Abstract
Vibrio cholerae, the causative agent of the severe diarrheal disease cholera, thrives in both marine environments and the human host. To do so, it must encode the tools necessary to acquire essential nutrients, including iron, under these vastly different conditions. A number of V. cholerae iron acquisition systems have been identified; however, the precise role of each system is not fully understood. To test the roles of individual systems, we generated a series of mutants in which only one of the four systems that support iron acquisition on unsupplemented LB agar, Feo, Fbp, Vct, and Vib, remains functional. Analysis of these mutants under different growth conditions showed that these systems are not redundant. The strain carrying only the ferrous iron transporter Feo grew well at acidic, but not alkaline, pH, whereas the ferric iron transporter Fbp promoted better growth at alkaline than at acidic pH. A strain defective in all four systems (null mutant) had a severe growth defect under aerobic conditions but accumulated iron and grew as well as the wild type in the absence of oxygen, suggesting the presence of an additional, unidentified iron transporter in V. cholerae. In support of this, the null mutant was only moderately attenuated in an infant mouse model of infection. While the null mutant used heme as an iron source in vitro, we demonstrate that heme is not available to V. cholerae in the infant mouse intestine.
INTRODUCTION
The Gram-negative bacterium Vibrio cholerae has an absolute requirement for iron (1). Despite the prevalence of iron within its environmental niches and human host, most of this iron is not readily available. Iron occurs naturally as oxidized (ferric) or reduced (ferrous) iron (2). Ferric iron(III), found in oxygenated environments at neutral to alkaline pH, has very low water solubility and forms large insoluble complexes. Studies of ocean water have found iron to be the limiting nutrient for microbial growth (3). Soluble, ferrous iron(II) is more prevalent under the anoxic conditions that V. cholerae may encounter while colonizing the human host. However, access to iron in the host is limited due to competition with other microbes on host surfaces, as well as sequestration by high-affinity iron-binding host proteins such as transferrin and lactoferrin (4–7). Despite the challenges of obtaining iron in these various environments, V. cholerae is proficient in growth in both marine and host environments.
The V. cholerae genome encodes multiple iron acquisition systems, which each transport a specific form of iron and may thus optimize iron acquisition in the various niches that V. cholerae inhabits (8–12). These include genes for the synthesis (vib) and utilization (viu) of vibriobactin, a siderophore that is secreted into the environment, where it binds ferric iron with extremely high affinity (9, 13–15). Ferrivibriobactin is bound by the outer membrane receptor ViuA (16, 17) and is transported across the outer membrane in a process that requires either of two energy transduction systems, TonB1 or TonB2 (18). Subsequent transport into the cytoplasm occurs through either ViuPDGC (19) or VctPDGC (11, 20), periplasmic binding protein-dependent, inner membrane catechol transport systems. Iron is removed from vibriobactin in the cytoplasm by ViuB (21). Heme, which also can be used as an iron source, is transported through an analogous TonB-dependent system involving the outer membrane receptor HutA (22–24), HutR (24), or HasR (24) and the inner membrane transport system HutBCD (18).
In addition to systems that allow transport of complexed iron, V. cholerae also encodes systems for the uptake of ferrous and ferric iron that is not bound to a siderophore or other high-affinity chelator. Ferrous iron can be transported through FeoABC (10, 25). FeoB is an integral inner membrane protein which is believed to form the pore through which iron is transported. The roles of FeoA and FeoC in V. cholerae remain unclear, but both are essential for Feo function (26). Unchelated ferric iron can be transported through FbpABC, an inner membrane system with homology to ABC transporters (10). In addition to its role in catechol siderophore transport, the VctPDGC system is capable of siderophore-independent transport of iron (11). Although the mechanism and substrate of the siderophore-free iron transport by Vct remain unknown, it supports moderate growth in the absence of functional Feo, Fbp, and Vib systems. It is not currently known how ferrous and ferric iron access the periplasmic space in V. cholerae; however, when expressed in Escherichia coli, these systems do not require TonB for iron acquisition, suggesting diffusion of unchelated iron through porins or similar proteins (10). The Vib, Feo, Fbp, and Vct systems all participate in iron uptake in V. cholerae growing under typical laboratory conditions, and genetic disruption of all four of these systems rendered V. cholerae unable to grow on LB agar, even with iron supplementation (11). This mutant strain, referred to as the null strain, retains the heme transport systems and could be maintained on medium containing heme. The relevant systems are listed in Table 1.
TABLE 1.
Known iron acquisition systems of V. cholerae
| Gene(s) | Function | Locus tag(s) | Reference(s) |
|---|---|---|---|
| feoABC | Transport of ferrous iron (Fe2+) | VC2078-VC2076 | 10 |
| fbpABC | Transport of ferric iron (Fe3+) | VC0608-VC0610 | 10 |
| vctPDGC | Inner membrane transport of vibriobactin, enterobactin, and unknown | VCA0227-VCA0230 | 11, 20 |
| viuPDGC | Inner membrane transport of vibriobactin and enterobactin | VC0776-VC0779 | 19 |
| viuA | Outer membrane vibriobactin receptor | VC2211 | 16, 17 |
| vibABCDEFH | Vibriobactin biosynthesis | VC0774, VC0771, VC0773, VC0780, VC0772, VC2209, VC0775 | 13, 15, 66, 67 |
| hutBCD | Inner membrane transport of heme | VCA0914-VCA0916 | 18 |
| hutA, hutR, hasR | Outer membrane heme receptors | VCA0576, VCA0064, VCA0625 | 22–24 |
| irgA, vctA | Outer membrane enterobactin receptors | VC0475, VCA0232 | 20 |
| fhuBCD | Inner membrane transport of ferrichrome | VC0203, VC0201, VC0202 | 68 |
| fhuA | Outer membrane ferrichrome receptor | VC0200 | 68 |
It is not clear what roles each of the iron transport systems play in the acquisition of iron during growth in the environment or in human infection. The presence of multiple systems suggests that V. cholerae might use different systems to optimally acquire iron under different conditions. To test this, we constructed a set of isogenic strains in which only one of the Feo, Fbp, Vct, and vibriobactin systems is functional. By assessing the growth of these strains under different environmental conditions, we demonstrate that these iron acquisition systems are not functionally redundant but differ in their ability to support growth as pH and oxygen levels are varied. Further, we present evidence for an additional, unidentified iron transport system that functions under anoxic growth conditions. This system may also function when the bacteria are growing in the intestine of a mammalian host, since the feo fbp vct vib viuA mutant displayed only a modest loss of fitness in the infant mouse model, despite its severe growth defect in vitro.
MATERIALS AND METHODS
Bacterial strains, plasmids, and media.
The bacterial strains and plasmids used in this study are listed in Table 2. Strains were maintained at −80°C in tryptic soy broth with 20% glycerol. Strains were routinely grown in Luria-Bertani (LB) broth (1% tryptone, 0.5% yeast extract, and 1% sodium chloride) or on LB agar (1.5% agar) at 37°C. Heme (5 μM) was used for the routine growth of iron transport-deficient strains. Antibiotics were used as follows for V. cholerae strains: 75 μg/ml streptomycin, 50 μg/ml kanamycin, 100 μg/ml ampicillin, 5 μg/ml chloramphenicol, and 5 μg/ml gentamicin. For E. coli strains, 50 μg/ml kanamycin and 100 μg/ml ampicillin were used.
TABLE 2.
Strains and plasmids
| Strain or plasmid | Description | Reference or source |
|---|---|---|
| Strains | ||
| V. cholerae | ||
| O395 | Classical biotype | 69 |
| ARM591 | O395 ΔvibB fbpA::cam | 10 |
| ARM592 | O395 ΔvibB fbpA::cam vctP::gent | This study |
| EPV6 | O395 ΔvibB fbpA::cam vctP::gent feoABC::kan | This study |
| EPV102 (null) | O395 ΔvibB fbpA::cam vctP::gent feoABC::kan ΔviuA | This study |
| EPV104 (vFeo) | O395 ΔvibB fbpA::cam vctP::gent viuA | This study |
| ALV101 | O395 ΔvibB | 10 |
| EPV105 | O395 ΔvibB ΔviuA | This study |
| EPV113 | O395 ΔvibB ΔviuA vctP::gent | This study |
| EPV115 (vFbp) | O395 ΔvibB ΔviuA vctP::gent feoABC::kan | This study |
| CFV1 | O395 feoB::tmp ΔvibB fbpA::cam | 10 |
| EPV103 (vVct) | O395 feoB::tmp ΔvibB fbpA::cam ΔviuA | This study |
| EPV111 | O395 feoABC::kan | This study |
| EPV114 | O395 feoABC::kan vctP::gent | This study |
| EPV126 (vVib) | O395 feoABC::kan vctP::gent fbpA::cam | This study |
| EPV134 | O395 ΔhemA | This study |
| N16961 | El Tor biotype | 70 |
| E. coli | ||
| DH5α | Cloning strain | 71 |
| DH5α λpir | Cloning strain | 71 |
| SM10 λpir | Conjugation strain | 72 |
| Plasmids | ||
| pWKS30 | Cloning vector | 73 |
| pHM5 | Suicide vector pGP704 carrying sacB | 74 |
| pCVD442N | pCVD442 with a NotI adapter inserted in the SacI site | 10 |
| pCGM120 | pCVD442N with V. cholerae sequences for vctP::gent | This study |
| pSfeoΔ::kan | pCVD442N with V. cholerae sequences for feoABC::kan | This study |
| pSviuA | pCVD442N with V. cholerae sequences for deleting viuA | This study |
| pShemA | pCVD442N with V. cholerae sequences for deleting hemA | This study |
| pSfbpA::cam | pCVD442N with V. cholerae sequences for fbpA::cam | 10 |
| pSvibBΔ | pCVD442N with V. cholerae sequences for deleting vibB | 10 |
| pHemA | V. cholerae hemA cloned into pWKS30 | This study |
The iron content of the LB broth was determined by a FerroZine-based assay. Iron sulfate was added to LB broth at final concentrations of 0, 5, 10, and 20 μM. The reducing agent ascorbate was added to a final concentration of 10 mM to reduce total iron in the samples to the ferrous state. Total iron was then quantified by the addition of 1 mM (final) FerroZine (Sigma), vortexed, and incubated at room temperature for 5 min prior to measuring the absorbance at 562 nm. This assay was performed on three separate LB broth samples prepared on separate days. Linear regression was used (GraphPad Prism), and the molar extinction coefficient of Fe-FerroZine (27.9 mM−1 cm−1 at 562 nm) was used to calculate iron in LB broth with no added iron. For growth assays, iron sulfate (FeSO4) and heme were added where indicated to final concentrations of 40 μM and 10 μM, respectively. LB medium at pH 6.5, 7.5, and 8.5 was buffered with 50 mM piperazine-N,N′-bis(2-ethanesulfonic acid) (PIPES), HEPES, or N-(2-hydroxy-1,1-bis(hydroxymethyl)ethyl)glycine (Tricine), respectively. Deferrated ethylene diamine diorthohydroxyphenyl acetic acid (EDDA) (27) was used at a final concentration of 20 μg/ml to induce iron starvation. Microaerobic environments were generated using the GasPak EZ Campy container system (BD Biosciences); anaerobic environments were generated using AnaeroGen 2.5L (Thermo Scientific). Sucrose supplementation was used at a final concentration of 0.4% (wt/vol) for inductively coupled plasma mass spectrometry (ICP-MS) sample preparation.
