Abstract
Despite the growing body of work on molecular components required for regenerative repair, we still lack a deep understanding of the ability of some animal species to regenerate their appropriate complex anatomical structure following damage. A key question is how regenerating systems know when to stop growth and remodeling – what mechanisms implement recognition of correct morphology that signals a stop condition? In this work, we review two conceptual models of pattern regeneration that implement a kind of pattern memory. In the first one, all cells communicate with each other and keep the value of the total signal received from the other cells. If a part of the pattern is amputated, the signal distribution changes. The difference fromthe original signal distribution stimulates cell proliferation and leads to pattern regeneration, in effect implementing an error minimization process that uses signaling memory to achieve pattern correction. In the second model, we consider a more complex pattern organization with different cell types. Each tissue contains a central (coordinator) cell that controls the tissue and communicates with the other central cells. Each of them keeps memory about the signals received from other central cells. The values of these signals depend on the mutual cell location, and the memory allows regeneration of the structure when it is modified. The purpose of these models is to suggest possible mechanisms of pattern regeneration operating on the basis of cell memory which are compatible with diverse molecular implementation mechanisms within specific organisms.
Keywords: target morphology, pattern regeneration, cell memory, morphogenesis, cell signaling, mathematical modeling, agent-based model
Cell Memory Can Regulate Morphogenesis and Regeneration
Many biological organisms can regenerate some of their tissues and organs. Some species, such as hydra and planaria, can regenerate the whole organism from its small parts (Birn-baum et al., 2008). Other organisms, such as salamanders, can regenerate limbs, eyes, portions of the brain, spinal cords, and jaws (McCusker and Gardiner, 2011; Tanaka and Reddien, 2011). Mammals have more limited regenerative potential that nevertheless includes not only wound healing (Baddour et al., 2012; Sousounis et al., 2014) but also regeneration of complex structures such as antlers (Li, 2012), liver (Mao et al., 2014), and children's fingertips (Illingworth, 1974).
Experiments on regeneration in numerous species suggest that tissues can keep some information about their former states, including anatomical location (Carlson, 1983; Kragl et al., 2009; Wang et al., 2009), and communicate this information to surrounding cells during repair processes. Specific cell types that exhibit instructive capacity for positional information include muscle cells (Witchley et al., 2013) and fibroblasts (Chang et al., 2002; Rinn et al., 2006). Another cell type that non-cell-autonomously directs cell movement and differentiation state is the glycine-gated chloride channel-expressing instructor cell, which regulates the behavior of other cell types via serotonergic signaling (Blackiston et al., 2011; Lobikin et al., 2012). Importantly, this information is not cell-autonomous, and often must coordinate activity of cells across large distances. For example, when bisected, the anterior end of a planarian flatworm grows a head, while the posterior piece grows a tail: radically different anatomical structures are produced by cells that were adjacent neighbors before the arbitrarily-placed cut (Salo et al., 2009; Lobo et al., 2012), and had been at the same position within the worm. Thus, it is clear that the decision of which body-parts to regenerate at a wound site cannot be made locally; the worms adult stem cells must integrate information from distant body regions that reveals what parts are missing and where the wound is located. The long-range communication within the body is at least partially mediated by physiological signaling through electrical synapses known gap junctions (Palacios-Prado and Bukauskas, 2009; Pereda et al., 2013), as revealed by the result of allowing worms to regenerate after being exposed to gap junctional inhibitors or being injected with RNAi that knock down innexin proteins (Oviedo et al., 2010). Upon modulation of normal electrical connectivity among its cells, a planarian with two heads is obtained from the bisection of a normal one-headed animal (Levin et al., 2012; Levin, 2014). Remarkably, when both heads are amputated in plain water (no more gap junctional blockers), in subsequent rounds of regeneration, pieces of the normal mid-body (gut) reliably regenerate 2 heads inperpetuity (Levin, 2014), revealing the ability of physiological networks to store patterning information on long time scales (a kind of long-term memory). Here, we review our recent work attempting to computationally model the events that control regeneration in planaria.
