Abstract
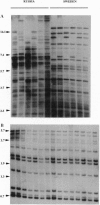
Loss of genetic variation due to population bottlenecks may be a severe threat for the survival of endangered species. Assessment and maintenance of genetic variability are thus crucial for conservation programs related to endangered populations. Scandinavian beavers went through an extensive bottleneck during the last century due to overhunting. In Sweden the species became extirpated but in Norway extinction was avoided by legal protection. Following reintroductions of small numbers of remaining Norwegian animals in 1922-1939, the Swedish population has increased tremendously, now harboring 100,000 animals. We show here that this viable population of beavers possesses extremely low levels of genetic variability at DNA fingerprinting loci and monomorphism at major histocompatibility complex (MHC) class I and class II loci. A similar pattern was also evident among Norwegian beavers but low levels of genetic variability were not a characteristic of the species since Russian conspecifics displayed substantial DNA fingerprinting polymorphism. However, the Russian animals were monomorphic at MHC loci, indicating that the European beaver is exceptional in its low level of MHC variability. The results demonstrate that a conservation program can be successful despite low levels of genetic variation in the founder population.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Andersson L., Lundberg C., Rask L., Gissel-Nielsen B., Simonsen M. Analysis of class II genes of the chicken MHC (B) by use of human DNA probes. Immunogenetics. 1987;26(1-2):79–84. doi: 10.1007/BF00345458. [DOI] [PubMed] [Google Scholar]
- Andersson L., Rask L. Characterization of the MHC class II region in cattle. The number of DQ genes varies between haplotypes. Immunogenetics. 1988;27(2):110–120. doi: 10.1007/BF00351084. [DOI] [PubMed] [Google Scholar]
- Andersson L., Sigurdardóttir S., Borsch C., Gustafsson K. Evolution of MHC polymorphism: extensive sharing of polymorphic sequence motifs between human and bovine DRB alleles. Immunogenetics. 1991;33(3):188–193. doi: 10.1007/BF01719239. [DOI] [PubMed] [Google Scholar]
- Bonnel M. L., Selander R. K. Elephant seals: genetic variation and near extinction. Science. 1974 May 24;184(4139):908–909. doi: 10.1126/science.184.4139.908. [DOI] [PubMed] [Google Scholar]
- Dunbar M. R., Jessup D. A., Evermann J. F., Foreyt W. J. Seroprevalence of respiratory syncytial virus in free-ranging bighorn sheep. J Am Vet Med Assoc. 1985 Dec 1;187(11):1173–1174. [PubMed] [Google Scholar]
- Ellegren H., Andersson L., Johansson M., Sandberg K. DNA fingerprinting in horses using a simple (TG)n probe and its application to population comparisons. Anim Genet. 1992;23(1):1–9. [PubMed] [Google Scholar]
- Ellegren H. DNA typing of museum birds. Nature. 1991 Nov 14;354(6349):113–113. doi: 10.1038/354113a0. [DOI] [PubMed] [Google Scholar]
- Gilbert D. A., Lehman N., O'Brien S. J., Wayne R. K. Genetic fingerprinting reflects population differentiation in the California Channel Island fox. Nature. 1990 Apr 19;344(6268):764–767. doi: 10.1038/344764a0. [DOI] [PubMed] [Google Scholar]
- Hedrick P. W., Thomson G. Evidence for balancing selection at HLA. Genetics. 1983 Jul;104(3):449–456. doi: 10.1093/genetics/104.3.449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedrick P. W., Whittam T. S., Parham P. Heterozygosity at individual amino acid sites: extremely high levels for HLA-A and -B genes. Proc Natl Acad Sci U S A. 1991 Jul 1;88(13):5897–5901. doi: 10.1073/pnas.88.13.5897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes A. L., Nei M. Pattern of nucleotide substitution at major histocompatibility complex class I loci reveals overdominant selection. Nature. 1988 Sep 8;335(6186):167–170. doi: 10.1038/335167a0. [DOI] [PubMed] [Google Scholar]
