT1 on cell invasion by transwell analysis. MALAT1 silencing resulted in a significant decrease in invasion (Figure 2C). Together, these data indicated that MALAT1 modulated cell migration and invasion.
Figure 2.
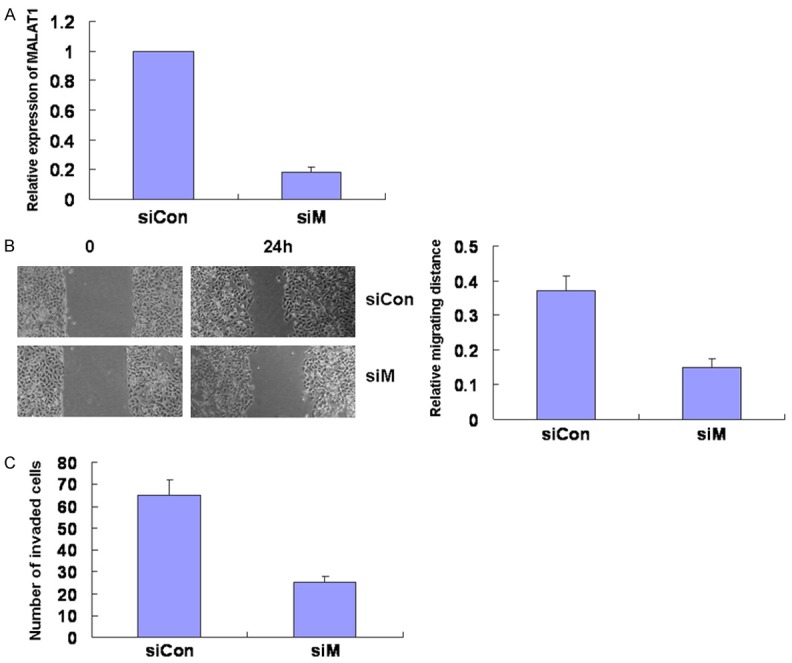
Knockdown of MALAT1 decreases cell migration and invasion. A549 cells were transfected with siRNA targeting for MALAT1 (siM) or control siRNA (siCon). A. After 24 h of transfection, cells were collected and MALAT1 expression was assessed by real time PCR. B. At 24 h after transfection, cells were scratched in 6-well plates. After 24 h, the scratch widths were measured under microscope and analyzed. C. After 24 h of transfection, cells were added onto cell culture inserts coated with matrigel inside matrigel invasion chambers. Cells invaded through matrigel to the bottom side of the inserts were stained and counted.
Knockdown of CXCL5 suppressed the migration and invasion
It is reported that MALAT1 knockout in lung cancer cells showed a significant deregulation of CXCL5 gene [15]. In this study, the MALAT1 silencing reduced the expression of CXCL5 in NSCLC (Figure 3A). To investigate the role of CXCL5 in NSCLC progression, the CXCL5 was knocked down in A549 cells and the cell migration and invasion was examined. The results showed that the knockdown of CXCL5 suppressed the cell migration and invasion ability (Figure 3B, 3C). These suggested that MALAT1 targeted CXCL5 and this interaction may mediate the effects of MALAT1 on the cell migration and invasion.
Figure 3.
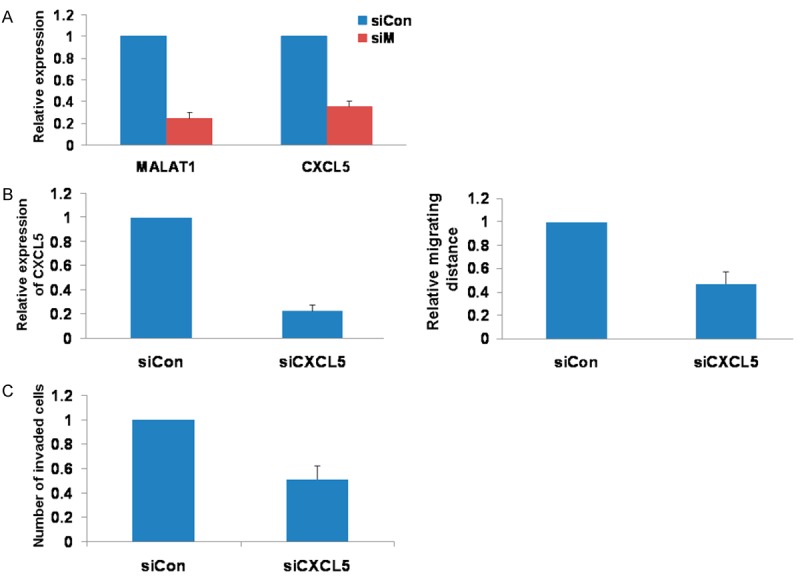
Knockdown of CXCL5 suppressed the migration and invasion. A. The knockdown of MALAT1 induced the decrease of CXCL5 expression. B. The knockdown of CXCL5 suppressed the cell migration ability. C. The knockdown of CXCL5 inhibited cell invasion.