During planarian regeneration, one or two heads are possible: the basic bodyplan of the worm can be altered by a brief perturbation of the endogenous bioelectric network guiding pattern formation (Levin, 2013; Tseng and Levin, 2013; Mustard and Levin, 2014), switching between the 2-headed and the 1-headed state (Oviedo et al., 2010; Beane et al., 2011). The choice between these different configurations of the planarian anatomy is not determined entirely at the genetic level, because the target morphology of the worm can be permanently altered by a transient physiological perturbation that does not alter genomic sequence. While chromatin modification (epigenetic remodeling) may be involved, this does not in itself resolve the question of how the patterning information is stored in a distributed manner. Since any reprogrammed (ectopic head) tissues are discarded at each cut, and the cut can be made anywhere, it is clear that every piece of the worm now knows that upon cutting, it is to make a 2-headed worm instead of the normal 1-headed pattern. However, side wounds made in such animals do not automatically make ectopic heads, showing that the worm's state has not simply been changed to implement a rule like “any wound forms a head”. The information about the number and location of heads that the stem cells must build is kept in the remaining tissue after the ectopic heads are removed. It is important to note the fundamental fact that even a normal worm is a system whose cells know to stop moving, differentiating, and proliferating when the correct pattern has been achieved (when for example are generated head is complete). Together, these observations suggest the existence of a set of properties and mechanisms that allow pieces of the worm to stably encode information that, upon cutting, guides cellular activity towards appropriate morphogenesis and also provides a sufficient stop condition when the correct shape has been restored.
Despite many recent high-resolution studies of the genetic pathways regulating stem cell differentiation (Reddien et al., 2005; Petersen and Reddien, 2011; Wagner et al., 2011; Agata et al., 2014), it is entirely unclear how altered bodyplan layouts can be re-written into the default planarian target morphology, or how the stem cells know to stop their remarkable activity once a correctly shaped and sized head(s) has been produced. Moreover, the development of conceptual models to understand information processing and control dynamics in large-scale pattern repair has not kept up with the rapid progress of mechanistic detail of pathways regulating events at the level of single cells. Thus, it is crucial to attempt to formulate computational models that serve as examples of the kinds of mechanisms that could exist (Reitz, 2012; Friston et al., 2015; Ogawa and Miyake, 2015). This is especially important for progress in regenerative medicine and developmental biology, as such models will be needed to guide strategies for repairing birth defects and traumatic injury, where growth must be induced towards a particular organ shape (not merely gene expression), and limited to avoid cancer in favor of regeneration of complex body structures (Ingber and Levin, 2007; Davies, 2008; Wang et al., 2009; Baddour et al., 2012; Levin, 2013). We analyzed a class of models of pattern regeneration that implements a kind of cell memory sufficient to explain the amazing properties of planarian regeneration.
In this work, we review two conceptual models of pattern regeneration (Bessonov et al., 2015; Tosenberger et al., 2015). In the first one (Section 2) all cells communicate with each other and keep the value of the total signal received from the other cells. If a part of the pattern is amputated, the signal distribution changes. The difference with the original signal distribution stimulates cell proliferation and leads to pattern regeneration, in effect implementing an error minimization process that uses signaling memory to implement pattern correction. In the second model (Section 3) we consider a more complex pattern organization with different cell types. Each tissue contains a central (coordinator) cell that controls the tissue and communicates with the other central cells. Each of them keeps memory about the signals received from other central cells. The values of these signals depend on the mutual cell location, and the memory allows regeneration of the structure when it is modified. The purpose of these models is to suggest possible mechanisms of pattern regeneration operating on the basis of cell memory which are compatible with diverse molecular implementation mechanisms within specific organisms.
