Abstract
Cerebral malaria (CM) is a neurological complication of infection with Plasmodium falciparum that is partly caused by cytokine-mediated inflammation. It is not known whether interleukin-17 (IL-17) cytokines, which regulate inflammation, control the development of CM. To evaluate the involvement of IL-17 cytokines in CM, we analyzed 46 common polymorphisms in IL17A, IL17F, and IL17RA (which encodes the common receptor chain of the members of the IL-17 family) in two independent African populations. A case-control study involving 115 Nigerian children with CM and 160 controls from the community (CC) showed that IL17F reference single nucleotide polymorphism (SNP) 6913472 (rs6913472) (P = 0.004; odds ratio [OR] = 3.12), IL17F rs4715291 (P = 0.004; OR = 2.82), IL17RA rs12159217 (P = 0.01; OR = 2.27), and IL17RA rs41396547 (P = 0.026; OR = 3.15) were independently associated with CM. A replication study was performed in 240 nuclear Malian family trios (two parents with one CM child). We replicated the association for 3 SNPs, IL17F rs6913472 (P = 0.03; OR = 1.39), IL17RA rs12159217 (P = 0.01; OR = 1.52), and IL17RA rs41396547 (P = 0.04; OR = 3.50). We also found that one additional SNP, IL17RA rs41433045, in linkage disequilibrium (LD) with rs41396547, was associated with CM in both Nigeria and Mali (P = 0.002; OR = 4.12 in the combined sample). We excluded the possibility that SNPs outside IL17F and IL17RA, in strong LD with the associated SNPs, could account for the observed associations. Furthermore, the results of a functional study indicated that the aggravating GA genotype of IL17F rs6913472 was associated with lower IL-17F concentrations. Our findings show for the first time that IL17F and IL17RA polymorphisms modulate susceptibility to CM and provide evidence that IL-17F protects against CM.
INTRODUCTION
Cerebral malaria (CM) is one of the most severe complications of infection with Plasmodium falciparum and occurs predominantly in young children under 5 years of age and in “nonimmune” adults. The clinical characteristics of CM are an unarousable coma lasting for at least 1 h, with or without generalized convulsions, and asexual P. falciparum parasitemia with normal cerebrospinal fluid and no other cause of encephalopathy. This reversible encephalopathy is characterized by the sequestration of infected red blood cells (IRBC) in the capillaries of the brain together with the accumulation of leukocytes, platelets, and uninfected red blood cells (URBC) causing mechanical obstruction of microvessels and excessive activation of immune cells. Other pathological consequences are brain edema, alterations of the integrity of the blood-brain barrier (BBB), microhemorrhages, and tissue necrosis (1). However, despite the large number of studies that have investigated CM, the orchestration of the pathogenic mechanisms leading to CM is not well understood.
Proinflammatory cytokines are thought to contribute to brain pathology in CM. Interleukin-17A (IL-17A) and IL-17F, the best-studied members of the IL-17 family (2, 3), control infections at epithelial barriers (4, 5); they were also shown to aggravate autoimmune diseases and inflammation, including in the brain (5–7). We tested here the hypothesis that IL-17A and IL-17F could aggravate CM in children. We performed in samples from two African populations a study of association between CM and common genetic variants in IL17A, IL17F, and IL17RA (encoding the chain common to all receptors of the IL-17), providing comprehensive coverage of these genes. Our data indicate that genetic variants in IL17F and IL17RA are associated with susceptibility to CM in the two study populations. Furthermore, our findings provide evidence that IL-17F protects against CM.
MATERIALS AND METHODS
Study participants.
The discovery cohort was recruited among children from Ibadan, Nigeria. This city has the second largest urban population in the Yoruba plateau, which is a region of holoendemic transmission. We used the Nigerian sample as the discovery cohort because the linkage disequilibrium (LD) map of the Yoruba is described as an African reference population in data banks. The replication cohort was recruited among children from Bamako (Mali) who were living under conditions of seasonal malaria.
The internationally recognized joint ethics committee of the College of Medicine of the University of Ibadan and the University College Hospital in Ibadan approved the Nigerian case-control study. Parents or guardians of children from the city of Ibadan gave written informed consent for their children to participate in the study. The Malian study was approved by the local ethic committees of the Faculty of Medicine of the University of Bamako; written informed consent was obtained from all parents.
All participating children from Ibadan were recruited under the direction of the Childhood Malaria Research Group (CMRG) at the Department of Pediatrics of the University College Hospital (UCH), Ibadan, Nigeria, as previously described (8–11). Briefly, children were 6 months to 13 years of age. WHO criteria were used to define severe malaria (12, 13). CM was defined as a state of unarousable coma (Blantyre coma score ≤ 2) lasting for at least 1 h accompanied by asexual Plasmodium falciparum parasitemia with normal cerebrospinal fluid results. Children with CM were also considered to have severe malarial anemia (SMA) if they had a packed cell volume (PCV) of less than 16%. The group of controls from the community (CC) included age-matched parasite-negative healthy children from the same community. Uncomplicated malaria (UM) cases were defined as febrile children with P. falciparum parasitemia and with a PCV greater than 20% who did not require hospital admission.
Malian children with CM were recruited as described previously (14–17). The children with CM were hospitalized between 1999 and 2003 in the pediatric department of the Gabriel Toure Hospital in Bamako (Mali). A total of 240 trios (two parents with one CM child) were recruited for the family-based association studies. All these nuclear families were prospectively recruited. The criteria used to define children with a CM phenotype were a coma with a Blantyre score of ≤2 and a thick blood smear positive for P. falciparum. Meningitis was ruled out by lumbar puncture. The two study populations are described in Table 1.
TABLE 1.
Demographics of genetic study participantsa
| Parameter | Result(s) |
|||
|---|---|---|---|---|
| Discovery cohort (city of Ibadan, Nigeria) | Replication cohort (city of Bamako, Mali) | |||
| Study design | Cases | Controls | Uncomplicated | Family based |
| Total no. of participants | 115 | 160 | 89 | 240 trios |
| Mean age (range) | 4.5 yrs (10 mo–13 yrs) | 6 yrs (6 mo–13 yrs) | 4.5 yrs (6 mo–13 yrs) | 6 yrs (10 mo–15 yrs) |
| No. (%) of females | 55 (47.8) | 77 (48.1) | 39 (43.8) | 119 (49.6) |
| No. with indicated coma score (Blantyre) | ≤2 | 5 | 5 | ≤2 |
| No. (%) with hematocrit: | ||||
| >15% | 112 (97.4) | 160 (100) | 89 (100) | 210 (87.5) |
| ≤15% | 3 (2.6) | 30 (12.5) | ||
Severe cases (“Cases”) were defined as consisting of children with cerebral malaria (CM, n = 112) and as cerebral malaria with severe malarial anemia (CMSMA, n = 3). Controls were defined as consisting of community controls (CC, n = 160). Uncomplicated malaria (UM, n = 89) cases were defined as consisting of febrile children with P. falciparum parasitemia.
