Multimodal spectroscopic imaging resolved controversies on biochemical changes associated with cerebral malaria pathology.
Keywords: Cerebral Malaria, Murine Model, Molecular Mechanism, Multi-modal Imaging, X-ray Fluorecence Microscopy, FTIR Imaging, resonance Raman Imaging, Nuclear Microprobe, Altered Cerebral Metabolism, Protein Oxidation
Abstract
Using a multimodal biospectroscopic approach, we settle several long-standing controversies over the molecular mechanisms that lead to brain damage in cerebral malaria, which is a major health concern in developing countries because of high levels of mortality and permanent brain damage. Our results provide the first conclusive evidence that important components of the pathology of cerebral malaria include peroxidative stress and protein oxidation within cerebellar gray matter, which are colocalized with elevated nonheme iron at the site of microhemorrhage. Such information could not be obtained previously from routine imaging methods, such as electron microscopy, fluorescence, and optical microscopy in combination with immunocytochemistry, or from bulk assays, where the level of spatial information is restricted to the minimum size of tissue that can be dissected. We describe the novel combination of chemical probe–free, multimodal imaging to quantify molecular markers of disturbed energy metabolism and peroxidative stress, which were used to provide new insights into understanding the pathogenesis of cerebral malaria. In addition to these mechanistic insights, the approach described acts as a template for the future use of multimodal biospectroscopy for understanding the molecular processes involved in a range of clinically important acute and chronic (neurodegenerative) brain diseases to improve treatment strategies.
INTRODUCTION
Malaria is a major global health and economical problem, and although annual fatalities are decreasing, approximately 584,000 global deaths (the majority of which are children younger than 5 years) were reported by the World Health Organization in the 2014 World Malaria Report (1). Cerebral malaria (CM) is a neurological complication of malarial infection and a large contributor to malarial fatalities, with a similar number of survivors retaining long-term neurological deficits (2). CM is most common in children younger than 5 years in sub-Saharan Africa and adults in parts of Southeast Asia. The disease always exhibits severe neurological symptoms, such as blurred vision, impaired consciousness, and convulsions, which may proceed to coma and death (3, 4). Poor understanding of the specific biochemical mechanisms that contribute to the development of disease pathogenesis currently hinders any improvement in patient therapy.
Although much remains unknown about the chemical pathways through which CM causes mortality, an increased level of lactate in the cerebrospinal fluid (CSF) has been identified as a clinical prognostic marker in both advanced human CM and murine experimental cerebral malaria (ECM) (5–10). The pathology of ECM shares many similarities with the human condition, and postmortem studies of human victims and ECM reveal characteristic histological features that include sequestration of parasitized red blood cells (PRBCs) and immune cells (leukocytes, monocytes) within the cerebral blood microvessels, decreased patency of the blood-brain barrier (BBB) accompanied by edema, and microhemorrhage (4, 11–13). These findings suggest that elevated cerebral lactate is a direct consequence of ischemia, which results from vascular obstruction and/or hemorrhage. However, contributions from direct effects of proinflammatory cytokines are possible (7).
It is known that hemorrhage results in substantially increased cerebral Fe, which, alongside ischemia, results in peroxidative stress in disorders, such as stroke (14). Because cerebral ischemia and microhemorrhage are components of CM pathogenesis, this led to the hypothesis that reactive oxygen species, peroxidative stress, and protein oxidation are key components of CM pathogenesis (15, 16). Indeed, antioxidant administration before presentation of clinical signs in ECM prevents the disease (17). However, antioxidant treatment administered after the onset of clinical symptoms does not prevent or reduce illness, and biochemical analysis of brain homogenates provides no evidence of increased protein oxidation during ECM (18). These results appear to indicate that peroxidative stress is not a key player in CM pathogenesis and that the positive effect observed by early administration of antioxidants is the result of actions on another pathway, such as the inflammatory response (17, 18). However, it is possible that peroxidative damage occurs at discrete foci around microhemorrhage, and the critical biochemical alterations responsible for altered brain function are too dilute in a tissue homogenate for detection. Thus, the critical question of whether peroxidative stress is a key component of CM pathogenesis has remained unanswered until now.