Model of Pattern Regeneration Based on Cell Memory
In this model, biological cell structures are considered as ensemble of mathematical points on the plane. Each cell produces a signal which propagates in space. It is received by other cells. The total signal received by each cell forms a signal distribution defined on the cell structure. This distribution characterizes the geometry of the cell structure. If a part of this structure is removed, then remaining cells have two signals. They keep the value of the signal which they had before the amputation (memory), and they receive a new signal produced after the amputation. Regeneration of the cell structure is stimulated by the difference between the old and the new signals. It is stopped when the two signals coincide.
Consider a 2D domain D filled by cells. Each cell produces a signal u which spreads in space. Its intensity decays with distance as some function f(d). If the distance between cell i (Ci) and cell j (Cj) is dij, then Cj receives signal f(dij) from Ci. We will assume here that each cell produces the same signal. Therefore, Ci receives from Cj a signal of the same intensity f(dij). For each cell Ci we can count the total signal which it receives from other cells:
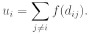
We will use also the notation u(x) assuming that x belong to the domain D, ui = u(xi), where xi is the coordinate of the ith cell.
Clearly, cells located in different part of the domain will receive different signals. For example, a cell located at the boundary receives signals only from one side and the value of the signal can be less than for a cell which is located inside the domain. Therefore the distribution u(x) represents some information about the geometry of the domain.
We suggest an algorithm for the placement of new cells, as one hypothesis of the policy guiding the migratory and differentiation behavior of neoblasts during regeneration. It is determined by the following three conditions:
1. All cells are placed in the nodes of the square grid. Each cell has 8 neighbors, 4 of them with a common side and 4 other cell with a common diagonal. Each new cell is placed in such a way that among its neighbors there is at least one cells from the cut (blastema) or another new cell. This condition provides continuity of growth of the regenerating domain, beginning from the place of the cut,
2. When we add a new cell we calculate the signal in every control cell. The new signal should be less than or equal to the old signal,

In numerical simulations this condition should be satisfied with certain accuracy.
3. Let us introduce total signals:
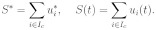
Among all cells, which satisfy conditions 1 and 2, at each time step we choose the cell for which the difference S*– S(t) is minimal (Figure 1).
Figure 1.

An example of pattern regeneration in the first model.
The pattern after amputation represents a rectangle. Regeneration reproduces exactly the original patterns before amputation. Different regenerated patterns are obtained because the memory remaining after amputation is different. Reprinted from Bessonov et al. (2015).
Tissues Exchange Signals and Control Their Mutual Location
We suppose in this model that not all cells possess memory and the ability to instruct surrounding cells, but only some special cells within each tissue (a reasonable supposition based on the positional memory and instructive capacity found in specific cell types). We call them central or coordinator cells. Moreover these cells exchange signals with other cells of this tissue and with the coordinating cells of other tissues. Thus, the organism is considered as an ensemble of tissues, while the growth of each individual tissue is directed by its centre of organisation. In the simplest model, we assume that the centre of organization is a single stem cell, which is able to generate its surrounding tissue through a series of asymmetric divisions. The question we pose here is whether the cell memory can be sufficient to restore the relative spatial position of individual tissues in the organism and thus characterize the organism's morphology. As was the case in the regeneration model, here we again assume that tissue centres can exchange signals and that each centre can possess a genetic or a temporary memory of signal intensity. Therefore, we consider that each centre Ci produces a signal si which decays exponentially in space. The signal si is then perceived by cell Cj as sji = f(dij), where dij is the distance between cells Ci and Cj and f is the exponential decay function. Each organizing cell Cj possesses amemory uji* of the ideal intensity of the signal produced by each other cell Ci. In a case of a perturbation of the spatial configuration of organizing cells, each organizing cell receives signals with intensities that differ from the memorized intensities. As a result of that difference cell Cj will produce a response to cell Ci:
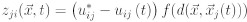
where 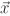 is the vector of position, t is the moment of time, and d(
is the vector of position, t is the moment of time, and d(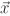 ,
, 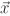 j(t)) is the distance between the spatial position
j(t)) is the distance between the spatial position 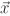 and the position of the cell Cj at the moment t. The response signals zji then direct the movement of the cell Ci along their gradient:
and the position of the cell Cj at the moment t. The response signals zji then direct the movement of the cell Ci along their gradient:
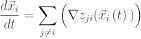
where 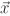 i(t) denotes the spatial position of the organizing cell Ci at the moment t.
i(t) denotes the spatial position of the organizing cell Ci at the moment t.