Preparation of genomic DNA and selection of single nucleotide polymorphisms (SNPs) for analysis.
Genomic DNA from the Nigerian cohort was extracted from peripheral blood leukocytes with a QIAamp blood kit (Qiagen). Genomic DNA from the Malian cohort was extracted as described previously (14).
For the discovery stage, SNPs in the Nigerian samples with a minor allele frequency (MAF) of ≥0.05 were selected within IL17A, IL17F, and IL17RA genes from the 1000 Genomes Yoruba (YRI) database (18).
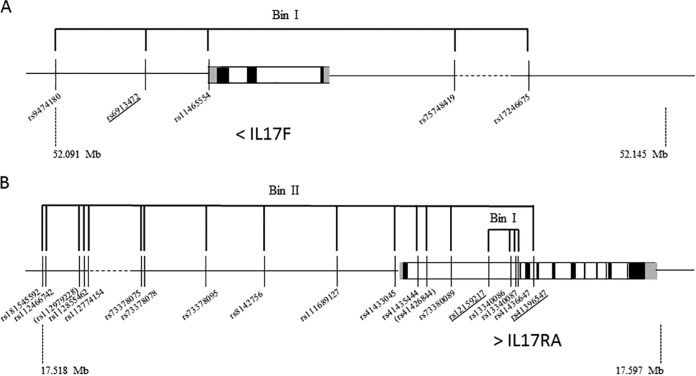
PLINK (19) was used to determine correlation bins (R2 ≥ 0.8) and TagSNPs (i.e., SNPs in a region of the genome with high linkage disequilibrium that represent a group of SNPs) within 5 kb upstream and downstream from the genes. We selected one SNP per correlation bin and a few singletons, providing comprehensive coverage of these genes (n = 48; see Table S1 in the supplemental material). For the replication stage in the Malian samples, SNPs associated with CM in the Nigerian samples (P < 0.05) were analyzed (n = 4). When the SNP was replicated, we also included additional SNPs that were in linkage disequilibrium (LD) with them (R2 > 0.6 [assessed by PLINK using the 1000 genome YRI database]) (18) and located 500 kb upstream or downstream from the associated SNP (n = 21, Fig. 1).
FIG 1.
Correlation bins for IL17F and IL17RA genes and flanking regions according to the 1000 Genomes YRI project (18). Pairwise R2 values from comparisons of SNPs associated with CM in the two study populations (underlined SNPs) and SNPs in a 1-Mb region were determined with PLINK software (19). Polymorphisms belonging to the same correlation group or “bin” (R2 > 0.6) are linked by a thick black line. (A) Chromosomal location of SNPs in the 6p region from 52.091 to 52.145 Mb that includes the IL17F gene (indicated by black vertical lines). The direction of gene transcription is indicated by the “<” sign. Correlation bins with R2 of >0.6 are as follows: bin I, SNPs rs6913472, rs11465554, rs9474180, rs75748419, and rs17246675. (B) Chromosomal location of SNPs in the 22q region from 17.518 to 17.597 Mb that includes the IL17RA gene (indicated by black vertical lines). The direction of gene transcription is indicated by the “>” sign. Correlation bins with R2 of >0.6 are as follows: bin I, rs12159217, rs13340087, rs13340086 and rs41436647; bin II, rs41396547 and 14 additional SNPs. The two SNPs whose designations are shown in parentheses, rs112979228 and rs41426844, were not informative and were excluded from the statistical analysis.
Genotyping and quality control.
For the discovery stage, genotyping was performed with 115 cases and 160 controls of the Nigerian sample using the custom designed Sequenom IPLEX assay. Genotyping was performed following the manufacturer's instructions. Two SNPs (reference SNP 12201582 [rs12201582] and rs115866730 of IL17F) had a low (<90%) call rate and were excluded from the analysis (see Table S1 in the supplemental material). Four SNPs associated with CM and 21 SNPs in LD with them (Fig. 1) were genotyped in the replication Malian sample using either the IPLEX assay (Sequenom) or the TaqMan SNP genotyping assays (Applied Biosystems). SNPs rs112979228 and rs41426844 in IL17RA were not informative and were excluded from the study.
Plasma collection and cytokine assays.
Plasma samples were collected from CM and UM children from Mali. For UM, the subjects had a thick blood film positive for P. falciparum, a Blantyre score of >4, and a hematocrit level of >21%. These children had never developed CM. They attended the outpatient clinic for an episode of febrile malaria. For CM, the criteria described previously were used.
IL-17A and IL-17F concentrations were determined by enzyme-linked immunosorbent assays (ELISAs) with pairs of cytokine-specific monoclonal antibodies according to the manufacturer's instructions. The detection thresholds were 8 pg/ml for IL-17A (R&D) and 15.6 pg/ml for IL-17F (eBiosciences).
Cell cultures and functional study.
Peripheral blood mononuclear cells (PBMCs) were isolated by blood centrifugation on Ficoll-Paque (GE Healthcare) (400 × g, 45 min), were washed two times in phosphate-buffered saline (PBS)–2 mM EDTA, and were resuspended in supplemented medium. We cultured 1 × 106 cells per well per 100-μl volume in a 24-well microplate in the presence of anti-CD3/CD28 antibodies or in the presence of medium alone. Plates were incubated at 37°C in a 5% CO2 atmosphere. Supernatants were collected at 3 days, centrifuged, and stored at −70°C for analysis of cytokine production. Cytokine levels of IL-17F were measured in supernatants as described above. Data are presented as arithmetic means of duplicate values.
Genomic DNA was extracted from peripheral blood leukocytes with a QIAamp blood kit (Qiagen). Genotyping of the IL17F rs6913472 SNP was assessed using TaqMan probe assays (Applied Biosystems).
Statistical analysis.
