Abstract
Background
Calcium, as a second messenger, transduces extracellular signals into cellular reactions. A rise in cytosolic calcium concentration is one of the first plant responses after exposure to microbe-associated molecular patterns (MAMPs). We reported previously the isolation of Arabidopsis thaliana mutants with a “changed calcium elevation” (cce) response to flg22, a 22-amino-acid MAMP derived from bacterial flagellin.
Results
Here, we characterized the cce2 mutant and its weaker allelic mutant, cce3. Besides flg22, the mutants respond with a reduced calcium elevation to several other MAMPs and a plant endogenous peptide that is proteolytically processed from pre-pro-proteins during wounding. Downstream defense-related events such flg22-induced mitogen-activated protein kinase activation, accumulation of reactive oxygen species and growth arrest are also attenuated in cce2/cce3. By genetic mapping, next-generation sequencing and allelism assay, CCE2/CCE3 was identified to be ALG3 (Asparagine-linked glycosylation 3). This encodes the α-1,3-mannosyltransferase responsible for the first step of core oligosaccharide Glc3Man9GlcNAc2 glycan assembly on the endoplasmic reticulum (ER) luminal side. Complementation assays and glycan analysis in yeast alg3 mutant confirmed the reduced enzymatic function of the proteins encoded by the cce2/cce3 alleles – leading to accumulation of M5ER, the immature five mannose-containing oligosaccharide structure found in the ER. Proper protein glycosylation is required for ER/Golgi processing and trafficking of membrane proteins to the plasma membrane. Endoglycosidase H-insensitivity of flg22 receptor, FLS2, in the cce2/cce3 mutants suggests altered glycan structures in the receptor.
Conclusion
Proper glycosylation of MAMP receptors (or other exported proteins) is required for optimal responses to MAMPs and is important for immune signaling of host plants.
Electronic supplementary material
The online version of this article (doi:10.1186/s12870-016-0718-3) contains supplementary material, which is available to authorized users.
Keywords: Calcium, Immune signaling, Pattern recognition receptors (PRRs), N-glycosylation, Defense
Background
For optimal growth, plants need to fend off pathogen attacks. Besides preformed barriers, one of the initial resistance mechanisms relies on the sensing of conserved microbe- or pathogen-associated molecular patterns (MAMPs/PAMPs) or of endogenous plant molecules released during the infection process (so-called damage-associated molecular patterns, DAMPs). This activates signaling events to coordinate local and systemic defense responses and mount so-called pattern-triggered immunity (PTI) [1]. MAMPs recognized by plants include microbe cell wall components such as lipopolysaccharides (LPS) [2], peptidoglycan [3] or chitin [4–8] or bacterial proteins such as flagellin [9] or EF-Tu [10], while DAMPs could be extracellular ATP [11], oligogalacturonides released from pectin [12] or plant peptides known as PEPs [13].
Sensing of MAMPs/DAMPs is achieved after binding to their cognate pattern-recognition receptors (PRRs), which are typically surface-localized proteins with an extracellular ectodomain for ligand-binding, a single-pass transmembrane domain and in some cases, intracellular signaling domains (see below) [14]. Different ectodomains (such as LysM, leucine-rich repeats (LRR), lectin and epidermal growth factor (EGF)-like domains) correspond to the different classes of molecules they bind (for a comprehensive review see [15]). In plants, most of the LRR-type receptors recognize peptidic MAMPs, whereas known LysM-, lectin- and EGF-like domain containing receptors bind different (lipo)glycan structures [16]. PRRs with an intracellular kinase domain are designated as receptor-like kinases (RLKs) while those lacking such signaling domain are classified as receptor-like proteins (RLPs) [17]. Both RLKs and RLPs form heterocomplexes with co-receptors or adapter proteins for full induction of immune signaling. Examples of such protein partners include members of the SERK (somatic embryogenesis receptor kinase) family and/or SOBIR1 (suppressor of BIR1) [18]. For LysM-domain PRRs such as CERK1 (chitin elicitor receptor kinase 1), two interacting kinases, LIK1 [19] and LYK5 [20], regulate chitin signaling. Contrary to earlier work showing chitin-induced CERK1 dimerization to form the optimal chitin-binding pocket [21], LYK5-CERK1 heterodimer is now thought to be the major chitin receptor with a higher affinity than CERK1 homodimers [20].
Upon MAMP/DAMP binding to the receptors, cis and trans phosphorylation between the receptor complex components [9] and phosphorylation of other intracellular targets are required for immune responses. One class of intracellular phosphorylation targets of PRR complexes are the receptor-like cytoplasmic kinases (RLCKs) such as BIK1 (Botrytis-induced kinase 1) and other members of the BIK1/PBL (avrPphB sensitive 1-like) family such as PBL1, PBL2 and PBL5, as well as BSK1 (Brassinosteroid receptor-signaling kinase1) [22–25]. These RLCKs, which are released from the receptor complex upon phosphorylation, can in turn phosphorylate downstream substrates. The NADPH oxidase RBOHD (respiratory burst oxidase homolog D), which is the main source of apoplastic reactive oxygen species (ROS) upon MAMP perception [26, 27], has been recently described as a target of BIK1, directly linking pattern recognition with a defense and signaling output [28, 29]. RBOHD is also targeted by the calcium dependent protein kinase, CPK5 [30], thus showing an interplay of ROS, phosphorylation and calcium signaling.
As integral membrane proteins, PRRs are subjected to endoplasmic reticulum (ER)-based quality control and the relevance of asparagine-linked glycosylation (N-glycosylation) for receptor function and PTI has been shown [31–34]. N-Glycosylation, the transfer of oligosaccharide moieties onto asparagine residues of proteins targeted to the secretory pathway, is a conserved protein modification [35]. In eukaryotes, this is achieved co- and post-translationally by the oligosaccharyl transferase (OST) complex in the ER lumen, which transfers a pre-formed tetradecasaccharide (the so-called core oligosaccharide) to the nascent polypeptide as it is being transferred through the translation translocon. The structurally conserved core oligosaccharide is pre-assembled on a dolichol-pyrophosphate (Dol-PP) carrier at both faces of the ER membrane by a series of glycosyltransferases termed ALGs (Asparagine-Linked Glycosylation). For a summary of the pathway, please see Additional file 1: Figure S1. Synthesis starts at the cytoplasmic face of the membrane by the sequential transfer of N-acetylglucosamine (GlcNAc) and mannose (Man) residues to the growing glycan structure. After flipping through the membrane, the final synthesis steps are carried out at the luminal face of the ER membrane by ER-localized ALGs through the transfer of Man and glucose (Glc) residues. The finished glycan (Glc3Man9GlcNAc2) is then transferred en bloc to growing polypeptides by the OST complex [36]. N-Glycosylation affects biological functions of the modified protein; one of which is in the ER quality control of protein folding during the calnexin/calreticulin (CNX/CRT) cycle. Briefly, unfolded proteins undergo a cycle of glycan trimming or reglycosylation by ER glucosidases or UDP- Glucose:glycoprotein glucosyltransferase, respectively, facilitating binding or dissociation with either CNX or CRT, which are lectins specific for monoglucosylated core oligosaccharides. This is repeated until proteins are correctly folded, whereby a final glycan trimming step releases the protein from recognition by CNX and CRT and allows the protein to exit the ER to the Golgi complex. If the protein remains unfolded, it is targeted for ER-associated degradation (ERAD) [37]. Therefore, N-glycosylation is important for the quality control of all secreted and membrane proteins. In the case of MAMP signaling, plants with non-functional calreticulin 3 (CRT3) or OST subunits accumulate the EF-Tu-receptor EFR at lower levels than the wild type and are more susceptible to infection by Pseudomonas syringae pv. tomato (Pto) DC3000. Surprisingly, function and protein levels of the FLS2 receptor appear to be less affected in crt3 and ost mutants [32, 34, 38].
After PRR activation, ROS and nitric oxide generation, MAPK activation, as well as ion fluxes (including an increase in cellular calcium concentration) constitute the early signaling steps that coordinately control MAMP/DAMP signal transduction. The molecular identity of the plasma-membrane calcium channels responsive to MAMPs/DAMPs is still unclear [18]. Isolation of calcium signaling mutants has suggested a very close association between receptor complexes and calcium (in)flux-mediating channels [39]. Although no direct interaction has been shown to date, genetic and inhibitor-based studies suggest channels may be activated by phosphorylation through the PRR complex components or their downstream target(s) [24]. Alternatively, as was proposed for the Pep1 (DAMP) receptor(PEPR1), a guanylyl cyclase activity of PEPR1 generates cGMP to activate the CNGC2 cyclic nucleotide gated calcium channel [40]. The Arabidopsis Ca2+-ATPase ACA8, which regulates MAMP responses, directly interacts with the flagellin receptor, FLS2 [41]. These findings support the hypothesis of close association and tight integration of calcium signaling with the receptor complexes in plant immune signaling.
To elucidate the MAMP-mediated calcium signaling pathway, we previously described the isolation of several mutants with a changed calcium elevation (cce) phenotype using the flagellin-derived flg22 peptide. Besides several new fls2 and bak1 (BRI1 receptor associated receptor kinase 1) alleles, cce mutants with enhanced or reduced calcium elevations were identified [39]. In this work, we show that the allelic cce2 and cce3 mutants are caused by mutations in AtALG3 (Asparagine-Linked Glycosylation 3), thereby resulting in underglycosylation of several PRRs and compromised MAMP/DAMP signaling.
Results
Genetic analysis of the two changed calcium elevation mutants, cce2 and cce3
We tested three of the previously reported mutants with reduced calcium elevation after flg22 treatment [39] for allelism. The lack of complementation in the F1 seedlings of the crosses between the cce2 and cce3 mutants indicates that these are allelic. By contrast, the cce1 mutant complemented cce2 or cce3 and is mutated in a different gene (Fig. 1a). For the rest of this report, we will concentrate on the characterization of the CCE2/CCE3 locus.
Fig. 1.