Liquid growth assays.
Liquid growth assays were carried out in 96-well plates (Corning) with automated readings using a Flexstation 3 (Molecular Devices). Cultures were grown overnight in LB supplemented with streptomycin and heme and were then diluted 1:100 or 1:200 into fresh LB medium with the indicated supplements. Optical density was assessed at 650 nm every 2.5 min over 10 h at 37°C; a reduced number of data points are depicted in the figures here. All liquid growth assays displayed are the means from three biological replicates.
Colony size assays.
Colony size assays were performed on LB agar. Overnight cultures of indicated strains were diluted and plated to obtain well-isolated colonies. After 24 h of incubation at 37°C, the diameters of 10 well-isolated colonies were measured using a reticle. The data shown are the means from single experiments and are representative of biological replicates.
Plasmid construction.
Primers used for PCR are listed in Table 3. For the generation of vctP mutations, the SmaI fragment of pGMΩ1 (28) was cloned into the blunted pCat120 (11) AvrII and Mlul sites. The SalI/NotI fragment of this plasmid was then cloned into the SalI/NotI sites of pCVD442N to result in pCGM120.
TABLE 3.
Primers used for this study
| Primer | Sequence (5′ to 3′) |
|---|---|
| Cloning | |
| feo1-sal | CAAAGTCGACAAACTGATTGAGC |
| feo2-sma | CGTTGCCAACACCCGGGAGAACGCC |
| feo3-sma | GGCGTTCTCCCGGGTGTTGGGAACG |
| feo4-RV | TGGATATCAATGAGCTAACAGGC |
| ViuA.776 | ATGCGCGGCCAATAGGGC |
| ViuA.1817.rev | CTCTAAATTAACCACTACACTGCG |
| ViuA.3912.SO | CGCAGTGTAGTGGTTAATTTAGAGGGACTGAAACAAAACAACGCT |
| ViuA.4941.rev | AATGGCGTAAATCTTATCTCTTGC |
| HemA.234 | TGCCATCGCGGCTTTGTAAATC |
| HemA.1233.rev | CTATAGAGGATTCTCAGATATGTG |
| HemA.2134.SO | CACATATCTGAGAATCCTCTATAGTTGCAGTCGATTGTCGATAGC |
| HemA.3133.rev | GTACAAGCGGACGTGTGGAT |
| HemA.934 | ATTGATAATGTTGAATTTGTTCTAGAG |
| HemA.2493.rev | TTAGTTCAGATCGTCAAGACCTA |
| Quantitative PCR | |
| feoB_RTFor | ACGATTTGCTGGCGAGTTTG |
| feoB_RTRev | TTCTTGCTCTTGCGCAACAG |
| fbpA_RTFor | AAACGCGCAGAAACTGATGG |
| fbpA_RTRev | TCCACCAATGCAGAACGTTG |
| vctP_RTFor | TGCGGTCGAAGCCAAAATTG |
| vctP_RTRev | TTGGCACCAAAAGTCGTCAG |
| vibB_RTFor | AAGCTTGCGAACCATGCAAC |
| vibB_RTRev | ACAACTGAATCGCGCGAATC |
| rpoZ_RTFor | TCTGCGCGAAATCGAAGAAG |
| rpoZ_RTRev | AACGGTTGTGCATGATGCTG |
The deletion of feoABC was created using pSfeoΔ::kan. Primers feo1-sal–feo2-sma and feo3-sma–feo4-RV were used to amplify V. cholerae N16961 sequence. Splice overlap PCR was performed to join these fragments together, and the resulting product was ligated into the SmaI site of pWKS30. Primers were designed to introduce a SmaI site at the junction of the splice overlap PCR product, and the SmaI site was used to introduce the kan cassette from pUC4K (Amersham). This construct was moved into pHM5 using SalI and EcoRV restriction sites to generate pSfeoΔ::kan.
Deletion of viuA was achieved using pSviuA. Regions up- and downstream of viuA (VC2211) were amplified from V. cholerae O395 using PCR with primers ViuA.776-ViuA.1817.rev and ViuA.3912.SO-ViuA.4941.rev. Splice overlap PCR was performed to join these fragments together, and the resultant product was ligated into the SmaI site of pCVD442N to generate pSviuA.
Mutations in hemA (VC2180) were generated using pShemA. PCR fragments were amplified from V. cholerae O395 using primers HemA.234-HemA.1233.rev and HemA.2134.SO-HemA.3133.rev. Splice overlap PCR was used to join these fragments together, and the resultant product was ligated into the SmaI site of pCVD442N to generate pShemA.
The V. cholerae O395 hemA gene locus was amplified using primers HemA.934-HemA.2493.rev by PCR. The blunt product was ligated into the SmaI site of pWKS30 to generate the plasmid pHemA.
Mutant construction.
Allelic exchange, as described previously (24), was used to generate all mutations in this study. Mutations in fbpA and vibB were created using pSfbpA::cam and pSvibBΔ as described previously (10). Mutations in vctP, feoABC, viuA, and hemA were introduced using plasmids pCGM120, pSfeoΔ::kan, pSviuA, and pShemA, respectively. Allelic exchange was performed using SM10 λpir, which carries the necessary genes for conjugation. Selection for sucrose-resistant exconjugants was carried out in the presence of 10% (wt/vol) sucrose with 5 μM heme, as needed.
(i) EPV102 (ΔviuA ΔvibB feoABC::kan vctP::gent fbpA::cam) (null).
The gent cassette was introduced into the vctP locus of ARM591 (10) to generate ARM592. The kan cassette was then used to replace feoABC to create EPV6. The unmarked deletion of viuA was then introduced into EPV6 to result in EPV102.
(ii) EPV104 (ΔviuA ΔvibB vctP::gent fbpA::cam) (vFeo).
The unmarked viuA deletion was introduced into ARM592 to generate EPV104.
(iii) EPV115 (ΔviuA ΔvibB feoABC::kan vctP::gent) (vFbp).
The unmarked viuA deletion was introduced into ALV101 (10) to generate EPV105. The gent insertion into vctP was then introduced to create EPV113. The replacement of the feoABC locus with the kan cassette was then introduced into EPV113 to generate EPV115.
(iv) EPV103 (ΔviuA ΔvibB feoB::tmp fbpA::cam) (vVct).
The unmarked viuA deletion was introduced into CFV1 (10) to generate EPV103.
(v) EPV126 (feoABC::kan vctP::gent fbpA::cam) (vVib).
The feoABC::kan mutation was introduced into wild-type O395 to generate EPV111. Next, the gent insertion into vctP was introduced into EPV111 to generate EPV114. Finally, the fbpA::cam mutation was introduced into EPV114 to generate EPV126.
(vi) EPV134 (ΔhemA).
EPV134 (ΔhemA) was created through the introduction of the unmarked hemA mutation into the wild-type O395 background, resulting in deletion of hemA.
qPCR.
Overnight cultures of O395 were diluted 1:200 into 25 ml LB buffered at pH 6.5, 7.5, or 8.5 and grown to mid-logarithmic phase. EDDA was then added to induce iron starvation. Fifteen minutes after the addition of EDDA, RNA was harvested from cells using RNA-Bee (Tel-Test, Inc.). Following DNase I (Life Technologies) treatment, Superscript III reverse transcriptase (Life Technologies) was used to generate cDNA. Power SYBR green (Life Technologies) chemistry was used for the amplification and detection of products. Samples were prepared and processed in biological triplicate. Primers used for quantitative PCR (qPCR) are listed in Table 3. Threshold cycle (CT) values were normalized against those for rpoZ. Analysis of values was done through the ΔΔCT approach.
ICP-MS.
Overnight cultures of O395 (wild type) or EPV102 (null) were diluted in LB and plated for single colonies on LB agar supplemented with sucrose. Cells were washed from plates with sterile saline after 24 h of incubation aerobically or anaerobically at 37°C, and 2.5 × 109 to 5 × 109 cells (determined by A650) were pelleted and washed with saline. Pellets were then resuspended in 1 ml of 35% (vol/vol) trace-metal-grade nitric acid (Fisher Scientific). Cell suspensions were transferred to 5-ml Savillex vials, sealed, and heated at 80°C for 3 to 6 h. Vials were cooled and allowed to evaporate to dryness at 60°C overnight. The residue was resuspended in 1 ml of 2% (vol/vol) nitric acid and incubated at 60°C overnight. Metal content was measured using the Agilent 7500ce inductively coupled plasma mass spectrometer and analyzed against defined standards at the quadrupole inductively coupled plasma mass spectrometry (ICP-MS) laboratory in the Jackson School of Geosciences at the University of Texas at Austin. Samples were prepared in biological triplicate.
In vivo and in vitro competition.
The in vivo competition assays in 5-day-old BALB/c mice were performed as described by Taylor et al. (29), using a protocol approved by the University of Texas Institutional Animal Use and Care Committee. A minimum of five mice per experiment were inoculated intragastrically with 50 μl saline containing 0.02% Evans blue dye and 105 CFU of each competing strain grown to mid-logarithmic phase in LB medium supplemented with 5 μM heme. The mice were sacrificed 18 h after inoculation, and the total intestine was homogenized in sterile saline. Serial dilutions of both the inoculum and the homogenate were plated on selective medium to allow determination of the ratios of the competing strains. For competition of the hemA mutant (EPV134) and its wild-type parental strain, quantification of strains was done by plating on streptomycin with heme (2.5 μM) and patching colonies onto streptomycin plates lacking heme to identify hemA mutants. In vitro competitions were performed by inoculating a 3-ml LB culture with 50 μl saline containing approximately 105 CFU of each strain in competition. For competition between the hemA mutant and wild type, heme was added to the medium at indicated amounts. Anaerobic competition assays were performed in an anaerobic chamber (Coy Laboratories). All cultures were incubated at 37°C with shaking. Cultures were serially diluted and plated at time zero and at 18 h on selective medium to allow quantification of each strain in competition.
Relative fitness was determined through calculating the competitive index (CI) as follows: CI = (mutantoutput/wild typeoutput)/(mutantinput/wild typeinput). A CI equal to 1 indicates no fitness defect, while a CI of <1 indicates a fitness defect in the mutant and a CI of >1 indicates a fitness advantage for the mutant.
RESULTS
Characterization of the roles of individual V. cholerae iron transport systems.
