Abstract
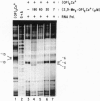
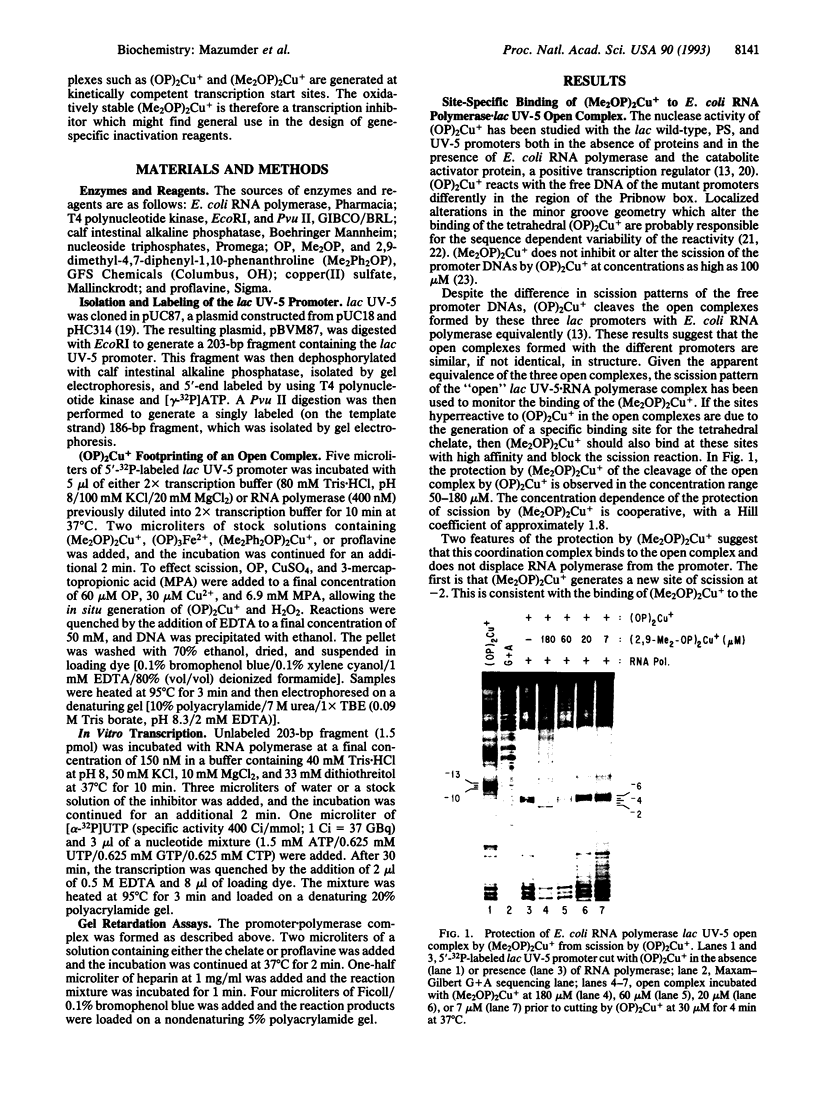
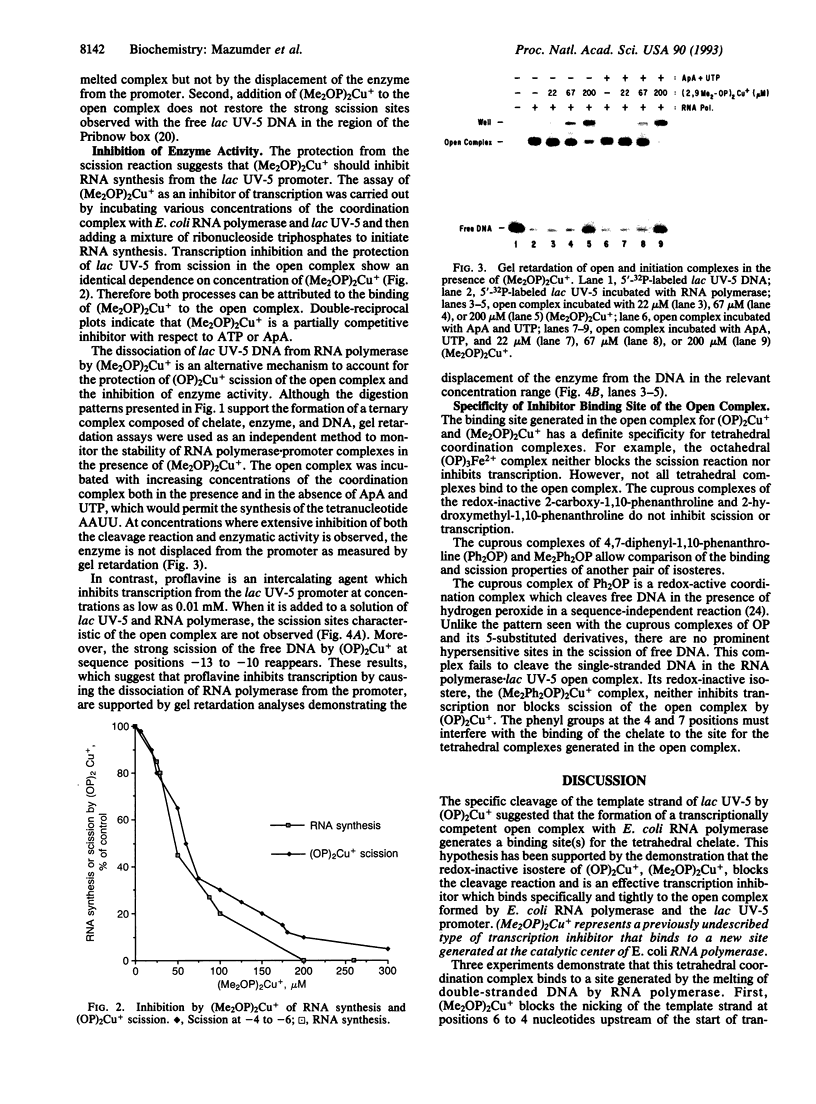
The kinetically component open complexes formed at prokaryotic and eukaryotic transcription start sites are efficiently nicked by the chemical nuclease activity of the 2:1 1,10-phenanthroline-copper(I) complex [(OP)2Cu+] and hydrogen peroxide. This reaction specificity has been attributed to the creation of a binding site(s) for redox-active tetrahedral (OP)2Cu+ when RNA polymerase form productive complexes with promoters. This proposal has been confirmed for the Escherichia coli lac UV-5 promoter by the demonstration that the 2:1 2,9-dimethyl-1,10-phenanthroline-copper(I) complex [(Me2OP)2Cu+], a redox-inactive isostere of (OP)2-Cu+, protects the transcription start site from scission by the chemical nuclease activity. (Me2OP)2Cu+ is also an effective inhibitor of transcription. The inhibition of transcription and the protection from scission of the open complex by (OP)2Cu+ exhibit the same dependence on the concentration of (Me2OP)2Cu+. This redox- and exchange-stable species is a previously undescribed transcription inhibitor that binds to a site generated by the interaction of RNA polymerase with the promoter. Unlike the intercalating agent proflavine, which is also an effective transcription inhibitor, it does not displace the enzyme from the promoter. The ability of (Me2OP)2Cu+ to inhibit transcription may be partially responsible for its potent cytotoxicity.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- BLANK F. In vitro fungistatic action of phenanthrolines against pathogenic fungi. Nature. 1951 Sep 22;168(4273):516–517. doi: 10.1038/168516a0. [DOI] [PubMed] [Google Scholar]
- Barton J. K. Metals and DNA: molecular left-handed complements. Science. 1986 Aug 15;233(4765):727–734. doi: 10.1126/science.3016894. [DOI] [PubMed] [Google Scholar]
- Buc H., McClure W. R. Kinetics of open complex formation between Escherichia coli RNA polymerase and the lac UV5 promoter. Evidence for a sequential mechanism involving three steps. Biochemistry. 1985 May 21;24(11):2712–2723. doi: 10.1021/bi00332a018. [DOI] [PubMed] [Google Scholar]
