Abstract
Interferon-induced transmembrane protein 1 (IFITM1) has recently been implicated in tumorigenesis. However, the prognostic value of IFITM1 in colorectal cancer remains unknown. The present study aimed to examine the expression and prognostic significance of IFITM1 in human colorectal cancer. IFITM1 expression was analyzed in 144 archived, paraffin-embedded colorectal cancer tissues and corresponding normal colorectal mucosa by immunohistochemistry. The correlation of IFITM1 with clinic-pathological features and overall survival of colorectal cancer patients was evaluated. IFITM1 was overexpressed in colonic cancer tissues but not in rectal cancer tissues, compared to control normal tissues. The expression of IFITM1 was significantly higher in patients with poor differentiation (P=0.031). The patients with higher IFITM1 expression had worse overall survival outcomes than those with lower IFITM1 expression in rectal cancer (P=0.037). Univariate Cox regression suggested that older age and poorly differentiation status predict shorter overall survival in colorectal cancer (P<0.05). However, IFITM1 expression was not a significant prognostic factor for survival by univariate or multivariate analyses. In conclusion, high expression of IFITM1 is associated with poor prognosis of rectal cancer. IFITM1 may serve as an independent prognostic biomarker for colorectal cancer.
Keywords: IFITM1, colorectal cancer, immunohistochemistry, prognosis
Introduction
Colorectal cancer is the third most commonly diagnosed cancer in males and the second in females. In 2008, there were over 1.2 million new cancer cases and 608,700 deaths worldwide [1]. Although the improvements in early detection and treatment decrease the incidence and death rates, colorectal cancer is still a great threat to human health [2]. Consequently, much effort has been made to identify reliable molecular biomarkers of colorectal cancer that can predict tumor progression and improve the survival rates of patients [3].
INF-induced transmembrane protein 1 (IFITM1) is a member of the IFN-inducible transmembrane protein family and a component of multimeric complex involved in the transduction of anti-proliferation and cell adhesion signals [4]. IFITM1 expression has been found to be highly increased in esophageal cancer, cervical cancer and ovarian cancer [5-7]. Furthermore, recent studies suggest that IFITM1 promotes the progression of malignancy by enhancing migration and invasion in gastric cancer, head and neck cancer and glioma cells [8-10]. In colorectal cancer, the up-regulation of IFITM1 expression is highly specific to human colorectal carcinogenesis and identified as a potential new marker for colorectal tumors [11]. However, few studies have investigated the correlation between the expression of IFITM1 and its prognostic value in colorectal cancer.
In this study, we used immunohistochemistry to detect the expression of IFITM1 in 144 colorectal cancer tissues. IFITM1 expression within diseased epithelium was compared with adjacent normal epithelium in patient-matched samples. Furthermore, we determine the correlation of IFITM1 expression with tumor progression and patient survival in patients with variable clinicopathological characteristics.
Materials and methods
Patients and tissue specimens
Patients with colorectal cancer who had undergone tumor surgical resection at Huaian First People’s Hospital Affiliated to Nanjing Medical University from July 2006 through August 2007 were involved in this study. Each patient had been diagnosed with colorectal cancer based on clinicopathological characteristics and none of them received radiotherapy or adjuvant chemotherapy before surgery. All patients have been prospectively followed up until death or when last seen alive on their clinical visit (August 2011), with the median follow-up of 51 months. A total of 144 paraffin-embedded colorectal cancer and adjacent normal epithelium in patient-matched tissues from tumor and adjacent tissues were collected for immunohistochemitry. The study protocols were approved by Ethics Committee of Nanjing Medical University. Every patient signed a written informed consent before surgery.
Immunohistochemistry
A total of 144 formalin-fixed paraffin-embedded colorectal tumor samples were cut into 4 µm sections and baked at 60°C. The slides were dewaxed in xylene and rehydrated in ethanol and then incubated in a 3% hydrogen peroxide to inhibit the endogenous peroxidase activity. Next the slides were boiled in antigen retrieval EDTA buffer for 10 min. After blocking with goat serum, the slides were incubated with a mouse polyclonal antibody against IFITM1 (1:100, Sigma) at 4°C overnight. After washing with phosphate-buffered saline (PBS), the sections were incubated with second antibody for 30 min, then washed with PBS. Finally, the section were visualized by diaminobenzidine solution and counterstained with hematoxylin. Negative controls were performed with the incubation with rabbit IgG instead of primary antibody.
