Abstract
Fibroblast growth factor 21 (FGF21) plays an important role in the treatment of disease associated with muscle insulin resistance which is characterized by various factors, such as intramuscular triglyceride (IMT) content. Studies have also shown that FGF21 inhibits triglyceride synthesis in vivo. However, the precise mechanism whereby FGF21 regulates triglyceride metabolism in intramuscular fat (IMF), which may influence the muscle insulin sensitivity, is not clearly understood. In order to understand the role of FGF21 in IMF deposition, we performed FGF21 overexpression in IMF cells by stable transfection. Our results showed that FGF21 inhibited the key adipogenesis gene mRNA expression of peroxisome proliferator-activated receptor gamma (PPARG), CCAAT/enhancer-binding protein (CEBP) family by reducing lysine-specific demethylase 1 (LSD1) expression which led to significant decline in lipid accumulation, and the result was confirmed by Western blot. Moreover, triggered by FGF21, parts of the adipokines—fatty acid-binding protein 4 (FABP4), glucose transporter 4 (GLUT4), adiponectin (ADIPOQ), and perilipin (PLIN1)—were also down-regulated. Furthermore, FGF21 gene expression was suppressed by transcription factor CEBP beta (CEBPB) which contributed strongly to triglyceride synthesis. Taken together, our study is the first to experimentally demonstrate FGF21 emerging as an efficient blockade of adipogenesis in IMF, thus also providing a new understanding of the mechanism whereby FGF21 improves insulin sensitivity.
Keywords: pig, FGF21, adipogenesis, IMT, CEBPB
1. Introduction
Intramuscular triglyceride (IMT), as an indispensable energy source for skeletal muscles, has been considered to be a robust indicator of muscle insulin sensitivity, which is an essential predictive factor for type 2 diabetes. Larson Meyer et al. [1,2,3,4] found that lipid accretion in skeletal muscles contributes to insulin resistance. In addition, ectopic fat accumulation in skeletal muscles is associated with the early pathogenesis of insulin resistance. This has inspired heightened efforts to be made to better understand the precise lipid metabolism in muscles.
Many molecules and pathways are involved in lipid metabolism. Wnt signal plays an important role in the regulation of adipocyte differentiation, which was first reported in the MacDougald laboratory [5]. By blocking induction of PPARG and CEBP alpha (CEBPα), Wnt signal shows ability in inhibiting adipogenesis. However, disruption of Wnt signal leads to spontaneous adipocyte differentiation [6,7,8]. The Wnt coreceptor LRP6 plays a very critical role in the reduction of body mass by reducing nuclear localization of β-catenin and inactivation of forkhead box O1 (FOXO1) which promotes adipocyte differentiation [9], resulting in down-regulation of genes involved in adipogenesis [10].
Kruppel-like factor (KLF) family encodes both transcriptional activator and repressor proteins which carry out important roles on differentiation of cells in mammals [11].
It has been known for years that histone deacetylases (HDACs) regulate a variety of processes, including growth arrest, differentiation, cytotoxicity, induction of apoptosis [12], and adipogenic transcription factor activity [13]. HDAC3 controls the circadian rhythm of hepatic lipogenesis. Mice with liver-specific depletion of HDAC3 reroute metabolic precursors towards lipid synthesis and storage within lipid droplets [14].
In the last decade, FGF21 as a novel metabolic regulator attracted much attention of scholars. Studies showed that FGF21 could enhance adipogenesis or attenuate lipolysis [15,16,17,18,19,20]. However, some conflicting data has also been reported. Coskun et al. [21] showed that FGF21 could correct obesity in mice via ameliorating insulin and leptin resistance. Furthermore, Chau demonstrated that FGF21 was a potential function as a therapy for obesity by activating AMPK-sirtuin 1 (SIRT1)-PGC-1α pathway [22]. In addition, FGF21 was reported to suppress the adipogenesis-related genes in liver, FGF21 had a benefit to fatty liver disease [23], and the treatment of recombinant FGF21 in 3T3-L1 could increase lipolysis [15]. What is more, Hotta [21] demonstrated FGF21 had opposite roles in different conditions.
Given the situation mentioned above, the functions of FGF21 in pharmacology and physiology are somewhat discordant. To understand whether and how FGF21 influences lipid metabolism in intramuscular fat cells, FGF21 gain-of-function by stable transfection in intramuscular preadipocyte was performed in this study. Our results showed FGF21 down-regulated the expression of LSD1 and resulted in the decrease of the adipogenesis-related key genes expression, which resulted in decline of the accumulation of triglyceride in muscles. It has contributed to our understanding of the mechanisms regulating triglyceride metabolism in IMF. Moreover, it also provides another perspective to interpret the pharmacological properties of FGF21 in the treatment of insulin resistant associated disease, such as type 2 diabetes.
2. Result
2.1. Low Expression of Fibroblast Growth Factor 21 (FGF21) in Fatty Tissue
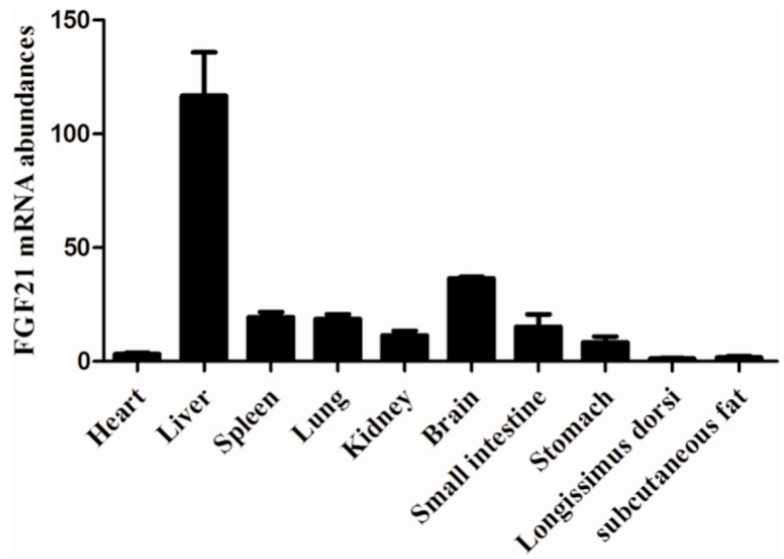
In general, the result of FGF21 expression profile analysis showed us that the higher expression of FGF21 was found in non-fatty tissues in spleen and brain, especially in liver, rather than in subcutaneous fat. Surprisingly, weaker expression in longissimus dorsi was observed (Figure 1).
Figure 1.
Expression profiles analysis of FGF21. The relative expression of FGF21 in heart, liver, spleen, lung, kidney, brain, small intestine, stomach, longissimus dorsi and subcutaneous fat in three-month Large White pigs and the mRNA level was normalized with glyceraldehydes phosphate dehydrogenase (GAPDH).
2.2. Establishment and Identification of the Intramuscular Preadipocyte Cell Line
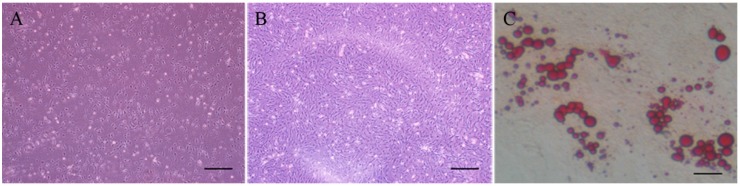
Cells isolated from a three day new born Large White pig were presented as spindle, which were similar to fibroblasts and there were no lipid droplets in the cytoplasm. Oil red staining was used to identify the cells and oil red could be dissolved by fat and oil red presented (jacinth). Oil red staining was performed after the cell differentiation was induced on the 8th day by IBMX + DEX + insulin. A significant amount of lipid droplets generated in the cytoplasm was observed under the inverted microscope which provided direct evidence that the cells we isolated were intramuscular preadipocyte cells (Figure 2).