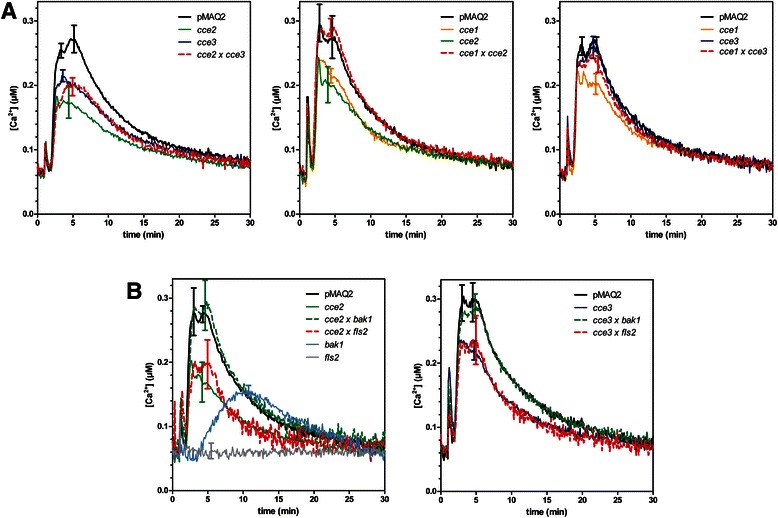
Allelic test of three cce mutants. a Flg22-induced calcium elevations were measured in 8-day-old seedlings of the F1 generation of the indicated crosses between the cce1, cce2 and cce3 mutants and compared to the parental lines. b cce2 or cce3 were crossed to bak1-4 or fls2 (SALK_062054) and flg22-induced calcium elevations determined as described above. Traces for the F1 crosses are always marked with broken lines. Error bars denote standard deviations (n > 10). pMAQ2 is the corresponding aequorin-expressing parental line of the cce mutants
Further crosses were made with bak1 or fls2 mutants to exclude that cce2/cce3 are weak mutant alleles of known flg22 signaling components that have been shown to affect calcium elevations. In the F1 crosses between bak1 and cce2 or cce3, the flg22-induced calcium elevation was restored to levels similar to that in the parental pMAQ2 line (Fig. 1b). By contrast, the cross with fls2 did not complement the cce2/cce3 phenotype. While an initial inference of this observation would be that cce2/cce3 is in fact mutated in FLS2, no mutations was detected in the sequence of a 4031 bp amplicon of the FLS2 gene from cce2/cce3 mutants [39]. In addition, the reduced calcium elevations in the cce2/cce3 mutants are also seen after treatment with other MAMPs/DAMPs such as elf18, chitin, LPS and AtPep1 (Fig. 2). This broad spectrum effect of the cce2/cce3 mutation to multiple MAMPs/DAMPs argues against a mutation in a specific PRR gene but for a common factor needed for receptor function. Also noteworthy from these measurements is that the reduction in MAMP/DAMP-induced calcium elevation is more pronounced for cce2 than cce3 (Fig. 2), indicating that cce3 is a weaker mutation compared to cce2.
Fig. 2.
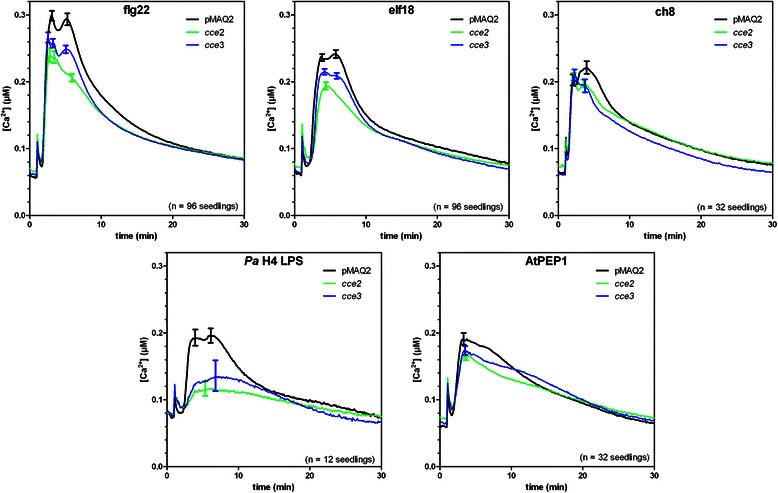
Calcium elevations induced by various MAMP/DAMPs. Eight-day-old seedlings were treated with 1 μM of the indicated peptide MAMPs, DAMP or chitin octamer (ch8) or 25 μg/ml lipopolysaccharide prepared from Pseudomonas aeruginosa strain H4 (Ps H4 LPS). Error bars depict the 95 % confidence intervals
Altered flg22-induced responses in the cce2/cce3 mutants
To test how other responses, in addition calcium elevations, are affected in the cce2/cce3 mutants, we looked at flg22-induced MAPK activation, reactive oxygen species (ROS) accumulation and growth arrest. Based on an immunoblot assay that detects phosphorylated (i.e. activated) forms of MAPKs, the flg22-induced MAPK activation is delayed and reduced in cce2 (Fig. 3a). For cce3, while there is little difference in the activated MAPK band intensities, there appears to be a marginal delay in the timing of MPK6 activation (that can be seen in three independent experiments). Flg22-stimulated ROS accumulation is another rapid plant response that can be measured within minutes after MAMP exposure. In agreement with the attenuated calcium response, the relative ROS accumulation is reduced in the cce2/cce3 mutants compared to the parental pMAQ2 line (Fig. 3b).
Fig. 3.
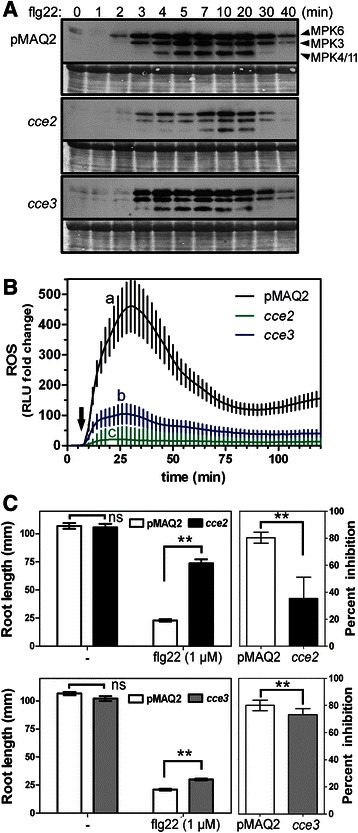
Some flg22-induced responses downstream of calcium transients are partially attenuated a Fourteen-day-old seedlings were collected at the indicated time points after flg22 (100 nM) treatment and proteins extracted for western blot analysis with an antibody that recognizes phosphorylated (i.e. activated forms of) MAPKs. The identities of the three MAPK bands are marked on the right. The experiment was repeated thrice with similar outcome. b Reactive oxygen species (ROS) accumulation was assessed in 24 leaf disks per genotype over 120 min. The basal luminescence (relative light units, RLU) of each disk was monitored for 6 min (before treatment) and used to normalize the fold change in RLU after flg22 treatment (marked with arrow). Error bars depict the 95 % confidence intervals. Statistical significance was analyzed using one-way ANOVA with Tukey’s multiple comparison post-test (The different alphabets denote groups that are considered statistically different, p < 0.05). c Growth inhibition of cce2/cce3 mutants compared to the pMAQ2 parental line. Seedlings were placed on agar media without (−) or containing flg22 (1 μM) and the root length measured after 14 days. Left panels show the absolute root lengths of a representative experiment (n = 22–23 seedlings). Right panels are the percent inhibition consolidated from 3 independently performed experiments (n = 67–69 seedlings). Statistical significance was analyzed using two-way ANOVA with Bonferroni post-test (** = p < 0.0001, ns = not significant)
Plants constantly exposed to MAMPs are typically stunted in growth. Based on root length differences of seedlings grown without or with flg22 (1 μM), it can be seen that the cce2 mutants are less inhibited than the pMAQ2 parents (Additional file 1: Figure S2). By comparison, the primary roots of the cce3 mutants are still strongly reduced in length when grown on flg22-containing media but are, nevertheless, significantly (p < 0.01) longer than the pMAQ2 parent. This growth arrest by flg22 is dependent on the presence of a functional FLS2 receptor [39]. A representative experiment of three independent experiments is shown in Fig. 3c (left panels). While there are variations in absolute root lengths between experiments, the percent growth inhibition consolidated from the three independent experiments (Fig. 3c, right panels) confirm that cce3 is less affected than the cce2 mutant. Taken together, all three tested flg22-induced reactions show attenuated responses in the cce2/cce3 mutants. Additionally, in agreement with the MAMP/DAMP-induced calcium elevation, the less dramatic impact of the cce3 allele supports the notion that cce3 is a weaker allele compared to cce2.
Mapping and next generation sequencing data reveal SNPs in At2g47760
To identify the CCE2 gene, the stronger cce2 mutant was crossed to the Arabidopsis accession Landsberg erecta (Ler-0). The mapping analysis of 44 F2 plants suggested that CCE2 is located on chromosome 2 between the INDEL marker CER452347 [42] at 18892029 bp (TAIR 10, 5 recombinants) and the end of the chromosome at 19698289 bp (TAIR 10). This region contains 233 gene loci. Illumina whole genome sequencing was performed with genomic DNA prepared from the cce2 mutant as well as from the pMAQ2 parental line. Comparison to the reference Col-0 genome revealed a total of 405 tentative single nucleotide polymorphisms (SNPs) in cce2, of which 77 SNPs led to missense mutations (Additional file 2: Table S1). Within the mapped region on chromosome 2, two SNPs (in gene loci At2g46060 and At2g47760) were detected, which could be re-confirmed by sequencing DNA amplicons encompassing the mutated regions from cce2. Since cce3 is allelic to cce2, the corresponding cce3 genomic region was also PCR-amplified and sequenced. No SNP was found for At2g46060 but a SNP was detected for the locus At2g47760 in the cce3 amplicons. At2g47760 encodes ASPARAGINE-LINKED GLYCOSYLATION 3 (ALG3), the dolichol pyrophosphate (Dol-PP)-mannose α-1,3-mannosyltransferase that catalyzes the first mannosylation of precursor glycans after flipping of the glycans from the cytosolic to the luminal side of the ER (See Additional file 1: Figure S1 for biosynthesis pathway) [43, 44].
The cce2 allele is a nonsense mutation that converts tryptophan 139 to a premature stop while the cce3 SNP leads to an A63V exchange. Semi-quantitative reverse-transcription-PCR showed that transcripts for the cce2/cce3-derived mRNAs were detectable; although, presumably due to nonsense-mediated mRNA decay, the expression in the cce2 background was much lower (Fig. 4b).