Evaluation of immunohistochemical staining
Two experienced pathologists who were blind to the clinical-pathological data evaluated immunohistochemistry staining independently. The score of immunohistochemical staining was based on the percentage of positive tumor cells and the intensity of the staining. The percentage of cells with staining was scored as: “0” (0%), “1” (1%-25%), “2” (26%-50%), “3” (51%-75%) and “4” (75%-100%). The staining intensity was scored as: “0” (negative), “1” (weak), “2” (moderate), “3” (strong). The immmunoreactivity score was classified by multiplying the two scores, and classified as three grades: weak (0-4), moderate (4-8), strong (8-12).
Statistical analysis
Differences between the grades of malignant and adjacent tissue samples were compared using Wilcoxon rank sum test. The relationship between IFITM1 expression and clinicopathological characteristics was performed by Pearson chi-square and Fisher’s exact tests. Survival curves were evaluated by the Kaplan-Meier method and compared by the log-rank test. Univariate and multivariate survival analysis were performed using Cox regression analysis. Statistical analysis was performed using SPSS software 18.0 (SPSS Inc., Chicago, IL, USA). All analyses were considered statistical significant when p values <0.05.
Results
Clinicopathological features
The median age of the 144 patients was 65 years (range 24 to 90 years). Among them, 65 patients had colon cancer and 79 patients had rectal cancer. The median follow-up was 51 months (range 1 to 61 months). The clinicopathological data of the patients were listed in Table 1.
Table 1.
Correlation between IFITM1 expression and clinic-pathological features of colorectal cancer patients
| Characteristic | IFITM1 expression | P value | ||
|---|---|---|---|---|
|
| ||||
| Weak | Moderate | Strong | ||
| Gender | 0.276 | |||
| Male | 45 | 37 | 3 | |
| Female | 38 | 18 | 3 | |
| Age | ||||
| <60 years | 30 | 16 | 2 | 0.688 |
| ≥60 years | 53 | 39 | 4 | |
| Location | 0.142 | |||
| Colon | 36 | 24 | 5 | |
| Rectum | 47 | 31 | 1 | |
| Differentiation | 0.031 | |||
| Well | 19 | 22 | 2 | |
| Moderately | 29 | 16 | 4 | |
| Poorly | 35 | 17 | 0 | |
| Depth of invasion | 0.605 | |||
| T1 | 3 | 2 | 0 | |
| T2 | 7 | 9 | 0 | |
| T3 | 67 | 39 | 5 | |
| T4 | 6 | 5 | 1 | |
| Lymph node | 0.865 | |||
| Negative | 45 | 32 | 3 | |
| Positive | 38 | 23 | 3 | |
| TNM stage | 0.638 | |||
| I | 8 | 9 | 0 | |
| II | 37 | 23 | 3 | |
| III | 37 | 21 | 3 | |
| IV | 1 | 2 | 0 | |
| Status at last review | 0.858 | |||
| Alive | 51 | 33 | 3 | |
| Dead | 32 | 22 | 3 | |
Comparison of IFITM1 expression in normal and malignant colorectal tissue
We performed mmunohistochemistry to assess IFITM1 expression in colorectal tissue samples and matched adjacent noncancerous tissue samples. The expression of IFITM1 was predominantly cytoplasmic in tumor cells and adjacent noncancerous epithelial cells (Figure 1). IFITM1 expression was significantly higher in malignant colon tissue than in non-malignant tissue (P=0.044), but there was no significant difference in IFITM1 expression between rectal cancer tissues and their precancerous counterparts. In addition, the difference in IFITM1 expression between colorectal cancer and normal mucosa was not statistically significant.
Figure 1.