- Buratowski S., Sopta M., Greenblatt J., Sharp P. A. RNA polymerase II-associated proteins are required for a DNA conformation change in the transcription initiation complex. Proc Natl Acad Sci U S A. 1991 Sep 1;88(17):7509–7513. doi: 10.1073/pnas.88.17.7509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow C. S., Barton J. K. Recognition of G-U mismatches by tris(4,7-diphenyl-1,10-phenanthroline)rhodium(III). Biochemistry. 1992 Jun 23;31(24):5423–5429. doi: 10.1021/bi00139a001. [DOI] [PubMed] [Google Scholar]
- Daube S. S., von Hippel P. H. Functional transcription elongation complexes from synthetic RNA-DNA bubble duplexes. Science. 1992 Nov 20;258(5086):1320–1324. doi: 10.1126/science.1280856. [DOI] [PubMed] [Google Scholar]
- Diffley J. F., Cocker J. H. Protein-DNA interactions at a yeast replication origin. Nature. 1992 May 14;357(6374):169–172. doi: 10.1038/357169a0. [DOI] [PubMed] [Google Scholar]
- Feig A. L., Thederahn T., Sigman D. S. Mutagenicity of the nuclease activity of 1,10-phenanthroline-copper ion. Biochem Biophys Res Commun. 1988 Aug 30;155(1):338–343. doi: 10.1016/s0006-291x(88)81090-8. [DOI] [PubMed] [Google Scholar]
- Gralla J. D. Rapid "footprinting" on supercoiled DNA. Proc Natl Acad Sci U S A. 1985 May;82(10):3078–3081. doi: 10.1073/pnas.82.10.3078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gralla J. D. Transcriptional control--lessons from an E. coli promoter data base. Cell. 1991 Aug 9;66(3):415–418. doi: 10.1016/0092-8674(81)90001-5. [DOI] [PubMed] [Google Scholar]
- Kirkegaard K., Buc H., Spassky A., Wang J. C. Mapping of single-stranded regions in duplex DNA at the sequence level: single-strand-specific cytosine methylation in RNA polymerase-promoter complexes. Proc Natl Acad Sci U S A. 1983 May;80(9):2544–2548. doi: 10.1073/pnas.80.9.2544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohindru A., Fisher J. M., Rabinovitz M. 2,9-Dimethyl-1,10-phenanthroline (neocuproine): a potent, copper-dependent cytotoxin with anti-tumor activity. Biochem Pharmacol. 1983 Dec 1;32(23):3627–3632. doi: 10.1016/0006-2952(83)90314-3. [DOI] [PubMed] [Google Scholar]