A chi-square test was used to determine whether the genotype distributions in parents and in controls conformed to the Hardy-Weinberg equilibrium. None of polymorphisms deviated from the Hardy-Weinberg equilibrium with a significance level of ≤0.05. The analysis was carried out with GenePop software (Web version 4.2, option 1). Univariate and multivariate analyses of SNPs were carried out with SPSS (statistical software version 10.1) to examine the association between SNPs and CM for unrelated Nigerian subjects. Differences were considered significant if the two-sided P value was <0.05. We also examined SNP-SNP interactions for independently associated SNPs within the Nigerian population with a two-locus test in PLINK (19). The family-based association test package (FBAT; version 1.7) (20, 21) was used for association tests in nuclear families from Mali. This analysis tests the transmission of the different alleles from heterozygous parents to affected children. A SNP was considered to be replicated if the association analysis yielded a one-tailed P value of <0.05 with the same risk allele as in the Nigerian population. A case-pseudocontrol data set analysis and a conditional logistic regression analysis were performed as described previously (16, 22, 23) to estimate odds ratio (OR) values for Malian subjects. Combined analyses that included both the discovery case-control sample and the replication family-based sample were also performed using the conditional logistic regression. Linear regression analysis was performed to test correlations between IL-17F concentrations and IL17F rs6913472 genotypes (SPSS statistical software).
In silico analysis.
The TFSEARCH program (24) was used to predict the potential transcription factor binding sites in IL17F and IL17RA genes. Associations of genotype and IL17F or IL17RA expression were examined with the GeneVar (GENe Expression Variation) database and Java application (25). The gene names and rs identifiers (ID) were entered in the “eQTL-SNP-Gene” option. GeneVar provides Spearman's correlation coefficient and P values for analytical comparisons between gene expression and genotype data from 8 different populations. Unfortunately, only the rs6913472 SNP was available.
RESULTS
Several common polymorphisms in IL17F and IL17RA genes are associated with an increased risk of CM in Nigerian children.
We first selected 48 representative TagSNPs within IL17A, IL17F, and IL17RA genes with 5 kb upstream and downstream from these genes. These SNPs were selected from the 1000 Genomes data bank (YRI reference population) (18), and all were common (MAF ≥ 0.05). Forty-six TagSNPs in IL17A, IL17F, and IL17RA were successfully genotyped in the Nigerian samples (see Table S1 in the supplemental material). We found no significant association (P > 0.05) between CM and polymorphisms in IL17A. In contrast, CM was associated with 3 SNPs in IL17F, IL17F rs6913472 (P = 0.039; OR = 1.95), IL17F rs9382083 (P = 0.004; OR = 2.63), and IL17F rs4715291 (P = 0.028; OR = 1.93) (Table 2), and with 2 SNPs in IL17RA, IL17RA rs12159217 (P = 0.009; OR = 2.20) and IL17RA rs41396547 (P = 0.028; OR = 2.76) (Table 2).
TABLE 2.
Results of the population-based association study in the Nigerian cohort among 115 cases with cerebral malaria and 160 healthy controlsa
| Analysis category | Gene | SNP | Positionb | Minor allele | MAFc | Genotype (risk) | % CC cases | % CM cases | OR | 95% CI | Pd |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Univariate | IL17F | rs6913472e | 52097072 | A | 0.10 (0.08) | GA | 13.1 | 22.7 | 1.95 | 1.03–3.69 | 0.039 |
| IL17F | rs9382083 | 52097773 | A | 0.07 (0.10) | GG | 74.5 | 88.5 | 2.63 | 1.33–5.18 | 0.004 | |
| IL17F | rs4715291 | 52113360 | T | 0.08 (0.12) | CC | 70.0 | 81.8 | 1.93 | 1.07–3.48 | 0.028 | |
| IL17RA | rs12159217 | 17573915 | T | 0.14 (0.16) | GG | 64.5 | 80.0 | 2.20 | 1.21–4.02 | 0.009 | |
| IL17RA | rs41396547 | 17579054 | C | 0.04 (0.05) | TT | 86.2 | 94.5 | 2.76 | 1.08–7.06 | 0.028 | |
| Multivariatef | IL17F | rs6913472 | A | GA | 3.12 | 1.43–6.82 | 0.004 | ||||
| IL17F | rs4715291 | A | GG | 2.82 | 1.39–5.75 | 0.004 | |||||
| IL17RA | rs12159217 | T | GG | 2.27 | 1.22–4.24 | 0.010 | |||||
| IL17RA | rs41396547 | C | TT | 3.15 | 1.15–8.70 | 0.026 |
Abbreviations: SNP, single-nucleotide polymorphism; OR, odds ratio; CI, confidence interval; MAF, minor allele frequency. All cases were either cerebral malaria (CM = 112) cases or cerebral malaria and severe malarial anemia (CMSMA = 3) cases. All controls were community controls (CC = 160).
Data represent the position on chromosome 6 for IL17F and on chromosome 22 for IL17RA according to human hg19 coordinates.
MAF values were estimated from Ensembl for the YRI population according to the 1000 Genomes project guidelines. MAF values for our Nigerian sample are shown in parentheses.
All P values represent results of two-sided analyses.
No AA genotype has been observed for rs6913472.
Data represent results of multivariate analysis combining the five significant SNPs (rs6913472, rs9382083, rs4715291, rs12159217 and rs41396547).
Multivariate analysis stepwise binary association test results confirmed the association of SNPs IL17F rs6913472 and IL17F rs4715291 as the best model, whereas IL17F rs9382083 was excluded. Thus, IL17F rs6913472 and IL17F rs4715291 were independently associated with CM. Likewise, the results of a multivariate analysis performed on the IL17RA polymorphisms indicated that IL17RA rs12159217 and IL17RA rs41396547 were also independently associated with disease. Finally, the best model revealed by the regression analysis of the SNPs of both genes included IL17F rs6913472 (P = 0.004; OR = 3.12), IL17F rs4715291 (P = 0.004; OR = 2.82), IL17RA rs12159217 (P = 0.01; OR = 2.27), and IL17RA rs41396547 (P = 0.026; OR = 3.15) (Table 2). Thus, these four SNPs are responsible for the observed associations and exert independent effects on CM. We did not find any evidence for SNP-SNP interactions between these polymorphisms with a two-locus test in PLINK (19).
Comparisons of CM children to UM children from Nigeria revealed evidence of an association between IL17RA rs12159217 and CM (P = 0.037; OR = 2.04), and a trend of association was obtained with IL17F rs6913472 (P = 0.059; OR = 2.09) (Table 3). Multivariate analysis combining the two SNPs (rs12159217 and rs6913472) confirmed the association of IL17RA rs12159217 with CM (P = 0.04; OR = 2.04) (Table 3). Finally, no significant association was detected in comparisons of CC to UM subjects from Nigeria (P > 0.202) (Table 3).
TABLE 3.