Figure 2.
Morphology of the primary cultured cells. (A) Morphology of primary cultured pig intramuscular fat cells on the 3rd day (bar = 200 μm); (B) Morphology of primary cultured pig intramuscular fat cells on the 8th day (bar = 200 μm); and (C) Lipid droplets became jacinth colored by oil red O (bar = 200 μm).
2.3. Cells Transfection and Stable FGF21 Cell Line Acquisition
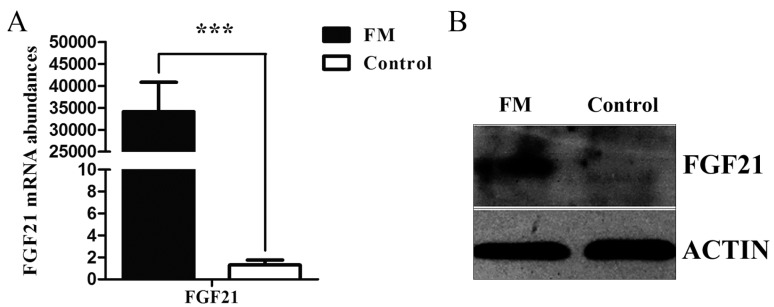
Because of the low transfection efficiency of pig IMF cells, we performed a stable transfection. After 10 days of screening, we acquired the FGF21 stable monoclone (FM). The shape of monoclone did not change too much from that of the initial cells. To investigate whether the FGF21 was up-regulated in FM, qRT-PCR was used to measure the mRNA expression of FGF21. Data showed that FGF21 was weakly expressed in the control, but strongly expressed in FM which was also verified by data of Western blot (Figure 3A,B).
Figure 3.
FGF21 expression in the FGF21 stable monoclone (FM) cells before cell differentiation. (A) qRT-PCR analysis revealed that FM cells had higher FGF21 mRNA level than that in the control; and (B) Western blot analysis showed that the FGF21 protein level was up-regulated significantly in FM cells. Gene expression was normalized to GAPDH. Data was presented as mean ± SD, *** for p < 0.001.
2.4. The Function of FGF21 in Adipogenesis
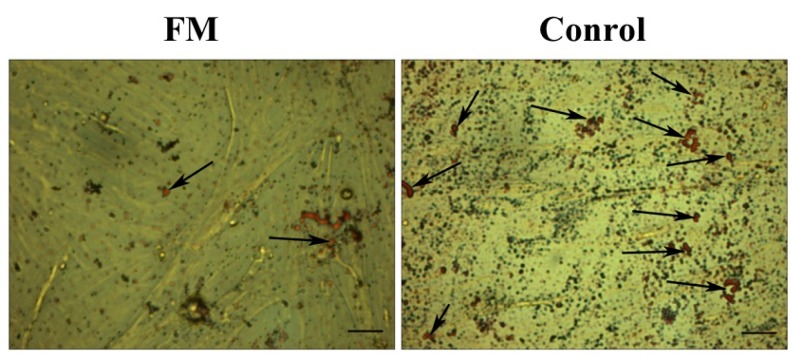
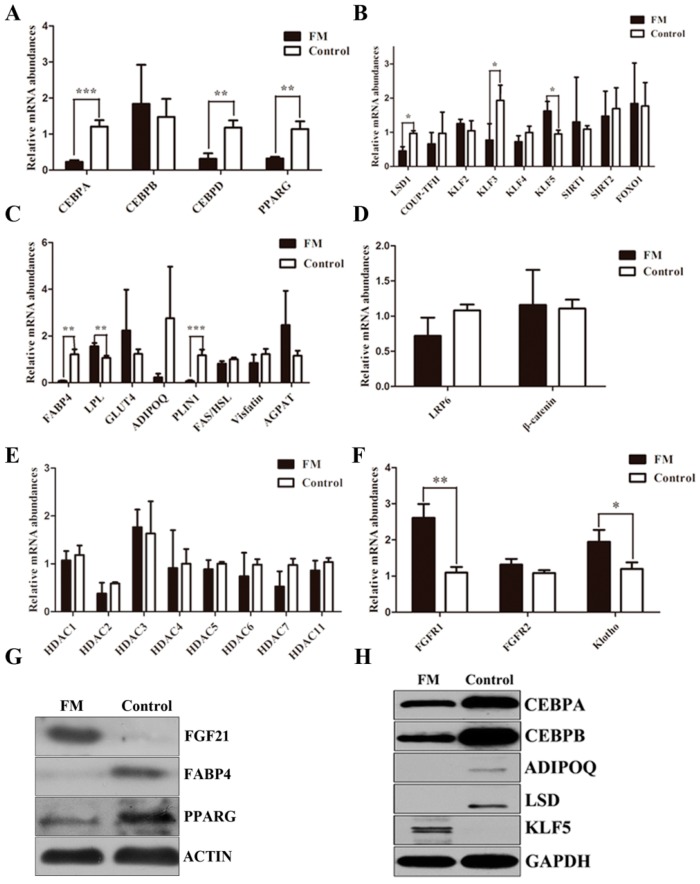
To specifically evaluate a potential role for FGF21 in adipogenesis, we examined the effect of FGF21 on the 8th day of adipocyte differentiation. We found that the mRNA expression of the key genes PPARG, CEBPA and CEBPD involved in adipogenesis was down-regulated dramatically in FM compared with the control, which was the same with down-regulation of the protein level. Interestingly, no change occurred in the CEBPB mRNA level, but the protein of CEBPB declined strongly. In short, FGF21 suppresses adipogenesis as assessed by oil red O staining (Figure 4 and Figure 5).
Figure 4.
FGF21 suppressed the accumulation of triglycerides. Oil red staining results showed less lipid droplets in FM cells (left) than that in the control (right) (bar = 100 μm). The arrows indicated the lipid.
Figure 5.
FGF21 reduced the adipogenesis-related genes expression on the cell differentiation 8th day. The qRT-PCR analysis of CEBP family and PPARG (A); The genes expression regulating CEBP family and PPARG (B); adipokines (C); Wnt signal (D); HDAC family (E) and FGFRs signal (F) on the cell differentiation 8th day. Gene expression was normalized with GAPDH. Data was presented as mean ± SD, * for 0.01 < p < 0.05, ** for 0.001 < p < 0.01 and *** for p < 0.001; Western blot analysis of the adipogenesis-related genes on the cell differentiation 8th day. The gene expression was normalized with ACTIN (G) and GAPDH (H).
To further explore the mechanism above, some molecules participating in the regulation of the key adipogenic genes were also analyzed. There were no significant changes in the mRNA levels of KLF2, SIRT1, SIRT2 and FOXO1, which could regulate PPARG expression. Chicken ovalbumin upstream promoter-transcription factor II (COUP-TF II), also known as NR2F2, played an important role in the function of CEBPA. There was no significant difference in the COUP-TF II expression between FM and control. The same level was found in mRNA of KLF4 which participated in CEBPB expression. Unexpectedly, there was a higher level in KLF3 mRNA and a lower level in KLF5 mRNA.
Compared with FGF21 expression of FM and control, FAS and hormone-sensitive lipase (HSL) mRNA levels were both up-regulated. However, the ratio of FAS and HSL was down-regulated although it did not reach a significant level. We also found that AGPAT2 and visfatin mRNA levels did not change.