MALAT1 expression was dependent on the methylation
DNA methylation is a dynamic and reversible process that governs gene expression during development and disease [16]. We want to explore if DNA methylation contributed to the expression of MALAT1 in lung cancer. We analyzed the CpG island in MALAT1 promoter with UCSC genome browser, MethPrimer and EMBOSS Cpgplot prediction software and found that CpG islands are located within 2 kb of the human MALAT1 promoters (Figure 4A). To analyze the methylation in regulation of MALAT1 in lung cancer, we evaluated the effect of the methyl donor S-Adenosylmethionine (SAM) on MALAT1 expression. We found that MALAT1 expression was suppressed by incubation with SAM in A549 cells (Figure 4B). And then we evaluated methylation status of the CpG island at the MALAT1 promoter in lung cancer cells by performing a methylation specific PCR (Figure 4C). Although normal lung cells and lung cancer cells have both the methylated and unmethylated forms, the methylated forms in lung cancer cells decrease compared with normal lung cells. Finally MSP of DNA from lung cancer and normal tissues showed a marked reduction of the methylated form in lung cancer compared with adjacent tissues (Figure 4D).
Figure 4.
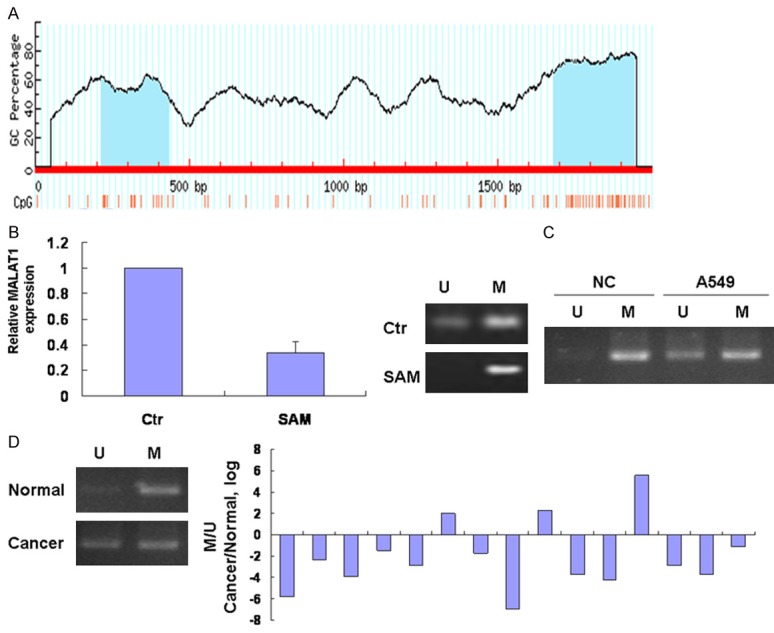
MALAT1 expression is dependent on the methylation. A. The CpG island in MALAT1 promoter was analyzed with UCSC genome browser prediction software. B. Cells were incubated with SAM 10 µM for 24 h and MALAT1 expression was assessed by real time PCR. Right panel was methylation level of MALAT1. C. DNA was extracted by using phenol-chloroform and treated with bisulfite using the EzDNA Methylation kit (Zymoresearch, Orange, CA, USA). Bisulfite-treated DNA was then used for methylation specific PCR (MSP) reaction. PCR products were identified by ethidium bromide staining after 2% agarose gel electrophoresis. For each sample a primer set for the methylated (M) and unmethylated (U) copies of MALAT1 gene was used. D. Methylation specific PCR analysis of DNA from lung cancer and normal tissues was performed.
Discussion
LncRNAs are found to play important roles in cancer [17]. We previously reported that inhibition of MALAT1 in CaSki human cervical cancer cells suppressed cell proliferation and invasion [18]. In this study, we also assessed the role of MALAT1 in NSCLC.