Fig. 4.
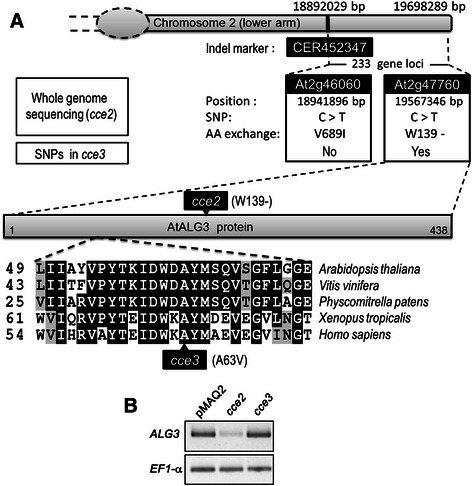
The cce2/cce3 mutants contain SNPs in the gene locus At2g47760 (ASPARAGINE-LINKED GLYCOSYLATION 3, ALG3). a Schematic representation summarizing the SNPs from whole genome re-sequencing of cce2 within the relevant lower arm of chromosome 2 identified by genetic mapping and amino acid (AA) exchanges in the predicted protein sequences of the cce2/cce3 alleles. CER452347 is the indel marker used for mapping the cce2 mutation onto the lower arm of chromosome 2. Sequence alignment of a segment of the ALG3 homologs from the indicated organisms is depicted to highlight that the amino acid exchange caused by the cce3 mutation lies in a conserved region. b Reverse-transcription-PCR analysis of the ALG3 transcripts. Elongation factor 1-α (EF1-α) was used as a loading control
Allelism assay by crossing with a T-DNA insertion mutant (alg3-T)
To confirm that the detected mutations in ALG3 are causal for the cce2/cce3 phenotype, crosses were made with a T-DNA alg3 insertion mutant, alg3-T [44]. The success of the cross was confirmed by PCR in the F1 generation and the calcium fluxes tested in the F2 generation. If ALG3 is not responsible for the reduced calcium signature in cce2/cce3, one would expect 75 % of the segregating F2 population to have a wild type signature. However, there is no complementation and the “calcium response” in the F2 population is reduced in amplitude compared to the pMAQ2 control (Fig. 5a). This suggests that alg3-T and cce2/cce3 are allelic. Furthermore, we genotyped F2 plants from the cce2 x alg3-T cross to identify aequorin-expressing plants that are homozygous for the alg3-T mutation and leaf disks from these plants were directly used for calcium measurements. As seen in Fig. 5b, flg22-induced calcium elevation in alg3-T is reduced compared to the pMAQ2 controls but not significantly different from the cce2 (1-way ANOVA, p < 0.01).
Fig. 5.
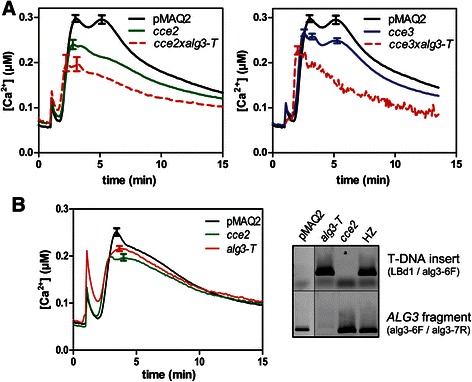
The cce2/cce3 mutants are allelic to ALG3. a A T-DNA insertion alg3 mutant (alg3-T, SALK_064006) was crossed to cce2 and cce3, successful crosses were validated by PCR in the F1, and the flg22-induced calcium response was monitored in the segregating F2 seedlings (marked with red broken lines). Error bars denote standard deviations (n =96 for pMAQ2, cce2 or cce3; n = 66 for cce2xalg3-T, n = 49 for cce3xalg3-T). The same pMAQ2 (“wild type”) reference calcium curve was used for both graphs. Calcium signatures were tested for statistical significance between genotypes by one-way ANOVA (with Tukey’s multiple comparison test, p < 0.05). b After identifying homozygous alg3-T individuals (from the cce2xalg3-T cross with the indicated primer pairs by PCR of genomic DNA, right panel), flg22-induced calcium response was monitored as above except leaf disks (Ø 4 mm) of 4-week-old plants were used (n = 96 for alg3-T; n =32 for pMAQ2 or cce2). Calcium elevations in cce2 and alg3-T are not different but are both statistically distinct from the pMAQ2 control (one-way ANOVA with Tukey’s multiple comparison test, p < 0.01)
The cce2/cce3 alleles fail to or only partially complement a yeast alg3 mutant
The A63V amino acid exchange in ALG3 protein encoded by the cce3 allele lies in a conserved region of ALG3 proteins of several taxa, ranging from lower/higher plants, amphibians to mammals (Fig. 4). However, alanine to valine is a “conservative” amino acid exchange. Thus, we were interested to see if this mutated variant is functional. Therefore, we cloned the ORFs from the cce2 and cce3 alleles and used them to complement a Saccharomyces cerevisiae alg3-deficient mutant strain (YG170) [44, 45]. These are under the control of a galactose-inducible promoter and reverse-transcription-PCR analysis showed that these are well expressed in the yeast cells (Fig. 6a, b). Under restrictive conditions (at 30 °C), the alg3-deficient strain (YG170) does not grow when transformed with an empty vector or with the cce2W139stop allele, but is able to grow when expressing wild type AtALG3 or the cce3A63V variant, albeit a little less growth is seen for the latter (Fig. 6c). When the galactose concentration used for inducing expression is lowered, reduced complementation is seen for cce3A63V (Fig. 6d). These results suggest that the ALG3A63V variant is enzymatically functional but is possibly less efficient.
Fig. 6.
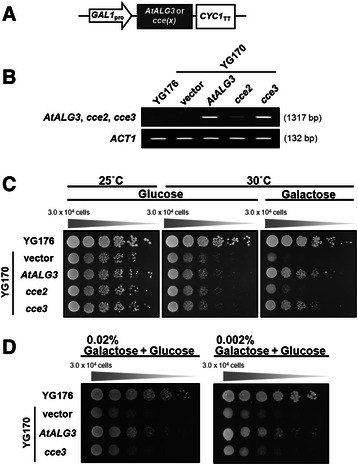
Complementation of yeast alg3-1 mutant strain (YG170) with the Arabidopsis ALG3 variants. The Arabidopsis ALG3 (AtALG3) open-reading frames (ORFs) were amplified from cDNAs of wild-type plants or the cce2/cce3 mutants, cloned into yeast expression vectors, and transformed into yeast strain YG170. a Scheme of expression vector constructs of the indicated ORFs under the control of a galactose-inducible promoter (GAL1pro) and a cytochrome c terminator (CYC1TT). b RT-PCR validation of gene expression from the transfected constructs. Actin 1 (ACT1) was used as an amplification/loading control. c Complementation of the alg3-1 mutant yeast strain YG170 was determined by comparing growth without or with galactose-induced expression of the indicated constructs under non-permissive temperature (30 °C). Growth at permissive temperature (25 °C) was used to show the viability of the yeast cells. Five serial dilutions were performed in all cases. d Complementation was further tested by reducing expression of the AtALG3 or the cce3 variant (by decreasing galactose concentrations from 2 % to 0.02/0.002 %)
Yeast glycan analysis confirms the enzymatic activity of ALG3
To confirm that the yeast growth is due to complementation of the alg3 deficiency in the yeast strain YG170, lipid-linked oligosaccharides were extracted from the various yeast strains grown in galactose-inducing media, hydrolyzed and labelled with 2-pyridylamine (PA) for glycan analysis. HPLC-based size fractionation showed distinct major peaks with retention times matching those of the Glc3M9 standards in the wild type (YG176) and the complemented strain. The alg3 mutant (YG170) and the cce2-complemented strains both have a major peak with retention time that corresponds to the 5-mannose-containing M5 standard. The cce3-complemented strain had both peaks (Fig. 7a).
Fig. 7.
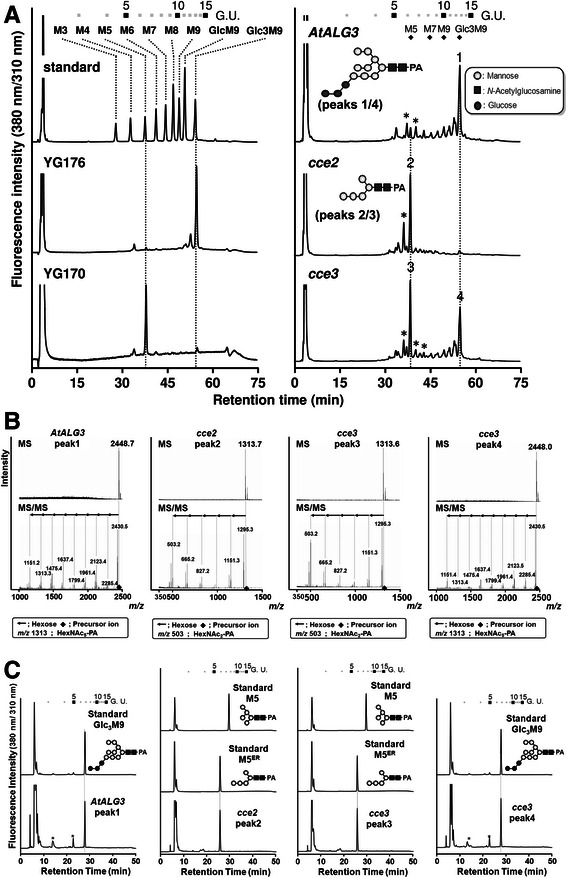
Analysis of dolichol-linked oligosaccharides. a HPLC-based size fractionation of 2-pyridylamine (PA)-labelled oligosaccharides after acid hydrolysis and purification of the lipid-linked oligosaccharides from the indicated yeast strains. Inserts depict the deduced structures of peaks 1/4 and peaks 2/3 (as described below in b and c). b Structure determination by LC-MS/MS of peaks 1–4. c RP-HPLC comparison of peaks 1–4 to known oligosaccharide standards. Symbols depicting oligosaccharide structures are as defined in A. *For A and C, low levels of contaminants in the HPLC traces are marked with asterisks. (Abbreviations: G.U. = Glucose oligomer unit, Glc = Glucose, GlcNAc = N-Acetylglucosamine)
Four samples (peak1 from the AtALG3 complemented strain; peak2 from the cce2-complemented strain; and peaks 3 and 4 from the cce3-complemented strain; see Fig. 7a) were collected. Structural determination by LC-MS/MS (Fig. 7b) indicated peaks1/4 to be Glc3Man9GlcNAc2-PA (or Hex12HexNAc2) while peaks 2/3 to be M5ER (i.e. the 5-mannose-containing Man5GlcNAc2-PA substrate of ALG3 found in the ER; or Hex5-PA). When rechromatographed by reverse-phase-HPLC, the elution time of peak 1 matched the Glc3M9 PA-sugar standard while peaks 2/3 differed from the M5 PA-sugar standard, which is the form typically generated in the eukaryotic Golgi apparatus, but co-migrated with the M5ER form (Fig. 7c). These findings confirm that the complemented AtALG3 or the ALG3A63V variants are indeed ER-localized α-1,3-mannosyltransferases in vivo. The cce3A63V-complemented yeast contained a mixture of both Glc3M9GlcNAc2 and M5ER forms, suggesting that the ALG3A63V protein has reduced enzyme activity or alternatively, it may be partially mis-localized in the yeast cells.