The described model of tissue centres organization has been shown to be sufficient for keeping their spatial configuration stable and resistant to non-extreme perturbations. Thus, for an organism with n organizing cells (n tissues) It is sufficient to memorise n(n − 1) different signal intensities and to be able to produce n2 different signal types. If we reduce this requirement to two different signal types (one initial signal type and the response signal type), the system becomes unstable even for small perturbations.
In order to demonstrate the possibility of the model to characterize spatial configuration of different tissues we apply a simple model of tissue growth and growth control. Each organizing cell is a stem cell which can divide in an asymmetric way, producing a new stem cell containing the memory of the mother cell and a differentiated cell of the corresponding tissue unable to divide. In order to limit tissue growth each stem cell produces a life support signal which decays in space. If a differentiated cell of that tissue perceives the intensity of the support signal which is lower than some threshold, it dies through apoptosis. Figure 2 shows a stable configuration of several bordering tissues representing a model organism. Firstly, the figure shows the formation of tissues and that the configuration remains stable when the tissues come in a contact with each other. Secondly, it is shown how the organism stays stable through the process of regeneration following an amputation of a part of the tissue.
Figure 2.
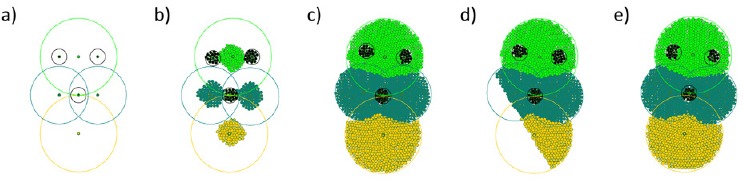
Numerical simulation of organism growth and regeneration with the second model.
There are several different tissues represented by different colors. a) The dots show equilibrium positions of central cells of each tissue, the circles around them the size of the corresponding tissue controlled by the central cells. b) Beginning of growth. c) Final form of the organism. d) A part of the organism is amputated. e) Regeneration to the original form.
Discussion
Information and communication
The models presented above suggest possible mechanisms of pattern regeneration on the basis of cell communication and memory. In the first model, we consider the case of horizontal communication. All cells are similar to each other, they produce the same type of signal and receive signals from all other cells. The information cost of this model for each particular cell is very low. Each cell memory consists of a single real number. On the other hand, since each cell participates in this signal exchange, this mechanism poses a limitation on the total number of cells and on the size of the pattern.
The two-level communication scheme suggested in the second model is more flexible. The pattern consists now of several tissues. Each tissue has some central or coordinating cells whose function is twofold. They communicate with the cells of this tissue with the purpose to control its form and size formandsize. On the other hand, they communicate with the coordinating cells of other tissues. This second signal exchange keeps the information about the global pattern organization: how different tissues are located with respect to each other. Here only coordinating cells possess memory, but it is now more extended than in the first model. It consists of several numbers, where “several” is the number of other coordinating cells.
These models suggest conceptual mechanisms of pattern regeneration. They do not take into account numerous specific features of biological organisms. Many mechanisms of cell communication and memory (including chemical, bioelectrical, and tensile forces) compatible with the signaling described in these models are currently an active area of study (Davies, 2008; Mustard and Levin, 2014).