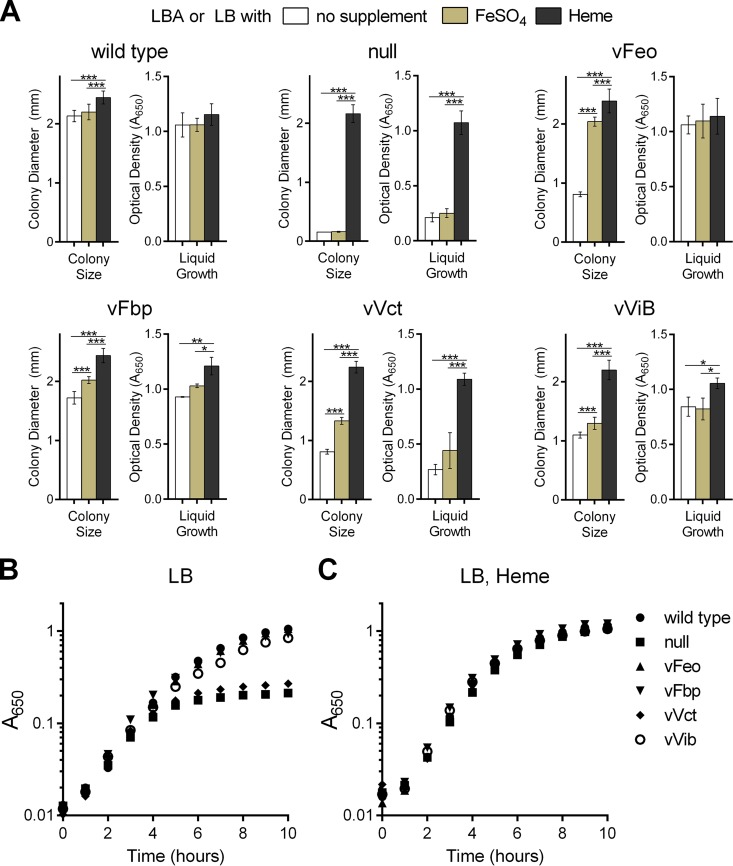
We previously demonstrated that mutation of four iron acquisition systems, feo, fbp, vct, and vib, in a single V. cholerae strain background eliminated growth on LB medium (11). The amount of iron in LB medium is 5.5 ± 0.2 μM, as determined by the FerroZine assay, which is sufficient to support growth of the wild-type strain. Failure of the mutant to grow in this medium indicates that even where iron is abundant, at least one of the iron transporters is needed. To investigate the roles of these systems, we constructed a series of isogenic strains in which only one of these four systems is functional. viuA, which encodes the vibriobactin receptor, was also deleted from vibriobactin biosynthesis mutant (vibB) strains to eliminate cross-feeding when a vibriobactin-producing strain was used in competition assays. The strain with all four systems disrupted is designated the null strain, and strains retaining only a single functional system are designated vFeo, vFbp, vVct, and vVib for their respective functional system (Table 2 and Fig. 1). As expected, the wild-type strain carrying the full complement of iron acquisition genes displayed robust growth on LB agar while the null strain failed to grow (Fig. 1A). The strains with individual systems varied in growth density on LB agar. Neither vFeo nor vVct appeared to grow as densely as the wild type, vFbp, or vVib in this assay, suggesting differences in the ability of these transport systems to acquire iron under this condition. All of these strains, including the null strain, retain the ability to acquire and utilize heme as an iron source, allowing strains to be routinely maintained on medium with heme. Growth was observed for the null strain on LB agar supplemented with heme, and differences in growth density among the other strains were reduced (Fig. 1B).
FIG 1.
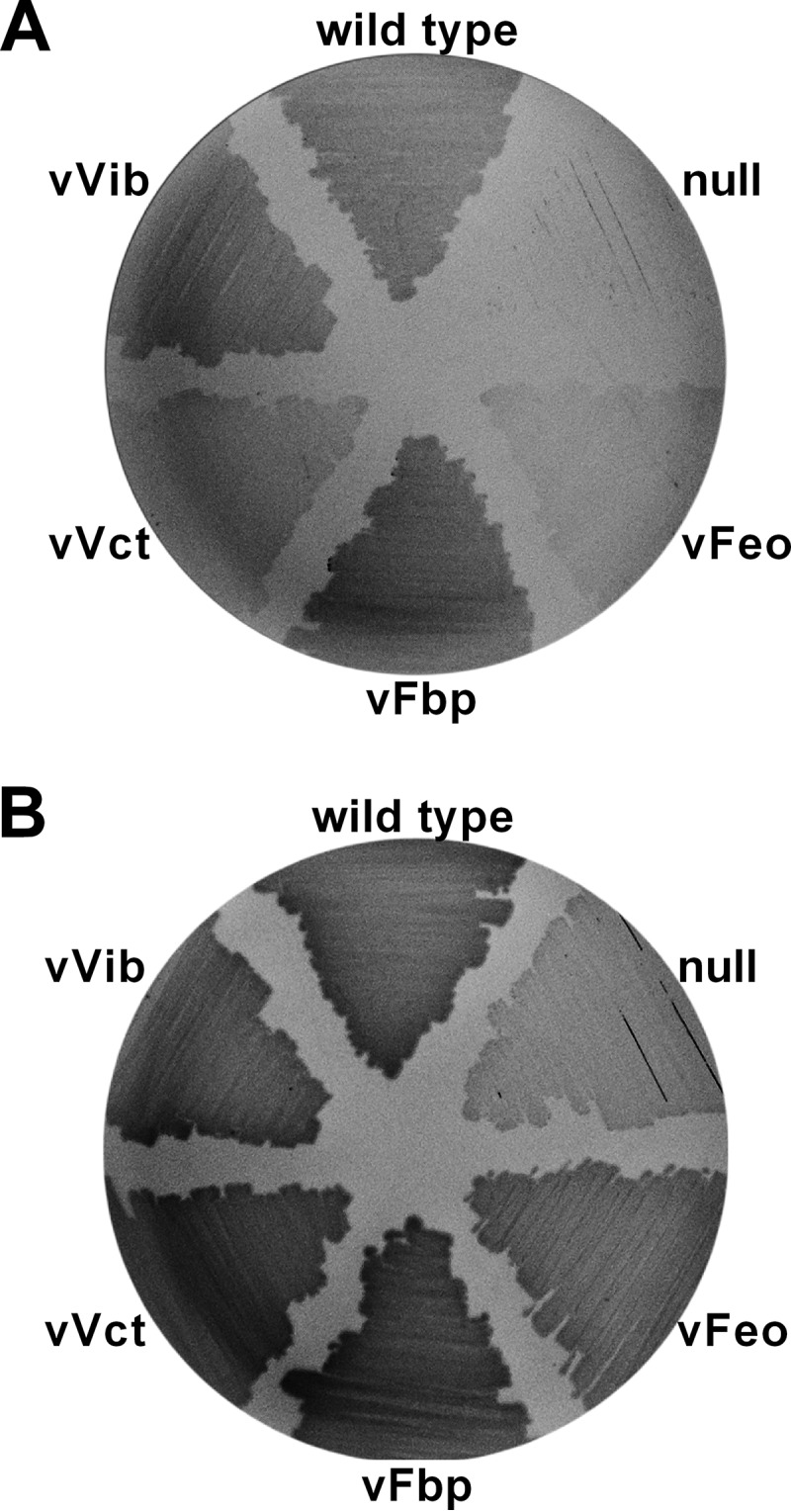
Single-system strains and null growth on LB agar plates. Overnight cultures of wild-type O395, null (EPV102), vFeo (EPV104), vFbp (EPV115), vVct (EPV103), and vVib (EPV126) were streaked on unsupplemented LB agar plates (A) or LB agar plates supplemented with heme (B). Plates were incubated overnight at 37°C.
Differential growth of single-system strains on solid or in liquid medium.
Because the initial screening on agar suggested that the single-system strains differed in their growth, quantitative assays were used to compare the strains. Colony sizes were determined for each strain growing on LB agar as described previously (11), and growth in liquid culture was used to measure differences in both final cell density and growth rate. The wild type grew well on solid medium and in liquid culture, and there was no significant difference between growth on unsupplemented medium and in the medium containing additional iron (Fig. 2A). The null strain formed microcolonies on LB agar in the absence of heme, and the colony size was not stimulated by the addition of iron to the medium (Fig. 2A). In LB broth, the null strain underwent 3 to 4 doublings, most likely the result of iron stores acquired during overnight growth in heme (Fig. 2B). As observed with the colony size assay, supplementation of the liquid medium with iron did not cause a significant increase in growth, suggesting that there is at most minimal iron acquisition during the course of the experiment. Supplementation with the usable iron source, heme, allowed the null mutant to grow to the same final density as the wild type. All of the single-system strains grew to near-wild-type levels when grown in medium containing heme in both assays (Fig. 2A and C), but there were distinct differences in their growth in unsupplemented or iron-supplemented medium (Fig. 2A and B).
FIG 2.
Differential growth of single-system strains on solid or in liquid medium. (A) For colony size assays (left), the diameters of 10 well-isolated colonies were measured using a reticle after 24 h of incubation at 37°C on unsupplemented LB agar or agar supplemented with FeSO4 or heme. The data indicate the means and standard deviations and are representative of biological replicates. For monitoring growth in liquid medium (right), overnight cultures of strains were diluted 1:100 into LB with supplements as indicated. The values are the means and standard deviations of optical density values (A650) measured after 10 h of growth for three independent experiments. The strains assessed in each panel are O395 (wild type), EPV102 (null), EPV104 (vFeo), EPV115 (vFbp), EPV103 (vVct), and EPV126 (vVib). In both assays, *, **, and *** indicate significance as determined by two-way analysis of variance, Holm-Šidák multiple-comparison test, with P values of <0.05, 0.01, and 0.0001, respectively. (B and C) The growth of the individual strains was assayed through the measurement of A650 over time in LB medium (B) and LB medium with heme supplementation (C). The means from three independent experiments are represented in both assays.
vFeo formed small colonies on LB agar (Fig. 2A). While the amount of iron found in LB agar was sufficient for robust growth of the wild-type strain, supplemental iron was necessary for the vFeo strain to achieve wild-type colony sizes, suggesting that ferrous iron is limiting. In contrast, growth of vFeo in liquid medium was similar to that of the wild type, independent of supplementation. This may be due to differences in the oxidation state of iron in these media due to aeration, increased access to iron ligands due to greater diffusion in liquid than solid media, or differences in expression of iron transport genes.
vFbp showed the most robust growth on LB agar of any of the single-system strains (Fig. 2A). This was most evident in growth on unsupplemented agar, although the colony size for vFbp was still smaller than the wild type, even with iron supplementation. In liquid medium, vFbp grew similarly to the wild type (Fig. 2A and B).
vVct had the poorest growth of the single-system strains in both assays (Fig. 2A and B). Although larger than the null mutant, the colonies were small on unsupplemented agar and were significantly smaller than the wild type, even in the presence of supplemental iron. The poor growth of vVct was most evident in liquid medium. In the absence of iron supplementation, its growth was not significantly better than the null mutant (Fig. 2B). However, unlike the null mutant, its growth was significantly stimulated by supplemental iron (Fig. 2A). The reason for the poor growth in liquid medium is not known, but it may reflect properties of this transporter's ligand, which has not been identified.
vVib produced small colonies and had a low growth rate in the absence of heme (Fig. 2A and B). This was unexpected, given the high iron-binding affinity of the vibriobactin system (9). The reduced growth and absence of stimulation by FeSO4 in either solid or broth medium suggest that vibriobactin, and not iron, is the limiting factor for the growth of this strain. In the absence of siderophore-independent systems, growth of vVib may be constrained until significant vibriobactin accumulates in the medium.