Various plant pattern recognition receptors are underglycosylated but appear to be still targeted to the plasma membrane
Since glycosylation is important for the ER quality control of PRRs [31, 32, 34], we analyzed the PRR proteins in the cce2/cce3 mutants by transiently expressing YFP/GFP-tagged PRRs in Arabidopsis protoplasts. As expected for compromised N-glycosylation, a higher mobility (in SDS-PAGE) of the tagged FLS2, EFR, CERK1, LORE (putative LPS receptor) and PEPR1 was observed when expressed in the cce2/cce3 background as compared to the parental pMAQ2 (Fig. 8a). The respective MAMPs/DAMPs all elicited a reduced calcium response in the cce2/cce3 background (Fig. 2). To know if this may be due to altered PRR localization, confocal microscopy was used to image the YFP/GFP-tagged PRRs. In agreement to plasma membrane localization, most of the YFP/GFP signals were at the periphery, and in some cases, in “patchy” spots along the membrane, of the protoplasts. No obvious difference in localization was seen between the mutants and the pMAQ2 background (Fig. 8b).
Fig. 8.
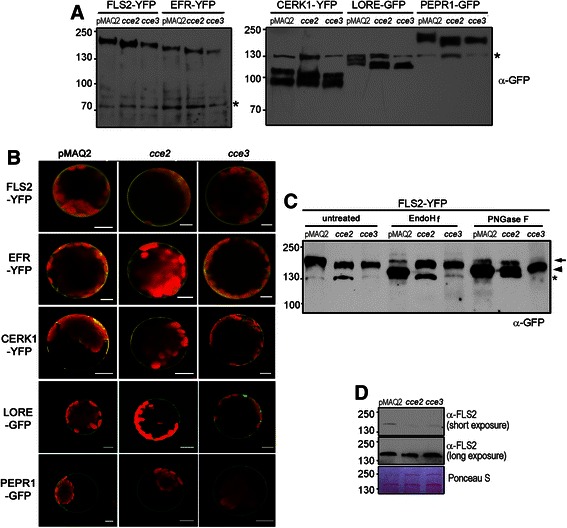
Glycosylation and localization of PRRs in the cce2/cce3 mutants. a Plasmid constructs for expressing p35S::PRR-YFP/GFP fusions were transfected into protoplasts isolated from the indicated genotypes and analyzed by western blotting with an α-GFP antibody. The asterisks mark unspecific bands recognized by the GFP antibody, which was used as an internal loading control. Note that different buffers (see methods section) were used for protein extraction in the right or left panels. Numbers on the left are the molecular size markers in kDa. b Confocal microscopical images of the GFP- or YFP- tagged PRRs in the pMAQ2 parental, cce2 or cce3 background. Arabidopsis protoplasts were transiently transfected with plasmids for expressing the indicated PRRs. Scale bars represent 10 μm. c FLS2-GFP was expressed in protoplasts as described above in A, and extracted membrane proteins were subjected to endoglycosidase H (EndoHf) or peptide:N-glycosidase F (PNGase F) digestion. An arrow marks the weak glycosidase-resistant FLS-YFP bands while the arrowhead marks the mobility shift of the digested proteins. In the cce mutants, Endo Hf-resistance results from altered N-glycan structure upon blockage of the ALG3 step in the pathway (see Additional file 1: Figure S1). In the PNGase-F digests, remaining weak bands of FLS2-YFP in both wildtype (pMAQ2) and cce lanes may be explained by about 10 % core fucose-decorated (and thus PNGase F-resistant) N-glycans with terminal GlcNAc residues (see Table II of Kajiura et al. [44]), indicative of passage through the Golgi apparatus after correct folding of FLS2-YFP in the ER. Note that the ~130 kDa bands (marked with asterisk) are unspecific signals that appear with the anti-GFP antibody. d Endogenous FLS2 levels in the cce2/cce3 mutants were compared to the pMAQ2 parental line. Western blotting (α-FLS2) was used to visualize FLS2 levels in microsomal proteins prepared from 8-day-old seedlings
To check if the PRRs are indeed glycosylated, we treated the membrane protein extracts of the transfected protoplasts with peptide-N-glycosidase F (PNGase F) to remove the N-glycan chains directly from the asparagine. FLS2 was used as a representative PRR for our assay. Most of the FLS2-YFP were digested and showed a mobility shift after PNGase F treatment for all the tested genotypes. This indicates that the FLS2 protein is N-glycosylated. By contrast, when treated with endoglycosidase H (EndoH) that cleaves the β-1,4-bond between the two GlcNAc moieties of the N,N’ diacetylchitobiose core linked to the modified asparagine residue, a mobility shift was seen for the FLS2-YFP expressed in the pMAQ2 but not for the FLS2-YFP expressed in cce2/cce3 background (Fig. 8c). Hence, the resistance to EndoH digestion shows that the N-glycan trees on FLS2-YFP occurring in the cce2/cce3 background have a different structure from that in the parental pMAQ2 background.
To exclude misinterpretation due to overexpression used for most of the above assays, we used anti-FLS2 to look at endogenous FLS2 levels in microsomal membrane protein preparations. While not as obvious as the YFP-tagged FLS2, a slight difference in mobility of the FLS2 band can be seen in the cce2/cce3 background. Furthermore, FLS2 protein levels were slightly reduced in the cce2/cce3 mutants (Fig. 8d). Longer exposure of the western blot revealed that substantial FLS2 is still expressed in the mutants, which would explain why these mutants have weakened response but are generally still responsive to flg22 (Fig. 2). Taken together, altered glycosylation led to a marginal reduction of FLS2 levels and presumably the attenuated responses to flg22. The same situation can be assumed for all the PRRs examined in this study, albeit it remains to be tested if altered glycosylation influences ligand binding in each individual case.
MAMP-induced resistance is not affected in the alg3 mutants
Since altered glycosylation of PRRs is reflected by a reduced calcium response, we tested if MAMP-induced resistance to pathogens is affected. Flg22 pretreatment activates resistance to subsequent infection, leading to reduced growth of pathogens e.g. Pseudomonas syringae pv. tomato (Pto) DC3000 [46]. As seen in Fig. 9, this PTI response is maintained in the cce2, cce3 and alg3-T mutants, as well as in the salicylic acid deficient sid2 mutant [47], which was used as a reference for hypersusceptibility. Hence, in line with the calcium measurements showing reduced but still substantial responsiveness to MAMPs (Fig. 2), we may conclude that sufficient functional FLS2 (see Fig. 8d) is present to trigger the MAMP-induced resistance pathway. In addition to the induced resistance, note that the basal resistance levels between the three alg3 mutants are different. While the cce3 and alg3-T mutants (like sid2) supported more bacterial growth, the cce2 plants are more resistant (see Additional file 1: Figure S3). These variations in susceptibility to Pto DC3000 may be due to additional mutations found in each mutant, especially for the EMS mutagenized cce mutants, and are unlikely to be linked to the alg3 mutation. Taken together, while the basal resistance levels differ between the tested alg3 alleles (due to still unknown reasons), the MAMP-induced resistance is mostly intact.
Fig. 9.
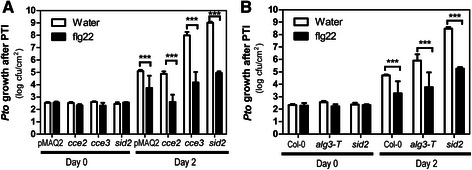
Pattern triggered immunity is not strongly affected in the cce2, cce3 or alg-T mutants. a, b For the pattern-triggered immunity (PTI) assay, plants were infiltrated with 1 μM flg22 for 24 h before syringe-inoculation with Pto DC3000 (at 10−5 cfu/ml). Error bars depict standard errors (n = 6). Two-way ANOVA (with Bonferroni post-test) was used to evaluate statistical significance (*** = p < 0.001). The sid2 mutant [47] was included as a hypersusceptible control but note that PTI appears to be intact in this mutant
Discussion
In this study, we show that CCE2/CCE3 encodes ALG3, the α-1,3-mannosyltransferase that catalyzes the first mannosylation of Man5GlcNAc2-PP-Dol (M5ER) glycan precursors within the ER after the flipping of the glycan precursor from the cytosol to the ER luminal side [43–45]. In wild type plants, the resulting Man6GlcNAc2-PP-Dol is further mannosylated by two other ER-luminal mannosyltransferases, ALG9 and ALG12, to form the nine-mannose-containing Man9GlcNAc2-PP-Dol. The fully assembled glycan, Glc3Man9GlcNAc2, is formed subsequently through consecutive triglucosylation by ALG6, ALG8 and ALG10 glucosyltransferases, respectively (For a summary of the lipid-linked oligosaccharide biosynthesis pathway, see Additional file 1: Figure S1). With the aid of oligosaccharide transferase, the glycan is transferred to polypeptides for co- and post-translational N-glycosylation, which can affect folding and modulate the biological function of the proteins [48]. For instance, association with CNX or CRT lectins is important for the folding of nascent polypeptides and ER-quality control processes where ER-associated degradation (ERAD) pathway is activated if incomplete or misfolded glycoproteins are formed [49].