In the circumstances that FGF21 suppresses adipogenesis, we would expect FGF21 could down-regulate the adipocytokines. So in order to find out the role of FGF21 on adipocytokines, those encoding FABP4 (also known as adipocyte protein 2, ap2), GLUT4, ADIPOQ, and PLIN1 were evaluated. We proved FABP4 and PLIN1 declined dramatically in mRNA level. However, there was no significant change in GLUT4 and ADIPOQ. Western blot analysis demonstrated FABP4 and ADIPOQ protein were rapidly decreased. To identify FGF21 candidate pathway with the potential to modulate adipogensis, we assessed the regulatory program governing inhibition of adipogenesis by the canonical Wnt/β-catenin signal. We analyzed the gene expression of LRP6 and β-catenin in mature adipocyte that was induced to differentiate on the 8th day. QRT-PCR analysis demonstrated FGF21 did not change the mRNA expression of LRP6 and β-catenin. Due to the important role of HDACs in adipogensis, we conducted the necessary test to determine whether FGF21 affects the mRNA expression of HDACs. However, all of the family of HDACs in our study did not change significantly. Yet, LSD1, a crucial regulator in histone methylation of CEBPA, decreased in both mRNA and protein levels (Figure 5).
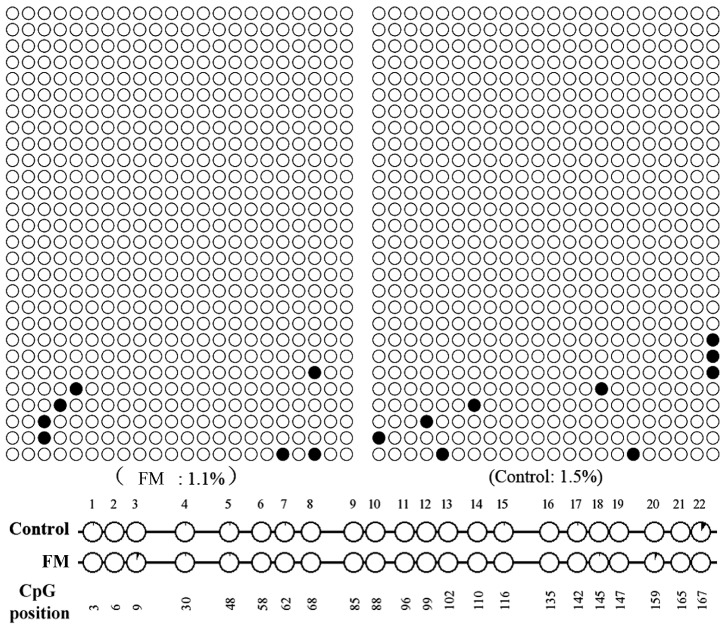
Given an important role of the gene promoter methylation for gene expression and to figure out whether FGF21 influenced the methylation level of CEBPA promoter to regulate CEBPA and further regulate PPARG expression, CEBPA promoter methylation level was also detected. The level of methylation of the CEBPA promoter did not change with the Quantification Tool for Methylation Analysis on line (Figure 6).
Figure 6.
Methylation analysis of CEBPA promoter. No significant difference of methylation level between FM and the control was found in the region from −172 to +2 bp in CEBPA promoter with the Quantification Tool for Methylation Analysis on line.
2.5. The Inhibitory Effect of the Transcription Factor CEBPB in FGF21 Expression
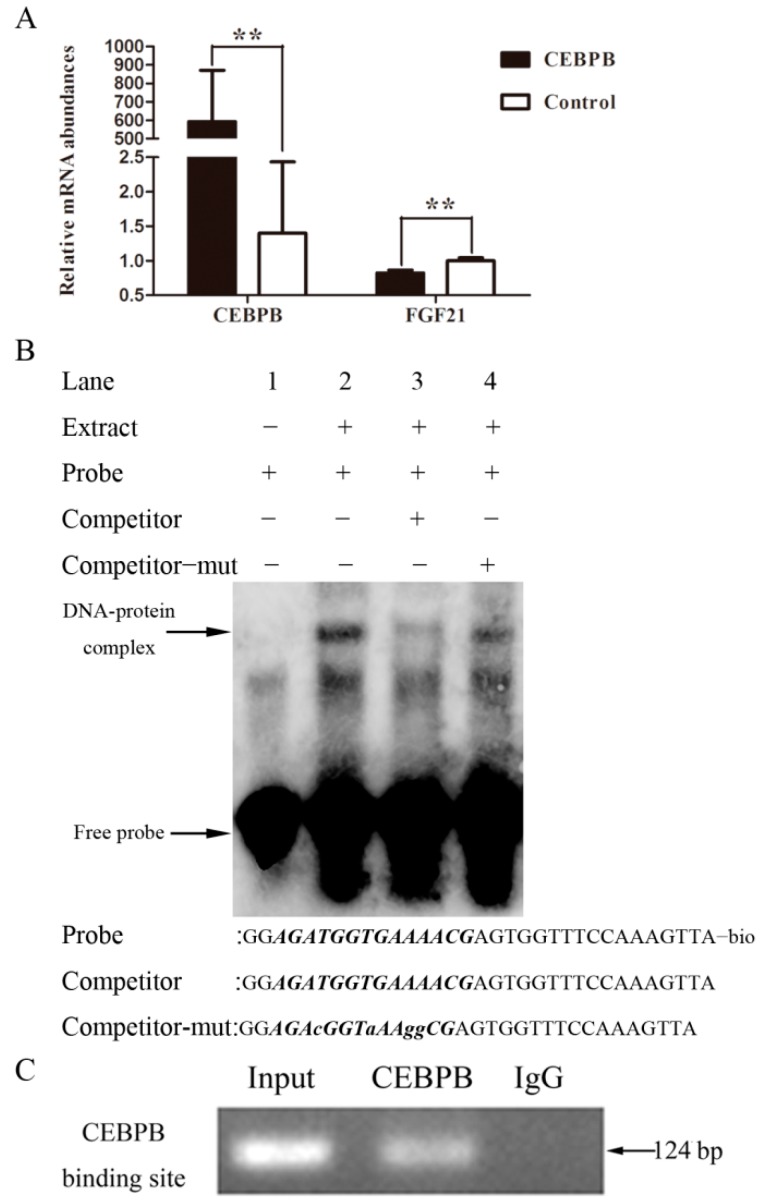
The porcine CEBPB cDNA fragment was inserted into plasmid pcDNA 3.1(+), named pcDNA3.1-CEBPB, which was transfected into PK15 cells using Lipofectamine 2000 (Invitrogen, New York, NY, USA), and then the total RNA was extracted and reversed. The result of qRT-PCR suggested CEBPB suppressed FGF21 expression in the mRNA level (Figure 7A).
Figure 7.
The transcription factor CEBPB suppressed FGF21 expression by binding to FGF21 promoter. QRT-PCR analysis showed CEBPB reduced FGF21 mRNA level. ** for 0.001 < p < 0.01 (A); The electrophoretic mobility shift assay (EMSA) was used to simulate the binding of CEBPB with FGF21 promoter in vitro (B); The chromatin immunoprecipitation (CHIP) assay was performed to detect the binding of CEBPB with FGF21 promoter in vivo (C), total chromatin and normal mouse IgG were used as the input and the negative control respectively.
To investigate whether the transcription factor CEBPB can bind to the promoter of FGF21 to regulate the gene expression, the electrophoretic mobility shift assay was carried out. Our results showed that a DNA-protein complex emerged when the nuclear extracts of IMF cells were incubated with CEBPB probe (lane 2). However the complex was weakened sharply in the presence of 50× cold probe (lane 3) compared with that in 50× mutation cold probe (Figure 7B).