These data show that these iron transport systems, Feo, Fbp, Vct, and Vib, differ in their ability to support growth in complex media. To characterize these differences in more detail, the single-system strains were grown under specific conditions to determine environmental effects on the ability of each system to provide iron for growth.
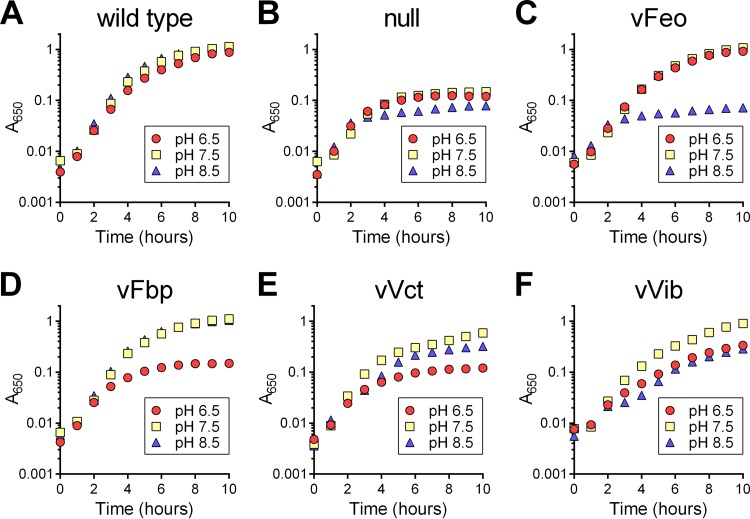
pH affects the function of the V. cholerae iron acquisition systems.
Because pH can affect the relative amounts of available ferrous and ferric iron, we wanted to test the function of these systems under differing pH conditions. V. cholerae encounters a wide range of pHs in infection of the human small intestine (pH 6.4 to 7.5) (30) and in marine environments (pH 7.5 to 8.4) (31). Growth of the single-system strains was assayed in LB buffered at pH 6.5, 7.5, or 8.5 (Fig. 3). While the wild type maintained robust growth under all three pH conditions (Fig. 3A), the null strain grew poorly under the tested conditions (Fig. 3B), indicating that at least one of the four systems is needed for growth at each pH. vFeo grew robustly at pH 6.5 and 7.5 but had the same phenotype as the null strain at pH 8.5 (Fig. 3C). This is consistent with the ligand for the Feo system, ferrous iron, being less abundant under more alkaline conditions. Conversely, the strain with only the ferric iron transporter Fbp grew robustly at pH 7.5 and 8.5, conditions under which most of the iron would be in the ferric form, and grew poorly at pH 6.5 (Fig. 3D). vVct grew best at pH 7.5. It is not known what nonsiderophore iron substrate the VctPDGC system transports, but given the poor growth at pH 6.5, it is unlikely that VctPDGC transports ferrous iron (Fig. 3E). vVib grew best at pH 7.5, and intermediate phenotypes were observed at both pH 6.5 and 8.5. Even at pH 7.5, vVib grew more slowly than the wild type. This may again suggest that the amount of vibriobactin synthesis or transport, rather than the amount or state of available iron, is the growth-limiting factor for this strain.
FIG 3.
pH affects growth of single-system strains. The growth of strains was assessed under different pH conditions. Overnight cultures of strains were diluted 1:200 into LB medium buffered at pH 6.5, 7.5, and 8.5. Growth of strains was assessed over 10 h at 37°C. The strains assessed in each panel are O395 (wild type) (A), EPV102 (null) (B), EPV104 (vFeo) (C), EPV115 (vFbp) (D), EPV103 (vVct) (E), and EPV126 (vVib) (F). The data represent the means from three biological replicates.
Effects of pH on growth are not solely due to changes in expression of iron acquisition genes.
Differences in growth at the different pHs may reflect not only the relative abundance of ferric and ferrous iron but may also be attributable to the effects of pH on expression of the genes encoding the transport systems. The transcriptional regulator Fur is known to repress each of these systems under high-iron conditions (32). Binding of iron to Fur allows Fur to repress transcription of genes with the Fur-box motif (32–34). However, it is not known whether expression of these genes is also controlled by regulators that respond to environmental pH. To assess regulatory effects on the expression of iron acquisition genes under the different pH conditions, wild-type O395 was grown in LB buffered at pH 6.5, 7.5, or 8.5, and iron starvation was induced by the addition of the iron chelator EDDA. RNA was isolated, and quantitative PCR was used to assess the relative levels of feoB, fbpA, vctP, and vibB mRNA. In each case, the level of mRNA was compared to the level in cells grown at pH 7.5. For vibB, the mRNA levels were nearly equivalent at all three pHs (Fig. 4), suggesting that the difference in growth as a function of pH was not due to differential expression of vib genes.
FIG 4.
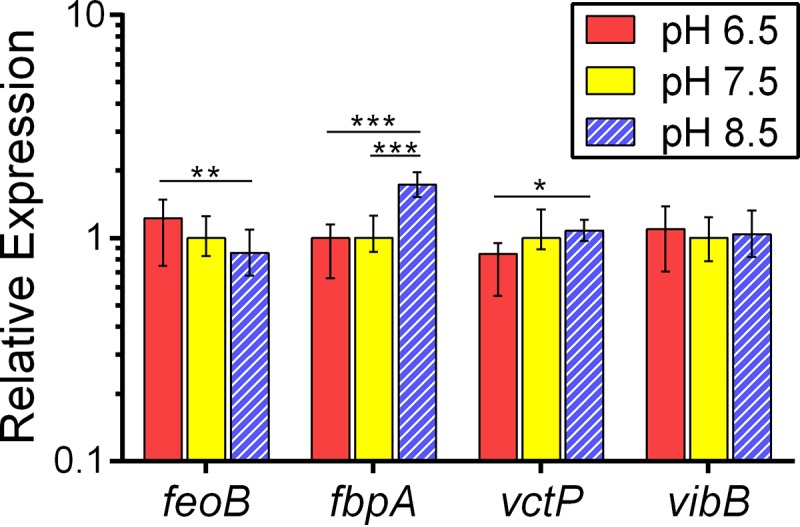
mRNA levels of iron transport genes in response to pH changes. Quantitative PCR was used to measure relative levels of mRNA isolated from wild-type bacteria grown to mid-logarithmic phase and exposed to iron starvation stress through the addition of the iron chelator EDDA. The level of feoB, fbpA, vctP, and vibB mRNAs was assessed. The data represent the means and standard deviations from biological triplicates. *, P < 0.05; **, P < 0.01; and ***, P < 0.0001, as determined by two-way analysis of variance, Holm-Šidák multiple-comparison test for ΔCT values.
While growth of vFeo was robust at pH 7.5 and poor at pH 8.5 (Fig. 3C), feoB expression was not different between pH 7.5 and pH 8.5 (Fig. 4), suggesting that the poor growth of vFeo at pH 8.5 was not a consequence of feoB transcriptional repression. However, a significant increase in mRNA levels at pH 6.5 was observed compared to levels at pH 8.5. Increased expression of feoB at lower pH may benefit V. cholerae, as ferrous iron, the substrate of the Feo system, is expected to be more prevalent at acidic pH.
For vctP, significantly higher expression was observed at pH 8.5 than pH 6.5, consistent with the reduced growth of vVct at lower pH (Fig. 3E).
fbpA mRNA levels were approximately 2-fold higher at pH 8.5 than at lower pH (Fig. 4). While the expression of fbp at higher pH may increase the amount of Fbp available for transport of ferric iron under conditions where this form of metal may be more abundant, growth at pH 7.5 was not impaired, indicating that Fbp is not rate-limiting for growth. While the growth of vFbp was reduced at pH 6.5 compared to pH 7.5 (Fig. 3D), relative expression of fbpA was not significantly different between cells grown under the two pH conditions (Fig. 4). Altogether, these data suggest that the differential growth of these strains as a function of pH was not solely the result of transcriptional repression.
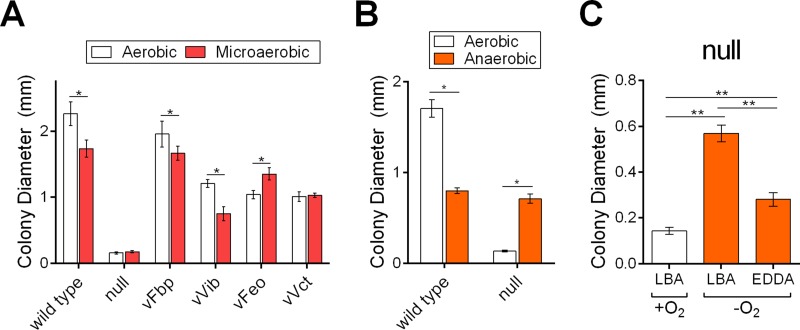
Effect of oxygen levels on iron acquisition.
Oxygen oxidizes ferrous iron to ferric iron. Under reduced-oxygen (microaerobic) conditions, more ferrous iron is expected to be present; as such, the amount of oxygen in an environment is expected to have an impact on the function of iron transport systems with ferrous and ferric iron substrates. When oxygen is absent from an environment, iron is expected to be predominantly in the reduced ferrous form. First, we tested the effects of reduced environmental oxygen on the growth of the single-system strains using the colony size assay, and plates were incubated under aerobic or microaerobic conditions. The wild-type strain displayed a modest reduction in colony size when oxygen was reduced, while the null strain grew equally poorly under both conditions (Fig. 5A). The strains that transport ferric iron, vFbp and vVib, had slightly reduced colony sizes, while vFeo, the strain with the ferrous iron transporter, formed larger colonies when growing microaerobically than aerobically. This is consistent with oxygen-limiting conditions increasing the availability of ferrous iron for transport through the Feo system while decreasing available ferric iron. vVct formed similarly sized colonies under both conditions, suggesting that its substrate is not significantly affected by reduced oxygen.
FIG 5.