The cce2 and cce3 alleles lead to a premature stop (W139-) and an A63V exchange, respectively. While the shortened ALG3W139- is non-functional, the ALG3A63V variant can partially replace the missing function in alg3 yeast but is apparently less efficient since it leads to the production of a mixture of fully assembled Glc3M9GlcNAc2 and M5ER forms in the complemented yeast cells. Alternatively, since ALG3 is ER-localized in both yeast and Arabidopsis [43] where its action takes place, it is also possible that some of the ALG3A63V may be partially mis-localized. Previous N-glycan profiling of total proteins from Arabidopsis alg3 mutants showed rare N-glycan structures that are typically not detected; these include M3, M4ER, M5ER and GlcM5ER (see Additional file 1: Figure S1 for structures). There are also, overall, lower levels of complex-type N-glycan than in wild type Col-0 plants. Surprisingly, despite protein N-glycosylation differences compared with wild type, alg3 showed no obvious phenotype under normal and high temperature or salt/osmotic stress conditions [44]. In an independent report where no high-mannose-type glycoproteins was detected in another alg3 mutant (alg3-2, SALK_040296), no growth phenotype under normal growth conditions was also observed. However, the glycosylation abnormalities resulted in activation of marker genes diagnostic of an unfolded protein response. [43]. These results indicate that while AtALG3 is critical for mature N-glycosylation of proteins, it is not essential for cell viability and growth in Arabidopsis but may perhaps affect certain stress related processes. In accordance, our cce2/cce3 mutants show normal growth (albeit the cce2 is a little smaller in rosette size, not shown) but are compromised in the calcium response to multiple MAMPs/DAMPs. However, PTI resistance to subsequent bacterial infection is intact in the cce2/cce3/alg3-T mutants (Fig. 9). Alternatively, since some level of complex-type N-glycans is detectable in the alg3 mutants, compensatory pathways to reduce impact of glycosylation defects probably exist in plants. One possibility is that after transport into the Golgi, the two α-1,2-mannose residues of M5ER may be cleavable by α-1,2-mannosidase I; and the resultant M3 structure may be subsequently processed by N-acetylglucosaminyltransferase I (GnTI) and enable eventual maturation of plant-specific N-glycans in the Golgi apparatus [44]. Since GnTI has a 20-fold lower affinity for M3 compared to M5 [50], this reaction is feasible but perhaps less efficient. This “salvage” mechanism is indicative of the importance of proper N-glycosylation of proteins and hence viability of the organism.
In humans, N-glycosylation of the extracellular domain (ECD) of G protein-coupled receptors is necessary for functions ranging from agonist binding, folding, maturation, stability, to internalization [51]. Our work here showed that several PRRs are underglycosylated (Fig. 8) and contribute to attenuated MAMP/DAMP-induced calcium responses in the cce2/cce3 (alg3) mutants (Fig. 2). In the human example mentioned above, the reduced localization of underglycosylated G-protein receptors to the cell surface may be, in part, compensated by the presence of receptor accessory proteins [51]. Whether members of the BAK1/SERK family that associate with FLS2 (or EFR or PEPR1) receptors may diminish the effects of underglycosylated plant PRRs remains to be tested. However, based on the failure to co-immunoprecipitate the ECDs of EFR and BAK1 after mutagenesis of a predicted N-glycosylation site (N590Q/S592T) in EFR, this does not seem to be the case – at least for EFR [52]. In another study where another conserved N-glycosylation site in the EFR ECD was mutated, this EFRN143Q variant accumulated at reduced levels, lost the ability to bind elf18 ligand and the corresponding transgenic plants lacked elf18-elicited oxidative burst. This would explain why, despite EFR being correctly targeted to the plasma membrane, EFR function is impaired in stt3a mutants (encoding the STT3A subunit of the ER resident oligosaccharyltransferase, OST, complex) [31]. Glycosylation sites within the binding pocket may contribute to ligand binding in some cases (such as EFR) [31]. FLS2, on the other hand, tolerated mild underglycosylation occurring in stt3a mutants. These findings are corroborated in recent work using mutants of other OST subunits, ost3 or ost6, where MAMP responses of EFR are more severely affected than the heavily glycosylated FLS2 [38]. Thus, distinct PRRs may be differentially affected by altered glycosylation. By contrast, we did not observe dramatic differences between the calcium responses to flg22 or elf18 in our cce2/cce3 mutants, but this may be because ALG3 acts in an earlier step than the OST subunits of the N-glycosylation pathway (Additional file 1: Figure S1). Hence the difference may be PRRs modified with smaller glycan structures versus complete underglycosylation, respectively.
To the best of our knowledge, our work on ALG3 is the first report of such an early step of N-glycosylation on PRRs’ role in plant immunity signaling, while other studies are based on the later OST oligosaccharide transferase step. For plants, ALGs have, so far, mostly been implicated in growth and development. In EMS-mutagenesis screens for suppressor of the brassinosteroid bri1-9 receptor dwarf mutant, ASPARAGINE-LINKED GLYCOSYLATION-9 and −12 (ALG9/ALG12) were isolated [53, 54]. These catalyze the two steps, after ALG3, of mannoysl-transfer to Man6GlcNAc2-PP-Dol in the ER (see Additional file 1: Figure S1). The bri1-9 phenotype results from ERAD-mediated removal of the brassinosteroid receptors. Altered glycosylation caused by the alg9 or alg12 mutations affects ER quality control and ERAD of BRI1, thereby allowing – despite the bri1-9 background – the accumulation of sufficient BRI1 receptors for brassinosteroid signaling. Surprisingly, although occurring in the same linear pathway, introducing the alg3 mutation has the opposite effect (i.e. causing reduced accumulation) on BRI1-9 abundance [54]. Likewise, we see a marginal reduction of FLS2 levels in our cce2/cce3 (alg3) mutants.
As discussed above, FLS2 may be more tolerant of alteration in glycosylation [38] and perhaps various PRRs are differentially affected. In our quantitative calcium measurements, the response to LPS seems proportionately more affected than the response to the other tested MAMPs/DAMPs in the cce2/cce3 mutants (Fig. 2). The ECD of the putative LPS receptor, LORE [2], contains a B-type lectin-like S-domain that belongs to the S-locus domain initially described from gametophytic “Self-Incompatibility (SI)” locus receptors. Like the S-RNases associated with SI, which are known to be glycosylated within the putative recognition sites, it is likely that the LORE ECD may also be glycosylated. Accordingly, we see an altered mobility of the LORE-GFP fusion protein in our cce2/cce3 mutants (Fig. 8a). N-glycosylation ensures the proper and efficient subcellular trafficking of S-locus receptors to the plasma membrane [55]. However, LORE localization to the plasma membrane appears to be unaffected in the cce2/cce3 mutants (albeit this is based on overexpression through the strong 35S promoter). Future studies may reveal if altered glycosylation of LORE may affect affinity for its ligand but unfortunately, LPS binding assay is still a challenging task currently.
Besides the host, N-glycosylation is also important for pathogens. Mutation of ALG3 in the rice blast fungus Magnaporthe oryzae causes underglycosylation and reduces the stability and chitin binding activity of the secreted SLP1 effector protein. SLP1’s inhibitory effect on chitin recognition by the rice chitin elicitor binding protein, CEBiP, is attenuated, resulting in a stronger immune response induction than that caused by wild type fungus. Consequently, the M. oryzae alg3 mutant is less virulent [56]. We show here that ALG3 is important for proper PRR glycosylation and compromises plant immunity-related signaling such as calcium fluxes. Interestingly, N-glycosylation directly affects the surface expression and function of low-voltage-activated T-type calcium channels and control neuron excitability during glucose stimulation [57]. Since most membrane proteins will have to pass through the secretory pathway to target the plasma membrane, it is tempting to speculate that the still elusive plant calcium channel(s) responsive to MAMPs/DAMPs might perhaps also be glycosylated and affected by the alg3 mutation. Finally, it should be mentioned that in animals/humans, N-glycosylation defects are often associated with diseases and especially congenital disorder [58]. Hence, N-glycosylation is highly conserved in eukaryotes. The lack of dramatic phenotypes for plant glycosylation mutants may be due to compensatory pathways to repair any deleterious effects of this important co- and post-translational protein modification pathway.
Conclusions
The calcium signaling mutants, cce2/cce3 are affected in ALG3 (Asparagine-linked glycosylation 3), encoding the α-1,3-mannosyltransferase responsible for the first step of core oligosaccharide glycan assembly in the E.R. Defective glycosylation of exported proteins (e.g. MAMP receptors) in alg3 mutants compromises immunity responses to MAMPs in plants. On the pathogen side, it has also been reported to affect virulence of fungal pathogens.
Methods
Plant material and growth
The alg3-T (SALK_064006, insertion in exon 6) and alg3-T2 (SALK_046061, insertion in exon 3) lines have been described previously [44]. For calcium and MAPK assays, seeds were surface-sterilized, stratified at 4 °C for >2 d and grown in liquid MS under long day conditions (16 h light, 8 h dark cycles) as described [39]. For adult plants, seedlings were transferred to soil and grown in climate chambers under short day conditions (8 h light, 16 h dark cycles) at 22 °C.
Mapping and identification of the mutation
The cce2 mutant was crossed to the Arabidopsis accession Landsberg erecta (Ler-0) and mapping performed as described [24]. Genomic DNA was prepared from young flower buds and sent to GATC Biotech (Constance, Germany) for Illumina whole genome sequencing.
Putative SNPs in cce2 were validated by amplifying the corresponding genomic fragments from cce2, cce3 and pMAQ2 plants with the primers listed in Additional file 3: Table S2. The amplified fragments were sent for Sanger sequencing to GATC Biotech. Sequence alignments were performed with BioEdit (Tom Hall, Ibis Biosciences, Carlsbad, California, USA).
Yeast complementation assays
The following yeast strains were used in this study: YG170 (MATα ade2-101 ade3 ura3-52 his3 alg3-1 stt3-3) and YG176 (MATa ade2-101 ade3 ura3-52 his3Δ200 leu2 tyr1 stt3-3) [59]. The ALG3 ORF was amplified from cDNA prepared from pMAQ2, cce2 or cce3 plants (using primers listed in Additional file 3: Table S2) and cloned into BamH I/Xho I-digested pYES2 vector (Invitrogen, Carlsbad, CA). All clones were verified by sequencing. The resulting pYES clones were introduced into YG170 strain by electroporation and complementation analysis carried out as described previously [44].