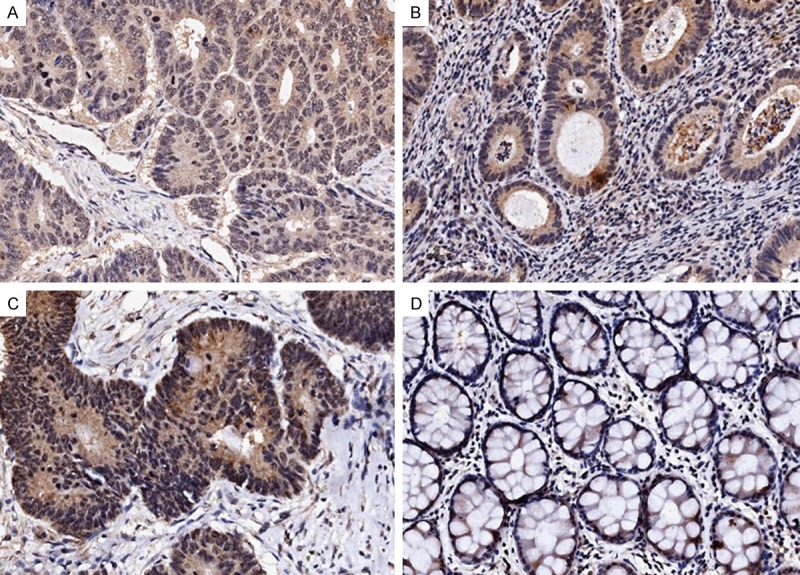
Immunohistochemical staining of IFITM1 in human colorectal cancers. Representative images from immunohistochemical staining of IFITM1 in colorectal cancer and matched adjacent noncancerous tissues. Weak IFITM1 staining (A), moderate IFITM1 staining (B), and strong IFITM1 staining (C). Negative staining of IFITM1 in matched noncancerous tissues adjacent to tumors (D). Magnification: ×200.
Association between IFITM1 expression and clinicpathological features of colorectal cancer
The association between IFITM1 expression and clinic-pathological features of colorectal tumor was shown in Table 2. IFITM1 expression was higher in histological differentiated colorectal carcinomas (P=0.031). However, gender, age, tumor location, depth of invasion, lymph node metastasis, TNM stage and survival had no significant relationship with IFITM1 expression (Table 2).
Table 2.
Univariate and multivariate analysis of factors related to overall survival in colorectal cancer patients
| Risk factor | Univariate analysis | P | Multivariate analysis | P |
|---|---|---|---|---|
|
|
|
|||
| HR (95% CI) | HR (95% CI) | |||
| Age | 1.964 (1.028-3.751) | 0.041 | 2.193 (1.183-4.066) | 0.013 |
| Gender | 0.810 (0.463-1.418) | 0.460 | ||
| Tumor location | 0.884 (0.508-1.538) | 0.662 | ||
| Tumor size | 1.391 (0.759-2.551) | 0.286 | ||
| Histological differentiation | 1.411 (1.018-1.956) | 0.038 | 1.337 (0.974-1.835) | 0.073 |
| TNM stage | 3.438 (2.081-5.680) | <0.001 | 3.282 (1.994-5.402) | <0.001 |
| IFITM1 expression | 1.178 (0.730-1.900) | 0.503 |
HR hazard ratio, CI confidence interval, P values in bold were statistically significant. Age, ≥60 or <60; Gender, male or female; Tumor location, rectum or colon; Tumor size, >4 cm or ≤4 cm; Histological differentiation, poorly differentiated, moderately differentiated or well differentiation; Tumor Node Metastasis, I+II, III or IV; IFITM1 expression, strongly expression, moderately expression or weakly expression.
Association between IFITM1 expression and clinical outcome
By analysis of Kaplan-Meier survival curves we found that the survival of the patients with moderate and strong staining of IFITM1 in colorectal tumor infiltrate was not significantly different from those with weak staining of IFITM1 (P=0.700, Figure 2). The overall survival rates in patients with high expression, moderate expression and low expression of IFITM1 were 55.4, 59.5 and 50.0% respectively.
Figure 2.