To better understand the roles of MALAT1 in NSCLC, we first measured the expression of MALAT1 in lung cancer and we found that the expression of MALAT1 was upregulated in lung cancer cells as compared with NC and the expression of MALAT1 in lung cancer tissues was remarkably increased than the adjacent normal tissues. Over-expression of MALAT1 has been reported in other multiple cancer types [19,20]. These suggested that MALAT1 plays an important role on tumor progression and may serve as a prognostic tumor biomarker.
MALAT1 has been related to cell migration and invasion of cancer [21]. It is reported that MALAT1 promoted colorectal cancer cell and esophageal squamous cell carcinoma proliferation and metastasis [22,23]. MALAT1 enhanced migration of glioblastoma cells [24]. MALAT1 induced neuroblastoma cell migration and invasion [25]. MALAT1 is also a critical regulator of the metastasis phenotype of lung cancer, gastric and bladder cancer cells [26-28]. Here knockdown of MALAT1 decreases NSCLC cell migration and invasion.
CXCL5 as an epithelial-derived neutrophil-activating peptide belongs to the CXC chemokine family. CXCL5 as a novel serum prognostic marker in patients with colorectal cancer promote cell proliferation, migration and invasion [29]. Furthermore, CXCL5 promotes prostate and bladder cancer progression [30,31]. CXCL5 overexpression was associated with late stage gastric cancer and high N stage [32]. In addition, CXCL5 enhanced proliferation and invasion of head and neck squamous cell carcinomas [33]. CXCL5 is a potential novel prognostic factor in early stage non-small cell lung cancer and passive immunization of NSCLC tumor-bearing mice with neutralizing anti-CXCL5 antibodies resulted in reduced tumor growth, tumor vascularity and metastases [34,35]. The present study showed that CXCL5 as a downstream gene of MALAT1 mediated the function of MALAT1 regulating the NSCLC migration and invasion.
Although there are lots of studies about tumor phenotype caused by MALAT1 abnormal expression, the mechanisms of MALAT1 expression is little known. Epigenetic changes such as DNA methylation act to regulate gene expression in normal mammalian development [36]. But promoter hypermethylation or hypomethylation also plays a major role in cancer by regulating gene expression. More attention is paid to the long non-coding RNA as a major regulator in cancer epigenetics. Long non-coding RNA HOX antisense intergenic RNA (HOTAIR) plays a critical role in gene regulation and chromatin dynamics in a variety of cancers [37]. TARID as a long non-coding RNA was found to direct demethylation and activation of the tumor suppressor TCF21 via GADD45A [38]. In this study the methylated forms of MALAT1 promoter in lung cancer cells or tissues decreased compared with normal lung cells or tissues. Treatment with SAM suppressed of MALAT1. These data suggested that MALAT1 expression can be regulated by DNA methylation. However yang et al reported that there are no effects of CpG island methylation status on MALAT1 expression in esophageal squamous cell carcinoma [23]. This might be the reason of different kinds of cancer.
In summary, we have identified the overexpression of MALAT1 in NSCLC, demonstrated functional effect and provided an epigenetic mechanism of regulation of MALAT1 expression in NSCLC. This study also extends the known role of MALAT1 in NSCLC and might help identify potential clinical application in NSCLC.
Acknowledgements
This work was supported by grants from Tianjin Education Committee Foundation (No. 20110101; to FJ Guo), the National Natural Science Foundation of China (No. 81201645; to FJ Guo), Doctoral Program Foundation of Institutions of Higher Education (No. 20121202120008; to FJ Guo), the National Natural Science Foundation of China (No. 30973383; to ZG Li), the Key Project from National Natural Science Foundation of China (No. 30430300); to QH Zhou, National 863 Program (No. 2006AAOZA401; to QH Zhou), National 973 Program (No. 2010CB529405; to QH Zhou) and China-Sweden Cooperative Foundation (No. 09ZCZDSF04100; to QH Zhou).
Disclosure of conflict of interest
None.