Although traditional biochemical approaches, such as assays, nuclear magnetic resonance (NMR) spectroscopy, high-performance liquid chromatography, and mass spectrometry, are crucial for the quantification of specific metabolites in biological samples, they provide this information at the whole-tissue level (7, 19). Small yet critical biochemical alterations at the cellular level may not be detected when analysis is performed on tissue homogenates. Advances in technology have provided mass spectroscopic imaging modalities, but these are not applicable at the cellular level (20). Traditional imaging methods, such as electron microscopy, fluorescence, and optical microscopy in combination with immunocytochemistry, suffer serious limitations for direct quantification of important markers of peroxidative stress. For example, histochemical stains for Fe do not detect heme Fe and display varied specificity and sensitivity between ferric and ferrous states (21). Qualitative localization of highly concentrated regions of β sheet aggregates, such as amyloid-β plaques, can be performed with thioflavin S/T, Congo red dyes, or immunocytochemistry with antibodies specific for amyloid-β (22). However, no method is available for semiquantitative localization of non–plaque-bound aggregated proteins that may be formed during protein oxidation (23, 24). Similarly, lactate, which is a marker of hypoxic metabolism and a mobile water-soluble metabolite, cannot be studied using histology or immunocytochemistry. Although lactate can be investigated in vivo using NMR spectroscopy, determination at spatial resolution relevant to microvasculature (~10 μm) is not possible (6, 7, 19, 25). Conventional microscopy techniques are well suited to studying alterations in cell structure as a consequence of disease or the location of specific enzymes implicated in cerebral metabolism [for example, lactate dehydrogenase (LDH)] and oxidative stress (glutathione reductase). However, the large discrepancy in spatial resolution between microscopy and techniques capable of direct quantification of metabolites and oxidation products means correlation between data obtained by both approaches is speculative, at best. This is an important reason behind the uncertainty in discerning aspects of the biochemical mechanisms that contribute to CM and other neurodegenerative diseases. Therefore, a novel multimodal imaging approach that provides the ability to image heme and nonheme Fe, aggregated proteins, and lactate, at the cellular level, would be of great benefit to studies of CM pathogenesis, and neurodegenerative diseases in general.
Several chemical probe–free spectroscopic modalities show great potential for application to the neuroimaging field: Fourier transform infrared microscopy (FTIRM), resonance Raman spectroscopy, x-ray fluorescence microscopy (XFM), particle-induced x-ray emission (PIXE) spectroscopy, scanning transmission ion microscopy, and Rutherford backscattering (26–33). FTIRM is highly sensitive to protein secondary structure and has found numerous applications in imaging increased aggregated β sheet proteins within degenerating neurons, lesions, and plaques in Alzheimer’s disease (34), Parkinson’s disease (35), multiple sclerosis (36), and prion-diseased tissue (37), and following seizures (38). The method has also been used to map locations of elevated lactate in cerebral tumors (39) and to image lactate within cerebellar Purkinje neurons (28).
Nondestructive FTIRM can be combined with complementary resonance Raman spectroscopy, PIXE, and XFM in multimodal imaging (26, 27, 34). XFM and PIXE elemental mapping have found widespread applications for monitoring changes in brain chemistry in an array of neurological conditions [for example, stroke (33), Alzheimer’s disease (34), Parkinson’s disease (40), prion disease (41), and epilepsy (42)]. In addition, resonance Raman spectroscopy, which takes advantage of the preferential enhanced sensitivity to Fe coordinated in a heme environment, has been used to image the distribution of hemoglobin in vascular tissue (43–45). Unfortunately, a large portion of these publications reported results that may be compromised by inappropriate sample preparation, such as formalin fixation and paraffin embedding. Although these methods preserve cellular architecture, they cause significant alterations in molecular and elemental distribution, and oxidation state, within tissues (30). Further, although it is common to use FTIRM and XFM methods in biochemical investigations (34), no previous study has used a suite of spectroscopic techniques in combination with traditional microscopy and biochemical methods to take full advantage of their complementary nature.
In this investigation, FTIRM, PIXE, XFM, and resonance Raman spectroscopy were used to map or image biochemical alterations at the cellular or near-cellular level within the cerebellum that resulted from murine CM and to correlate the location of these alterations with the classical histological features of CM. This multimodal spectroscopic approach was performed in combination with traditional immunohistochemistry and biochemical techniques [enzymatic assays and real-time polymerase chain reaction (PCR)] and forms the first study to reveal a wealth of biochemical information at sufficient spatial resolution to test the hypothesis that protein oxidation and anaerobic metabolism occur at the sites of microhemorrhage and elevated cerebral Fe content within the cerebellum during murine CM and, by inference, in human diseases.