Results of the population-based association study in the Nigerian cohort, with cerebral malaria compared to uncomplicated malaria and uncomplicated malaria compared to community controlsa
| Analysis category | Gene | SNP | Genotype (risk) | % CM cases | % UM cases | OR (95% CI) | Pb | % CC cases | % UM cases | Pb |
|---|---|---|---|---|---|---|---|---|---|---|
| Univariate | IL17F | rs6913472 | GA | 22.7 | 12.4 | 2.09 (1.04–4.17) | 0.059 | 13.1 | 12.4 | 1 |
| rs9382083 | GG | 88.5 | 82.6 | 0.304 | 74.5 | 82.6 | 0.202 | |||
| rs4715291 | CC | 81.8 | 77.5 | 0.481 | 70.0 | 77.5 | 0.236 | |||
| IL17RA | rs12159217 | GG | 80.0 | 66.2 | 2.04 (1.04–4.00) | 0.037 | 64.5 | 66.2 | 0.883 | |
| rs41396547 | TT | 94.5 | 92.1 | 0.570 | 86.2 | 92.1 | 0.217 | |||
| Multivariatec | IL17F | rs6913472 | GA | 2.09 (1.08–4.74) | 0.077 | |||||
| IL17RA | rs12159217 | GG | 2.04 (1.03–4.03) | 0.040 |
Abbreviations: SNP, single-nucleotide polymorphism; OR, odds ratio; CI, confidence interval; CC, community controls; UM, uncomplicated malaria; CM, cerebral malaria. All uncomplicated malaria cases were febrile children with P. falciparum parasitemia (UM = 89).
All P values represent results of two-sided analyses.
Data represent results of multivariate analysis combining the two SNPs (rs6913472 and rs12159217).
IL17F and IL17RA SNPs are also associated with CM in Malian trios.
We then attempted to confirm these associations of IL17F and IL17RA SNPs in independent Malian trios. Hence, only the four SNPs significantly associated with CM in the multivariate analysis were selected for genotyping in the familial Malian replication sample. We detected a significant association (P = 0.03; OR = 1.39) of IL17F rs6913472 with CM (Table 4); the A allele was overtransmitted to children with CM. However, we were not able to replicate the association with rs4715291 in this study population (Table 4). For IL17RA polymorphisms, we replicated the same direction of association for rs12159217 (P = 0.01; OR = 1.52) and rs41396547 (P = 0.04; OR = 3.5). The G allele of rs12159217 was more frequently transmitted to CM children (n = 127) than expected from the null hypothesis (n = 116), and this association was also significant in permutation testing (P = 0.01). For the rs41396547 polymorphism, the same risk allele identified in the Nigerian sample (the T allele) was overtransmitted to children with CM (Table 4).
TABLE 4.
Results of family-based association study in the Malian cohorta
| Gene | SNPb | Minor allele/major allele | Risk allele | Freqc (risk allele) | OR (95% CI) | Pd |
|---|---|---|---|---|---|---|
| IL17F | rs6913472 | A/G | A | 0.109 | 1.39 (1.13–1.76) | 0.03 |
| rs4715291 | T/C | T | 0.181 | 0.17 | ||
| IL17RA | rs12159217 | T/G | G | 0.885 | 1.52 (1.03–2.37) | 0.01 |
| rs41396547 | C/T | T | 0.976 | 3.50 (1.06–13.04) | 0.04 |
Abbreviations: SNP, single-nucleotide polymorphism; OR, odds ratio; CI, confidence interval; Freq, frequency.
Data represent TagSNPs selected for their association with CM in the Nigerian sample and tested for association with CM in the Malian sample.
Risk allele frequency values were estimated from Malian parents.
All P values represent results of 1-sided tests.
SNP IL17RA rs41433045 in LD with rs41396547 is also associated with CM in the two African populations.
To take into account the possibility that additional polymorphisms in LD with associated SNPs may be involved in CM susceptibility, we also included SNPs that correlate (R2 > 0.6) with SNPs IL17F rs6913472, IL17RA rs12159217, and IL17RA rs41396547 on the basis of the data in the 1000 Genomes YRI database (18). Four SNPs in IL17F were correlated in Yoruba with rs6913472 in bin I (Fig. 1A). These SNPs were located close to the 3′ or 5′ end of the IL17F gene. Three IL17RA SNPs were correlated with rs12159217 in bin I, and 14 SNPs were correlated with rs41396547 in bin II (Fig. 1B). All 17 of these SNPs were located within or close to the IL17RA gene in a region of 61 kb. These SNPs were genotyped in the 240 Malian trios. Two of them (rs112979228 and rs41426844) were not informative in our study population and were excluded from the statistical analysis.
No other significant association with SNPs in bin I of IL17F and in bin I of IL17RA was found. The testing of the 14 SNPs in bin II of IL17RA revealed a significant association of rs41433045 (P = 0.03 [2-sided test]) with CM that was also significant in the permutation test (P = 0.04). The T allele of rs41433045 is overtransmitted to CM children. We thus examined association results between this polymorphism and CM in Nigerian samples, and we confirmed its association (P = 0.02) with an OR of developing CM for TT homozygous subjects versus those with a TC genotype, estimated at 3.1 (1.1 to 9.4).
We investigated the LD pattern of the SNPs rs6913472, rs12159217, rs41396547, and rs41433045 in a 1-Mb region, and we found no SNP with an R2 value of >0.6 in others surrounding genes that could account for the observed associations.
Association results for IL17F and IL17RA polymorphisms in combined samples.
Finally, we performed association comparisons between CM and the 4 SNPs in the whole sample set by combining the genotype data of the Nigerian case-control and the Malian nuclear families. The 3 SNPs of IL17RA (rs12159217, rs41396547, and rs41433045) were significantly associated (P < 0.05) (Table 5), whereas rs6913472 of IL17F indicated a trend of association (P = 0.059) (Table 5). The results of multivariate regression analysis performed on the four SNPs showed that IL17RA rs12159217 and rs41396547 polymorphisms remained significantly independently associated with CM as follows: for rs12159217, P = <0.0001 and OR (95% confidence interval [CI]) = 3.40 (2.22 to 5.21); for rs41396547, P = <0.0001 and OR (95% CI) = 4.71 (1.99 to 11.17) (Table 5).
TABLE 5.
Results of combined analyses that included the case-control and familial dataa
| Analysis category | Gene | SNP | Minor allele/major allele | Risk allele | Freqb (risk) | OR (95% CI) | Pc |
|---|---|---|---|---|---|---|---|
| Univariate | IL17F | rs6913472 | A/G | A | 0.093 | 1.36 (1.06–1.97) | 0.059 |
| IL17RA | rs12159217 | T/G | G | 0.882 | 1.79 (1.24–2.52) | 0.001 | |
| rs41396547 | C/T | T | 0.956 | 3.17 (1.48–6.78) | 0.001 | ||
| rs41433045 | C/T | T | 0.973 | 4.12 (1.54–11.00) | 0.002 | ||
| Multivariated | IL17RA | rs12159217 | T/G | G | 0.882 | 3.40 (2.22–5.21) | <0.0001 |
| rs41396547 | C/T | T | 0.956 | 4.71 (1.99–11.17) | <0.0001 |
Abbreviations: SNP, single-nucleotide polymorphism; OR, odds ratio; CI, confidence interval; Freq, frequency.