References
- 1.Mercer TR, Dinger ME, Mattick JS. Long noncoding RNAs: insights into functions. Nat Rev Genet. 2009;10:155–159. doi: 10.1038/nrg2521. [DOI] [PubMed] [Google Scholar]
- 2.Johnsson P, Lipovich L, Grandér D, Morris KV. Evolutionary conservation of long non-coding RNAs; sequence, structure, function. Biochim Biophys Acta. 2014;1840:1063–1071. doi: 10.1016/j.bbagen.2013.10.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.He X, Chen X, Zhang X, Duan X, Pan T, Hu Q, Zhang Y, Zhong F, Liu J, Zhang H, Luo J, Wu K, Peng G, Luo H, Zhang L, Li X, Zhang H. An Lnc RNA (GAS5)/SnoRNA-derived piRNA induces activation of TRAIL gene by site-specifically recruiting MLL/COMPASS-like complexes. Nucleic Acids Res. 2015;43:3712–3725. doi: 10.1093/nar/gkv214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Satpathy AT, Chang HY. Long noncoding RNA in hematopoiesis and immunity. Immunity. 2015;42:792–804. doi: 10.1016/j.immuni.2015.05.004. [DOI] [PubMed] [Google Scholar]
- 5.Shi SJ, Wang LJ, Yu B, Li YH, Jin Y, Bai XZ. LncRNA-ATB promotes trastuzumab resistance and invasion-metastasis cascade in breast cancer. Oncotarget. 2015;6:11652–11663. doi: 10.18632/oncotarget.3457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ji P, Diederichs S, Wang W, Böing S, Metzger R, Schneider PM, Tidow N, Brandt B, Buerger H, Bulk E, Thomas M, Berdel WE, Serve H, Müller-Tidow C. MALAT-1, a novel noncoding RNA, and thymosin beta4 predict metastasis and survival in early-stage non-small cell lung cancer. Oncogene. 2003;22:8031–8041. doi: 10.1038/sj.onc.1206928. [DOI] [PubMed] [Google Scholar]
- 7.Hutchinson JN, Ensminger AW, Clemson CM, Lynch CR, Lawrence JB, Chess A. A screen for nuclear transcripts identifies two linked noncoding RNAs associated with SC35 splicing domains. BMC Genomics. 2007;8:39. doi: 10.1186/1471-2164-8-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Szymanski M, Barciszewska MZ, Erdmann VA, Barciszewski J. A new frontier for molecular medicine: noncoding RNAs. Biochim Biophys Acta. 2005;1756:65–75. doi: 10.1016/j.bbcan.2005.07.005. [DOI] [PubMed] [Google Scholar]
- 9.Wilusz JE, Freier SM, Spector DL. 3’ end processing of a long nuclear-retained noncoding RNA yields a tRNA-like cytoplasmic RNA. Cell. 2008;135:919–932. doi: 10.1016/j.cell.2008.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lin R, Maeda S, Liu C, Karin M, Edgington TS. A large noncoding RNA is a marker for murine hepatocellular carcinomas and a spectrum of human carcinomas. Oncogene. 2007;26:851–858. doi: 10.1038/sj.onc.1209846. [DOI] [PubMed] [Google Scholar]
- 11.Wang X, Li M, Wang Z, Han S, Tang X, Ge Y, Zhou L, Zhou C, Yuan Q, Yang M. Silencing of long noncoding RNA MALAT1 by miR-101 and miR-217 inhibits proliferation, migration, and invasion of esophageal squamous cell carcinoma cells. J Biol Chem. 2015;290:3925–3935. doi: 10.1074/jbc.M114.596866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.West JA, Davis CP, Sunwoo H, Simon MD, Sadreyev RI, Wang PI, Tolstorukov MY, Kingston RE. The long noncoding RNAs NEAT1 and MALAT1 bind active chromatin sites. Mol Cell. 2014;55:791–802. doi: 10.1016/j.molcel.2014.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang L, Lin C, Liu W, Zhang J, Ohgi KA, Grinstein JD, Dorrestein PC, Rosenfeld MG. ncRNAand Pc2 methylation-dependent gene relocation between nuclear structures mediates gene activation programs. Cell. 2011;147:773–788. doi: 10.1016/j.cell.2011.08.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guil S, Soler M, Portela A, Carrère J, Fonalleras E, Gómez A, Villanueva A, Esteller M. Intronic RNAs mediate EZH2 regulation of epigenetic targets. Nat Struct Mol Biol. 2012;19:664–670. doi: 10.1038/nsmb.2315. [DOI] [PubMed] [Google Scholar]