RESULTS
Demonstration of characteristic histological features of murine CM
Immunohistochemical detection of the serum protein fibrinogen and routine hematoxylin and eosin (H&E) histology were used to demonstrate the characteristic histological features of ECM, such as immune cell sequestration, BBB compromise, and microhemorrhage. A representative example of immunohistochemical localization of fibrinogen that demonstrates increased BBB permeability to protein in ECM is presented in Fig. 1.
Fig. 1. Immunohistochemistry shows compromised BBB and increased fibrinogen content in the brain parenchyma of CM mice.
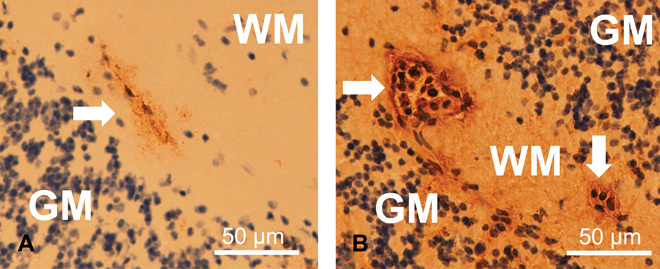
(A and B) Immunohistochemical localization of the plasma protein fibrinogen in the cerebellum of control (A) and CM (B) mice. Brown DAB staining, indicating increased BBB compromise and leakage of the plasma protein fibrinogen into the brain parenchyma, was observed in mice with CM. Counterstaining with hematoxylin revealed numerous leukocyte nuclei within the microvessels (arrows) in CM mice. WM, white matter; GM, gray matter.
Malaria parasites are not the source of elevated cerebral lactate during murine CM
Lactate and parasite DNA content were determined via enzymatic assay (lactate) and real-time PCR (DNA) in lung and brain homogenates (Fig. 2L). A significant increase in brain lactate was observed in ECM relative to controls. ECM brain lactate was significantly greater than ECM lung lactate. No difference in lung lactate was observed between ECM and controls. However, parasite DNA content displayed the reverse trend to lactate, with significantly more parasite DNA present in the ECM lung than in ECM brain tissue. Thus, there was no correlation between the lactate concentration and the number of malaria parasites (as indicated by parasite DNA) in these tissues, indicating that parasites are not the source of increased brain lactate during ECM.
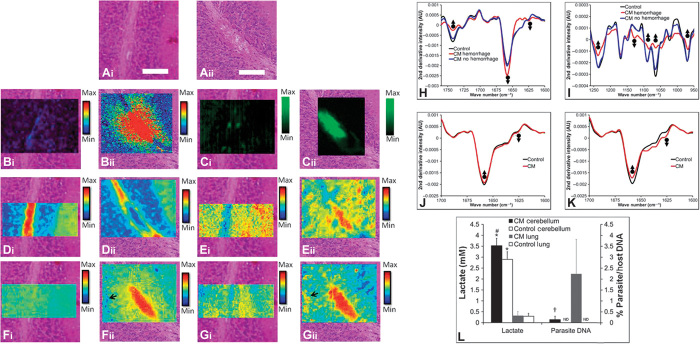
Fig. 2. Multimodal biospectroscopic approach to studying the biochemical mechanisms of CM.
(A to G) Biospectroscopic imaging in combination with histology of cerebellum tissue in CM and control mice. (A) Representative example of H&E histology of cerebellum tissue from control (i) and CM (ii) mice, highlighting the appearance of tissue edema at the site of vascular hemorrhage. (B) XFM elemental mapping of the distribution of Fe in control (i) and surrounding hemorrhaged tissue in CM (ii) cerebellum tissue. (C) Resonance Raman mapping of the distribution of hemoglobin in control (i) and hemorrhaged tissue in CM (ii) cerebellum tissue. (D) FTIR imaging of the lipid (second-derivative band intensity at 1742 cm−1) distribution in control (i) and hemorrhaged tissue in CM (ii) cerebellum tissue. (E) FTIR imaging of the lactate (second-derivative band intensity at 1127 cm−1) distribution in control (i) and hemorrhaged tissue in CM (ii) cerebellum tissue. (F) FTIR imaging of the α helix protein (second-derivative band intensity at 1656 cm−1) distribution in control (i) and hemorrhaged tissue in CM (ii) cerebellum tissue. (G) FTIR imaging of the aggregated β sheet protein (second-derivative band intensity at 1627 cm−1) distribution in control (i) and hemorrhaged tissue in CM (ii) cerebellum tissue. Black arrows in (Fii) and (Gii) highlight the location of elevated aggregated β sheet/α helix protein ratio hotspots. Scale bar in (A), 50 μm. (H to K) Representative second derivatives of the average FTIR spectra from control and CM mouse cerebellum tissue. (H) Lipid and protein alterations in the spectra from hemorrhaged and nonhemorrhaged inner white matter from CM mice and from inner white matter of control mice. AU, arbitrary units. (I) Differences in lactate in the spectra from hemorrhaged and nonhemorrhaged inner white matter from CM mice and from inner white matter of control mice. (J) Protein alterations in the spectra from granular layer tissue in CM mice and control mice. (K) Protein alterations in aggregated protein “hotspots” in the spectra from granular layer tissue of CM and control mice. Upward-facing arrows highlight spectral locations of decreased second-derivative band intensity, and downward facing arrows highlight spectral locations of increased second-derivative band intensity. (L) Brain and lung lactate levels in CM diseased and control mice (n = 5). Parasite DNA was not detected (ND) in the brain or lung of control mice. #Significant difference in brain lactate between CM and control mice. *Significant difference in lactate between the brain and lung of CM mice, or between the brain and lung of control mice. †Significant difference in parasite DNA between the brain and lung of CM mice.