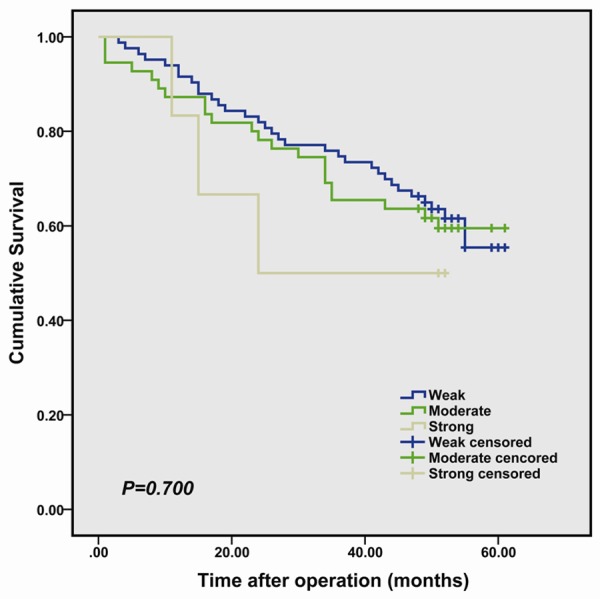
Kaplan-Meier survival curves in colorectal cancer according to IFITM1 staining. The log rank test was used to calculate P values.
With the use of univariate analysis, we found that age, histological differentiation and TNM stage were the significant prognostic factors for predicting overall survival. Gender, the primary location of tumor and the size of tumor were not prognostic factor in univariate analysis. By multivariate analysis, we still found that age and TNM stage had prognostic value. IFITM1 expression was not a significant prognostic factor for survival by either univariate or multivariate analysis.
To further assess the prognostic value of IFITM1 expression, we performed a stratified analysis. In the subgroup of patients, there was a significant association between higher IFITM1 expression in the rectal cancer and poorer overall survival (P=0.037, Figure 3B). No association was found between IFITM1 expression and the prognostic significance with patients with colonic cancer (P=0.873, Figure 3A).
Figure 3.
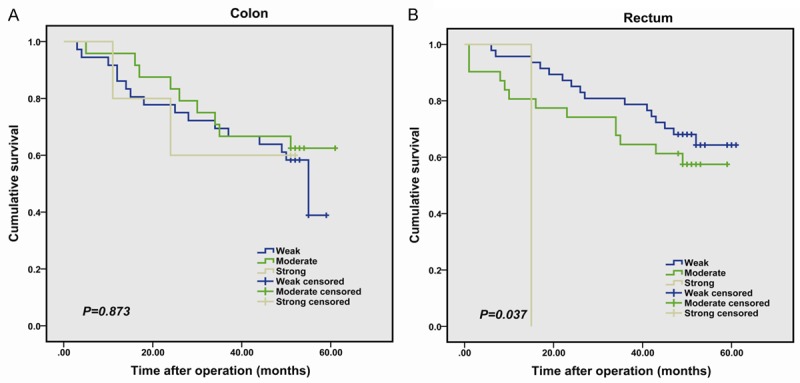
Kaplan-Meier analysis of the overall survival of colorectal cancer patients according to tumor location, colon cancer (A) and rectum cancer (B). The log rank test was used to calculate P values.
Discussion
In the present study, we analyzed the expression of IFITM1 in colorectal cancer tissues by immunohistochemistry. We found that IFITM1 protein was significantly up-regulated in colon cancer tissues compared to their corresponding non-tumor colonic tissues. Furthermore, the level of IFITM1 expression was inversely correlated with histological differentiation of colorectal cancer. Patients with high IFITM1 expression had a shorter survival time than those with low IFITM1 expression in rectal cancer.
Interferon-induced transmembrane protein 1 (IFITM1) is a cell surface 17-KDa membrane protein that is encoded on chromosome arm 11p15.5. Loss of heterozygosity in this region was observed in several solid tumors, including breast cancer and lung cancer [12,13]. Consistent with this, downregulated IFITM1 expression has been reported in hepatoma, astrocytomas and breast cancer [14-16]. These findings indicate that IFITM1 as a tumor suppressor. On the other hand, several studies have established the role of IFITM1 in tumorigenesis. IFITM1 is upregulated in early and late intestinal neoplasm’s. Moreover, IFITM1 expression is upregulated by the Wnt/β-catenin signaling and its upregulation seems to be an early event in intestinal tumorigenesis [11]. In gastric cancer cells, IFITM1 modulates the susceptibility to natural killer cells and invasiveness of gastric cancer cells [17]. In addition, overexpression of IFITM1 has shown to promote invasion and migration in glioma cells and head and neck cancer cells [9,10].