- Mohindru A., Fisher J. M., Rabinovitz M. Bathocuproine sulphonate: a tissue culture-compatible indicator of copper-mediated toxicity. Nature. 1983 May 5;303(5912):64–65. doi: 10.1038/303064a0. [DOI] [PubMed] [Google Scholar]
- Murakawa G. J., Chen C. H., Kuwabara M. D., Nierlich D. P., Sigman D. S. Scission of RNA by the chemical nuclease of 1,10-phenanthroline-copper ion: preference for single-stranded loops. Nucleic Acids Res. 1989 Jul 11;17(13):5361–5375. doi: 10.1093/nar/17.13.5361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pope L. E., Sigman D. S. Secondary structure specificity of the nuclease activity of the 1,10-phenanthroline-copper complex. Proc Natl Acad Sci U S A. 1984 Jan;81(1):3–7. doi: 10.1073/pnas.81.1.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roe J. H., Burgess R. R., Record M. T., Jr Temperature dependence of the rate constants of the Escherichia coli RNA polymerase-lambda PR promoter interaction. Assignment of the kinetic steps corresponding to protein conformational change and DNA opening. J Mol Biol. 1985 Aug 5;184(3):441–453. doi: 10.1016/0022-2836(85)90293-1. [DOI] [PubMed] [Google Scholar]
- Sasse-Dwight S., Gralla J. D. KMnO4 as a probe for lac promoter DNA melting and mechanism in vivo. J Biol Chem. 1989 May 15;264(14):8074–8081. [PubMed] [Google Scholar]
- Sasse-Dwight S., Gralla J. D. Probing the Escherichia coli glnALG upstream activation mechanism in vivo. Proc Natl Acad Sci U S A. 1988 Dec;85(23):8934–8938. doi: 10.1073/pnas.85.23.8934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro A., Nathan H. C., Hutner S. H., Garofalo J., McLaughlin S. D., Rescigno D., Bacchi C. J. In vivo and in vitro activity by diverse chelators against Trypanosoma brucei brucei. J Protozool. 1982 Feb;29(1):85–90. doi: 10.1111/j.1550-7408.1982.tb02885.x. [DOI] [PubMed] [Google Scholar]
- Sigman D. S. Chemical nucleases. Biochemistry. 1990 Oct 2;29(39):9097–9105. doi: 10.1021/bi00491a001. [DOI] [PubMed] [Google Scholar]