Identification and validation of spectroscopic markers of protein oxidation and altered metabolism for FTIRM
The use of spectroscopic marker bands centered at 1742 and 1656 cm−1 to image the relative distribution of lipids and α helix proteins, respectively, in cerebellum tissue is well established (28–30, 36). Here, we conclusively demonstrate that the second-derivative intensity of the bands at 1627 and 1127 cm−1 from FTIR spectra collected from brain tissue is suitable for the analysis of relative aggregated β sheet protein (a marker of protein oxidation) and lactate content (a marker of disturbed metabolism), respectively. The methods used to validate these markers are presented in fig. S1.
FTIRM demonstrates elevated lactate and protein aggregates at the site of vascular hemorrhage
Relative distributions of lipids, α helix proteins, aggregated β sheet proteins, and lactate in the cerebellum were determined from second-derivative intensities at 1742, 1656, 1625, and 1127 cm−1, respectively. To demonstrate that molecular alterations observed in the cerebellum during ECM were not a consequence of terminal illness, or a response to generalized parasite infection, brain tissues from three sets of control animals were imaged: saline-injected mice, mice with mild malarial anemia, and mice with terminal severe malarial anemia without cerebral complications (see Materials and Methods for a detailed description of the animal models).
To correlate molecular images with brain anatomy and CM pathology, the false-color functional group images were overlayed with visible light images of the H&E-stained adjacent tissue section (Fig. 2, A to G, and fig. S3). The average spectrum was determined from regions of interest that corresponded to specific tissue regions: molecular layer, granular layer, inner white matter, and hemorrhaged tissue (Fig. 2). A representative example of these regions is shown in fig. S3. An alternative approach, using multivariate analysis, yielded similar scientific conclusions (figs. S4 and S5). Comparison of average spectra from each tissue region for each treatment group was used for statistical evaluation of significant differences in the average relative lipid, α helix and aggregated β sheet protein, and lactate contents (Fig. 2 and Table 1).
Table 1. Differences in relative content of lipids, α helix protein, aggregated β sheet protein, and lactate as determined from second derivatives of FTIR band intensities for hemorrhaged inner white matter (WM HM), nonhemorrhaged inner white matter (WM), granular layer (GL), and molecular layer (ML) from CM and sham, mild malaria anemia (MMA), and severe malaria anemia (SMA) mice.
Bold indicates a statistically significant difference (P < 0.05), relative to sham animals.