IFITM1 is frequently overexpressed in several solid tumors [11]. One study indicated that higher IFITM1 expression correlates with improved survival in chronic myeloid leukemia [18]. However, the prognostic significance of IFITM1 in colorectal cancer remains unknown. In the present study, we analyzed IFITM1 expression level using immunohistochemistry. The results showed that IFITM1 protein level was significantly higher in colonic cancer tissues than in adjacent noncancerous tissues but not in rectal cancer tissues, which were not completely consistent with previous findings. This study also found higher IFITM1 expression of histologically differentiated colorectal cancer (P=0.031). Our data indicate that IFITM1 is an oncogene in colorectal cancer. Moreover, patients with rectal cancer with moderate and strong expression of IFITM1 had worse outcome, and IFITM1 expression was not an independent prognostic factor for colorectal cancer among these patients. The reason for the difference in expression and clinical outcome between colonic and rectal cancer remains unknown. One possible explanation is that IFITM1 expression tends to associate with tumor location, although not statistically significant.
One of limitations of our study is the relatively small number of patients analyzed. This may reduce the power to assess statistical association and significantly affect survival analyses. In addition, scoring with immunohistochemistry is a semiquantitative method, which represents a weakness of the study.
In conclusion, the overexpression of IFITM1 tends to be associated with poor prognosis in colorectal patients and may represent a potential prognosis factor for colorectal cancer. Further investigations are needed to confirm that IFITM1 is an independent prognostic factor for colorectal cancer patients.
Acknowledgements
This study was supported by grants from Nature Science Fund of Jiangsu Province (No. BK2008192) and Project Fund of Health Bureau of Jiangsu Province (No. H201358).
Disclosure of conflict of interest
None.
References
- 1.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 2.Siegel R, Ward E, Brawley O, Jemal A. Cancer statistics, 2011: the impact of eliminating socioeconomic and racial disparities on premature cancer deaths. CA Cancer J Clin. 2011;61:212–36. doi: 10.3322/caac.20121. [DOI] [PubMed] [Google Scholar]
- 3.Arienti C, Tesei A, Verdecchia GM, Framarini M, Virzì S, Grassi A, Scarpi E, Turci L, Silvestrini R, Amadori D, Zoli W. Role of conventional chemosensitivity test and tissue biomarker expression in predicting response to treatment of peritoneal carcinomatosis from colon cancer. Clin Colorectal Cancer. 2013;12:122–7. doi: 10.1016/j.clcc.2012.11.006. [DOI] [PubMed] [Google Scholar]
- 4.Deblandre GA, Marinx OP, Evans SS, Majjaj S, Leo O, Caput D, Huez GA, Wathelet MG. Expression cloning of an interferon-inducible 17-kDa membrane protein implicated in the control of cell growth. J Biol Chem. 1995;270:23860–6. doi: 10.1074/jbc.270.40.23860. [DOI] [PubMed] [Google Scholar]
- 5.Chattopadhyay I, Phukan R, Singh A, Vasudevan M, Purkayastha J, Hewitt S, Kataki A, Mahanta J, Kapur S, Saxena S. Molecular profiling to identify molecular mechanism in esophageal cancer with familial clustering. Oncol Rep. 2009;21:1135–46. doi: 10.3892/or_00000333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pan Z, Chen S, Pan X, Wang Z, Han H, Zheng W, Wang X, Li F, Qu S, Shao R. Differential gene expression identified in Uigur women cervical squamous cell carcinoma by suppression subtractive hybridization. Neoplasma. 2010;57:123–8. doi: 10.4149/neo_2010_02_123. [DOI] [PubMed] [Google Scholar]