- Sigman D. S., Chen C. H. Chemical nucleases: new reagents in molecular biology. Annu Rev Biochem. 1990;59:207–236. doi: 10.1146/annurev.bi.59.070190.001231. [DOI] [PubMed] [Google Scholar]
- Sigman D. S., Spassky A., Rimsky S., Buc H. Conformational analysis of lac promoters using the nuclease activity of 1,10-phenanthroline-copper ion. Biopolymers. 1985 Jan;24(1):183–197. doi: 10.1002/bip.360240115. [DOI] [PubMed] [Google Scholar]
- Spassky A., Sigman D. S. Nuclease activity of 1,10-phenanthroline-copper ion. Conformational analysis and footprinting of the lac operon. Biochemistry. 1985 Dec 31;24(27):8050–8056. doi: 10.1021/bi00348a032. [DOI] [PubMed] [Google Scholar]
- Thederahn T., Spassky A., Kuwabara M. D., Sigman D. S. Chemical nuclease activity of 5-phenyl-1,10-phenanthroline-copper ion detects intermediates in transcription initiation by E. Coli RNA polymerase. Biochem Biophys Res Commun. 1990 Apr 30;168(2):756–762. doi: 10.1016/0006-291x(90)92386-e. [DOI] [PubMed] [Google Scholar]
- Wang W., Carey M., Gralla J. D. Polymerase II promoter activation: closed complex formation and ATP-driven start site opening. Science. 1992 Jan 24;255(5043):450–453. doi: 10.1126/science.1310361. [DOI] [PubMed] [Google Scholar]
- Yoon C., Kuwabara M. D., Law R., Wall R., Sigman D. S. Sequence-dependent variability of DNA structure. Influence of flanking sequences and fragment length on digestion by conformationally sensitive nucleases. J Biol Chem. 1988 Jun 15;263(17):8458–8463. [PubMed] [Google Scholar]
- Yoon C., Kuwabara M. D., Spassky A., Sigman D. S. Sequence specificity of the deoxyribonuclease activity of 1,10-phenanthroline-copper ion. Biochemistry. 1990 Feb 27;29(8):2116–2121. doi: 10.1021/bi00460a022. [DOI] [PubMed] [Google Scholar]
- von Hippel P. H., Bear D. G., Morgan W. D., McSwiggen J. A. Protein-nucleic acid interactions in transcription: a molecular analysis. Annu Rev Biochem. 1984;53:389–446. doi: 10.1146/annurev.bi.53.070184.002133. [DOI] [PubMed] [Google Scholar]
- von Hippel P. H., Yager T. D. The elongation-termination decision in transcription. Science. 1992 Feb 14;255(5046):809–812. doi: 10.1126/science.1536005. [DOI] [PubMed] [Google Scholar]