Data represent risk allele frequencies.
All P values represent the results of 1-sided tests in reference to the risk allele.
Data represent results of multivariate analysis combining the four SNPs (rs6913472, rs12159217, rs41396547, and rs41433045).
In silico analysis of the possible functional effects of the associated SNPs.
Our analysis identified several SNPs within or surrounding IL17F and IL17RA that were independently associated with CM. In silico analysis of these SNPs using TFSEARCH (24) showed that several of them may alter the binding sites of transcription factor: alleles rs6913472 A, rs12159217 T, and rs41433045 C create new binding sites for SRY, Oct-1, and NIT2, respectively. Moreover, the major risk allele, rs12159217 G, created a new binding site for ADR1. Then, we used genotype data from HapMap (26) and gene expression data from GeneVar (25) to perform a quantitative trait locus (eQTL) analysis and assess whether polymorphisms associated with CM are correlated with mRNA abundance. This analysis did not show significant correlations between the transcription levels of IL17F rs6913472 genotypes. Data for other SNPs were not available.
Levels of IL-17A and IL-17F were not delectable in plasma.
We measured the IL-17A and IL-17F levels by ELISAs of the plasma (diluted by a factor of 2) of 77 children with CM and 45 children with UM from Mali. Levels of IL-17F in the plasma were not detectable in either the CM and the UM children. For IL-17A, the levels were detectable in only 17% of UM and 10% of CM children. Hence, the number of subjects with detectable IL-17A levels was too low to perform statistical analysis.
Aggravating genotype IL17F rs6913472 GA was correlated with lower IL-17F concentration.
To further explore the potential functional effect of the rs6913472 polymorphism, we sought to determine whether genotypes may be correlated with the level of the IL-17F protein. Hence, we analyzed the IL-17F production in cultures of PBMCs from healthy subjects from the Marseille blood bank stimulated with anti-CD3/anti-CD28 antibodies. We found that the at-risk GA genotype of rs6913472 was significantly associated with lower levels of IL-17F production (P = 0.02) (Fig. 2).
FIG 2.
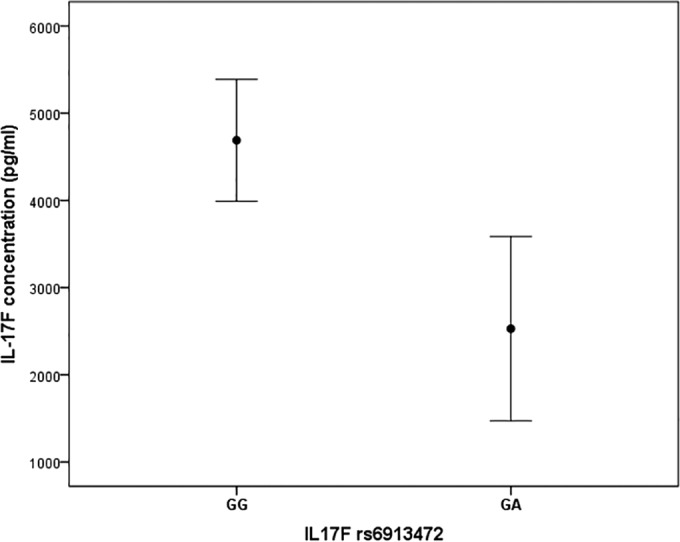
Concentration of IL-17F according to the IL17F rs6913472 genotypes. The GA genotype was associated (P = 0.02) with the lower levels of IL-17F production in supernatants from the cell cultures of 50 healthy subjects. The concentrations are represented as arithmetic means of duplicate values (± standard errors of the mean).
DISCUSSION
The aim of this study was to evaluate whether genetic variants in IL-17A and IL-17F influence a child's risk of developing cerebral malaria. We thoroughly analyzed genetic variants of IL17A and IL17F and of their common receptor, IL17RA, in two independent African populations. We found that CM was significantly associated with SNPs in IL17F and IL17RA in a population of children from Nigeria. We then tested these findings in a cohort of nuclear families from Mali.
Among polymorphisms in IL17F, the rs6913472 and rs4715291 SNPs showed the strongest association with CM in Nigerian children and rs6913472 was replicated in the Malian sample. The rs12159217 polymorphism of the IL17RA gene was associated with CM in both the Nigerian and Malian populations (P < 0.05 in both cohorts) with the same G risk allele. Similarly, IL17RA rs41396547 and rs41433045 (in strong LD; R2 = 0.67, D′ = 1) were associated in both populations with the major T allele as a risk for CM. Our findings showed that all SNPs associated with CM are located near or within the IL17F or IL17RA genes. The regional LD patterns indicated that no SNPs in the 1-Mb region and in strong LD with the associated SNPs are located in genes other than IL17F and IL17RA, supporting the idea of the involvement of these genes in the development of CM.
The association observed between IL17F rs4715291 and CM in the Nigerian sample was not replicated in the Malian population. This SNP is in strong LD (R2 = 0.76) with the nonsynonymous SNP rs763780, which did not show convincing results in our two study populations. The rs763780 was previously described as a functional polymorphism (27) and has found to be associated with susceptibility to tuberculosis (28) and resistance to asthma (27). This result may have been due to the poor conservation of LD blocks among African populations, which is widely recognized. Indeed, this may explain in part the limited success of studies carried out with a pool of samples from various African ethnic groups, because microarrays include TagSNPs that have been defined in a few African populations. This problem occurs less frequently in other world populations (i.e., European, Asian, and American populations) that diverged much later than African populations. This underlines the importance of carefully determining LD patterns before attempting validation studies in different African populations.
Using TFSEARCH (24), which provides an in silico prediction model for transcription factor binding, we found that transcription factors bound to the DNA sequence overlapping several associated SNPs (rs6913472, rs12159217, and rs41433045) when a particular allele was present. These results indicate that these SNPs may play a functional role by acting on regulatory functions. Unfortunately, plasma levels of IL-17F were not detectable in either UM and CM Malian subjects. Hence, no correlation could be established between the genotype and the levels of IL-17F. We thus performed functional studies to investigate the potential functional effect of the IL17F rs6913472 polymorphism on cultures of PBMCs. The rs6913472 GA genotype was associated with a lower level of IL-17F production in anti-CD3/CD28-stimulated cells. Thus, rs6913472 GA is associated with (i) downregulation of IL-17F, (ii) modification of transcription factor binding, and (iii) an increased risk of CM.