| Sample | 1742 cm−1 (lipid) | 1656 cm−1 (α helix protein) | 1625 cm−1 (aggregated β sheet protein) | 1127 cm−1 (lactate) | |
| CM WM HM | Average | −2.5 × 10−4 | −2.6 × 10−3 | 2.3 × 10−4 | 1.3 × 10−5 |
| SD | 1.1 × 10−5 | 3.5 × 10−4 | 3.0 × 10−5 | 2.0 × 10−6 | |
| CM WM | Average | −6.6 × 10−4 | −1.8 × 10−3 | 3.0 × 10−4 | 6.6 × 10−5 |
| SD | 4.5 × 10−5 | 3.5 × 10−4 | 9.8 × 10−6 | 3.3 × 10−5 | |
| Control (sham) WM | Average | −7.2 × 10−4 | −1.8 × 10−3 | 3.6 × 10−4 | 9.1 × 10−5 |
| SD | 5.2 × 10−5 | 1.6 × 10−4 | 5.9 × 10−5 | 1.9 × 10−5 | |
| Control (MMA) WM | Average | −6.9 × 10−4 | −1.8 × 10−3 | 3.3 × 10−4 | 8.7 × 10−5 |
| SD | 2.5 × 10−5 | 2.6 × 10−4 | 2.7 × 10−5 | 1.4 × 10−5 | |
| Control (SMA) WM | Average | −7.0 × 10−4 | −1.8 × 10−3 | 3.4 × 10−4 | 8.6 × 10−5 |
| SD | 3.5 × 10−5 | 3.0 × 10−4 | 4.3 × 10−5 | 1.3 × 10−5 | |
| CM GL | Average | −2.4 × 10−4 | −2.0 × 10−3 | 2.5 × 10−4 | 6.0 × 10−5 |
| SD | 2.2 × 10−5 | 1.7 × 10−4 | 1.9 × 10−5 | 6.6 × 10−6 | |
| Control (sham) GL | Average | −2.4 × 10−4 | −2.1 × 10−3 | 3.0 × 10−4 | 5.0 × 10−5 |
| SD | 2.5 × 10−5 | 2.7 × 10−4 | 8.8 × 10−6 | 2.0 × 10−5 | |
| Control (MMA) GL | Average | −2.4 × 10−4 | −2.1 × 10−3 | 2.9 × 10−4 | 6.0 × 10−5 |
| SD | 3.0 × 10−5 | 2.7 × 10−4 | 1.5 × 10−5 | 3.3 × 10−5 | |
| Control (SMA) GL | Average | −2.6 × 10−4 | −2.1 × 10−3 | 2.7 × 10−4 | 5.5 × 10−5 |
| SD | 2.9 × 10−5 | 2.7 × 10−4 | 9.8 × 10−6 | 2.5 × 10−5 | |
| CM ML | Average | −1.6 × 10−4 | −2.1 × 10−3 | 2.7 × 10−4 | 4.0 × 10−5 |
| SD | 4.0 × 10−4 | 1.9 × 10−4 | 2.5 × 10−5 | 2.9 × 10−6 | |
| Control (sham) ML | Average | −3.5 × 10−5 | −2.1 × 10−3 | 2.9 × 10−4 | 4.1 × 10−5 |
| SD | 3.4 × 10−4 | 1.9 × 10−4 | 4.2 × 10−5 | 6.2 × 10−6 | |
| Control (MMA) GL | Average | −4.1 × 10−4 | −2.1 × 10−3 | 3.0 × 10−4 | 4.1 × 10−5 |
| SD | 7.5 × 10−5 | 2.0 × 10−4 | 4.2 × 10−5 | 3.0 × 10−6 | |
| Control (SMA) ML | Average | −2.3 × 10−4 | −2.2 × 10−3 | 2.7 × 10−4 | 3.8 × 10−5 |
| SD | 3.9 × 10−4 | 1.6 × 10−4 | 1.1 × 10−5 | 5.7 × 10−6 |
A significant decrease in lipid content (second-derivative intensity of the band at 1742 cm−1) was observed at the site of hemorrhaged tissue within cerebellar white matter during ECM, relative to control white matter (Fig. 2 and Table 1). This decrease was colocalized with an increased relative content of α helix (second-derivative intensity of the band at 1656 cm−1), greater aggregated β sheet proteins (second-derivative intensity of the band at 1627 cm−1), and higher lactate (second-derivative intensity of the band at 1127 cm−1), as shown in Fig. 2 and Table 1. Although additional lipid bands were not quantified, the decreased intensity in the lipid carbonyl band at 1742 cm−1 in hemorrhaged white matter occurred concomitant with reduced intensity of characteristic phospholipid bands, such as νas(PO22−), νs(PO22−), ν(C–O), and ν(P–O–C) at 1242, 1085, 1065, and 970 cm−1, respectively. In addition, on average, there was a subtle, yet significant, increase in the relative content of aggregated β sheet proteins and a decrease in the relative content of α helix proteins in the granular layer tissue surrounding hemorrhaged white matter during ECM (Fig. 2 and Table 1). Differences were not uniform across the granular layer but rather occurred in discrete “hotspots” (representative spectra from the hotspots are shown in Fig. 2). Of significance, these hotspots occurred at the periphery of elevated brain Fe microdomains, which were not hemoglobin, as determined from XFM and resonance Raman mapping (see below). These results from univariate spectral analysis are in agreement with, and are further supported by, multivariate analysis of the data (principal components analysis), presented in figs. S4 and S5. Additional analyses showed that the scientific conclusions drawn from these results were not influenced by spectral artifacts caused by resonant Mie scattering or electric field standing wave artifacts (fig. S6).