Glycan analysis
Extraction of lipid-linked glycans from yeast, 2-pyridylamine labelling, size-fractionation HPLC and MS/MS analysis were performed as described previously [44].
Protein extraction and western blotting
For protein extraction from mature plants or seedlings, frozen leaf discs or seedlings were homogenized in a Precellys 24 device (Bertin Technologies, Montigny-le-Bretonneux, France) in the presence of ~10–15 Zircosil® micro-milling beads (Ø1.2–1.7 mm) (Mühlmeier Mahltechnik, Bärnau, Germany) at 6500 rpm for 20 s. If necessary, the procedure was repeated until the tissues were thoroughly homogenized and to prevent premature thawing, samples were kept in liquid N2 between homogenization rounds. The transiently transformed protoplasts (in 1.5 ml tubes) were harvested by short pulse centrifugation in a Mikro120 microcentrifuge (Hettich, Tuttlingen, Germany), the incubation buffer was removed and the pellet was snap-frozen in liquid N2. Total protein was extracted from protoplasts or ground tissues by thawing the samples in 50 μL of extraction buffer (either Buffer A: 25 mM Tris–HCl pH 8.0, 150 mM NaCl, 1 % (v/v) 2-[2-[4-(2,4,4-trimethylpentan-2-yl)phenoxy]ethoxy]ethanol (Nonidet P-40), 1 % (v/v) Serva Protease Inhibitor mix P or Buffer B: 50 mM Tris pH 9.6, 100 mM NaCl, 20 mM DTT, 1 % (v/v) Serva protease inhibitor mix P, 0.1 % Triton X-100 and 0.1 % SDS) under constant vortexing for 15 min. Insoluble debris was pelleted by centrifugation at 20.000 x g for 10 min at 4 °C. Proteins were separated on 8 % SDS-PAGE gels and transferred onto PVDF membranes (Carl Roth, Karlsruhe, Germany). The blots were incubated with α-GFP (Life Technologies, Carlsbad, USA, 1:2000 dilution) in case of GFP- and YFP-tagged proteins or with α-FLS2 [60] for endogenous untagged FLS2 and the immune-decorated proteins detected with goat-anti-rabbit peroxidase-conjugated antibodies (Life Technologies, Carlsbad, USA) and enhanced chemiluminescence (ECL Prime, GE Healthcare, Freiburg, Germany).
Deglycosylation assays
Total protein was extracted from protoplasts expressing FLS2-YFP as described above. Extracted proteins were boiled at 100 °C for 10 min in 1 x glycoprotein denaturing buffer (i.e. 0.5 % SDS, 40 mM DTT). Samples were allowed to cool to room temperature and 10 μL of each sample were either treated with 1 μL Peptide: N-glycosidase F in G7 reaction buffer (50 mM sodium phosphate pH 7.5, 1 % NP-40) or with 1 μL Endo Hf (Endoglycosidase H fused to maltose binding protein) in G5 reaction buffer (50 mM sodium citrate pH 5.5). Glycosidase treatment was performed for 1 h at 37 °C. All buffers and enzymes were obtained from New England Biolabs, Ipswich, Massachusetts, USA.
Protoplast transfection and confocal microscopy
Protoplasts were isolated from 5-week-old plants from the pMAQ2/cce1/cce2/cce3 genotype as described [61]. Protoplasts were transformed with pXCSG-FLS2-YFP, pXCSG-EFR-YFP [31], pAMPAT-PEPR-GFP, pUBC-CERK1-GFP (which was generated by LR-cloning recombination using a pDONR-CERK1 clone provided by Yusuke Saijo) or pGGInA-224C-LORE-eGFP (plasmid for 35S promoter-mediated expression of LORE-eGFP fusion; S.R., unpublished) [2]. After an overnight incubation to allow protein expression, microscopy of transformed protoplasts was performed with a LSM710 confocal laser-scanning microscope (Carl Zeiss, Jena, Germany). GFP signals were detected by excitation with a 488 nm argon laser and YFP molecules with a 514 nm argon laser. Images were acquired with the Zen 2012 software (Carl Zeiss) and processed with ImageJ software (National Institutes of Health, Bethesda, Maryland, USA) equipped with the Fiji plugin bundle [62] and the Biovoxxel toolbox (Jan Borcher/Biovoxxel; www.biovoxxel.de).
Plant assays to assess responses to MAMPs (ROS, growth inhibition and pathoassays)
Detection of early MAMP-triggered responses such as MAPK activation and reactive oxygen species (ROS) accumulation and growth inhibition assays were performed as previously described [27].
Availability of data
The datasets supporting the conclusions of this article are available in the LabArchives repository (https://mynotebook.labarchives.com/share/JLee%2520IPB%2520lab/MzUuMXwxNDcxNTUvMjcvVHJlZU5vZGUvMjY2OTg4MzE2OHw4OS4x) or DOI: 10.6070/H4RX9932. This includes (1) the numerical datasets used to plot Figs. 1, 2, 3, 4, 5, 6, 7, 8 and 9 and (2) the SNP/Indel lists generated from the Illumina Whole Genome Re-sequencing Data.
Acknowledgements
We thank the following for providing material: Birgit Kemmerling (bak1-4 and fls2 mutants), Marc Knight (apoaequorin transgenic line pMAQ2), Silke Robatzek (pXCSG-FLS2-YFP and pXCSG-EFR-YFP plasmids) and Yusuke Saijo (pAMPAT-PEPR-GFP and pDONR-CERK1 plasmids). For evaluating the whole genome sequencing data, Christian Ullrich (from the group of Marcel Quint, IPB) wrote the R-script used in assigning SNPs to gene loci and amino acid exchange. This work is supported by the German Research Foundation through Priority Program SPP1212 (to JL and DS), Collaborative Research Centre SFB648/TP-B1 “Molecular mechanisms of information processing in plants” (to JL and DS) and SFB924/TP-B10 “Molecular mechanisms regulating yield and yield stability in plants” to S.R.
Abbreviations
- ACA8
arabidopsis-autoinhibited Ca2+-ATPase, isoform 8
- ACT1
actin 1
- ALG
asparagine-linked glycosylation
- ALG3
asparagine-linked glycosylation 3
- BAK1
BRI1-associated receptor kinase 1
- BIR1
BAK1-interacting receptor-like kinase 1
- BRI1
brassinosteroid insensitive 1
- BSK1
brassinosteroid receptor-signaling kinase1
- cce
changed calcium elevation
- CERK1
chitin elicitor receptor kinase 1
- CNGC2
cyclic nucleotide gated channel 2
- CNX
calnexin
- CPK5
calcium-dependent protein kinase 5
- CRT
calreticulin
- CYC1TT
cytochrome c terminator
- DAMP
damage-associated molecular pattern
- Dol-PP
dolichol-pyrophosphate
- DPM1
dolichol phosphate mannose synthase 1
- ECD
extracellular domain
- ECL
enhanced chemiluminescence
- EF1-α
elongation factor 1-α
- EFR
EF-TU receptor
- EF-Tu
elongation factor thermo unstable
- EGF
epidermal growth factor
- elf18
18-residue peptide from EF-TU
- EMS
ethane methylsulfonate
- Endo H
endoglycosidase H
- ERAD
ER-associated degradation
- flg22
22-residue peptide from flagellin
- FLS2
flagellin sensitive 2
- G.U.
glucose oligomer unit
- GAL1pro
galactose-inducible yeast promotor
- GFP
green fluorescent protein
- Glc
glucose
- GlcM5ER
unusual glycan structure, monoglucosylated M5ER
- GlcN
glucosamine
- GlcNAc
N-Acetylglucosamine
- GnTI
N-acetylglucosaminyltransferase
- HexNAc
hexose-acetylgucosamine structure
- LIK1
LysM RLK1-interacting kinase 1
- LPS
lipopolysaccharide
- LRR
leucine-rich repeat
- LYK5
LysM-containing receptor-like kinase 5
- LysM
lysin motif
- M3
glycan synthesis intermediate with 3 mannose residues, product of ALG2
- M4ER
glycan synthesis intermediate with 4 mannose residues, product of ALG11
- M5
trimmed glycan tree with 5 mannose residues, reaction catalyzed in Golgi apparatus
- M5ER
glycan synthesis intermediate with 5 mannose residues, product of ALG11
- MAMP
microbe-associated molecular pattern
- Man
mannose
- MAPK/MPK
mitogen activated protein kinase
- MS
murashige and skoog
- OST
oligosaccharyltransferase
- PA
2-pyridylamine
- PAMP
pathogen-associated molecular pattern
- PBL
avrPphB sensitive 1-like
- PEP
Arabidopsis thaliana peptide
- PEPR1
PEP1 receptor 1
- PNGaseF
peptide-N-glycosidase F
- PRR
pattern recognition receptor
- Ps H4
Pseudomonas aeruginosa strain H4
- PTI
pattern-triggered immunity
- Pto
Pseudomonas syringae pathovar tomato
- pv
pathovar
- RBOHD
respiratory burst oxidase homolog D
- RLCK
receptor-like cytoplasmic kinase
- RLK
receptor-like kinase
- RLP
receptor-like protein
- RLU
relative light unit
- ROS
reactive oxygen species
- SEC59
secretory 59
- SERK
somatic embryogenesis receptor kinase
- SI
self incompatibility
- sid2
salicylic acid induction deficient 2
- SLP1
secreted lysM protein1
- SNP
single nucleotide polymorphism
- SOBIR1
suppressor of BIR1
- S-RNAse
S-locus encoded ribonuclease
- STT3A
staurosporin and temperature sensitive 3-like A
- YFP
yellow fluorescent protein
- α-FLS2
anti-FLS2
- α-GFP
anti-GFP
Additional files
N-linked glycosylation model in yeast (A) and the N-linked glycan structures mentioned in the text (B). (A) Model is redrawn after Aebi [35] and Kajiura et al. [44]; and the Saccharomyces cerevisiae names of the enzymes or subunits are used. (B) The glycan structures mentioned in the text are schematically drawn using the same codes depicted in (A) above, which are based on the recommended Consortium for Functional Glycomics glycan structure nomenclature (http://www.functionalglycomics.org/static/consortium/Nomenclature.shtml). Figure S2: MAMP-induced seedling arrest assay. Representative photos showing root lengths of Arabidopsis seedlings grown on agar plates with or without flg22 (1 μM) for 14 days. Figure S3: Resistance to Pseudomonas syringae pv. tomato DC3000 (Pto DC3000) in the cce2, cce3 or alg3-T mutants. A representative experiment (of three independently performed experiments) showing quantification of Pto DC3000 colony forming units (cfu) in leaf tissues after spray inoculation. Different alphabets indicate the distinct statistical groupings according to one-way ANOVA with Bonferroni’s multiple comparison test (p < 0.05). The hypersusceptible sid2 mutant [47] was used as a control. (PDF 416 kb)
List of single nucleotide polymorphisms (SNPs) found in the cce2 mutant after Illumina-based genome resequencing. (XLSX 163 kb)
Primers used in this work. (PDF 94 kb)
Footnotes
Competing interests
The author(s) declare(s) that they have no competing interests.