IL-17F stimulates the production of both antimicrobial peptides and molecules capable of recruiting or stimulating immune cells (granulocyte colony-stimulating factor [G-CSF], granulocyte-macrophage colony-stimulating factor [GM-CSF], IL-6) and chemokines such as CXCL1, CXCL2, and CXCL5 that promote neutrophil recruitment. Both IL-17A and IL-17F stimulate matrix metalloproteases, thus increasing cell mobility. Studies in mice showed that IL-17A, IL-17E, IL-17F, and IL17RA are required for host resistance to Trypanosoma cruzi (29); furthermore, human resistance to kala azar caused by Leishmania donovani was associated with high levels of IL-17 and IL-22 responses (30). IL-17A and IL-17F are also crucial in the clearance of extracellular bacteria, such as Staphylococcus aureus, Citrobacter rodentium, and Klebsiella pneumoniae (31, 32).
However, several studies indicate that Th17 cells aggravate neurological disorders such as multiple sclerosis (MS) and experimental autoimmune encephalomyelitis (EAE) (7). The pathophysiology of MS involves a neuroinflammatory reaction and disruption of the blood-brain barrier (BBB). These pathophysiological changes are also found in CM, suggesting that Th17 cells may play a similar role in the development of cerebral manifestations associated with P. falciparum. Cytokines secreted by Th17 cells, including IL-17 and IL-22, may alter the permeability of the human BBB to soluble molecules and circulating CD4+ lymphocytes (33). IL-17A is also required for the initiation of EAE, whereas IL-17F plays only a minor role and maintains inflammation in the central nervous system (CNS) (34). Indeed, anti-IL-17F treatment does not improve the course of EAE whereas the onset of EAE is delayed and its progression is slow in IL-17 knockout (KO) mice and wild-type mice treated with anti-IL-17 (35, 36). Our data, however, do not support the hypothesis that IL-17A and IL-17F could aggravate CM by increasing inflammation in particular because (i) we found no association between CM and SNPs in IL17A, whose product is more proinflammatory than IL-17F, and (ii) subjects with the at-risk GA genotype associated with IL17F rs6913472 had significantly lower IL-17F production than genotype GG subjects.
In the experimental CM mouse model infected with P. berghei ANKA (PbA) (sharing certain characteristics with human CM), mice deficient in IL-23 or IL-17A develop neurological symptoms and die, similarly to wild-type mice (37), suggesting that these cytokines do not aggravate CM. Thus, in spite of the observation that IL-17A aggravates inflammation associated with various neurophysiological disorders, our data from children and observations in mice do not support the view that IL-17A increases the risk of CM. In fact, we suggest that the presence of IL-17A and IL-17F increases antiparasite immunity during infection by P. falciparum and then protects against severe disease.
All IL-17 family members use receptors that share the common IL-17RA chain. IL-17A and IL-17F signal through IL-17RA or through the dimeric receptor IL-17RA/IL-17RC (38); however, these two cytokines interact differently with the two chains. IL-17F is highly dependent on IL-17RC, whereas IL-17A can signal through IL-17RA alone (39, 40). The expression of IL-17RC is restricted to nonhematopoietic tissue-resident cells, suggesting that the selective action of IL-17F on nonhematopoietic cells may be due to the presence of IL-17RC. IL-17RA is widely expressed by innate cells such as macrophages and neutrophils and by nonhematopoietic cells such as epithelial cells and fibroblasts (38). Our finding that polymorphisms in IL17RA are associated with CM confirms that cytokines belonging to the IL-17 cytokine family are implicated in malaria. Given that IL-17F relies on IL-17RC more than on any other IL-17-related cytokine, we are currently examining genetic variants of this receptor gene and their associations with CM.
In conclusion, we used discovery and replication cohorts of childhood P. falciparum malaria to show that polymorphisms in IL17F and IL17RA significantly contribute to susceptibility to CM. Functional studies provide evidence that lower production of IL-17F leads to CM. Further investigation of this promising association is required to decipher the underlying mechanisms of the involvement of these genes in the pathogenesis of CM. Finally, our validated findings expand current knowledge of the complex host genetic factors that predispose to childhood cerebral malaria. Our results are relevant for the development of diagnostic algorithms to identify children who have a high risk of developing CM. Furthermore, such diagnostic algorithms will also guide the design of functional studies aimed at the development and deployment of severe malaria prevention strategies. Our results emphasize the importance of deciphering the role of this key immunological pathway in the pathogenesis of severe childhood malaria. More importantly, our results may contribute to the development of adjunct therapies aimed at reducing mortality and neurological sequelae in children presenting with cerebral malaria.
Supplementary Material
ACKNOWLEDGMENTS
We thank all children, guardians, and parents for participating in the study. We thank the staff of the pediatric wards at the Gabriel Toure Hospital in Bamako, Mali, for their help. We also thank all the consultants, registrars, nurses, and administrative staff at the department of Pediatrics, University College Hospital, Ibadan, Nigeria, for all the support. We thank Christophe Chevillard for helpful advice.
Part of the genotyping presented in the present publication was performed at the Genomic and Sequencing Facility of Bordeaux (grants from the Conseil Regional d'Aquitaine [no. 20030304002FA and 20040305033FA] and from the European Union Fondul European de Dezvoltare Regională [FEDR] [no. 2003227] and from Investissements d'avenir, Convention attributive d'aide [no. ANR-10-EQPX-16-01]).
We declare that we have no conflicts of interest.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/IAI.00671-15.