Oxygen level affects colony formation. To assess the effect of oxygenation on the growth of strains, overnight cultures were diluted and plated for isolated colonies on LB agar with supplements as indicated, and colony diameters were measured after 24 h at 37°C under the indicated oxygen condition. (A) Comparison of growth in aerobic and microaerobic environments for the wild-type, null, and single-system strains. (B) Comparison of growth in aerobic and anaerobic environments for the wild-type and null strains. (C) Comparison of null strain growth in aerobic and anaerobic environments with the iron chelator EDDA (20 μg/ml) as indicated. The data are means and standard deviations and are representative of biological replicates. * indicates significance as determined by two-way analysis of variance, Šidák multiple-comparison test, P value of <0.0001. ** indicates significance as determined by one-way analysis of variance, Holm-Šidák multiple-comparison test, P value of <0.0001. LBA, LB agar.
We next assessed the growth of these strains in the absence of oxygen. The wild-type strain growing anaerobically formed smaller colonies than those with either aerobic or microaerobic growth. In contrast, the null strain grown anaerobically formed colonies that were larger than those under aerobic or microaerobic conditions and, in fact, were similar in size to wild-type colonies (Fig. 5B). Since the wild-type and null strains had nearly identical colony sizes, the strains with single functional systems were not assessed in this assay.
The ability of the null strain to form larger colonies in the absence of oxygen may indicate that a reduced amount of iron is necessary for anaerobic growth, such that iron stored from growth in the presence of heme prior to the start of the assay was sufficient. Alternatively, there may be an additional iron acquisition system that specifically functions under anaerobic conditions. If the iron requirement anaerobically can be fulfilled through iron stores, iron should not be required in the growth medium during the assay. To test this, colony sizes of the null strain were measured after anaerobic growth with or without the addition of the iron chelator EDDA to the agar (Fig. 5C). The addition of iron chelator resulted in a significant reduction in colony size, suggesting that the intracellular iron stores are insufficient for growth anaerobically, and thus, the growth of the null strain is dependent upon acquiring iron from the medium.
To confirm that the null strain was accumulating iron when grown anaerobically, ICP-MS was used to quantify the metal ions in bacterial cells growing aerobically and anaerobically. The wild-type and null strains were grown overnight in the presence of heme and plated onto LB agar for isolated colonies. After 24 h of incubation in either the presence or absence of atmospheric oxygen, bacterial cells were harvested from the plates to assess metal content. While the null strain accumulated only a very low level of iron when grown aerobically, a 5-fold-higher level of iron was found in cells grown anaerobically (Table 4). This suggests that there is an iron uptake system, separate from the four systems deleted from the null strain, that allows the cells to accumulate iron when growing in the absence of oxygen. The wild-type strain also showed a modest increase in intracellular iron levels when growing anaerobically. This result is distinct from observations in E. coli, where differences in intracellular iron levels were not seen when comparing cells grown aerobically and anaerobically (35). Zinc contents were not significantly different among samples, indicating that there was not a general impairment of metal acquisition in response to iron starvation or oxygen presence.
TABLE 4.
Metal content of cells growing aerobically and anaerobically
| Strain | Condition | Content (ng/109 cells ± SD) |
||
|---|---|---|---|---|
| Iron | Manganese | Zinc | ||
| Wild type | Aerobic | 35.7 ± 5.8a | 1.3 ± 0.2 | 25.1 ± 5.4d |
| Anaerobic | 50.4 ± 3.0a | 0.4 ± 0.3 | 23.1 ± 5.0d | |
| Null | Aerobic | 6.2 ± 4.5b | 13.6 ± 0.7c | 28.2 ± 2.1d |
| Anaerobic | 29.6 ± 8.4b | 2.8 ± 0.3c | 28.1 ± 4.8d | |
The difference in iron concentrations found between indicated sample sets is statistically significant as determined by two-way analysis of variance, Holm-Šidák multiple-comparison test, P value of <0.001.
The difference in iron concentrations found between indicated sample sets is statistically significant as determined by two-way analysis of variance, Holm-Šidák multiple-comparison test, P value of <0.0001.
The difference in manganese concentrations found between indicated samples sets is statistically significant as determined by two-way analysis of variance, Holm-Šidák multiple-comparison test, P value of <0.05.
Differences in zinc concentrations among all samples are not statistically significant as determined by two-way analysis of variance, Holm-Šidák multiple-comparison test, P value of >0.05.
The manganese content in null bacteria grown aerobically was significantly higher than that found in cells grown anaerobically or in wild-type cells under either condition. This is consistent with misregulation of Mn uptake as a consequence of iron starvation, as has been found in other bacteria (36, 37). Under iron-replete conditions, Fe-Fur represses not only iron acquisition genes but also a manganese import system (VC1688) (38) and the small RNA RyhB (32). When expressed, RyhB represses the genes sodB and fumC (39), which encode the manganese-containing superoxide dismutase and the noniron form of fumarate hydratase, respectively. Under low-iron conditions, the expression levels of VC1688, sodB, and fumC increase, suggesting that manganese and manganese enzymes fulfill some of the roles that would otherwise be performed by their iron-containing equivalents.
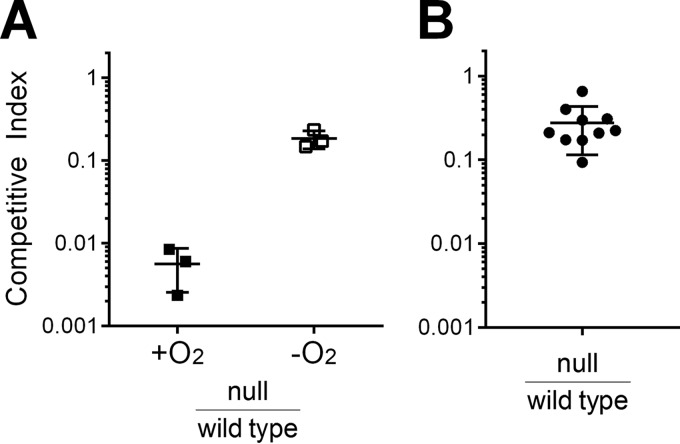
Iron acquisition by V. cholerae in the infant mouse.
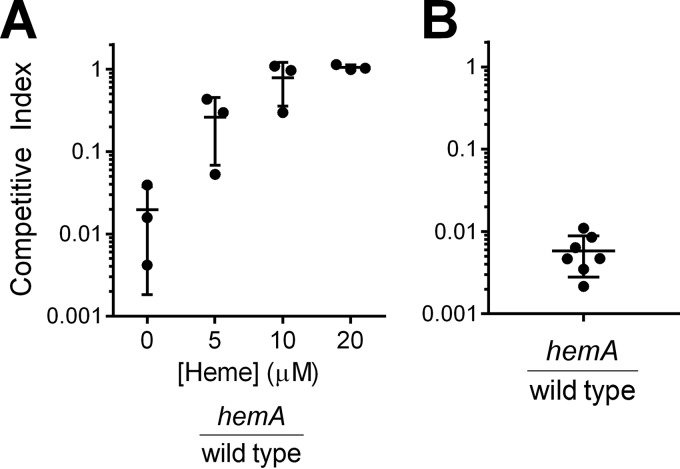
Oxygen is low or absent in the intestine, and we hypothesize that a V. cholerae iron transport system that supports sufficient iron acquisition for anaerobic growth in vitro could allow the bacteria to obtain iron in the intestine. Therefore, we assessed the ability of the null mutant to compete with the wild type for colonization and growth in the infant mouse intestinal infection model (29, 40). In prior studies, the infant mouse infection model has been employed to explore the function of iron acquisition systems in vivo, but most of the iron transport mutants tested thus far were only moderately attenuated or fully virulent (12). To determine whether the null mutant (vib viuA feo fbp vct) has an iron transport system capable of supporting growth in the host, infant mice were inoculated with equal numbers of the null and wild-type strains. The mean competitive index of the null strain in this assay was 0.28 (Fig. 6). This equates to a 3- to 4-fold loss of fitness relative to the wild type. This is a larger effect than seen with the vib feo fbp mutant (12), and this may indicate a role for vctP in vivo. However, this decrease in fitness is relatively modest compared with the loss of fitness of the null strain grown in vitro in the presence of oxygen (mean competitive index, 0.006). In fact, it is more consistent with the relative fitness observed in vitro in the absence of oxygen (mean competitive index, 0.19), suggesting that V. cholerae growing in the infant mouse intestine behaves similarly to cells growing anaerobically in vitro. The moderate defect of the null strain in the mouse infection model, in addition to the demonstrated anaerobic iron accumulation in this strain (Table 4), suggests that there is an iron transport system in V. cholerae that functions in the absence of oxygen and supports growth of the bacteria in vivo. Identification of this system will be crucial to understanding the interactions of this pathogen with its host.
FIG 6.
In vitro and in vivo competition between null and wild-type strains. The competitive index (CI) was calculated by normalizing the output ratio to the input ratio of the two competing strains. (A) Relative fitness of the wild-type and null strains was assessed in vitro in the presence of oxygen (atmospheric oxygen conditions) and absence of oxygen (anaerobic chamber). Each point represents an individual culture. The lines indicate the means and standard deviations. (B) Relative fitness of the wild-type and null strains was assessed in the infant mouse infection model. Each circle indicates a separate infant mouse, and horizontal lines indicate the means and standard deviations. A competitive index of <1 indicates a fitness defect for the null mutant under the condition.
Heme is not a source of iron for V. cholerae in the infant mouse.
Because the null mutant retains the ability to use heme as an iron source, it was possible that heme could serve as an iron source for V. cholerae in the infant mouse intestine. Previous studies indicated that the heme transporters are not essential for growth of V. cholerae in the infant mouse intestine, but the presence of additional iron transport systems did not allow determination of whether heme is available as a potential iron source within the host (24). To determine whether heme is present in the infant mouse intestine, a hemA deletion mutant in the wild-type O395 background was generated (EPV134). HemA catalyzes the first committed step of heme biosynthesis (41, 42), and a V. cholerae O139 hemA mutant was reported to have a significant growth defect (43). Because the heme transport systems allow V. cholerae to utilize available heme, we reasoned that growth of a hemA mutant could serve as a sensor for the presence of heme in the environment. Indeed, EPV134 grew as well as the wild type in LB medium supplemented with heme but poorly in the absence of supplementation (Fig. 7A). Supplying the wild-type copy of hemA on a low-copy-number vector allowed normal growth in the absence of heme supplementation (Fig. 7B), confirming that the defect in EPV134 is in the hemA gene. An in vitro competition assay was used to assess the growth of the hemA mutant relative to the wild type at different heme concentrations. The hemA mutant competed poorly with the wild type in the absence of heme in vitro, but relative fitness increased in the presence of increasing concentrations of supplemental heme (Fig. 8A). Heme concentrations of ≥10 μM eliminated the competitive advantage of the wild type over the hemA mutant. This concentration of heme supported growth of the null strain (Fig. 1B). Thus, if the hemA mutant is able to acquire sufficient heme for growth in the intestine, the level of heme available for growth should also be sufficient to sustain growth of the null mutant.