A possible suggestion of molecular basis of the phenomenon of cell memory implies that the signals between cells, included in our model, can depend on a set of epigenetic information, comprised by specific types and locations of cell surface molecules, distributed during cell divisions according to a set of laws, preserving some of them in each cell (Morozova and Shubin, 2012). Thus, the distribution of these molecules of the epigenetic code which has rested on the membranes of the control cells after amputation, will influence a process of new signals formation and distribution, as during the process of regeneration, so in the normal development. Then this additional level of molecular information can be considered as the epigenetic code for the target morphology of an organism.
Perspectives
Our model reveals a scheme which explains how a stable pattern can be stored, and re-written, in a cellular network. Future efforts will extend the model to other examples of target morphology change, such as crab claws and trophic memory in deer antlers (reviewed in (Lobo et al., 2014)). Fleshing out the model with additional realistic parameters will enable derivation of testable predictions, which can readily be tested in planaria. Convergence of agent-based models of pattern memory (Bessonov et al., 2015; Tosenberger et al., 2015) with other algorithmic (Slack, 1980; Friston et al., 2015) and continuous gradient models (Meinhardt, 2008, 2009; Werner et al., 2015) will provide the field with much-needed theory to guide experiments on pattern memory.
While much of the regeneration field is searching for ways to induce regeneration, there are two other crucial components. One is the possibility of programming growth at the level of organs: providing signals that trigger complex downstream patterning cascades (developmental modules) without needing to micromanage the process directly. This will allow us to defeat the complexity barrier that stymies efforts to rebuild organs such as the hand or the eye even if all stem cell derivatives were readily available (Levin, 2011). The second is the issue of how such growth, once triggered, stops after a correctly shaped and sized body-part is rebuilt; this is crucial if regenerative therapies are to avoid carcinogenesis. Cancers have often been called “wounds that do not heal” (Dvorak, 1986; Riss et al., 2006; Byun and Gardner, 2013), underlying the critical nature of patterning information that separates regenerative wound repair with uncontrolled and lethal tumor growth. The dynamics of this patterning information must be understood, if we are to develop effective strategies for guiding patterning by rewriting, in vivo, the parameters that regulate target anatomical outcomes.
Aside from regenerative repair, three other areas will benefit from similar approaches. First, the repair of birth defects during embryogenesis likewise depends on knowledge of the dynamics that implement particular shapes (in this case, self-organized from the fertilized egg stage). Second, evolutionary biology may face significant implications of genomic alterations that regulate key aspects of this kind of cell-to-cell communication; molecular studies in this field could ask which cells mutations and which epigenetic influences could rewrite the cellular parameters toward adaptive outcomes of the pattern regulation. Finally, synthetic bioengineering will need models such as this to guide efforts to build self-assembling hybrid constructs, the so-called biobots implemented with synthetic morphology (Doursat, 2006; Davies, 2008; Doursat et al., 2012, 2013; Doursat and Sanchez, 2014; Kamm and Bashir, 2014).
Footnotes
Funding: ML is grateful for the support of the G. Harold and Leila Y. Mathers Charitable Foundation, the Templeton World Charity Foundation (TWCF0089/AB55), and the W.M. Keck Foundation.