Elemental mapping of Fe distribution around hemorrhaged microvasculature shows spread of Fe from hemorrhage into brain parenchyma
After FTIRM, elemental maps (P, S, Cl, K, Ca, Fe, Cu, and Zn) were obtained from XFM mapping of the same sections to determine whether FTIR spectroscopic markers of protein oxidation observed in the CM mice colocalized with elevated tissue Fe. A preliminary study used XFM and PIXE elemental mapping to validate the ability to resolve Fe distribution in healthy and hemorrhaged cerebral blood microvessels (see figs. S7 and S8). A representative example of the elemental distribution across ECM and control cerebellum (5-μm step size), together with high-resolution maps (0.5-μm step size) of healthy cerebral blood microvessels and microhemorrhage, is presented in Fig. 3. Visual examination of the elemental maps provided discrimination between the different cerebellar tissue layers. The average elemental concentrations for each tissue region (molecular layer, granular layer, inner layer, and hemorrhaged tissue) were quantified from XFM elemental maps (Fig. 4). As can be seen from the elemental maps and average elemental concentrations, significantly increased Fe and significantly decreased P content were observed at the site of hemorrhaged white matter tissue in ECM relative to controls (Figs. 3 and 4). Further, as evident in the Fe map, the elevated Fe was not confined to the region identified as “hemorrhaged” tissue based on histology but extended into the surrounding granular layer tissue. Notably, the highly localized regions of elevated aggregated β sheet protein content were observed at the periphery of the Fe microdomain (Fig. 3).
Fig. 3. XFM analysis of the elemental alterations in the cerebellum during murine CM.
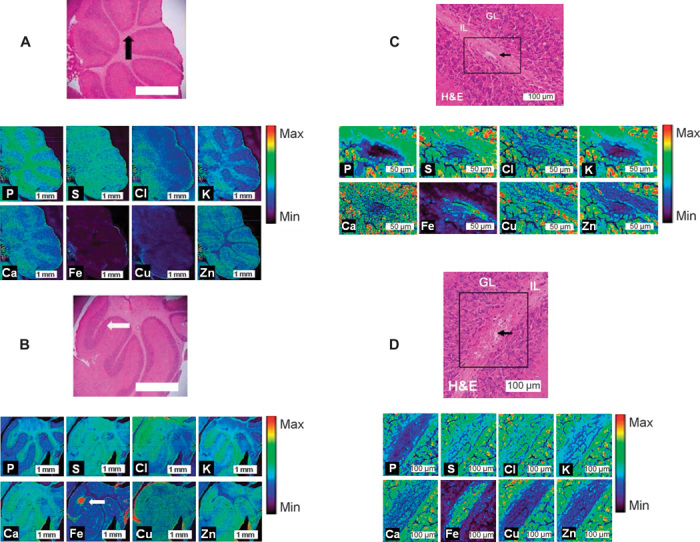
(A and B) XFM elemental maps (P, S, Cl, K, Ca, Fe, Cu, and Zn) collected with a 5-μm step size of the cerebellum of control (A) and CM (B) mice. H&E histology is presented for the entire cerebellum and at sites of healthy and hemorrhaged vasculature. IL, inner layer (white matter); GL, granular layer (gray matter). Arrows indicate the location of healthy vasculature in controls (black) and hemorrhaged (white) vasculature in ECM. (C and D) High–spatial resolution XFM elemental maps (P, S, Cl, K, Ca, Fe, Cu, and Zn) collected with a 0.5-μm step size of the cerebellum of control (C) and CM (D) mice. H&E histology is presented for the sites of healthy and hemorrhaged vasculature, indicated by black arrows. Note that high relative Fe content is only observed within the microvessel in the sham animal (similar to the preliminary investigation, fig. S8), whereas high Fe is observed throughout the white matter in the CM mouse, with low Fe observed in the white matter surrounding the vessel. IL, inner layer (white matter); GL, granular layer (gray matter).
Fig. 4. XFM elemental concentrations showing a significant difference in the P and Fe concentrations observed in hemorrhaged white matter tissue in CM mice, relative to healthy molecular layer, granular layer, and white matter tissue in control mice (n = 5).