REFERENCES
- 1.Coltel N, Combes V, Hunt NH, Grau GE. 2004. Cerebral malaria—a neurovascular pathology with many riddles still to be solved. Curr Neurovasc Res 1:91–110. doi: 10.2174/1567202043480116. [DOI] [PubMed] [Google Scholar]
- 2.Basso AS, Cheroutre H, Mucida D. 2009. More stories on Th17 cells. Cell Res 19:399–411. doi: 10.1038/cr.2009.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chang SH, Dong C. 2009. IL-17F: regulation, signaling and function in inflammation. Cytokine 46:7–11. doi: 10.1016/j.cyto.2008.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Iwakura Y, Nakae S, Saijo S, Ishigame H. 2008. The roles of IL-17A in inflammatory immune responses and host defense against pathogens. Immunol Rev 226:57–79. doi: 10.1111/j.1600-065X.2008.00699.x. [DOI] [PubMed] [Google Scholar]
- 5.Iwakura Y, Ishigame H, Saijo S, Nakae S. 2011. Functional specialization of interleukin-17 family members. Immunity 34:149–162. doi: 10.1016/j.immuni.2011.02.012. [DOI] [PubMed] [Google Scholar]
- 6.Hirota K, Duarte JH, Veldhoen M, Hornsby E, Li Y, Cua DJ, Ahlfors H, Wilhelm C, Tolaini M, Menzel U, Garefalaki A, Potocnik AJ, Stockinger B. 2011. Fate mapping of IL-17-producing T cells in inflammatory responses. Nat Immunol 12:255–263. doi: 10.1038/ni.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jadidi-Niaragh F, Mirshafiey A. 2011. Th17 cell, the new player of neuroinflammatory process in multiple sclerosis. Scand J Immunol 74:1–13. doi: 10.1111/j.1365-3083.2011.02536.x. [DOI] [PubMed] [Google Scholar]
- 8.Ajetunmobi WA, Orimadegun AE, Brown BJ, Afolabi NK, Olabiyi FA, Anetor JI, Omokhodion S, Osinusi K, Akinbami FO, Shokunbi WA, Sodeinde O, Fernandez-Reyes D. 2012. Haemoglobinuria among children with severe malaria attending tertiary care in Ibadan, Nigeria. Malar J 11:336. doi: 10.1186/1475-2875-11-336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Burté F, Brown BJ, Orimadegun AE, Ajetunmobi WA, Battaglia F, Ely BK, Afolabi NK, Athanasakis D, Akinkunmi F, Kowobari O, Omokhodion S, Osinusi K, Akinbami FO, Shokunbi WA, Sodeinde O, Fernandez-Reyes D. 2012. Severe childhood malaria syndromes defined by plasma proteome profiles. PLoS One 7:e49778. doi: 10.1371/journal.pone.0049778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Burté F, Brown BJ, Orimadegun AE, Ajetunmobi WA, Afolabi NK, Akinkunmi F, Kowobari O, Omokhodion S, Osinusi K, Akinbami FO, Shokunbi WA, Sodeinde O, Fernandez-Reyes D. 2013. Circulatory hepcidin is associated with the anti-inflammatory response but not with iron or anemic status in childhood malaria. Blood 121:3016–3022. doi: 10.1182/blood-2012-10-461418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bachmann J, Burte F, Pramana S, Conte I, Brown BJ, Orimadegun AE, Ajetunmobi WA, Afolabi NK, Akinkunmi F, Omokhodion S, Akinbami FO, Shokunbi WA, Kampf C, Pawitan Y, Uhlen M, Sodeinde O, Schwenk JM, Wahlgren M, Fernandez-Reyes D, Nilsson P. 2014. Affinity proteomics reveals elevated muscle proteins in plasma of children with cerebral malaria. PLoS Pathog 10:e1004038. doi: 10.1371/journal.ppat.1004038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.World Health Organization. 2000. Severe falciparum malaria. World Health Organization, Communicable Diseases Cluster. Trans R Soc Trop Med Hyg 94(Suppl 1):S1–S90. [PubMed] [Google Scholar]
- 13.World Health Organization. 2014. Severe malaria. Trop Med Int Health 19(Suppl 1):7–131. doi: 10.1111/tmi.12313_2. [DOI] [PubMed] [Google Scholar]
- 14.Cabantous S, Poudiougou B, Traore A, Keita M, Cisse MB, Doumbo O, Dessein AJ, Marquet S. 2005. Evidence that interferon-gamma plays a protective role during cerebral malaria. J Infect Dis 192:854–860. doi: 10.1086/432484. [DOI] [PubMed] [Google Scholar]
- 15.Cabantous S, Poudiougou B, Oumar AA, Traore A, Barry A, Vitte J, Bongrand P, Marquet S, Doumbo O, Dessein AJ. 2009. Genetic evidence for the aggravation of Plasmodium falciparum malaria by interleukin 4. J Infect Dis 200:1530–1539. doi: 10.1086/644600. [DOI] [PubMed] [Google Scholar]
- 16.Marquet S, Doumbo O, Cabantous S, Poudiougou B, Argiro L, Safeukui I, Konate S, Sissoko S, Chevereau E, Traore A, Keita MM, Chevillard C, Abel L, Dessein AJ. 2008. A functional promoter variant in IL12B predisposes to cerebral malaria. Hum Mol Genet 17:2190–2195. doi: 10.1093/hmg/ddn118. [DOI] [PubMed] [Google Scholar]
- 17.Ranque S, Poudiougou B, Traore A, Keita M, Oumar AA, Safeukui I, Marquet S, Cabantous S, Diakite M, Mintha D, Cisse MB, Keita MM, Dessein AJ, Doumbo OK. 2008. Life-threatening malaria in African children: a prospective study in a mesoendemic urban setting. Pediatr Infect Dis J 27:130–135. [DOI] [PubMed] [Google Scholar]
- 18.Abecasis GR, Altshuler D, Auton A, Brooks LD, Durbin RM, Gibbs RA, Hurles ME, McVean GA. 2010. A map of human genome variation from population-scale sequencing. Nature 467:1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, Sham PC. 2007. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Horvath S, Xu X, Laird NM. 2001. The family based association test method: strategies for studying general genotype–phenotype associations. Eur J Hum Genet 9:301–306. doi: 10.1038/sj.ejhg.5200625. [DOI] [PubMed] [Google Scholar]
- 21.Wilk JB, Volcjak JS, Myers RH, Maher NE, Knowlton BA, Heard-Costa NL, Demissie S, Cupples LA, DeStefano AL. 2001. Family-based association tests for qualitative and quantitative traits using single-nucleotide polymorphism and microsatellite data. Genet Epidemiol 21(Suppl 1):S364–369. [DOI] [PubMed] [Google Scholar]
- 22.Schaid DJ, Rowland C. 1998. Use of parents, sibs, and unrelated controls for detection of associations between genetic markers and disease. Am J Hum Genet 63:1492–1506. doi: 10.1086/302094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cordell HJ, Barratt BJ, Clayton DG. 2004. Case/pseudocontrol analysis in genetic association studies: a unified framework for detection of genotype and haplotype associations, gene-gene and gene-environment interactions, and parent-of-origin effects. Genet Epidemiol 26:167–185. doi: 10.1002/gepi.10307. [DOI] [PubMed] [Google Scholar]