References
- 1.Agata K, Tasaki J, Nakajima E, Umesono Y. Recent identification of an ERK signal gradient governing planarian regeneration. Zoology. 2014;117:161–162. doi: 10.1016/j.zool.2014.04.001. [DOI] [PubMed] [Google Scholar]
- 2.Baddour JA, Sousounis K, Tsonis PA. Organ repair and regeneration: an overview. Birth Defects Res C Embryo Today. 2012;96:1–29. doi: 10.1002/bdrc.21006. [DOI] [PubMed] [Google Scholar]
- 3.Beane WS, Morokuma J, Adams DS, Levin M. A chemical genetics approach reveals H, K-ATPase-mediated membrane voltage is required for planarian head regeneration. Chem Biol. 2011;18:77–89. doi: 10.1016/j.chembiol.2010.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bessonov N, Levin M, Morozova N, Reinberg N, Tosenberger A, Volpert V. 2015 On a model of pattern regeneration based on cell memory. PLoS One. 2015;10:e0118091. doi: 10.1371/journal.pone.0118091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Birnbaum KD, Alvarado AS. Slicing across kingdoms: regeneration in plants and animals. Cell. 2008;132:697–710. doi: 10.1016/j.cell.2008.01.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blackiston D, Adams DS, Lemire JM, Lobikin M, Levin M. Transmembrane potential of GlyCl-expressing instructor cells induces a neoplastic-like conversion of melanocytes via a serotonergic pathway. Dis Model Mech. 2011;4:67–85. doi: 10.1242/dmm.005561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Byun JS, Gardner K. Wounds that will not heal: pervasive cellular reprogramming in cancer. Am J Pathol. 2013;182:1055–1064. doi: 10.1016/j.ajpath.2013.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carlson BM. Limb Development and Regeneration. New York: Alan R. Liss; 1983. Positional memory invertebrate limb development and regeneration; pp. 433–443. [PubMed] [Google Scholar]
- 9.Chang HY, Chi JT, Dudoit S, Bondre C, vandeRijn M, Botstein D, Brown PO. Diversity topographic differentiation and positional memory in human fibroblasts. Proc Natl Acad Sci U S A. 2002;99:12877–12882. doi: 10.1073/pnas.162488599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Davies JA. Synthetic morphology: prospects for engineered self-constructing anatomies. J Anat. 2008;212:707–719. doi: 10.1111/j.1469-7580.2008.00896.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Doursat R. The growing canvas of biological development: Multiscale pattern generation on an expanding lattice of gene regulatory networks. InterJournal: Complex Systems, 1809. 2006 [Google Scholar]
- 12.Doursat R, Sanchez C. Growing fine-grained multi cellular robots. Soft Robot. 2014;1:110–121. [Google Scholar]
- 13.Doursat R, Sayama H, Michel O. Morphogenetic engineering: reconciling self-organization and architecture. Underst Complex Syst. 2012:1–24. [Google Scholar]
- 14.Doursat R, Sayama H, Michel O. A review of morphogenetic engineering. Nat Comput. 2013;12:517–535. [Google Scholar]
- 15.Dvorak HF. Tumors: wounds that do not heal. Similarities between tumor stroma generation and wound healing. N Engl J Med. 1986;315:1650–1659. doi: 10.1056/NEJM198612253152606. [DOI] [PubMed] [Google Scholar]
- 16.Friston K, Levin M, Sengupta B, Pezzulo G. Knowing one's place: a free-energy approach to pattern regulation. J R Soc Interface 12. 2015 doi: 10.1098/rsif.2014.1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Illingworth CM. Trapped fingers and amputated finger tips in children. J Pediatr Surg. 1974;9:853–858. doi: 10.1016/s0022-3468(74)80220-4. [DOI] [PubMed] [Google Scholar]
- 18.Ingber DE, Levin M. What lies at the interface of regenerative medicine and developmental biology? Development. 2007;134:2541–2547. doi: 10.1242/dev.003707. [DOI] [PubMed] [Google Scholar]
- 19.Kamm RD, Bashir R. Creating living cellular machines. Ann Biomed Eng. 2014;42:445–459. doi: 10.1007/s10439-013-0902-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kragl M, Knapp D, Nacu E, Khattak S, Maden M, Epperlein HH, Tanaka EM. Cells keep a memory of their tissue origin during axolotl limb regeneration. Nature. 2009;460:60–65. doi: 10.1038/nature08152. [DOI] [PubMed] [Google Scholar]