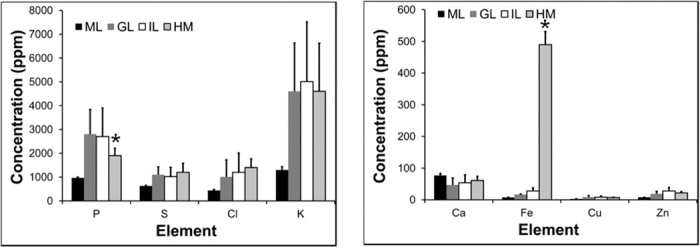
*Significant difference relative to control tissue. IL, inner layer (white matter); GL, granular layer (gray matter); ML, molecular layer; HM, hemorrhaged tissue.
Resonance Raman mapping of the distribution of hemoglobin around hemorrhaged microvasculature reveals elevated nonheme-bound Fe in brain parenchyma surrounding hemorrhaged tissue
The highest total Fe content in ECM brain tissue can be found at the center of the microhemorrhage, colocalized with the highest concentration of hemoglobin (Fig. 2). Further, the resonance Raman map of hemoglobin showed a similar distribution to hemorrhaged tissue, as seen by the loss of lipid and P from the white matter and from histological examination of the H&E-stained tissue section. No resonance Raman signal from hemoglobin or any other heme moiety colocalized with the regions of elevated Fe was observed in the granular layer tissue, which surrounded the hemorrhaged white matter tissue in ECM. A representative example of a Raman spectrum from the hemorrhaged tissue that shows the characteristic hemoglobin bands is presented in fig. S9.
DISCUSSION
Long-debated mechanistic controversies on the biochemical mechanisms of CM pathogenesis
Classical histological features of CM, common to human and ECM, are (i) sequestration of PRBCs and immune cells (leukocytes and monocytes) within cerebral blood microvessels, (ii) compromised BBB, and (iii) microhemorrhage (4, 12, 13). These pathologies suggest that reduction or loss of blood flow due to vascular obstruction caused by sequestered cells or hemorrhage would result in hypoxia/ischemia and contribute to disease pathogenesis (8, 15, 16, 18, 46–48). In addition, release of Fe into the brain parenchyma following microhemorrhage may mediate free radical production and peroxidative stress through classic Fenton chemistry and may contribute to disease morbidity and mortality as proposed, but not proven, by others (8, 16–18, 46–48). As the disease pathology is localized to the microvasculature, direct biochemical proof of disturbed energy metabolism and peroxidative stress has been difficult to obtain with traditional methods (7, 8, 18, 19, 47). For example, although there is a known net increase in cerebral lactate during CM, it is not known whether this is a uniform global change or a site-specific increase. Likewise, although administration of antioxidants before hemorrhage prevented ECM, increased levels of oxidized proteins or other oxidation products have not been detected in bulk assays (8, 17, 18, 47). Further, direct biochemical markers of altered metabolism and peroxidative stress, such as lactate and high–molecular weight protein aggregates, are essentially “invisible” to the traditionally used imaging modalities such as electron microscopy, visible light microscopy, and fluorescence microscopy. Consequently, the exact location and role of ischemia and peroxidative stress during CM have long remained controversial and unresolved.
Elevated brain lactate during CM is of host, not parasite, origin and is colocalized at the site of tissue hemorrhage
Occurrence of elevated brain lactate is well established in CM, yet the exact location remains unknown (5–10, 19, 25, 46). Elevated lactate observed previously in brain tissue homogenates and CSF may have been due to a low-level uniform increase or to localized regions of significant elevation. Recent studies in ECM suggest the latter, with reduced brain oxygen and hypoxic conditions identified at discrete foci (49). Several factors may contribute to elevated brain lactate during CM, including lactate production by malaria parasites or ischemic brain tissue, response to cytopathic hypoxia and inflammatory cytokines, or leakage from serum due to increased BBB permeability and microhemorrhage (4, 7, 8, 12, 13, 19).