CHIP analysis was preformed to verify whether CEBPB binds to the promoter of FGF21. Our results demonstrated one single DNA band was obtained on agarose gel electrophoresis from PCR product when PK15 cells were incubated with anti-CEBPB, but no band appeared with Normal Mouse IgG (Figure 7C).
3. Discussion
Given the essential role of IMT content in insulin resistant associated disease, the complicated and orderly expression of genes in muscles had been shown to be involved in triglyceride metabolism and their underlying action mechanism in muscles was beginning to be elucidated. Our new finding that FGF21 suppressed adipogenesis in muscles, however, provides a new insight for the treatment of insulin resistant associated disease.
FGF21 was known as a metabolic regulator taking part in glucose metabolism and lipid metabolism and had the ability to treat insulin resistant associated disease [23,24,25,26,27]. However, little was known about its effect on triglyceride metabolism in intramuscular triglycerides, which emerged as a hallmark of insulin resistant associated disease [28,29,30]. In order to understand the effect on IMF, we performed FGF21 overexpression in IMF cells and evaluated the expression of the relative genes participating in triglyceride metabolism.
PPARG was a key gene in adipocyte differentiation, activated most genes involved in adipogenesis directly [31] and played a critical role in fat deposition in mammals [32]. Considering the key role of this molecule, we first evaluated the expression of PPARG. The sharp decline of PPARG was observed, demonstrating FGF21 suppressed adipogenesis by decreasing the expression of PPARG directly or indirectly. As is well known, many factors influencing adipogenesis ultimately also affected the activity of PPARG [13]. CEBPB and CEBPD, induced by adipogenic induction, targeted the promoters of the genes encoding PPARG and CEBPA directly. Then, the interactive activation between PPARG and CEBPA induced a range of genes expressed in mature adipocytes [13]. In order to figure out how FGF21 decreased PPARG expression whether or not through down regulating the expression of CEBP family, the mRNA levels of these genes were measured. The result suggested the CEBPB and CEBPD protein levels were decreased significantly by the action of FGF21, although the mRNA of CEBPB did not change significantly, which together suggested that the decline of the CEBP family decreased the expression of PPARG expression.
Unexpectedly, the interaction between FGF21 and CEBPB was mutual. FGF21 down-regulated the expression of the transcription factor CEBPB as mentioned above. CEBPB led to the reduction of FGF21 expression by binding to the promoter of FGF21 based on the EMSA and CHIP analysis, thus demonstrating the biological function of FGF21 was constrained by CEBPB. This also explained the phenomenon of the low level of FGF21, but the high level of CEBPB in fatty tissue.
To figure out the deep interaction relation of FGF21 and CEBP family which contributed to the declined expression of PPARG, we also measured the expression of the genes involved in the key genes expression in triglyceride metabolism mentioned above. SIRT1, acting as a PPARG co-repressor, impaired adipogenesis [33]. SIRT2 reduced FOXO1 acetylation and phosphorylation, led to an increase in the nuclear localization of FOXO1, and resulted in inhibiting PPARG indirectly [34]. We demonstrated FGF21 indulged in the action of sirtuin during adipocyte differentiation. COUP-TF II, activated by Wnt/β-catenin, recruited the SMRT corepressor complex to PPARG promoter to repress PPARG gene expression [35]. We showed that there were no obvious changes observed in expression of LRP6, β-catenin and COUP-TF II, which suggested FGF21 appeared to have no effect on the Wnt/β-catenin-COUP-TF ii-PPARG pathway.
KLF2 inhibited PPARG and CEBPA expression in the process of 3T3-L1 cell differentiation [36]. By binding to CEBPA promoter in vivo, KLF3 repressed the transcription of CEBPA [37]. KLF4, together with Krox20, bound directly to the CEBPB promoter and transactivated CEBPB reporter cooperatively [38]. KLF5 regulated adipocyte differentiation by activating PPARG promoter [39]. Our data showed there were no significant changes in KLF2 and KLF4. The decrease of PPARG and CEBPA was not altered although an increase in KLF5 and decrease in KLF3 were observed.It was possible that the sharp decline of CEBPB and CEBPD attenuated the activity of KLF3 and KLF5.
It is worth noting that LSD1 expression was dramatically reduced compared with the control The result, together with the reduction of CEBPA in both mRNA and protein, was consistent with a previous finding that LSD1 is necessary for adipogenesis by regulating adipogenic transcription factor CEBPA [40]. It was possible that FGF21 realized its biological function in triglyceride metabolism by reducing LSD1.
In addition, ADIPOQ was believed to be produced exclusively by mature adipocytes and induced by PPARG and CEBPA [13,41]. FABP4 was also a major player involved in fatty acid metabolism and appeared to be necessary for the differentiation of IMF accretion in pigs [42]. PLIN1 was established for a distinct role in regulating both TAG storage and lipolysis in adipocytes and was regarded as a candidate gene contributing to human obesity [43]. In our research, FABP4 and PLIN1, except GLUT4, were down-regulated in FM significantly which was likely to be caused by the reduction of PPARG and CEBPA.
FAS played an important role in fat deposition, which could catalyze acetyl CoA and malonyl CoA synthesis of fatty acid [44,45]. HSL is a limiting velocity enzyme in fat hydrolysis, playing an opposite role to FAS [46]. Our results demonstrated that HSL mRNA level was up-regulated and Visfatin mRNA level was down-regulated, which indicated FGF21 promotes lipolysis by strengthening the fat hydrolysis process. Several studies have reported that the ratio of FAS and HSL was positively correlated with intramuscular fat content [47]. In our study, the ratio of FAS and HSL was down-regulated in FM, but did not reach a significant level. AGPAT2 converted lysophosphatidic acid to phosphatidic acid, a crucial step in synthesis of triglycerides [48]. However, the level of AGPAT2 mRNA did not change as obtained from our data.
In order to better understand the mechanism of FGF21 affected adipogenesis in IMF cells, we analyzed relative gene expression in FGF21 signal. FGF21 did not bind FGFRs directly. So an interaction partner is required for FGF21 to perform its function [25]. Klotho was a crucial cofactor in FGF21 signals. Firstly, FGF21 binds to Klotho via its C-terminus. Next FGF21 contacts FGFRs through its N-terminal [49]. In our study, FGFR1 and Klotho were significantly up-regulated, and the mRNA level of FGFR2 was also increased to some extent. Our results suggested that FGF21 may be a critical factor affecting IMF by inhibiting gene expression involved in lipogenesis and activating FGF21 signal, which well matched the result of FGF21 expression profiles analysis and explained the low expression level of FGF21 in fatty tissues and the high expression level in non-fatty tissues. As for the low expression in muscle tissues, such as longissimus dorsi, the method of modern breeding towards a high content of intramuscular fat may lead to the occurrence of this phenomenon. In addition, our results were consistent with the report by Alexei et al. [25], which further confirmed the role of FGF21 in adipogenesis.