- 15.Gutschner T, Hämmerle M, Eissmann M, Hsu J, Kim Y, Hung G, Revenko A, Arun G, Stentrup M, Gross M, Zörnig M, MacLeod AR, Spector DL, Diederichs S. The noncoding RNA MALAT1 is a critical regulator of the metastasis phenotype of lung cancer cells. Cancer Res. 2013;73:1180–1189. doi: 10.1158/0008-5472.CAN-12-2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Arab K, Park YJ, Lindroth AM, Schäfer A, Oakes C, Weichenhan D, Lukanova A, Lundin E, Risch A, Meister M, Dienemann H, Dyckhoff G, Herold-Mende C, Grummt I, Niehrs C, Plass C. Long noncoding RNA TARID directs demethylation and activation of the tumor suppressor TCF21 via GADD45A. Mol Cell. 2014;55:604–614. doi: 10.1016/j.molcel.2014.06.031. [DOI] [PubMed] [Google Scholar]
- 17.Pandey GK, Mitra S, Subhash S, Hertwig F, Kanduri M, Mishra K, Fransson S, Ganeshram A, Mondal T, Bandaru S, Ostensson M, Akyürek LM, Abrahamsson J, Pfeifer S, Larsson E, Shi L, Peng Z, Fischer M, Martinsson T, Hedborg F, Kogner P, Kanduri C. The risk-associated long noncoding RNA NBAT-1 controls neuroblastoma progression by regulating cell proliferation and neuronal differentiation. Cancer Cell. 2014;26:722–737. doi: 10.1016/j.ccell.2014.09.014. [DOI] [PubMed] [Google Scholar]
- 18.Guo F, Li Y, Liu Y, Wang J, Li Y, Li G. Inhibition of metastasis-associated lung adenocarcinoma transcript 1 in CaSki human cervical cancer cells suppresses cell proliferation and invasion. Acta Biochim Biophys Sin (Shanghai) 2010;42:224–229. doi: 10.1093/abbs/gmq008. [DOI] [PubMed] [Google Scholar]
- 19.Zhang HM, Yang FQ, Chen SJ, Che J, Zheng JH. Upregulation of long non-coding RNA MALAT1 correlates with tumor progression and poor prognosis in clear cell renal cell carcinoma. Tumour Biol. 2015;36:2947–2955. doi: 10.1007/s13277-014-2925-6. [DOI] [PubMed] [Google Scholar]
- 20.Ma KX, Wang HJ, Li XR, Li T, Su G, Yang P, Wu JW. Long noncoding RNA MALAT1 associates with the malignant status and poor prognosis in glioma. Tumour Biol. 2015;36:3355–3359. doi: 10.1007/s13277-014-2969-7. [DOI] [PubMed] [Google Scholar]
- 21.Gutschner T, Diederichs S. The hallmarks of cancer: a long non-coding RNA point of view. RNA Biol. 2012;9:703–719. doi: 10.4161/rna.20481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang MH, Hu ZY, Xu C, Xie LY, Wang XY, Chen SY, Li ZG. MALAT1 promotes colorectal cancer cell proliferation/migration/invasion via PRKA kinase anchor protein 9. Biochim Biophys Acta. 2015;1852:166–174. doi: 10.1016/j.bbadis.2014.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hu L, Wu Y, Tan D, Meng H, Wang K, Bai Y, Yang K. Up-regulation of long noncoding RNA MALAT1 contributes to proliferation and metastasis in esophageal squamous cell carcinoma. J Exp Clin Cancer Res. 2015;34:7. doi: 10.1186/s13046-015-0123-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vassallo I, Zinn P, Lai M, Rajakannu P, Hamou MF, Hegi ME. WIF1 re-expression in glioblastoma inhibits migration through attenuation of non-canonical WNT signaling by downregulating the lncRNA MALAT1. Oncogene. 2016;35:12–21. doi: 10.1038/onc.2015.61. [DOI] [PubMed] [Google Scholar]