- Jeffreys A. J., Wilson V., Thein S. L. Hypervariable 'minisatellite' regions in human DNA. Nature. 1985 Mar 7;314(6006):67–73. doi: 10.1038/314067a0. [DOI] [PubMed] [Google Scholar]
- Kuhnlein U., Zadworny D., Dawe Y., Fairfull R. W., Gavora J. S. Assessment of inbreeding by DNA fingerprinting: development of a calibration curve using defined strains of chickens. Genetics. 1990 May;125(1):161–165. doi: 10.1093/genetics/125.1.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindberg P. G., Andersson L. Close association between DNA polymorphism of bovine major histocompatibility complex class I genes and serological BoLA-A specificities. Anim Genet. 1988;19(3):245–255. doi: 10.1111/j.1365-2052.1988.tb00813.x. [DOI] [PubMed] [Google Scholar]
- O'Brien S. J., Roelke M. E., Marker L., Newman A., Winkler C. A., Meltzer D., Colly L., Evermann J. F., Bush M., Wildt D. E. Genetic basis for species vulnerability in the cheetah. Science. 1985 Mar 22;227(4693):1428–1434. doi: 10.1126/science.2983425. [DOI] [PubMed] [Google Scholar]
- O'Brien S. J., Wildt D. E., Bush M., Caro T. M., FitzGibbon C., Aggundey I., Leakey R. E. East African cheetahs: evidence for two population bottlenecks? Proc Natl Acad Sci U S A. 1987 Jan;84(2):508–511. doi: 10.1073/pnas.84.2.508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'brien S. J., Wildt D. E., Goldman D., Merril C. R., Bush M. The cheetah is depauperate in genetic variation. Science. 1983 Jul 29;221(4609):459–462. doi: 10.1126/science.221.4609.459. [DOI] [PubMed] [Google Scholar]
- Ralls K., Brugger K., Ballou J. Inbreeding and juvenile mortality in small populations of ungulates. Science. 1979 Nov 30;206(4422):1101–1103. doi: 10.1126/science.493997. [DOI] [PubMed] [Google Scholar]
- Reeve H. K., Westneat D. F., Noon W. A., Sherman P. W., Aquadro C. F. DNA "fingerprinting" reveals high levels of inbreeding in colonies of the eusocial naked mole-rat. Proc Natl Acad Sci U S A. 1990 Apr;87(7):2496–2500. doi: 10.1073/pnas.87.7.2496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahata N., Nei M. Allelic genealogy under overdominant and frequency-dependent selection and polymorphism of major histocompatibility complex loci. Genetics. 1990 Apr;124(4):967–978. doi: 10.1093/genetics/124.4.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trowsdale J., Groves V., Arnason A. Limited MHC polymorphism in whales. Immunogenetics. 1989;29(1):19–24. doi: 10.1007/BF02341609. [DOI] [PubMed] [Google Scholar]
- Vassart G., Georges M., Monsieur R., Brocas H., Lequarre A. S., Christophe D. A sequence in M13 phage detects hypervariable minisatellites in human and animal DNA. Science. 1987 Feb 6;235(4789):683–684. doi: 10.1126/science.2880398. [DOI] [PubMed] [Google Scholar]
- Vrijenhoek R. C., Douglas M. E., Meffe G. K. Conservation genetics of endangered fish populations in Arizona. Science. 1985 Jul 26;229(4711):400–402. doi: 10.1126/science.229.4711.400. [DOI] [PubMed] [Google Scholar]
- Watkins D. I., Garber T. L., Chen Z. W., Toukatly G., Hughes A. L., Letvin N. L. Unusually limited nucleotide sequence variation of the expressed major histocompatibility complex class I genes of a New World primate species (Saguinus oedipus). Immunogenetics. 1991;33(2):79–89. doi: 10.1007/BF00210819. [DOI] [PubMed] [Google Scholar]
- Yuhki N., O'Brien S. J. DNA variation of the mammalian major histocompatibility complex reflects genomic diversity and population history. Proc Natl Acad Sci U S A. 1990 Jan;87(2):836–840. doi: 10.1073/pnas.87.2.836. [DOI] [PMC free article] [PubMed] [Google Scholar]