- 21.Levin M. The wisdom of the body: future techniques and approaches to morphogenetic fields in regenerative medicine, developmental biology and cancer. Regen Med. 2011;6:667–673. doi: 10.2217/rme.11.69. [DOI] [PubMed] [Google Scholar]
- 22.Levin M. Morphogenetic fields in embryo genesis, regeneration and cancer: non-local control of complex patterning. Biol Syst. 2012;109:243–261. doi: 10.1016/j.biosystems.2012.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Levin M. Reprogramming cells and tissue patterning via bioelectrical pathways: molecular mechanisms and biomedical opportunities. Wiley Interdiscip Rev Syst Biol Med. 2013;5:657–676. doi: 10.1002/wsbm.1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Levin M. Endogenous bioelectrical networks store non-genetic patterning information during development and regeneration. J Physiol. 2014;592:2295–2305. doi: 10.1113/jphysiol.2014.271940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li C. Deer antler regeneration: A stem cell-based epimorphic process. Birth Defects Res C Embryo Today. 2012;96:51–62. doi: 10.1002/bdrc.21000. [DOI] [PubMed] [Google Scholar]
- 26.Lobikin M, Chernet B, Lobo D, Levin M. Resting potential, oncogene-induced tumorigenesis, and metastasis: the bioelectric basis of cancer in vivo. Phys Biol. 2012;9:065002. doi: 10.1088/1478-3975/9/6/065002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lobo D, Beane WS, Levin M. Modeling planarian regeneration: a primer for reverse-engineering the worm. PLoS Comput Biol. 2012;8:e1002481. doi: 10.1371/journal.pcbi.1002481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lobo D, Solano M, Bubenik GA, Levin M. A linear-encoding model explains the variability of the target morphology in regeneration. J R Soc Interface. 2014;11:20130918. doi: 10.1098/rsif.2013.0918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mao SA, Glorioso JM, Nyberg SL. Liver regeneration. Transl Res. 2014;163:352–362. doi: 10.1016/j.trsl.2014.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McCusker C, Gardiner DM. The axolotl model for regeneration and aging research: a mini-review. Gerontology. 2011;57:565–571. doi: 10.1159/000323761. [DOI] [PubMed] [Google Scholar]
- 31.Meinhardt H. Models of biological pattern formation: from elementary steps to the organization of embryonic axes. Curr Top Dev Biol. 2008;81:1–63. doi: 10.1016/S0070-2153(07)81001-5. [DOI] [PubMed] [Google Scholar]
- 32.Meinhardt H. Beta-catenin and axis formation in planarians. Bioessays. 2009;31:5–9. doi: 10.1002/bies.080193. [DOI] [PubMed] [Google Scholar]
- 33.Morozova N, Shubin M. The geometry of morphogenesis and the morphogenetic field concept. in: pattern formation in morphogenesis - problems and mathematical issues. Springer Proc Math. 2012;15:255–282. [Google Scholar]
- 34.Mustard J, Levin M. bioelectrical mechanisms for programming growth and form: taming physiological networks for soft body robotics. Soft Robot. 2014;1:169–191. [Google Scholar]
- 35.Ogawa K, Miyake Y. Robust patterning of gene expression based on internal co-ordinate system of cells. Bio Syst. 2015;132(133):6–12. doi: 10.1016/j.biosystems.2015.04.004. [DOI] [PubMed] [Google Scholar]