FIG 7.
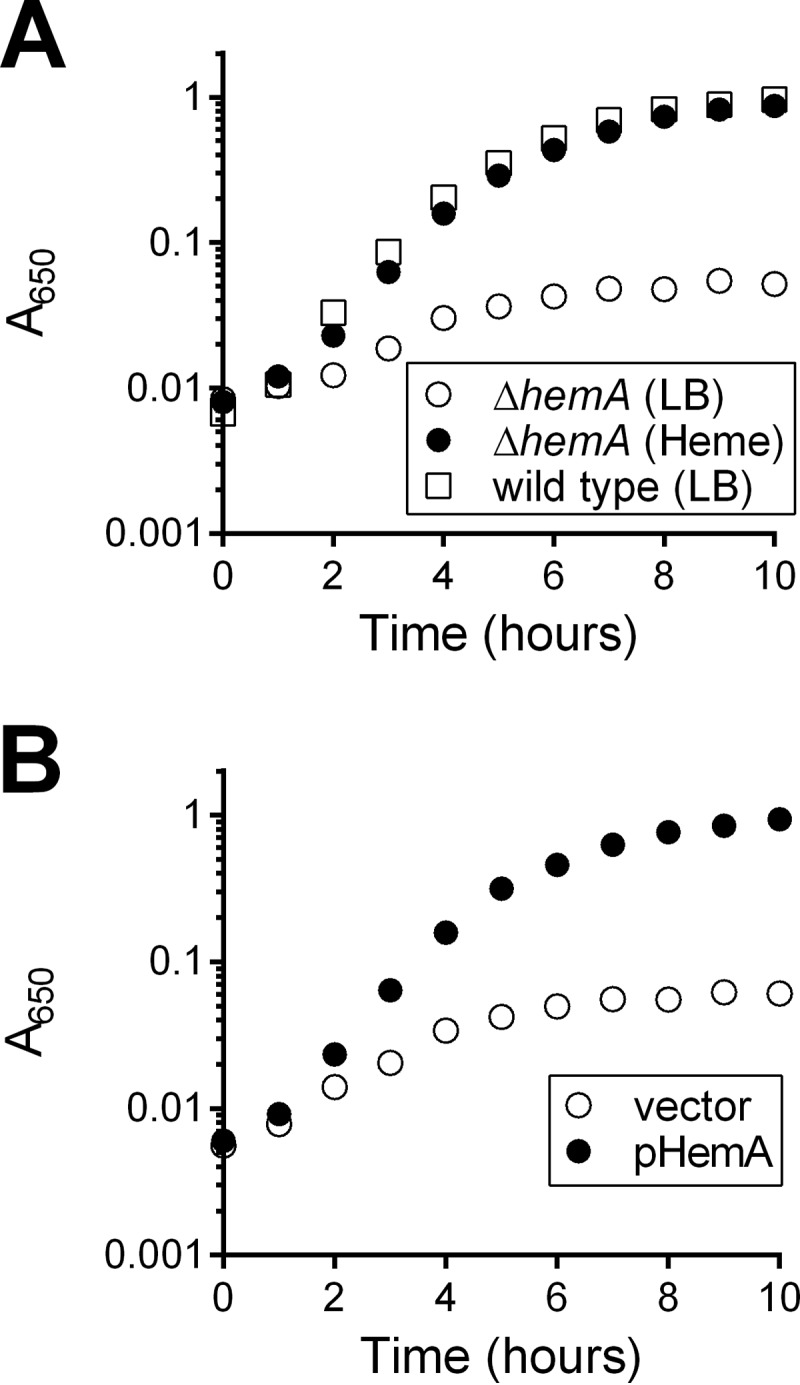
Growth of the V. cholerae O395 hemA mutant. (A) Overnight cultures of EPV134 (O395 ΔhemA) and wild-type O395 were diluted 1:200 in LB or LB supplemented with heme. Growth was determined by monitoring A650 over the course of 10 h. (B) Overnight cultures of EPV134 bearing empty vector (pWKS30) or pHemA were diluted 1:200 in LB, and growth was monitored as described above. The data represent the means from biological triplicates.
FIG 8.
Heme is not available as an iron source to V. cholerae in the infant mouse intestinal tract. (A) Relative fitnesses of wild-type O395 and EPV134 (O395 ΔhemA) were determined in vitro in LB supplemented with the indicated concentrations of heme. Each point represents a separate culture; lines indicate the means and standard deviations. (B) Relative fitness of wild-type O395 and EPV134 (O395 ΔhemA) was determined in vivo. Each circle indicates a separate infant mouse. Circles indicate the calculated competitive index in mice from which EPV134 was isolated (n = 7); EPV134 was not isolated from an additional 16 mice (not depicted). The competitive index (CI) was calculated by normalizing the output ratio to the input ratio of the two competing strains. Each data point represents one mouse, and the mean CI and standard deviation are represented by horizontal lines. A competitive index of <1 indicates a fitness defect for the hemA mutant under the condition.
When in vivo competition of the hemA strain against the wild type was performed, few hemA mutants were recovered from the infant mice after 18 h of infection (Fig. 8B). Of 23 infected mice, hemA mutants were recovered from only 7 animals (mean competitive index, 0.006), while the recovery of hemA mutants was below the level of detection in the remaining 16 mice (Fig. 8B). The mean competitive index observed in the mice was similar to that observed in the absence of heme in vitro (mean competitive index, 0.02) (Fig. 8A). The severe attenuation of the hemA mutant suggests that there is minimal heme available to V. cholerae in the infant mouse intestine, and growth of V. cholerae in this environment must depend on a source of iron other than heme. The null strain likely acquires this iron via an uncharacterized system that functions in anoxic environments.
DISCUSSION
V. cholerae has a large number of iron transport systems (11, 12). These include siderophore synthesis (Vib) and transport (Viu), transporters for xenosiderophores (IrgA, Vct, and Fhu), ferric (Fbp) and ferrous (Feo) iron transporters, and heme uptake systems (Hut and Has). Vct can also transport iron in the absence of siderophore (11). The maintenance of all these genes within the genome suggests that, while there may be some redundancy, each system confers a selective advantage during some stage of the V. cholerae life cycle. Having multiple iron acquisition systems may allow V. cholerae to acquire iron efficiently from the diverse sources available in the different environments that it inhabits. The need for multiple iron transport systems for optimal growth in different environments has been noted for other pathogens. In Yersinia pestis, the siderophore yersiniabactin is essential for the early stages of bubonic plague but is not needed for septicemic infection (44). Y. pestis has multiple ferrous iron transporters, including Feo, Yfe, and Fet. These do not appear to be redundant, as mutations in either feo or yfe resulted in a significant growth defect (45). Similarly, Bordetella pertussis uses different systems at different times during infection (46). The alcaligin and enterobactin siderophore systems are expressed early in the course of infection, while heme transport is induced late, reflecting the availability of different iron sources during the course of infection.
In V. cholerae, the presence of multiple systems, some with potentially overlapping functions, makes it difficult to assess the role of individual systems. To more fully characterize the role of each system, we constructed and analyzed strains that have only one of the four systems that promote growth in standard, unsupplemented LB medium. Our data indicate that the different iron acquisition systems are not functionally redundant but are needed to optimize iron acquisition under a variety of conditions. For example, the Feo system appears to function best at low pH or reduced oxygen, where its substrate ferrous iron is expected to be present. Conversely, the Fbp system supports robust growth at alkaline pH or normal atmospheric oxygen levels where more ferric iron is expected. However, when strains with either system are grown in a pH environment where their respective substrate is less prevalent, these systems do not appear to be capable of supporting growth, growing as poorly as the strain deficient in all four of the iron transport systems.
Vibriobactin is often considered the most effective iron acquisition system in V. cholerae, because of the high affinity of the siderophore for iron. Therefore, it was surprising that vVib did relatively poorly in growth assays, especially as individual colonies on a solid medium (Fig. 2A), compared to vFbp, which also transports ferric iron. While siderophores are often considered the primary iron transport systems, their role is generally assessed in assays that are different from those used here (5, 47, 48). Usually, assays measure the ability of the siderophore to compete with a soluble chelator, such as dipyridyl or EDDA, and frequently there is at least one nonsiderophore transport system, such as Feo, in the genetic background of the strain. In the experiments presented here, there is very limited siderophore available at the time when the strain is spread on the plate or diluted into fresh medium due to dilution at the beginning of the assays. The only way for the strain to obtain iron is to produce siderophore such that it would accumulate to a threshold concentration in the medium. We speculate that the growth defect is more pronounced in the colony size assay (Fig. 2A), since cells can easily use siderophore produced by other cells in liquid culture, while on solid medium, this will be limited by the rate of diffusion of siderophore in the agar.
Anaerobic growth of the null mutant in the absence of heme appears to be due to the presence of an additional, unidentified iron transport system. Both the null and the wild-type strains accumulate iron when grown anaerobically, and growth is significantly reduced when an iron chelator is added to the medium. This indicates that the ability of the null strain to grow anaerobically is dependent upon its ability to acquire iron. Growth of the null strain anaerobically, but not aerobically, suggests that ferrous iron is the likely ligand for the system. Feo also transports only ferrous iron, but unlike the null mutant, vFeo grows in the presence of oxygen, indicating that ferrous iron is available aerobically, possibly due to the presence of the VciB protein, which was proposed to act as a ferric iron reductase under aerobic conditions (49). This suggests that it is not a lack of ferrous iron that prevents the growth of the null mutant in oxygen. It is more likely that the system is expressed only under anoxic conditions. V. cholerae has both the Fnr and ArcAB oxygen-sensing transcriptional regulators, and regulation of feo expression by the presence of oxygen has been reported (50). One, or both, of these regulators may control expression of iron transport genes. Having oxygen, as well as iron, regulate iron uptake systems can help protect the cells from oxidative stress and damaging radicals associated with the Fenton reaction in the presence of iron (51). While iron is essential, excess intracellular iron in the presence of oxygen can be lethal (52), and the cell may repress some systems in aerobic environments to reduce potential toxic effects of iron. An effect of oxygen on expression of V. cholerae iron transport systems is suggested by the observation that the wild type accumulated higher levels of iron anaerobically than under aerobic conditions.
Although the growth and metal accumulation data strongly suggest the presence of an additional mechanism of iron acquisition, the identity of the system is unknown at this time. No clear homologues of the characterized ferrous iron transport systems Shigella flexneri SitABCD (53), S. flexneri MntH (54), Yersinia pestis YfeABCD (55, 56), E. coli EfeUOB (57), E. coli CorA (58), E. coli FetM (59), E. coli FecA (60), or Bordetella FtrABCD (61) are present within the V. cholerae genome, and mutation of other candidate iron-regulated iron transporter genes (32, 34) has, thus far, not been successful in abrogating anaerobic growth. It is possible that a transporter for another metal, such as manganese, can also transport iron, as has been noted for the Salmonella Sit system (36) and S. flexneri MntH (54).