- 25.Tee AE, Ling D, Nelson C, Atmadibrata B, Dinger ME, Xu N, Mizukami T, Liu PY, Liu B, Cheung B, Pasquier E, Haber M, Norris MD, Suzuki T, Marshall GM, Liu T. The histone demethylase JMJD1A induces cell migration and invasion by up-regulating the expression of the long noncoding RNA MALAT1. Oncotarget. 2014;5:1793–1804. doi: 10.18632/oncotarget.1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gutschner T, Hämmerle M, Eissmann M, Hsu J, Kim Y, Hung G, Revenko A, Arun G, Stentrup M, Gross M, Zörnig M, MacLeod AR, Spector DL, Diederichs S. The noncoding RNA MALAT1 is a critical regulator of the metastasis phenotype of lung cancer cells. Cancer Res. 2013;73:1180–1189. doi: 10.1158/0008-5472.CAN-12-2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Okugawa Y, Toiyama Y, Hur K, Toden S, Saigusa S, Tanaka K, Inoue Y, Mohri Y, Kusunoki M, Boland CR, Goel A. Metastasis-associated long non-coding RNA drives gastric cancer development and promotes peritoneal metastasis. Carcinogenesis. 2014;35:2731–2739. doi: 10.1093/carcin/bgu200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fu X, Liu Y, Zhuang C, Liu L, Cai Z, Huang W. Synthetic artificial microRNAs targeting UCA1-MALAT1 or c-Myc inhibit malignant phenotypes of bladder cancer cells T24 and 5637. Mol Biosyst. 2015;11:1285–1289. doi: 10.1039/c5mb00127g. [DOI] [PubMed] [Google Scholar]
- 29.Kawamura M, Toiyama Y, Tanaka K, Saigusa S, Okugawa Y, Hiro J, Uchida K, Mohri Y, Inoue Y, Kusunoki M. CXCL5, a promoter of cell proliferation, migration and invasion, is a novel serum prognostic marker in patients with colorectal cancer. Eur J Cancer. 2012;48:2244–2251. doi: 10.1016/j.ejca.2011.11.032. [DOI] [PubMed] [Google Scholar]
- 30.Begley LA, Kasina S, Mehra R, Adsule S, Admon AJ, Lonigro RJ, Chinnaiyan AM, Macoska JA. CXCL5 promotes prostate cancer progression. Neoplasia. 2008;10:244–254. doi: 10.1593/neo.07976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gao Y, Guan Z, Chen J, Xie H, Yang Z, Fan J, Wang X, Li L. CXCL5/CXCR2 axis promotes bladder cancer cell migration and invasion by activating PI3K/AKT-induced upregulation of MMP2/MMP9. Int J Oncol. 2015;47:690–700. doi: 10.3892/ijo.2015.3041. [DOI] [PubMed] [Google Scholar]
- 32.Park JY, Park KH, Bang S, Kim MH, Lee JE, Gang J, Koh SS, Song SY. CXCL5 overexpression is associated with late stage gastric cancer. J Cancer Res Clin Oncol. 2007;133:835–840. doi: 10.1007/s00432-007-0225-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Miyazaki H, Patel V, Wang H, Edmunds RK, Gutkind JS, Yeudall WA. Down-regulation of CXCL5 inhibits squamous carcinogenesis. Cancer Res. 2006;66:4279–4284. doi: 10.1158/0008-5472.CAN-05-4398. [DOI] [PubMed] [Google Scholar]
- 34.Kowalczuk O, Burzykowski T, Niklinska WE, Kozlowski M, Chyczewski L, Niklinski J. CXCL5 as a potential novel prognostic factor in early stage non-small cell lung cancer: results of a study of expression levels of 23 genes. Tumour Biol. 2014;35:4619–4628. doi: 10.1007/s13277-014-1605-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Arenberg DA, Keane MP, DiGiovine B, Kunkel SL, Morris SB, Xue YY, Burdick MD, Glass MC, Iannettoni MD, Strieter RM. Epithelial-neutrophil activating peptide (ENA-78) is an important angiogenic factor in non-small cell lung cancer. J Clin Invest. 1998;102:465–472. doi: 10.1172/JCI3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Baylin SB. DNA methylation and gene silencing in cancer. Nat Clin Pract Oncol. 2005;2(Suppl 1):S4–11. doi: 10.1038/ncponc0354. [DOI] [PubMed] [Google Scholar]
- 37.Bhan A, Mandal SS. LncRNA HOTAIR: A master regulator of chromatin dynamics and cancer. Biochim Biophys Acta. 2015;1856:151–164. doi: 10.1016/j.bbcan.2015.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Arab K, Park YJ, Lindroth AM, Schäfer A, Oakes C, Weichenhan D, Lukanova A, Lundin E, Risch A, Meister M, Dienemann H, Dyckhoff G, Herold-Mende C, Grummt I, Niehrs C, Plass C. Long noncoding RNA TARID directs demethylation and activation of the tumor suppressor TCF21 via GADD45A. Mol Cell. 2014;55:604–614. doi: 10.1016/j.molcel.2014.06.031. [DOI] [PubMed] [Google Scholar]