- 7.Gyorffy B, Dietel M, Fekete T, Lage H. A snapshot of microarray-generated gene expression signatures associated with ovarian carcinoma. Int J Gynecol Cancer. 2008;18:1215–33. doi: 10.1111/j.1525-1438.2007.01169.x. [DOI] [PubMed] [Google Scholar]
- 8.Lee J, Goh SH, Song N, Hwang JA, Nam S, Choi IJ, Shin A, Kim IH, Ju MH, Jeong JS, Lee YS. Overexpression of IFITM1 has clinicopathologic effects on gastric cancer and is regulated by an epigenetic mechanism. Am J Pathol. 2012;181:43–52. doi: 10.1016/j.ajpath.2012.03.027. [DOI] [PubMed] [Google Scholar]
- 9.Hatano H, Kudo Y, Ogawa I, Tsunematsu T, Kikuchi A, Abiko Y, Takata T. IFN-induced transmembrane protein 1 promotes invasion at early stage of head and neck cancer progression. Clin Cancer Res. 2008;14:6097–105. doi: 10.1158/1078-0432.CCR-07-4761. [DOI] [PubMed] [Google Scholar]
- 10.Yu F, Ng SS, Chow BK, Sze J, Lu G, Poon WS, Kung HF, Lin MC. Knockdown of interferon-induced transmembrane protein 1 (IFITM1) inhibits proliferation, migration, and invasion. J Neurooncol. 2011;103:187–95. doi: 10.1007/s11060-010-0377-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Andreu P, Colnot S, Godard C, Laurent-Puig P, Lamarque D, Kahn A, Perret C, Romagnolo B. Identification of the IFITM family as a new molecular marker in human colorectal tumors. Cancer Res. 2006;66:1949–55. doi: 10.1158/0008-5472.CAN-05-2731. [DOI] [PubMed] [Google Scholar]
- 12.Negrini M, Rasio D, Hampton GM, Sabbioni S, Rattan S, Carter SL, Rosenberg AL, Schwartz GF, Shiloh Y, Cavenee WK, et al. Definition and refinement of chromosome 11 regions of loss of heterozygosity in breast cancer: identification of a new region at 11q23.3. Cancer Res. 1995;55:3003–7. [PubMed] [Google Scholar]
- 13.Catchpoole D, Lam WW, Valler D, Temple IK, Joyce JA, Reik W, Schofield PN, Maher ER. Epigenetic modification and uniparental inheritance of H19 in Beckwith-Wiedemann syndrome. J Med Genet. 1997;34:353–9. doi: 10.1136/jmg.34.5.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang G, Xu Y, Chen X, Hu G. IFITM1 plays an essential role in the antiproliferative action of interferon-gamma. Oncogene. 2007;26:594–603. doi: 10.1038/sj.onc.1209807. [DOI] [PubMed] [Google Scholar]
- 15.Huang H, Colella S, Kurrer M, Yonekawa Y, Kleihues P, Ohgaki H. Gene expression profiling of low-grade diffuse astrocytomas by cDNA arrays. Cancer Res. 2000;60:6868–74. [PubMed] [Google Scholar]
- 16.Abba MC, Drake JA, Hawkins KA, Hu Y, Sun H, Notcovich C, Gaddis S, Sahin A, Baggerly K, Aldaz CM. Transcriptomic changes in human breast cancer progression as determined by serial analysis of gene expression. Breast Cancer Res. 2004;6:R499–513. doi: 10.1186/bcr899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang Y, Lee JH, Kim KY, Song HK, Kim JK, Yoon SR, Cho D, Song KS, Lee YH, Choi I. The interferon-inducible 9-27 gene modulates the susceptibility to natural killer cells and the invasiveness of gastric cancer cells. Cancer Lett. 2005;221:191–200. doi: 10.1016/j.canlet.2004.08.022. [DOI] [PubMed] [Google Scholar]
- 18.Akyerli CB, Beksac M, Holko M, Frevel M, Dalva K, Ozbek U, Soydan E, Ozcan M, Ozet G, Ilhan O, Gürman G, Akan H, Williams BR, Ozçelik T. Expression of IFITM1 in chronic myeloid leukemia patients. Leuk Res. 2005;29:283–6. doi: 10.1016/j.leukres.2004.07.007. [DOI] [PubMed] [Google Scholar]