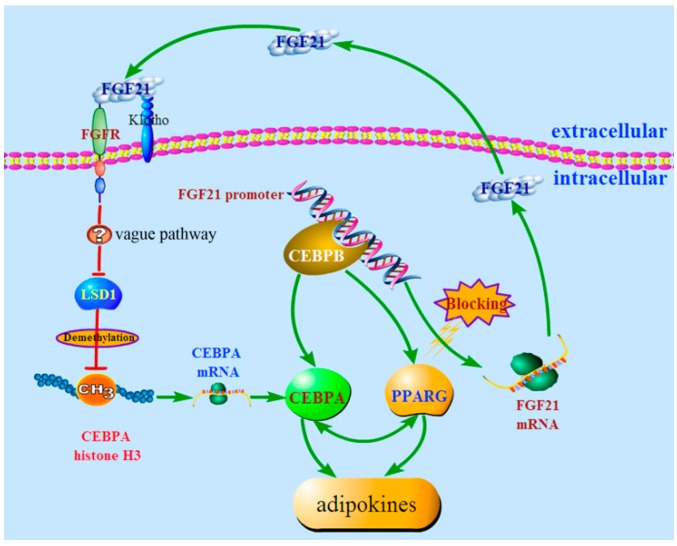
Herein, we believe our study to be the first to elaborate on the clear biological function of FGF21 on triglyceride metabolism in IMF. In brief, triggered by FGF21, the FGF21 signal was activated, and then LSD1 was attenuated, which led to the reduction of PPARG and CEBPA without changing the methylation level of CEBPA and resulted in down-regulation of adipokines and the accumulation of triglycerides in IMF (Figure 8). Our results offer new understanding on FGF21 as a candidate in the treatment of insulin resistant associated diseases.
Figure 8.
The proposed mechanism of FGF21 action in triglyceride metabolism. Briefly, FGF21 suppressed adipogenesis by reducing LSD1 expression, and in the meantime, FGF21 expression was repressed by the transcription factor CEBPB. The red line represented for the action of inhibition and the green line represented for the action of activation.
4. Experimental Section
4.1. Ethics Statement
All animal experiments were carried out according to Huazhong Agricultural University Animal Care and Use Committee Guidelines.
4.2. Expression Profile Analysis of FGF21
The total mRNA from heart, liver, spleen, lung, kidney, brain, small intestine, stomach, Longissimus dorsi, and subcutaneous fat of three-month Large White pigs was extracted by trizol (Invitrogen, New York, NY, USA) and reversely transcribed by Prime Script RT Reagent Kit with gDNA Eraser (TAKARA, Dalian, China). The qRT-PCR was used to analyze FGF21 mRNA expression levels.
4.3. Isolation and Identification of Intramuscular Preadipocyte
A three-day new born Large White pig was washed with 0.3% benzalkonium bromide and then the tissue longissimus dorsi (LD) was taken out. The tissue was digested by collagenase type II for 2 h, then passed through a 400 screen mesh filter and centrifuged 5 min at 1500 r/min. Next the precipitate was resuspended by DMEM/F12 with 15% fetal bovine serum (complete medium), and then the medium was changed after 2 h. After confluence, the medium was changed to the complete medium with 0.5 mmol/L 3-isobutyl-1-methyl xanthine (IBMX), 1 μmol/L dexamethasone (DEX), 10 mg/L insulin for 48 h, and then was changed to the complete medium with 10 mg/L insulin for 48 h. Finally, the complete medium was changed with no drug mentioned above for 8 days.
When the preadipocytes were induced into mature adipocyte, the complete media was discarded and the preadipocytes were washed 3 times with phosphate buffered saline (PBS). Subsequently, the cells were fixed by 10% paraformal dehyde for 1h at room temperature, then moderate oil red O was added for 1h at room temperature after washing 3 times with PBS, finally the cells were washed with PBS and observed under an inverted microscope Observer.A1 (ZEISS, Oberkochen, Germany).
4.4. Stable Transfection
A 627bp cDNA fragment of FGF21 from pigs was cloned and inserted into plasmid pcDNA 3.1(+), named pcDNA3.1-FGF21. Lipofectamine 2000 (Invitrogen, New York, NY, USA) was used to perform cell transfection and G418 was also used in order to get a stable cell line. Firstly, a set of gradients (100, 200, 300, 400, 500 and 600 ng/mL) of G418 were set up to determine the optimal concentration (300 ng/mL). Cell transfection was performed when cells extended to the third generation. Cells were digested by 0.25% trypsin after 6 h, and then inoculated into 96-well plates and only one cell was put into one well. After the cells were adherent, a certain amount of G418 was added into the media to 300 ng/mL. The screening medium was changed every two days until the monoclone appeared.
4.5. RNA Extraction and qRT-PCR
Total RNA was extracted using DNA/RNA/protein isolation kit (OMEGA, Norcross, GA, USA) according to the protocol of the manufacturer. The integrity of the total RNA was detected by 1.2% agarose gel electrophoresis and the concentration was determined using NanoDrop 2000 (Thermo, New York, NY, USA). Then Prime Script RT Reagent Kit with gDNA Eraser (TAKARA, Dalian, China) was used to perform RT-PCR and next CFX96 Real-Time System (Bio-Rad, Berkeley, CA, USA) was used to perform qRT-PCR. The primers for qRT-PCR were designed by Primer Premier 5, which are listed in Table 1.
Table 1.
Primer Sequences for qRT-PCR.
| Genes | Primer Sequences (5′–3′) | Length of Amplicon (bp) |
|---|---|---|
| FGF21 | F: ACTGTGGGTCCCTGTGCTG R: ATCCGTGTAGAGGTATCGTTGG |
118 |
| PPARG | F: TCCCGCTGACCAAAGCAAAGGC R: CCACGGAGCGAAACTGACACCC |
195 |
| FABP4 | F: GAAAGTCAAGAGCACGATAACC R: CAAGATACATTCCACCACCAAC |
124 |
| FAS | F: TTACACCTCCCTCAACTTCCG R: GGCACCATTCCCATCACG |
153 |
| HSL | F: GTCTTTGCGGGTATTCGG R: GAGTTCGGCCAGGTTGTG |
228 |
| Visfatin | F: TGCCTTTGGTTCTGGTGG R: GCGTAATGAGGTGCTGCTTC |
354 |
| LPL | F: TAACGAACCCGACCAGCATC R: TACACCACCGCCACAGCAA |
140 |
| FGFR1 | F: GAATCGGAGGCTACAAGGTC R: GATAGAGTTACCCGCCAAGC |
305 |
| FGFR2 | F: CTGCCGCCAACTCTGTCA R: CGGATGGAACCACGCTTT |
165 |
| Klotho | F: TCATCCTGTCACCGTTTATTC R: CCTCCACCTGAAATGCTCC |
169 |
| CEBPA | F: CGGTGCGTCTAAGATGAGG R: AGCGGTGAGTTTGCGTTT |
120 |
| CEBPB | F: AGCCTGTCCACATCCTCG R: CACGGTCTTCTTGGTCTTACTC |
135 |
| CEBPD | F: TCAAACACGCCGAACTACAC R: GAGCAAAGGGAAAGCAAATC |
207 |
| GLUT4 | F: TGTTGCGGATGCTATGGG R: GGGTTTCACCTCCTGCTCTAA |
169 |
| ADIPOQ | F: CTGGCGAGAAGAGTGAGA R: TGCTGAACGGTAGACATAGGC |
158 |
| AGPAT2 | F: GTTCGTCCGCTCCTACAA R: CAGGATGCTCTGGTGGTTG |
112 |
| PLIN1 | F: GTGCCAGGAACAGCAACAG R: GGGCTCTACCACCTTCTCATC |
194 |
| FOXO1 | F: AAGACCGCTTTACAAGTGCC R: TCTCCATCCATGAGGTCGTT |
200 |
| LSD1 | F: AGCCAGTTGACAGTGAGGAAT R: CTGAAGCGGTGTAGCGAAC |
109 |
| LRP6 | F: GCAGGGTGGAATGAATGTG R: GAGCAGGAAAGTAGTTGGAGC |
150 |
| β -catenin | F: AAGCAGGTGGATCTATTTCATG R: AGCATTGTATCACAGCAGGTTA |
159 |
| GAPDH | F: GGGCATGAACCATGAGAAGT R: AAGCAGGGATGATGTTCTGG124 |
230 |
| KLF2 | F: GGCGAGAAGCCCTAACACT R: CGCACAGATGGCATTGGA |
121 |
| KLF3 | F: CGCACAGATGGCATTGGA R: CAGCACATTCCTCCCAGTTA |
184 |
| KLF4 | F: CGGAGGGAGACGGAGGAGTT R: TGAAGCCGACGAGGACACG |
108 |
| KLF5 | F: GAAGGGTGCGACTGGAGGTT R: TGTGCTGGGCGAGGTGAT |
133 |
| COUP-TF ii | F: CAAGGCCATAGTCCTGTTCACC R: CGTACTCTTCCAAAGCACACTGG |
101 |
4.6. CEBPA Promoter Methylation Analysis
Genomic DNA from the control and FM induced cell differentiation for the 8th day was extracted using DNA/RNA/protein isolation kit (OMEGA), and then treated with sulfate using a Methylation-Gold Kit (ZYMO, Orange, CA, USA) according to the operation manual. The primers used for amplification of bisulfite-treated DNA were designed and the sequences were forward primer: AGAGGAGAGGGTTTTATTAGGGAT and reverse primer: CTCCATAAAAAAACTAAAATCCTCC. The region of CEBPA promoter from −172 to +2 bp was amplified for sequencing. The PCR amplification profile was: 94 °C for 4 min followed by 35 cycles of 94 °C for 30 s, 58 °C for 30 s, and 72 °C for 30 s, with a final extension at 72 °C for 5 min. The Quantification Tool for Methylation Analysis on line was used for CEBPA promoter methylation analysis.