It is of particular interest to identify the system(s) that enables V. cholerae to acquire iron during colonization of the mammalian intestine. Previous studies using single or multiple gene knockouts failed to identify any one iron transport system as critical for in vivo colonization and pathogenesis. Mutations in vibriobactin synthesis had reduced multiplication but still caused fluid accumulation in the mouse (62, 63) and a vibB fbpA feoB triple mutant competed well with its parent (10). The most severe defect observed in the mouse model was seen in a mutant in which vibriobactin synthesis as well as the genes encoding both TonB systems was mutated, but interpretation of this is complicated by the fact that TonB may have effects in addition to iron transport (63), and toxic peptides may have resulted from the manner in which the genes were disrupted. In competition assays, the null strain, lacking Vib, Feo, Fbp, and Vct, had a small but consistent defect in colonization. This indicates that one or more of these systems contribute to wild-type growth in the mouse. However, the null strain competed surprisingly well and could be recovered in relatively high numbers from the infected animals. This suggests that the null strain is able to acquire iron in vivo, most likely using the same system that allows it to grow and compete with the wild type under anaerobic conditions in vitro. Attempts to identify a candidate gene by bioinformatics approaches have, thus far, been unsuccessful. We have not excluded the possibility of a reduced iron requirement for V. cholerae during growth in vivo. However, the fact that the bacteria accumulate iron and are sensitive to an iron chelator when growing anaerobically suggests that iron is needed for growth under conditions that are similar to those in the intestine.
Heme is a potential iron source in the intestine and was reported to be present within the cecal fluid of the rabbit intestine (64, 65); however, to our knowledge, this has not been assessed in infant mice. Disruption of the three heme receptors in V. cholerae, which prevented heme transport, did not result in a loss of fitness during in vivo competition with the wild type (24). This indicated that heme transport was not essential for V. cholerae virulence but did not establish whether heme is available to serve as an iron source in vivo. The hemA mutant, which cannot synthesize heme but can grow in the presence of exogenous heme, was unable to acquire sufficient heme to colonize and compete with the wild-type strain, suggesting that little or no heme is available to V. cholerae growing in the mouse intestine. This indicates that the system used by the null mutant to acquire iron in the infant mouse model is not heme.
The data presented in this study support a model whereby multiple iron acquisition systems contribute to the growth of V. cholerae in different environments. These systems have optimal conditions for iron acquisition, but there is overlap, ensuring some redundancy for acquiring this essential element. More than one system contributes to iron acquisition in vivo. The null strain had a reduction in fitness, indicating that at least one of the four systems missing in this strain contributes to colonization; however, the defect was modest, suggesting the presence of an additional system. Identifying this system, which is likely the same one supplying iron in anaerobiosis, is important to understanding the pathogenicity of this organism.
ACKNOWLEDGMENTS
We thank Bryan Davies, Mary Lozano, and Jaquelin Dudley for technical assistance and advice. We also thank Shanzhong Gong for technical assistance. Additionally, we thank Marvin Whiteley for generously allowing use of his anaerobic chamber. We are grateful to Nathaniel Miller for helpful discussions and processing ICP-MS samples.
REFERENCES
- 1.Weinberg ED. 1989. Cellular regulation of iron assimilation. Q Rev Biol 64:261–290. doi: 10.1086/416359. [DOI] [PubMed] [Google Scholar]
- 2.Hem JD. 1972. Chemical factors that influence the availability of iron and manganese in aqueous systems. Geol Soc Am Spec Pap 140:17–24. [Google Scholar]
- 3.Martin JH, Gordon M, Fitzwater SE. 1991. The case for iron. Limnol Oceanogr 36:1793–1802. doi: 10.4319/lo.1991.36.8.1793. [DOI] [Google Scholar]
- 4.Deriu E, Liu JZ, Pezeshki M, Edwards RA, Ochoa RJ, Contreras H, Libby SJ, Fang FC, Raffatellu M. 2013. Probiotic bacteria reduce Salmonella typhimurium intestinal colonization by competing for iron. Cell Host Microbe 14:26–37. doi: 10.1016/j.chom.2013.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Caza M, Kronstad JW. 2013. Shared and distinct mechanisms of iron acquisition by bacterial and fungal pathogens of humans. Front Cell Infect Microbiol 3:80. doi: 10.3389/fcimb.2013.00080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cassat JE, Skaar EP. 2013. Iron in infection and immunity. Cell Host Microbe 13:509–519. doi: 10.1016/j.chom.2013.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Becker KW, Skaar EP. 2014. Metal limitation and toxicity at the interface between host and pathogen. FEMS Microbiol Rev 38:1235–1249. doi: 10.1111/1574-6976.12087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Payne SM, Finkelstein RA. 1978. Siderophore production by Vibrio cholerae. Infect Immun 20:310–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Griffiths GL, Sigel SP, Payne SM, Neilands JB. 1984. Vibriobactin, a siderophore from Vibrio cholerae. J Biol Chem 259:383–385. [PubMed] [Google Scholar]
- 10.Wyckoff EE, Mey AR, Leimbach A, Fisher CF, Payne SM. 2006. Characterization of ferric and ferrous iron transport systems in Vibrio cholerae. J Bacteriol 188:6515–6523. doi: 10.1128/JB.00626-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wyckoff EE, Payne SM. 2011. The Vibrio cholerae VctPDGC system transports catechol siderophores and a siderophore-free iron ligand. Mol Microbiol 81:1446–1458. doi: 10.1111/j.1365-2958.2011.07775.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wyckoff EE, Mey AR, Payne SM. 2007. Iron acquisition in Vibrio cholerae. Biometals 20:405–416. doi: 10.1007/s10534-006-9073-4. [DOI] [PubMed] [Google Scholar]
- 13.Wyckoff EE, Stoebner JA, Reed KE, Payne SM. 1997. Cloning of a Vibrio cholerae vibriobactin gene cluster: identification of genes required for early steps in siderophore biosynthesis. J Bacteriol 179:7055–7062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Keating TA, Marshall CG, Walsh CT. 2000. Vibriobactin biosynthesis in Vibrio cholerae: VibH is an amide synthase homologous to nonribosomal peptide synthetase condensation domains. Biochemistry 39:15513–15521. doi: 10.1021/bi001651a. [DOI] [PubMed] [Google Scholar]
- 15.Keating TA, Marshall CG, Walsh CT. 2000. Reconstitution and characterization of the Vibrio cholerae vibriobactin synthetase from VibB, VibE, VibF, and VibH. Biochemistry 39:15522–15530. doi: 10.1021/bi0016523. [DOI] [PubMed] [Google Scholar]
- 16.Butterton JR, Stoebner JA, Payne SM, Calderwood SB. 1992. Cloning, sequencing, and transcriptional regulation of viuA, the gene encoding the ferric vibriobactin receptor of Vibrio cholerae. J Bacteriol 174:3729–3738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stoebner JA, Butterton JR, Calderwood SB, Payne SM. 1992. Identification of the vibriobactin receptor of Vibrio cholerae. J Bacteriol 174:3270–3274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Occhino DA, Wyckoff EE, Henderson DP, Wrona TJ, Payne SM. 1998. Vibrio cholerae iron transport: haem transport genes are linked to one of two sets of TonB, ExbB, ExbD genes. Mol Microbiol 29:1493–1507. doi: 10.1046/j.1365-2958.1998.01034.x. [DOI] [PubMed] [Google Scholar]
- 19.Wyckoff EE, Valle AM, Smith SL, Payne SM. 1999. A multifunctional ATP-binding cassette transporter system from Vibrio cholerae transports vibriobactin and enterobactin. J Bacteriol 181:7588–7596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mey AR, Wyckoff EE, Oglesby AG, Rab E, Taylor RK, Payne SM. 2002. Identification of the Vibrio cholerae enterobactin receptors VctA and IrgA: IrgA is not required for virulence. Infect Immun 70:3419–3426. doi: 10.1128/IAI.70.7.3419-3426.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Butterton JR, Calderwood SB. 1994. Identification, cloning, and sequencing of a gene required for ferric vibriobactin utilization by Vibrio cholerae. J Bacteriol 176:5631–5638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Henderson DP, Payne SM. 1993. Cloning and characterization of the Vibrio cholerae genes encoding the utilization of iron from haemin and haemoglobin. Mol Microbiol 7:461–469. doi: 10.1111/j.1365-2958.1993.tb01137.x. [DOI] [PubMed] [Google Scholar]
- 23.Henderson DP, Payne SM. 1994. Characterization of the Vibrio cholerae outer membrane heme transport protein HutA: sequence of the gene, regulation of expression, and homology to the family of TonB-dependent proteins. J Bacteriol 176:3269–3277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mey AR, Payne SM. 2001. Haem utilization in Vibrio cholerae involves multiple TonB-dependent haem receptors. Mol Microbiol 42:835–849. [DOI] [PubMed] [Google Scholar]
- 25.Kammler M, Schön C, Hantke K. 1993. Characterization of the ferrous iron uptake system of Escherichia coli. J Bacteriol 175:6212–6219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Weaver EA, Wyckoff EE, Mey AR, Morrison R, Payne SM. 2013. FeoA and FeoC are essential components of Vibrio cholerae ferrous iron uptake system and FeoC interacts with FeoB. J Bacteriol 195:4826–4835. doi: 10.1128/JB.00738-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rogers HJ. 1973. Iron-binding catechols and virulence in Escherichia coli. Infect Immun 7:445–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schweizer HD. 1993. Small broad-host-range gentamycin resistance gene cassettes for site-specific insertion and deletion mutagenesis. Biotechniques 15:831–834. [PubMed] [Google Scholar]
- 29.Taylor RK, Miller VL, Furlong DB, Mekalanos JJ. 1987. Use of phoA gene fusions to identify a pilus colonization factor coordinately regulated with cholera toxin. Proc Natl Acad Sci U S A 84:2833–2837. doi: 10.1073/pnas.84.9.2833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Evans DF, Pye G, Bramley R, Clark AG, Dyson TJ, Hardcastle JD. 1988. Measurement of gastrointestinal pH profiles in normal ambulant human subjects. Gut 29:1035–1041. doi: 10.1136/gut.29.8.1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chester R, Jickells TD. 2012. Marine geochemistry, 3rd ed Wiley-Blackwell, Chichester, West Sussex, United Kingdom. [Google Scholar]