- 36.Oviedo NJ, Morokuma J, Walentek P, KemaI P, GuM B, Ahn JM, Hwang JS, Gojobori T, Levin M. Long-range neural and gap junction protein-mediated cues control polarity during planarian regeneration. Dev Biol. 2010;339:188–199. doi: 10.1016/j.ydbio.2009.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Palacios-Prado N, Bukauskas FF. Hetero typic gap junction channels as voltage-sensitive valves for intercellular signaling. Proc Natl Acad Sci U S A. 2009;106:14855–14860. doi: 10.1073/pnas.0901923106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pereda AE, Curti S, Hoge G, Cachope R, Flores CE, Rash JE. Gap junction mediated electrical transmission: regulatory mechanisms and plasticity. Biochim Biophys Acta. 2013;1828:134–146. doi: 10.1016/j.bbamem.2012.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Petersen CP, Reddien PW. Polarized notum activation at wounds inhibits Wnt function to promote planarian head regeneration. Science. 2011;332:852–855. doi: 10.1126/science.1202143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Reddien PW, Bermange AL, Murfitt KJ, Jennings JR, Sanchez Alvarado A. Identification of genes needed for regeneration, stem cell function and tissue homeostasis by systhematic gene perturbation inplanaria. Dev Cell. 2005;8:635–649. doi: 10.1016/j.devcel.2005.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Reitz FB. A “lookup table” schema for synthetic biological patterning. Theory Biosci. 2012;131:43–47. doi: 10.1007/s12064-012-0150-7. [DOI] [PubMed] [Google Scholar]
- 42.Rinn JL, Bondre C, Gladstone HB, Brown PO, Chang HY. Anatomic demarcation by positional variation in fibroblast gene expression programs. PLoS Genet. 2006;2:e119. doi: 10.1371/journal.pgen.0020119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Riss J, Khanna C, Koo S, Chandramouli GV, Yang HH, Hu Y, Kleiner DE, Rosenwald A, Schaefer CF, Ben-Sasson SA, Yang L, Powell J, Kane DW, Star RA, Aprelikova O, Bauer K, Vasselli JR, Maranchie JK, Kohn KW, Buetow KH, et al. Cancers as wounds that do not heal: differences and similarities between renal regeneration/repair and renal cell carcinoma. Cancer Res. 2006;66:7216–7224. doi: 10.1158/0008-5472.CAN-06-0040. [DOI] [PubMed] [Google Scholar]
- 44.Salo E, Abril JF, Adell T, Cebria F, Eckelt K, Fernandez-Taboada E, Handberg-Thorsager M, Iglesias M, Molina MD, Rodriguez-Esteban G. Planarian regeneration: achievements and future directions after 20 years of research. IntJ Dev Biol. 2009;53:1317–1327. doi: 10.1387/ijdb.072414es. [DOI] [PubMed] [Google Scholar]
- 45.Sousounis K, Baddour JA, Tsonis PA. Aging and regeneration invertebrates. Curr Top Dev Biol. 2014;108:217–246. doi: 10.1016/B978-0-12-391498-9.00008-5. [DOI] [PubMed] [Google Scholar]
- 46.Slack JM. A serial threshold theory of regeneration. J Theor Biol. 1980;82:105–140. doi: 10.1016/0022-5193(80)90092-2. [DOI] [PubMed] [Google Scholar]
- 47.Tanaka EM, Reddien PW. The cellular basis for animal regeneration. Dev Cell. 2011;21:172–185. doi: 10.1016/j.devcel.2011.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tosenberger A, Bessonov N, Levin M, Reinberg N, Volpert V, Morozova N. A conceptual model of morphogenesis and regeneration. Acta Biotheor. 2015 doi: 10.1007/s10441-015-9249-9. DOI:10.1007/s10441-015-9249-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tseng A, Levin M. Cracking the bioelectric code: Probing endogenous ionic controls of pattern formation. Commun Integr Biol. 2013;6:1–8. doi: 10.4161/cib.22595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wagner DE, Wang IE, Reddien PW. Clonogenic neoblasts are pluripotent adult stem cells that underlie planarian regeneration. Science. 2011;332:811–816. doi: 10.1126/science.1203983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang KC, Helms JA, Chang HY. Regeneration, repair and remembering identity: the three Rs of Hox gene expression. Trends Cell Biol. 2009;19:268–275. doi: 10.1016/j.tcb.2009.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Werner S, Stuckemann T, Beiran Amigo M, Rink JC, Julicher F, Friedrich BM. Scaling and regeneration of self-organized patterns. Phys Rev Lett. 2015;114:138101. doi: 10.1103/PhysRevLett.114.138101. [DOI] [PubMed] [Google Scholar]
- 53.Witchley JN, MayerM, Wagner DE, Owen JH, Reddien PW. Muscle cells provide instructions for planarian regeneration. Cell Rep. 2013;4:633–641. doi: 10.1016/j.celrep.2013.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]