4.7. Western Blot
The protein was extracted from the positive monoclone using DNA/RNA/protein isolation kit (OMEGA). A total of 40 micrograms of protein for each sample was loaded into 12% SDS-polyacrylamide gel lanes. Proteins were transferred to PVDF membranes after electrophoresis, and then the membranes were blocked by 5% skimmed milk (1g skimmed milk in 20 mL TBST). The blocked membranes were incubated with primary antibodies anti-FGF21 (1:1000; Abcam, Cambridge, UK), anti-PPARG (1:1000; Abcam, UK), anti-FABP4 (1:1000; Abcam, UK), anti-KLF5 (1:1000; Abcam, UK), anti-CEBPA (1:1000; Santa Cruz, CA, USA), anti-CEBPB (1:1000; Santa Cruz), anti-KDM1A (1:1000; Abcam, UK), anti-ADIPOQ (1:1000; Santa Cruz), anti-GAPDH (1:1000; Abcam, UK) and anti-beta-actin (ACTIN) (1:1000; Santa Cruz) overnight at 4 °C, then treated with labeled-horseradish peroxidase-conjugated (HRP) secondary antibody. Finally, photographic plate was used to expose and develop films.
4.8. Electrophoretic Mobility Shift Assay (EMSA)
Commercially-available Light Shift Chemiluminescent EMSA Kit (Thermo, New York, NY, USA) was used to perform electrophoretic mobility shift assay. Nuclear protein of pig IMF cells was extracted with Nucleoprotein Extraction Kit (Active Motif, Carlsbad, CA, USA). Oligonucleotides (Sangon, Shanghai, China) corresponding to the CEBPB binding sites of the FGF21 promoter were annealed into double strands. All the reagents in every group were added according to the manufacturer’s instructions. After being incubated for 15 min at room temperature, 20 μL of each mixture was loaded onto the 6% polyacrylamide gel until the bromophenol blue dye had migrated approximately 3/4 down the length of the gel. The transfer time was 45 min at 380 mA (~100 V). Finally, the nylon membrane was scanned with Automated BioSpectrum Imaging System (UVP).
4.9. Chromatin Immunoprecipitation (CHIP) Assay
CHIP assay was carried out using a commercially available CHIP Assay Kit (Beyotime, Shanghai, China). After incubating 1% formaldehyde at 37 °C for 10 min, PK15 cells were washed and harvested. Then, the nuclear lysates were sonicated at a setting of 20 times for 10 s pulses using the Scientz-IID (Scientz, Ningbo, China). The chromatin was immunoprecipitated with CEBPB antibody (Santa) at 4 °C overnight and the control, under the same experimental condition, was immunoprecipitated with Normal Mouse IgG (Millipore, Billerica, MA, USA). Finally, the immunoprecipitated DNA was purified by the AxyPrep PCR Cleanup Kit (Axygen, Hangzhou, China). One pair of oligonucleotide primers was synthesized to amplify the DNA fragments of CEBPB binding sites in the FGF21 promoter.
4.10. Statistical Analysis
Student t-test was used for qRT-PCR statistical analysis and all the data were presented as mean ± SD. Fisher’s exact test was used for Methylation Analysis. 0.01 < p < 0.05 was defined as significance, 0.001 < p < 0.01 was defined as very significant and p < 0.001 was defined as of great significance.
5. Conclusions
In this study, we isolated intramuscular preadipocytes from pig longissimus dorsi, and constructed stable transfection intramuscular preadipocytes cell line of pig FGF21. We evaluated the expression of the genes involved in adipogenesis when the cell induced adipogenic differentiation on the 8th day. Our final analysis conclusions can be summarized as follow:
FGF21 suppressed adipogenesis by reducing LSD1 expression.
The transcription factor CEBPB can bind to the promoter of FGF21 directly, and attenuates the expression of FGF21.
Acknowledgments
The authors are grateful for the help from all the members of their laboratory and kindly thank Hong Xiang for the technical support of cell isolation. This work was supported by grants from the National Key Basic Research Program of China (2012CB124702) and grants from the National Project for Breeding of Transgenic Pig (2012ZX08006-002, 2013ZX08006-002).