- 32.Mey AR, Wyckoff EE, Kanukurthy V, Fisher CR, Payne SM. 2005. Iron and fur regulation in Vibrio cholerae and the role of fur in virulence. Infect Immun 73:8167–8178. doi: 10.1128/IAI.73.12.8167-8178.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bagg A, Neilands JB. 1987. Ferric uptake regulation protein acts as a repressor, employing iron(II) as a cofactor to bind the operator of an iron transport operon in Escherichia coli. Biochemistry 26:5471–5477. doi: 10.1021/bi00391a039. [DOI] [PubMed] [Google Scholar]
- 34.Davies BW, Bogard RW, Mekalanos JJ. 2011. Mapping the regulon of Vibrio cholerae ferric uptake regulator expands its known network of gene regulation. Proc Natl Acad Sci U S A 108:12467–12472. doi: 10.1073/pnas.1107894108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Niehaus F, Hantke K, Unden G. 1991. Iron content and FNR-dependent gene regulation in Escherichia coli. FEMS Microbiol Lett 84:319–323. doi: 10.1111/j.1574-6968.1991.tb04617.x. [DOI] [PubMed] [Google Scholar]
- 36.Kehres DG, Janakiraman A, Slauch JM, Maguire ME. 2002. Regulation of Salmonella enterica serovar Typhimurium mntH transcription by H2O2, Fe2+, and Mn2+. J Bacteriol 184:3151–3158. doi: 10.1128/JB.184.12.3151-3158.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Puri S, Hohle TH, O'Brian MR. 2010. Control of bacterial iron homeostasis by manganese. Proc Natl Acad Sci U S A 107:10691–10695. doi: 10.1073/pnas.1002342107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Green RT, Todd JD, Johnston AWB. 2013. Manganese uptake in marine bacteria; the novel MntX transporter is widespread in Roseobacters, Vibrios, Alteromonadales and the SAR11 and SAR116 clades. ISME J 7:581–591. doi: 10.1038/ismej.2012.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mey AR, Craig SA, Payne SM. 2005. Characterization of Vibrio cholerae RyhB: the RyhB regulon and role of ryhB in biofilm formation. Infect Immun 73:5706–5719. doi: 10.1128/IAI.73.9.5706-5719.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Klose KE. 2000. The suckling mouse model of cholera. Trends Microbiol 8:189–191. doi: 10.1016/S0966-842X(00)01721-2. [DOI] [PubMed] [Google Scholar]
- 41.Săsărman A, Surdeanu M, Szégli G, Horodniceanu T, Greceanu V, Dumitrescu A. 1968. Hemin-deficient mutants of Escherichia coli K-12. J Bacteriol 96:570–572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Avissar YJ, Beale SI. 1989. Identification of the enzymatic basis for delta-aminolevulinic acid auxotrophy in a hemA mutant of Escherichia coli. J Bacteriol 171:2919–2924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ravichandran M, Ali SA, Rashid NHA, Kurunathan S, Yean CY, Ting LC, Bakar ASA, Lalitha P, Zainuddin ZF. 2006. Construction and evaluation of a O139 Vibrio cholerae vaccine candidate based on a HemA gene mutation. Vaccine 24:3750–3761. doi: 10.1016/j.vaccine.2005.07.016. [DOI] [PubMed] [Google Scholar]
- 44.Perry RD, Bobrov AG, Fetherston JD. 2015. The role of transition metal transporters for iron, zinc, manganese, and copper in the pathogenesis of Yersinia pestis. Metallomics 7:965–978. doi: 10.1039/C4MT00332B. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fetherston JD, Mier I, Truszczynska H, Perry RD. 2012. The Yfe and Feo transporters are involved in microaerobic growth and virulence of Yersinia pestis in bubonic plague. Infect Immun 80:3880–3891. doi: 10.1128/IAI.00086-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Brickman TJ, Hanawa T, Anderson MT, Suhadolc RJ, Armstrong SK. 2008. Differential expression of Bordetella pertussis iron transport system genes during infection. Mol Microbiol 70:3–14. doi: 10.1111/j.1365-2958.2008.06333.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Holden VI, Bachman MA. 2015. Diverging roles of bacterial siderophores during infection. Metallomics 7:986–995. doi: 10.1039/C4MT00333K. [DOI] [PubMed] [Google Scholar]
- 48.Wyckoff EE, Allred BE, Raymond KN, Payne SM. 2015. Catechol siderophore transport by Vibrio cholerae. J Bacteriol 197:2840–2849. doi: 10.1128/JB.00417-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mey AR, Wyckoff EE, Hoover LA, Fisher CR, Payne SM. 2008. Vibrio cholerae VciB promotes iron uptake via ferrous iron transporters. J Bacteriol 190:5953–5962. doi: 10.1128/JB.00569-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Helton EA. 2010. FeoA, FeoB, and FeoC encode essential components of the Vibrio cholerae ferrous iron transport system. Ph.D. dissertation. University of Texas at Austin, Austin, TX. [Google Scholar]
- 51.Carpenter C, Payne SM. 2014. Regulation of iron transport systems in Enterobacteriaceae in response to oxygen and iron availability. J Inorg Biochem 133:110–117. doi: 10.1016/j.jinorgbio.2014.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Frawley ER, Fang FC. 2014. The ins and outs of bacterial iron metabolism. Mol Microbiol 93:609–616. doi: 10.1111/mmi.12709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Runyen-Janecky LJ, Reeves SA, Gonzales EG, Payne SM. 2003. Contribution of the Shigella flexneri Sit, Iuc, and Feo iron acquisition systems to iron acquisition in vitro and in cultured cells. Infect Immun 71:1919–1928. doi: 10.1128/IAI.71.4.1919-1928.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Runyen-Janecky L, Dazenski E, Hawkins S, Warner L. 2006. Role and regulation of the Shigella flexneri Sit and MntH systems. Infect Immun 74:4666–4672. doi: 10.1128/IAI.00562-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bearden SW, Staggs TM, Perry RD. 1998. An ABC transporter system of Yersinia pestis allows utilization of chelated iron by Escherichia coli SAB11. J Bacteriol 180:1135–1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bearden SW, Perry RD. 1999. The Yfe system of Yersinia pestis transports iron and manganese and is required for full virulence of plague. Mol Microbiol 32:403–414. doi: 10.1046/j.1365-2958.1999.01360.x. [DOI] [PubMed] [Google Scholar]
- 57.Cao J, Woodhall MR, Alvarez J, Cartron ML, Andrews SC. 2007. EfeUOB (YcdNOB) is a tripartite, acid-induced and CpxAR-regulated, low-pH Fe2+ transporter that is cryptic in Escherichia coli K-12 but functional in E. coli O157:H7. Mol Microbiol 65:857–875. doi: 10.1111/j.1365-2958.2007.05802.x. [DOI] [PubMed] [Google Scholar]
- 58.Hantke K. 1997. Ferrous iron uptake by a magnesium transport system is toxic for Escherichia coli and Salmonella typhimurium. J Bacteriol 179:6201–6204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Koch D, Chan ACK, Murphy MEP, Lilie H, Grass G, Nies DH. 2011. Characterization of a dipartite iron uptake system from uropathogenic Escherichia coli strain F11. J Biol Chem 286:25317–25330. doi: 10.1074/jbc.M111.222745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wagegg W, Braun V. 1981. Ferric citrate transport in Escherichia coli requires outer membrane receptor protein FecA. J Bacteriol 145:156–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Brickman TJ, Armstrong SK. 2012. Iron and pH-responsive FtrABCD ferrous iron utilization system of Bordetella species. Mol Microbiol 86:580–593. doi: 10.1111/mmi.12003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sigel SP, Stoebner JA, Payne SM. 1985. Iron-vibriobactin transport system is not required for virulence of Vibrio cholerae. Infect Immun 47:360–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Henderson DP, Payne SM. 1994. Vibrio cholerae iron transport systems: roles of heme and siderophore iron transport in virulence and identification of a gene associated with multiple iron transport systems. Infect Immun 62:5120–5125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mandlik A, Livny J, Robins WP, Ritchie JM, Mekalanos JJ, Waldor MK. 2011. RNA-Seq-based monitoring of infection-linked changes in Vibrio cholerae gene expression. Cell Host Microbe 10:165–174. doi: 10.1016/j.chom.2011.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kamp HD, Patimalla-Dipali B, Lazinski DW, Wallace-Gadsden F, Camilli A. 2013. Gene fitness landscapes of Vibrio cholerae at important stages of its life cycle. PLoS Pathog 9:e1003800. doi: 10.1371/journal.ppat.1003800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wyckoff EE, Smith SL, Payne SM. 2001. VibD and VibH are required for late steps in vibriobactin biosynthesis in Vibrio cholerae. J Bacteriol 183:1830–1834. doi: 10.1128/JB.183.5.1830-1834.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Butterton JR, Choi MH, Watnick PI, Carroll PA, Calderwood SB. 2000. Vibrio cholerae VibF is required for vibriobactin synthesis and is a member of the family of nonribosomal peptide synthetases. J Bacteriol 182:1731–1738. doi: 10.1128/JB.182.6.1731-1738.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Rogers MB, Sexton JA, DeCastro GJ, Calderwood SB. 2000. Identification of an operon required for ferrichrome iron utilization in Vibrio cholerae. J Bacteriol 182:2350–2353. doi: 10.1128/JB.182.8.2350-2353.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Mekalanos JJ, Swartz DJ, Pearson GD, Harford N, Groyne F, de Wilde M. 1983. Cholera toxin genes: nucleotide sequence, deletion analysis and vaccine development. Nature 306:551–557. doi: 10.1038/306551a0. [DOI] [PubMed] [Google Scholar]
- 70.Heidelberg JF, Eisen JA, Nelson WC, Clayton RA, Gwinn ML, Dodson RJ, Haft DH, Hickey EK, Peterson JD, Umayam L, Gill SR, Nelson KE, Read TD, Tettelin H, Richardson D, Ermolaeva MD, Vamathevan J, Bass S, Qin H, Dragoi I, Sellers P, McDonald L, Utterback T, Fleishmann RD, Nierman WC, White O, Salzberg SL, Smith HO, Colwell RR, Mekalanos JJ, Venter JC, Fraser CM. 2000. DNA sequence of both chromosomes of the cholera pathogen Vibrio cholerae. Nature 406:477–483. doi: 10.1038/35020000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hanahan D. 1983. Studies on transformation of Escherichia coli with plasmids. J Mol Biol 166:557–580. doi: 10.1016/S0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- 72.Miller VL, Mekalanos JJ. 1988. A novel suicide vector and its use in construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires ToxR. J Bacteriol 170:2575–2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wang RF, Kushner SR. 1991. Construction of versatile low-copy-number vectors for cloning, sequencing and gene expression in Escherichia coli. Gene 100:195–199. doi: 10.1016/0378-1119(91)90366-J. [DOI] [PubMed] [Google Scholar]
- 74.Runyen-Janecky LJ, Hong M, Payne SM. 1999. The virulence plasmid-encoded impCAB operon enhances survival and induced mutagenesis in Shigella flexneri after exposure to UV radiation. Infect Immun 67:1415–1423. [DOI] [PMC free article] [PubMed] [Google Scholar]