Authors’ contributions
FT, SR and JG characterized the cce mutants, performed allelism tests, the next generation sequencing analysis and mapped the gene together with LW. HK and KF performed the yeast complementation and glycan analysis. CZ provided material essential for this work. CZ, DS, KF and JL supervised the work, conceived of the study, participated in its design, coordination and helped to draft the manuscript. All authors have read and approved the manuscript.
Contributor Information
Fabian Trempel, Email: Fabian.Trempel@ipb-halle.de.
Hiroyuki Kajiura, Email: kajiura@hitzbio.com.
Stefanie Ranf, Email: ranf@wzw.tum.de.
Julia Grimmer, Email: julia.grimmer@biochemtech.uni-halle.de.
Lore Westphal, Email: Lore.Westphal@ipb-halle.de.
Cyril Zipfel, Email: Cyril.Zipfel@sainsbury-laboratory.ac.uk.
Dierk Scheel, Email: dscheel@ipb-halle.de.
Kazuhito Fujiyama, Email: fujiyama@icb.osaka-u.ac.jp.
Justin Lee, Email: jlee@ipb-halle.de.
References
- 1.Boller T, Felix G. A renaissance of elicitors: perception of microbe-associated molecular patterns and danger signals by pattern-recognition receptors. Annu Rev Plant Biol. 2009;60:379–406. doi: 10.1146/annurev.arplant.57.032905.105346. [DOI] [PubMed] [Google Scholar]
- 2.Ranf S, Gisch N, Schäffer M, Illig T, Westphal L, Knirel YA, et al. A lectin S-domain receptor kinase mediates lipopolysaccharide sensing in Arabidopsis thaliana. Nat Immunol. 2015;16(4):426–436. doi: 10.1038/ni.3124. [DOI] [PubMed] [Google Scholar]
- 3.Willmann R, Lajunen HM, Erbs G, Newman MA, Kolb D, Tsuda K, et al. Arabidopsis lysin-motif proteins LYM1 LYM3 CERK1 mediate bacterial peptidoglycan sensing and immunity to bacterial infection. Proc Natl Acad Sci U S A. 2011;108(49):19824–19829. doi: 10.1073/pnas.1112862108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kaku H, Nishizawa Y, Ishii-Minami N, Akimoto-Tomiyama C, Dohmae N, Takio K, et al. Plant cells recognize chitin fragments for defense signaling through a plasma membrane receptor. Proc Natl Acad Sci U S A. 2006;103(29):11086–11091. doi: 10.1073/pnas.0508882103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Miya A, Albert P, Shinya T, Desaki Y, Ichimura K, Shirasu K, et al. CERK1, a LysM receptor kinase, is essential for chitin elicitor signaling in Arabidopsis. Proc Natl Acad Sci U S A. 2007;104(49):19613–19618. doi: 10.1073/pnas.0705147104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Petutschnig EK, Jones AM, Serazetdinova L, Lipka U, Lipka V. The LysM-RLK CERK1 is a major chitin binding protein in Arabidopsis thaliana and subject to chitin-induced phosphorylation. J Biol Chem. 2010;285(37):28902–28911. doi: 10.1074/jbc.M110.116657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shimizu T, Nakano T, Takamizawa D, Desaki Y, Ishii-Minami N, Nishizawa Y, et al. Two LysM receptor molecules, CEBiP and OsCERK1, cooperatively regulate chitin elicitor signaling in rice. Plant J. 2010;64(2):204–214. doi: 10.1111/j.1365-313X.2010.04324.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wan J, Zhang XC, Neece D, Ramonell KM, Clough S, Kim SY, et al. A LysM receptor-like kinase plays a critical role in chitin signaling and fungal resistance in Arabidopsis. Plant Cell. 2008;20(2):471–481. doi: 10.1105/tpc.107.056754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chinchilla D, Boller T, Robatzek S. Flagellin signalling in plant immunity. Adv Exp Med Biol. 2007;598:358–371. doi: 10.1007/978-0-387-71767-8_25. [DOI] [PubMed] [Google Scholar]
- 10.Zipfel C, Kunze G, Chinchilla D, Caniard A, Jones JD, Boller T, et al. Perception of the bacterial PAMP EF-Tu by the receptor EFR restricts Agrobacterium-mediated transformation. Cell. 2006;125(4):749–760. doi: 10.1016/j.cell.2006.03.037. [DOI] [PubMed] [Google Scholar]
- 11.Choi J, Tanaka K, Cao Y, Qi Y, Qiu J, Liang Y, et al. Identification of a plant receptor for extracellular ATP. Science. 2014;343(6168):290–294. doi: 10.1126/science.343.6168.290. [DOI] [PubMed] [Google Scholar]
- 12.Brutus A, Sicilia F, Macone A, Cervone F, De Lorenzo G. A domain swap approach reveals a role of the plant wall-associated kinase 1 (WAK1) as a receptor of oligogalacturonides. Proc Natl Acad Sci U S A. 2010;107(20):9452–9457. doi: 10.1073/pnas.1000675107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Krol E, Mentzel T, Chinchilla D, Boller T, Felix G, Kemmerling B, et al. Perception of the Arabidopsis danger signal peptide 1 involves the pattern recognition receptor AtPEPR1 and its close homologue AtPEPR2. J Biol Chem. 2010;285(18):13471–13479. doi: 10.1074/jbc.M109.097394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Monaghan J, Zipfel C. Plant pattern recognition receptor complexes at the plasma membrane. Curr Opin Plant Biol. 2012;15(4):349–357. doi: 10.1016/j.pbi.2012.05.006. [DOI] [PubMed] [Google Scholar]
- 15.Bohm H, Albert I, Fan L, Reinhard A, Nurnberger T. Immune receptor complexes at the plant cell surface. Curr Opin Plant Biol. 2014;20:47–54. doi: 10.1016/j.pbi.2014.04.007. [DOI] [PubMed] [Google Scholar]
- 16.Beck M, Heard W, Mbengue M, Robatzek S. The INs and OUTs of pattern recognition receptors at the cell surface. Curr Opin Plant Biol. 2012;15(4):367–374. doi: 10.1016/j.pbi.2012.05.004. [DOI] [PubMed] [Google Scholar]
- 17.Maekawa T, Kufer TA, Schulze-Lefert P. NLR functions in plant and animal immune systems: so far and yet so close. Nat Immunol. 2011;12(9):818–826. doi: 10.1038/ni.2083. [DOI] [PubMed] [Google Scholar]
- 18.Seybold H, Trempel F, Ranf S, Scheel D, Romeis T, Lee J. Ca2+ signalling in plant immune response: from pattern recognition receptors to Ca2+ decoding mechanisms. New Phytol. 2014;204:782–790. doi: 10.1111/nph.13031. [DOI] [PubMed] [Google Scholar]
- 19.Le MH, Cao Y, Zhang XC, Stacey G. LIK1, a CERK1-interacting kinase, regulates plant immune responses in Arabidopsis. PLoS One. 2014;9(7):e102245. doi: 10.1371/journal.pone.0102245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cao Y, Liang Y, Tanaka K, Nguyen CT, Jedrzejczak RP, Joachimiak A, et al.: The kinase LYK5 is a major chitin receptor in Arabidopsis and forms a chitin-induced complex with related kinase CERK1. Elife 2014;3:e03766 [DOI] [PMC free article] [PubMed]
- 21.Liu T, Liu Z, Song C, Hu Y, Han Z, She J, et al. Chitin-induced dimerization activates a plant immune receptor. Science. 2012;336(6085):1160–1164. doi: 10.1126/science.1218867. [DOI] [PubMed] [Google Scholar]
- 22.Liu Z, Wu Y, Yang F, Zhang Y, Chen S, Xie Q, et al. BIK1 interacts with PEPRs to mediate ethylene-induced immunity. Proc Natl Acad Sci U S A. 2013;110(15):6205–6210. doi: 10.1073/pnas.1215543110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lu D, Wu S, Gao X, Zhang Y, Shan L, He P. A receptor-like cytoplasmic kinase, BIK1, associates with a flagellin receptor complex to initiate plant innate immunity. Proc Natl Acad Sci U S A. 2010;107(1):496–501. doi: 10.1073/pnas.0909705107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ranf S, Eschen-Lippold L, Fröhlich K, Westphal L, Scheel D, Lee J. Microbe-associated molecular pattern-induced calcium signaling requires the receptor-like cytoplasmic kinases, PBL1 and BIK1. BMC Plant Biol. 2014;14(1):374. doi: 10.1186/s12870-014-0374-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang J, Li W, Xiang T, Liu Z, Laluk K, Ding X, et al. Receptor-like cytoplasmic kinases integrate signaling from multiple plant immune receptors and are targeted by a Pseudomonas syringae effector. Cell Host Microbe. 2010;7(4):290–301. doi: 10.1016/j.chom.2010.03.007. [DOI] [PubMed] [Google Scholar]
- 26.Mersmann S, Bourdais G, Rietz S, Robatzek S. Ethylene signalling regulates accumulation of the FLS2 receptor and is required for the oxidative burst contributing to plant immunity. Plant Physiol. 2010;154(1):391–400. doi: 10.1104/pp.110.154567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ranf S, Eschen-Lippold L, Pecher P, Lee J, Scheel D. Interplay between calcium signalling and early signalling elements during defence responses to microbe- or damage-associated molecular patterns. Plant J. 2011;68(1):100–113. doi: 10.1111/j.1365-313X.2011.04671.x. [DOI] [PubMed] [Google Scholar]
- 28.Kadota Y, Sklenar J, Derbyshire P, Stransfeld L, Asai S, Ntoukakis V, et al. Direct regulation of the NADPH oxidase RBOHD by the PRR-associated kinase BIK1 during plant immunity. Mol Cell. 2014;54(1):43–55. doi: 10.1016/j.molcel.2014.02.021. [DOI] [PubMed] [Google Scholar]
- 29.Li L, Li M, Yu L, Zhou Z, Liang X, Liu Z, et al. The FLS2-associated kinase BIK1 directly phosphorylates the NADPH Oxidase RbohD to control plant immunity. Cell Host Microbe. 2014;15(3):329–338. doi: 10.1016/j.chom.2014.02.009. [DOI] [PubMed] [Google Scholar]