- 24.Heinemeyer T, Wingender E, Reuter I, Hermjakob H, Kel AE, Kel OV, Ignatieva EV, Ananko EA, Podkolodnaya OA, Kolpakov FA, Podkolodny NL, Kolchanov NA. 1998. Databases on transcriptional regulation: TRANSFAC, TRRD and COMPEL. Nucleic Acids Res 26:362–367. doi: 10.1093/nar/26.1.362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang TP, Beazley C, Montgomery SB, Dimas AS, Gutierrez-Arcelus M, Stranger BE, Deloukas P, Dermitzakis ET. 2010. Genevar: a database and Java application for the analysis and visualization of SNP-gene associations in eQTL studies. Bioinformatics 26:2474–2476. doi: 10.1093/bioinformatics/btq452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Frazer KA, Ballinger DG, Cox DR, Hinds DA, Stuve LL, Gibbs RA, Belmont JW, Boudreau A, Hardenbol P, Leal SM, Pasternak S, Wheeler DA, Willis TD, Yu F, Yang H, Zeng C, Gao Y, Hu H, Hu W, Li C, Lin W, Liu S, Pan H, Tang X, Wang J, Wang W, Yu J, Zhang B, Zhang Q, Zhao H, Zhou J, Gabriel SB, Barry R, Blumenstiel B, Camargo A, Defelice M, Faggart M, Goyette M, Gupta S, Moore J, Nguyen H, Onofrio RC, Parkin M, Roy J, Stahl E, Winchester E, Ziaugra L, Altshuler D, Shen Y, Yao Z, et al. . 2007. A second generation human haplotype map of over 3.1 million SNPs. Nature 449:851–861. doi: 10.1038/nature06258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kawaguchi M, Takahashi D, Hizawa N, Suzuki S, Matsukura S, Kokubu F, Maeda Y, Fukui Y, Konno S, Huang SK, Nishimura M, Adachi M. 2006. IL-17F sequence variant (His161Arg) is associated with protection against asthma and antagonizes wild-type IL-17F activity. J Allergy Clin Immunol 117:795–801. doi: 10.1016/j.jaci.2005.12.1346. [DOI] [PubMed] [Google Scholar]
- 28.Peng R, Yue J, Han M, Zhao Y, Liu L, Liang L. 2013. The IL-17F sequence variant is associated with susceptibility to tuberculosis. Gene 515:229–232. doi: 10.1016/j.gene.2012.11.017. [DOI] [PubMed] [Google Scholar]
- 29.Tosello Boari J, Amezcua Vesely MC, Bermejo DA, Ramello MC, Montes CL, Cejas H, Gruppi A, Acosta Rodriguez EV. 2012. IL-17RA signaling reduces inflammation and mortality during Trypanosoma cruzi infection by recruiting suppressive IL-10-producing neutrophils. PLoS Pathog 8:e1002658. doi: 10.1371/journal.ppat.1002658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pitta MG, Romano A, Cabantous S, Henri S, Hammad A, Kouriba B, Argiro L, el Kheir M, Bucheton B, Mary C, El-Safi SH, Dessein A. 2009. IL-17 and IL-22 are associated with protection against human kala azar caused by Leishmania donovani. J Clin Invest 119:2379–2387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Happel KI, Dubin PJ, Zheng M, Ghilardi N, Lockhart C, Quinton LJ, Odden AR, Shellito JE, Bagby GJ, Nelson S, Kolls JK. 2005. Divergent roles of IL-23 and IL-12 in host defense against Klebsiella pneumoniae. J Exp Med 202:761–769. doi: 10.1084/jem.20050193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ye P, Garvey PB, Zhang P, Nelson S, Bagby G, Summer WR, Schwarzenberger P, Shellito JE, Kolls JK. 2001. Interleukin-17 and lung host defense against Klebsiella pneumoniae infection. Am J Respir Cell Mol Biol 25:335–340. doi: 10.1165/ajrcmb.25.3.4424. [DOI] [PubMed] [Google Scholar]
- 33.Kebir H, Kreymborg K, Ifergan I, Dodelet-Devillers A, Cayrol R, Bernard M, Giuliani F, Arbour N, Becher B, Prat A. 2007. Human TH17 lymphocytes promote blood-brain barrier disruption and central nervous system inflammation. Nat Med 13:1173–1175. doi: 10.1038/nm1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yang XO, Chang SH, Park H, Nurieva R, Shah B, Acero L, Wang YH, Schluns KS, Broaddus RR, Zhu Z, Dong C. 2008. Regulation of inflammatory responses by IL-17F. J Exp Med 205:1063–1075. doi: 10.1084/jem.20071978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Langrish CL, Chen Y, Blumenschein WM, Mattson J, Basham B, Sedgwick JD, McClanahan T, Kastelein RA, Cua DJ. 2005. IL-23 drives a pathogenic T cell population that induces autoimmune inflammation. J Exp Med 201:233–240. doi: 10.1084/jem.20041257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Park H, Li Z, Yang XO, Chang SH, Nurieva R, Wang YH, Wang Y, Hood L, Zhu Z, Tian Q, Dong C. 2005. A distinct lineage of CD4 T cells regulates tissue inflammation by producing interleukin 17. Nat Immunol 6:1133–1141. doi: 10.1038/ni1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ishida H, Matsuzaki-Moriya C, Imai T, Yanagisawa K, Nojima Y, Suzue K, Hirai M, Iwakura Y, Yoshimura A, Hamano S, Shimokawa C, Hisaeda H. 2010. Development of experimental cerebral malaria is independent of IL-23 and IL-17. Biochem Biophys Res Commun 402:790–795. doi: 10.1016/j.bbrc.2010.10.114. [DOI] [PubMed] [Google Scholar]
- 38.Gaffen SL. 2009. Structure and signalling in the IL-17 receptor family. Nat Rev Immunol 9:556–567. doi: 10.1038/nri2586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hu Y, Ota N, Peng I, Refino CJ, Danilenko DM, Caplazi P, Ouyang W. 2010. IL-17RC is required for IL-17A- and IL-17F-dependent signaling and the pathogenesis of experimental autoimmune encephalomyelitis. J Immunol 184:4307–4316. doi: 10.4049/jimmunol.0903614. [DOI] [PubMed] [Google Scholar]
- 40.Kuestner RE, Taft DW, Haran A, Brandt CS, Brender T, Lum K, Harder B, Okada S, Ostrander CD, Kreindler JL, Aujla SJ, Reardon B, Moore M, Shea P, Schreckhise R, Bukowski TR, Presnell S, Guerra-Lewis P, Parrish-Novak J, Ellsworth JL, Jaspers S, Lewis KE, Appleby M, Kolls JK, Rixon M, West JW, Gao Z, Levin SD. 2007. Identification of the IL-17 receptor related molecule IL-17RC as the receptor for IL-17F. J Immunol 179:5462–5473. doi: 10.4049/jimmunol.179.8.5462. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.