Author Contributions
Yongliang Wang and Yuanzhu Xiong designed the experiments. Yongliang Wang and Xinyi Liu performed the expriments. Yongliang Wang, Xinyi Liu, Wangjun Wu, Liming Hou and Yuanzhu Xiong analyzed the data. Yongliang Wang and Shuhong Zhao wrote and revised the manuscript. All authors read and approved the manuscript.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Muoio D.M. Revisiting the connection between intramyocellular lipids and insulin resistance: A long and winding road. Diabetologia. 2012;55:2551–2554. doi: 10.1007/s00125-012-2597-y. [DOI] [PubMed] [Google Scholar]
- 2.Guo Z. Intramyocellular lipids: Maker vs. marker of insulin resistance. Med. Hypotheses. 2008;70:625–629. doi: 10.1016/j.mehy.2007.03.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cesar A.S., Regitano L.C., Koltes J.E., Fritz-Waters E.R., Lanna D.P., Gasparin G., Mourao G.B., Oliveira P.S., Reecy J.M., Coutinho L.L. Putative regulatory factors associated with intramuscular fat content. PLoS ONE. 2015;10:e0128350. doi: 10.1371/journal.pone.0128350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Larson-Meyer D.E., Newcomer B.R., Ravussin E., Volaufova J., Bennett B., Chalew S., Cefalu W.T., Sothern M. Intrahepatic and intramyocellular lipids are determinants of insulin resistance in prepubertal children. Diabetologia. 2011;54:869–875. doi: 10.1007/s00125-010-2022-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Christodoulides C., Lagathu C., Sethi J.K., Vidal-Puig A. Adipogenesis and WNT signalling. Trends Endocrinol. Metab. 2009;20:16–24. doi: 10.1016/j.tem.2008.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ross S.E. Inhibition of Adipogenesis by Wnt Signaling. Science. 2000;289:950–953. doi: 10.1126/science.289.5481.950. [DOI] [PubMed] [Google Scholar]
- 7.Bennett C.N., Ross S.E., Longo K.A., Bajnok L., Hemati N., Johnson K.W., Harrison S.D., MacDougald O.A. Regulation of Wnt signaling during adipogenesis. J. Biol. Chem. 2002;277:30998–31004. doi: 10.1074/jbc.M204527200. [DOI] [PubMed] [Google Scholar]
- 8.Waki H., Park K.W., Mitro N., Pei L., Damoiseaux R., Wilpitz D.C., Reue K., Saez E., Tontonoz P. The small molecule harmine is an antidiabetic cell-type-specific regulator of PPARγ expression. Cell Metab. 2007;5:357–370. doi: 10.1016/j.cmet.2007.03.010. [DOI] [PubMed] [Google Scholar]
- 9.Nakae J., Kitamura T., Kitamura Y., Biggs W.H., III, Arden K.C., Accili D. The forkhead transcription factor Foxo1 regulates adipocyte differentiation. Dev. Cell. 2003;4:119–129. doi: 10.1016/S1534-5807(02)00401-X. [DOI] [PubMed] [Google Scholar]
- 10.Liu W., Singh R., Choi C.S., Lee H.Y., Keramati A.R., Samuel V.T., Lifton R.P., Shulman G.I., Mani A. Low density lipoprotein (LDL) receptor-related protein 6 (LRP6) regulates body fat and glucose homeostasis by modulating nutrient sensing pathways and mitochondrial energy expenditure. J. Biol. Chem. 2012;287:7213–7223. doi: 10.1074/jbc.M111.286724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Oates A.C. The zebrafish klf gene family. 2001;98:1792–1801. doi: 10.1182/blood.V98.6.1792. [DOI] [PubMed] [Google Scholar]
- 12.De Ruijter A.J.M., van Gennip A.H., Caron H.N., Kemp S., van Kuilenburg A.B.P. Histone deacetylases (HDACs): Characterization of the classical HDAC family. Biochem. J. 2003;370:737–749. doi: 10.1042/BJ20021321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lowe C.E., O’Rahilly S., Rochford J.J. Adipogenesis at a glance. J. Cell Sci. 2011;124:2681–2686. doi: 10.1242/jcs.079699. [DOI] [PubMed] [Google Scholar]
- 14.Sun Z., Miller R.A., Patel R.T., Chen J., Dhir R., Wang H., Zhang D., Graham M.J., Unterman T.G., Shulman G.I., et al. Hepatic Hdac3 promotes gluconeogenesis by repressing lipid synthesis and sequestration. Nat. Med. 2012;18:934–942. doi: 10.1038/nm.2744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Inagaki T., Dutchak P., Zhao G., Ding X., Gautron L., Parameswara V., Li Y., Goetz R., Mohammadi M., Esser V., et al. Endocrine regulation of the fasting response by PPARα-mediated induction of fibroblast growth factor 21. Cell Metab. 2007;5:415–425. doi: 10.1016/j.cmet.2007.05.003. [DOI] [PubMed] [Google Scholar]
- 16.Arner P., Pettersson A., Mitchell P.J., Dunbar J.D., Kharitonenkov A., Ryden M. FGF21 attenuates lipolysis in human adipocytes—A possible link to improved insulin sensitivity. FEBS Lett. 2008;582:1725–1730. doi: 10.1016/j.febslet.2008.04.038. [DOI] [PubMed] [Google Scholar]
- 17.Li X., Ge H., Weiszmann J., Hecht R., Li Y.S., Veniant M.M., Xu J., Wu X., Lindberg R., Li Y. Inhibition of lipolysis may contribute to the acute regulation of plasma FFA and glucose by FGF21 in ob/ob mice. FEBS Lett. 2009;583:3230–3234. doi: 10.1016/j.febslet.2009.09.012. [DOI] [PubMed] [Google Scholar]
- 18.Chen W., Hoo R.L., Konishi M., Itoh N., Lee P.C., Ye H.Y., Lam K.S., Xu A. Growth hormone induces hepatic production of fibroblast growth factor 21 through a mechanism dependent on lipolysis in adipocytes. J. Biol. Chem. 2011;286:34559–34566. doi: 10.1074/jbc.M111.285965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Murata Y., Konishi M., Itoh N. FGF21 as an endocrine regulator in lipid metabolism: From molecular evolution to physiology and pathophysiology. J. Nutr. Metab. 2011;2011 doi: 10.1155/2011/981315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Canto C., Auwerx J. Cell biology. FGF21 takes a fat bite. Science. 2012;336:675–676. doi: 10.1126/science.1222646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Coskun T., Bina H.A., Schneider M.A., Dunbar J.D., Hu C.C., Chen Y., Moller D.E., Kharitonenkov A. Fibroblast growth factor 21 corrects obesity in mice. Endocrinology. 2008;149:6018–6027. doi: 10.1210/en.2008-0816. [DOI] [PubMed] [Google Scholar]
- 22.Chau M.D., Gao J., Yang Q., Wu Z., Gromada J. Fibroblast growth factor 21 regulates energy metabolism by activating the AMPK-SIRT1-PGC-1α pathway. Proc. Natl. Acad. Sci. USA. 2010;107:12553–12558. doi: 10.1073/pnas.1006962107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xu J., Lloyd D.J., Hale C., Stanislaus S., Chen M., Sivits G., Vonderfecht S., Hecht R., Li Y.S., Lindberg R.A., et al. Fibroblast growth factor 21 reverses hepatic steatosis, increases energy expenditure, and improves insulin sensitivity in diet-induced obese mice. Diabetes. 2009;58:250–259. doi: 10.2337/db08-0392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hotta Y., Nakamura H., Konishi M., Murata Y., Takagi H., Matsumura S., Inoue K., Fushiki T., Itoh N. Fibroblast growth factor 21 regulates lipolysis in white adipose tissue but is not required for ketogenesis and triglyceride clearance in liver. Endocrinology. 2009;150:4625–4633. doi: 10.1210/en.2009-0119. [DOI] [PubMed] [Google Scholar]