- 30.Dubiella U, Seybold H, Durian G, Komander E, Lassig R, Witte CP, et al. Calcium-dependent protein kinase/NADPH oxidase activation circuit is required for rapid defense signal propagation. Proc Natl Acad Sci U S A. 2013;110(21):8744–8749. doi: 10.1073/pnas.1221294110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Häweker H, Rips S, Koiwa H, Salomon S, Saijo Y, Chinchilla D, et al. Pattern recognition receptors require N-glycosylation to mediate plant immunity. J Biol Chem. 2010;285(7):4629–4636. doi: 10.1074/jbc.M109.063073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li J, Zhao-Hui C, Batoux M, Nekrasov V, Roux M, Chinchilla D, et al. Specific ER quality control components required for biogenesis of the plant innate immune receptor EFR. Proc Natl Acad Sci U S A. 2009;106(37):15973–15978. doi: 10.1073/pnas.0905532106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nekrasov V, Li J, Batoux M, Roux M, Chu ZH, Lacombe S, et al. Control of the pattern-recognition receptor EFR by an ER protein complex in plant immunity. EMBO J. 2009;28(21):3428–3438. doi: 10.1038/emboj.2009.262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Saijo Y, Tintor N, Lu X, Rauf P, Pajerowska-Mukhtar K, Haweker H, et al. Receptor quality control in the endoplasmic reticulum for plant innate immunity. EMBO J. 2009;28(21):3439–3449. doi: 10.1038/emboj.2009.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Aebi M. N-linked protein glycosylation in the ER. Biochim Biophys Acta. 2013;1833(11):2430–2437. doi: 10.1016/j.bbamcr.2013.04.001. [DOI] [PubMed] [Google Scholar]
- 36.Burda P, Aebi M. The dolichol pathway of N-linked glycosylation. Biochim Biophys Acta. 1999;1426(2):239–257. doi: 10.1016/S0304-4165(98)00127-5. [DOI] [PubMed] [Google Scholar]
- 37.Anelli T, Sitia R. Protein quality control in the early secretory pathway. EMBO J. 2008;27(2):315–327. doi: 10.1038/sj.emboj.7601974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Farid A, Malinovsky FG, Veit C, Schoberer J, Zipfel C, Strasser R. Specialized roles of the conserved subunit OST3/6 of the oligosaccharyltransferase complex in innate immunity and tolerance to abiotic stresses. Plant Physiol. 2013;162(1):24–38. doi: 10.1104/pp.113.215509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ranf S, Grimmer J, Pöschl Y, Pecher P, Chinchilla D, Scheel D, et al. Defense-related calcium signaling mutants uncovered via a quantitative high-throughput screen in Arabidopsis thaliana. Mol Plant. 2012;5(1):115–130. doi: 10.1093/mp/ssr064. [DOI] [PubMed] [Google Scholar]
- 40.Qi Z, Verma R, Gehring C, Yamaguchi Y, Zhao Y, Ryan CA, et al. Ca2+ signaling by plant Arabidopsis thaliana Pep peptides depends on AtPepR1, a receptor with guanylyl cyclase activity, and cGMP-activated Ca2+ channels. Proc Natl Acad Sci U S A. 2010;107(49):21193–21198. doi: 10.1073/pnas.1000191107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Frei dit Frey N, Mbengue M, Kwaaitaal M, Nitsch L, Altenbach D, Häweker H, et al. Plasma membrane calcium ATPases are important components of receptor-mediated signaling in plant immune responses and development. Plant Physiol. 2012;159(2):798–809. doi: 10.1104/pp.111.192575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Salathia N, Lee HN, Sangster TA, Morneau K, Landry CR, Schellenberg K, et al. Indel arrays: an affordable alternative for genotyping. Plant J. 2007;51(4):727–737. doi: 10.1111/j.1365-313X.2007.03194.x. [DOI] [PubMed] [Google Scholar]
- 43.Henquet M, Lehle L, Schreuder M, Rouwendal G, Molthoff J, Helsper J, et al. Identification of the gene encoding the alpha1,3-mannosyltransferase (ALG3) in Arabidopsis and characterization of downstream n-glycan processing. Plant Cell. 2008;20(6):1652–1664. doi: 10.1105/tpc.108.060731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kajiura H, Seki T, Fujiyama K. Arabidopsis thaliana ALG3 mutant synthesizes immature oligosaccharides in the ER and accumulates unique N-glycans. Glycobiology. 2010;20(6):736–751. doi: 10.1093/glycob/cwq028. [DOI] [PubMed] [Google Scholar]
- 45.Aebi M, Gassenhuber J, Domdey H, Heesen ST. Cloning and characterization of the ALG3 gene of Saccharomyces cerevisiae. Glycobiology. 1996;6(4):439–444. doi: 10.1093/glycob/6.4.439. [DOI] [PubMed] [Google Scholar]
- 46.Zipfel C, Robatzek S, Navarro L, Oakeley EJ, Jones JD, Felix G, et al. Bacterial disease resistance in Arabidopsis through flagellin perception. Nature. 2004;428(6984):764–767. doi: 10.1038/nature02485. [DOI] [PubMed] [Google Scholar]
- 47.Nawrath C, Metraux JP. Salicylic acid induction-deficient mutants of Arabidopsis express PR-2 and PR-5 and accumulate high levels of camalexin after pathogen inoculation. Plant Cell. 1999;11(8):1393–1404. doi: 10.1105/tpc.11.8.1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Strasser R. Biological significance of complex N-glycans in plants and their impact on plant physiology. Front Plant Sci. 2014;5:363. doi: 10.3389/fpls.2014.00363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Meusser B, Hirsch C, Jarosch E, Sommer T. ERAD: the long road to destruction. Nat Cell Biol. 2005;7(8):766–772. doi: 10.1038/ncb0805-766. [DOI] [PubMed] [Google Scholar]
- 50.Strasser R, Stadlmann J, Svoboda B, Altmann F, Glossl J, Mach L. Molecular basis of N-acetylglucosaminyltransferase I deficiency in Arabidopsis thaliana plants lacking complex N-glycans. Biochem J. 2005;387(Pt 2):385–391. doi: 10.1042/BJ20041686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Roy S, Perron B, Gallo-Payet N. Role of asparagine-linked glycosylation in cell surface expression and function of the human adrenocorticotropin receptor (melanocortin 2 receptor) in 293/FRT cells. Endocrinology. 2010;151(2):660–670. doi: 10.1210/en.2009-0826. [DOI] [PubMed] [Google Scholar]
- 52.Koller T, Bent AF. FLS2-BAK1 extracellular domain interaction sites required for defense signaling activation. PLoS One. 2014;9(10):e111185. doi: 10.1371/journal.pone.0111185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hong Z, Jin H, Fitchette AC, Xia Y, Monk AM, Faye L, et al. Mutations of an alpha1,6 mannosyltransferase inhibit endoplasmic reticulum-associated degradation of defective brassinosteroid receptors in Arabidopsis. Plant Cell. 2009;21(12):3792–3802. doi: 10.1105/tpc.109.070284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hong Z, Kajiura H, Su W, Jin H, Kimura A, Fujiyama K, et al. Evolutionarily conserved glycan signal to degrade aberrant brassinosteroid receptors in Arabidopsis. Proc Natl Acad Sci U S A. 2012;109(28):11437–11442. doi: 10.1073/pnas.1119173109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yamamoto M, Tantikanjana T, Nishio T, Nasrallah ME, Nasrallah JB. Site-specific N-glycosylation of the S-locus receptor kinase and its role in the self-incompatibility response of the brassicaceae. Plant Cell. 2014;26(12):4749–4762. doi: 10.1105/tpc.114.131987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chen XL, Shi T, Yang J, Shi W, Gao XS, Chen D, et al. N-Glycosylation of Effector Proteins by an alpha-1,3-Mannosyltransferase Is Required for the Rice Blast Fungus to Evade Host Innate Immunity. Plant Cell. 2014;26(3):1360–1376. doi: 10.1105/tpc.114.123588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Weiss N, Black SA, Bladen C, Chen L, Zamponi GW. Surface expression and function of Cav3.2 T-type calcium channels are controlled by asparagine-linked glycosylation. Pflugers Arch - Eur J Physiol. 2013;465(8):1159–1170. doi: 10.1007/s00424-013-1259-3. [DOI] [PubMed] [Google Scholar]
- 58.Barone R, Fiumara A, Jaeken J. Congenital disorders of glycosylation with emphasis on cerebellar involvement. Semin Neurol. 2014;34(3):357–366. doi: 10.1055/s-0034-1387197. [DOI] [PubMed] [Google Scholar]
- 59.Zufferey R, Knauer R, Burda P, Stagljar I, te Heesen S, Lehle L, et al. STT3, a highly conserved protein required for yeast oligosaccharyl transferase activity in vivo. EMBO J. 1995;14(20):4949–4960. doi: 10.1002/j.1460-2075.1995.tb00178.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Boutrot F, Segonzac C, Chang KN, Qiao H, Ecker JR, Zipfel C, et al. Direct transcriptional control of the Arabidopsis immune receptor FLS2 by the ethylene-dependent transcription factors EIN3 and EIL1. Proc Natl Acad Sci U S A. 2010;107(32):14502–14507. doi: 10.1073/pnas.1003347107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yoo SD, Cho YH, Sheen J. Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat Protoc. 2007;2(7):1565–1572. doi: 10.1038/nprot.2007.199. [DOI] [PubMed] [Google Scholar]
- 62.Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, et al. Fiji: an open-source platform for biological-image analysis. Nat Methods. 2012;9(7):676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]