- 25.Kharitonenkov A., Shiyanova T.L., Koester A., Ford A.M., Micanovic R., Galbreath E.J., Sandusky G.E., Hammond L.J., Moyers J.S., Owens R.A., et al. FGF-21 as a novel metabolic regulator. J. Clin. Investig. 2005;115:1627–1635. doi: 10.1172/JCI23606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huang Z., Wang H., Lu M., Sun C., Wu X., Tan Y., Ye C., Zhu G., Wang X., Cai L., et al. A better anti-diabetic recombinant human fibroblast growth factor 21 (rhFGF21) modified with polyethylene glycol. PLoS ONE. 2011;6:e20669. doi: 10.1371/journal.pone.0020669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kharitonenkov A., Wroblewski V.J., Koester A., Chen Y.F., Clutinger C.K., Tigno X.T., Hansen B.C., Shanafelt A.B., Etgen G.J. The metabolic state of diabetic monkeys is regulated by fibroblast growth factor-21. Endocrinology. 2007;148:774–781. doi: 10.1210/en.2006-1168. [DOI] [PubMed] [Google Scholar]
- 28.Muoio D.M. Intramuscular triacylglycerol and insulin resistance: Guilty as charged or wrongly accused? Biochim. Biophys. Acta. 2010;1801:281–288. doi: 10.1016/j.bbalip.2009.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Muoio D.M., Newgard C.B. Molecular and metabolic mechanisms of insulin resistance and β-cell failure in type 2 diabetes. Nat. Rev. Mol. Cell Biol. 2008;9:193–205. doi: 10.1038/nrm2327. [DOI] [PubMed] [Google Scholar]
- 30.McGarry J. Dysregulation of fatty acid metabolism in the etiology of type 2 diabetes. Diabetes. 2002;51:17–18. doi: 10.2337/diabetes.51.1.7. [DOI] [PubMed] [Google Scholar]
- 31.Siersbaek R., Nielsen R., Mandrup S. PPARγ in adipocyte differentiation and metabolism—Novel insights from genome-wide studies. FEBS Lett. 2010;584:3242–3249. doi: 10.1016/j.febslet.2010.06.010. [DOI] [PubMed] [Google Scholar]
- 32.Lehrke M., Lazar M.A. The many faces of PPARγ. Cell. 2005;123:993–999. doi: 10.1016/j.cell.2005.11.026. [DOI] [PubMed] [Google Scholar]
- 33.Picard F., Kurtev M., Chung N., Topark-Ngarm A., Senawong T., de Machado Oliveira R., Leid M., McBurney M.W., Guarente L. Sirt1 promotes fat mobilization in white adipocytes by repressing PPAR-(gamma) Nature. 2004;429:771–776. doi: 10.1038/nature02583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jing E., Gesta S., Kahn C.R. SIRT2 Regulates Adipocyte differentiation through FoxO1 acetylation/deacetylation. Cell Metabol. 2007;6:105–114. doi: 10.1016/j.cmet.2007.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Okamura M., Kudo H., Wakabayashi K., Tanaka T., Nonaka A., Uchida A., Tsutsumi S., Sakakibara I., Naito M., Osborne T.F., et al. COUP-TFII acts downstream of Wnt/β-catenin signal to silence PPARγ gene expression and repress adipogenesis. Proc. Natl. Acad. Sci. USA. 2009;106:5819–5824. doi: 10.1073/pnas.0901676106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Banerjee S.S., Feinberg M.W., Watanabe M., Gray S., Haspel R.L., Denkinger D.J., Kawahara R., Hauner H., Jain M.K. The Kruppel-like factor KLF2 inhibits peroxisome proliferator-activated receptor-gamma expression and adipogenesis. J. Biol. Chem. 2003;278:2581–2584. doi: 10.1074/jbc.M210859200. [DOI] [PubMed] [Google Scholar]
- 37.Sue N., Jack B.H., Eaton S.A., Pearson R.C., Funnell A.P., Turner J., Czolij R., Denyer G., Bao S., Molero-Navajas J.C., et al. Targeted disruption of the basic Kruppel-like factor gene (Klf3) reveals a role in adipogenesis. Mol. Cell. Biol. 2008;28:3967–3978. doi: 10.1128/MCB.01942-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Birsoy K., Chen Z., Friedman J. Transcriptional regulation of adipogenesis by KLF4. Cell Metab. 2008;7:339–347. doi: 10.1016/j.cmet.2008.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kinoshita M., Ono K., Horie T., Nagao K., Nishi H., Kuwabara Y., Takanabe-Mori R., Hasegawa K., Kita T., Kimura T. Regulation of adipocyte differentiation by activation of serotonin (5-HT) receptors 5-HT2AR and 5-HT2CR and involvement of microRNA-448-mediated repression of KLF5. Mol. Endocrinol. 2010;24:1978–1987. doi: 10.1210/me.2010-0054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Musri M.M., Carmona M.C., Hanzu F.A., Kaliman P., Gomis R., Parrizas M. Histone demethylase LSD1 regulates adipogenesis. J. Biol. Chem. 2010;285:30034–30041. doi: 10.1074/jbc.M110.151209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Esteve E., Ricart W., Fernandez-Real J.M. Adipocytokines and Insulin Resistance: The possible role of lipocalin-2, retinol binding protein-4, and adiponectin. Diabetes Care. 2009;32:S362–S367. doi: 10.2337/dc09-S340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gerbens F., Jansen A., van Erp A.J.M., Harders F., Meuwissen T.H.E., Rettenberger G., Veerkamp J.H., te Pas M.F.W. The adipocyte fatty acid-binding protein locus: Characterization and association with intramuscular fat content in pigs. Mamm. Genome. 1998;9:1022–1026. doi: 10.1007/s003359900918. [DOI] [PubMed] [Google Scholar]
- 43.Jenkins N.T., McKenzie J.A., Damcott C.M., Witkowski S., Hagberg J.M. Endurance exercise training effects on body fatness, VO2max, HDL-C subfractions, and glucose tolerance are influenced by a PLIN haplotype in older Caucasians. J. Appl. Physiol. 2010;108:498–506. doi: 10.1152/japplphysiol.01018.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen Y., Mu P., He S., Tang X., Guo X., Li H., Xu H., Woo S.L., Qian X., Zeng L., et al. Gly482Ser mutation impairs the effects of peroxisome proliferator—Activated receptor γ coactivator-1α on decreasing fat deposition and stimulating phosphoenolpyruvate carboxykinase expression in hepatocytes. Nutr. Res. 2013;33:332–339. doi: 10.1016/j.nutres.2013.02.003. [DOI] [PubMed] [Google Scholar]
- 45.Rodriguez J.A., Ben Ali Y., Abdelkafi S., Mendoza L.D., Leclaire J., Fotiadu F., Buono G., Carrière F., Abousalham A. In vitro stereoselective hydrolysis of diacylglycerols by hormone-sensitive lipase. Biochim. Biophys. Acta (BBA) Mol. Cell Biol. Lipids. 2010;1801:77–83. doi: 10.1016/j.bbalip.2009.09.020. [DOI] [PubMed] [Google Scholar]
- 46.Yan X., Chen J., Zhang C., Zhou S., Zhang Z., Chen J., Feng W., Li X., Tan Y. FGF21 deletion exacerbates diabetic cardiomyopathy by aggravating cardiac lipid accumulation. J. Cell. Mol. Med. 2015;19:1557–1568. doi: 10.1111/jcmm.12530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cortés V.A., Cautivo K.M., Rong S., Garg A., Horton J.D., Agarwal A.K. Leptin ameliorates insulin resistance and hepatic steatosis in Agpat2−/− lipodystrophic mice independent of hepatocyte leptin receptors. J. Lipid Res. 2014;55:276–288. doi: 10.1194/jlr.M045799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kharitonenkov A., Larsen P. FGF21 reloaded: Challenges of a rapidly growing field. Trends Endocrinol. Metabol. 2011;22:81–86. doi: 10.1016/j.tem.2010.11.003. [DOI] [PubMed] [Google Scholar]
- 49.Jin D., Tan H.J., Lei T., Chen X.D., Long Q.Q., Feng B., Yang Z.Q. Molecular cloning and characterization of porcine sirtuin genes. Comp. Biochem. Phys. B. 2009;153:348–358. doi: 10.1016/j.cbpb.2009.04.004. [DOI] [PubMed] [Google Scholar]
- 50.Hou L., Ma F.H., Yang J.Z., Riaz H., Wang Y.L., Wu W.J., Xia X.L., Ma Z.Y., Zhou Y., Zhang L., et al. Effects of histone deacetylase inhibitor oxamflatinon in vitro porcine somatic cell nuclear transfer embryos. Cell. Reprogram. 2014;16:253–265. doi: 10.1089/cell.2013.0058. [DOI] [PMC free article] [PubMed] [Google Scholar]