This investigation demonstrated that malaria parasites are not the dominant source of elevated lactate during CM. We cannot rule out a lactate contribution derived from PRBCs sequestered within brain blood microvessels. However, this study showed a large number of parasites within the lung during CM but no accompanying increase in lactate relative to control mice, whereas greater lactate was observed in CM brain tissue where parasite numbers were lower than those in the lung (Fig. 2). Therefore, elevated brain lactate during ECM must have been of host and not parasite origin and was likely to have resulted from hypoxic and/or ischemic brain tissue. This result was in strong agreement with two previous investigations that demonstrated reduced oxygen content in tissue surrounding microhemorrhages (49) and increased occurrence of crystalline creatine deposits in cerebellum tissue, a characteristic marker of ischemia, during ECM (29). Further, we demonstrate that elevated lactate was not uniform but occurred at the site of microhemorrhage (Fig. 2). This result highlighted that generation of lactate by brain cells in response to hypoxia, ischemia, or inflammatory cytokines, or serum-derived lactate, may contribute to elevated brain lactate. Although the methods used in this study did not discriminate between these possibilities, the highly localized nature of elevated lactate at the site of tissue hemorrhage demonstrated that brain ischemia and altered metabolism were not uniformly present in ECM brains but, rather, were likely to colocalize with damaged vasculature. Previous studies have shown a direct correlation between brain lactate and disease severity (5, 9). Likewise, declining brain function has been shown to correlate with the number of hemorrhagic events that occurred during CM (50). This study has demonstrated a direct correlation between elevated cerebral lactate and microhemorrhage, thus providing a link between the previous observations.
Fe-mediated Fenton chemistry, peroxidative stress, and protein oxidation are components of CM pathogenesis
A novel multimodal biospectroscopic imaging approach allowed the distribution of important biological parameters to be assessed at micrometer spatial resolution, in relation to classical histological features of murine CM. This confirmed lipid loss at the location of edema and microhemorrhage within white matter as demonstrated in the FTIRM lipid (Fig. 2) and elemental P maps (Fig. 3). The appearance of serum proteins in brain parenchyma due to BBB leakiness was confirmed by immunohistochemistry for the serum protein fibrinogen (Fig. 1). Because of the high content of α helix secondary structure in most serum proteins, it is not surprising that FTIRM revealed a large increase in the relative content of proteins with this feature at the site of tissue microhemorrhage.
Of greater interest in this investigation was an increase in the relative content of aggregated β sheet proteins colocalized at the site of tissue microhemorrhage and at the periphery of parenchymal microdomains containing Fe (Fig. 2). Two physiological sites of protein oxidation, one at the site of tissue hemorrhage and another at the furthest extent of elevated parenchymal Fe, may initially seem counterintuitive. However, this agrees with the known pathology of vascular damage, red blood cell lysis, hemoglobin degradation, and Fenton chemistry. First, because of substantial inflammation and immune cell sequestration within the blood microvessels, free radical production is likely to be increased because of localized ischemia or cytokine-mediated release of reactive oxygen species from immune cells (15, 18, 47, 51, 52). Therefore, one would expect oxidation of proteins and the formation of high–molecular weight protein aggregates at the site of microhemorrhage. Following hemorrhage, most of the hemoglobin released into the brain tissue may not be immediately damaging; however, upon hemoglobin breakdown, liberation of redox-active free Fe(II) may saturate native antioxidant mechanisms through classical Fenton chemical pathways (53, 54). We hypothesize that Fe, which has spread furthest into the brain from the site of hemorrhage, has spent the longest time in the parenchyma, most likely originates from degraded hemoglobin, is redox-active, and may have reached tissue that is sufficiently distant from the vasculature that is yet to up-regulate levels of defense mechanisms to combat Fe-mediated oxidative stress. The results from this investigation provide strong support for this theory: Elemental mapping revealed elevated Fe at the site of hemorrhage as well as spreading into the surrounding brain parenchyma; resonance Raman mapping demonstrated that only Fe at the site of tissue hemorrhage, but not Fe in the surrounding brain parenchyma, was heme-bound; and FTIRM showed that there was a significant increase in the relative content of aggregated β sheet proteins colocalized with the boundary of elevated parenchymal nonheme Fe. Hence, this is the first study to provide direct evidence for Fe-mediated Fenton chemistry, peroxidative stress, and protein oxidation following microhemorrhage during CM. These results may help explain some of the success of deferoxamine Fe chelation therapy in reducing coma in children with CM (55). However, the exact mechanism of action of Fe chelation therapy is not fully understood, and it is not known whether Fe chelation acts via reduction of free Fe in the brain parenchyma, thus reducing oxidative stress, or whether Fe chelation acts to reduce parasitemia through Fe starvation of the parasite (55, 56). Although some success in minimizing patient coma with Fe chelation therapy has been achieved, other studies have shown no effect on patient morbidity (56). Therefore, the results of this study indicate that new therapies specifically targeted to chelate free Fe, minimize oxidative stress, and minimize BBB breakdown may provide substantial patient benefit.
The combination of resonance Raman spectroscopy and XFM allowed identification of elevated nonheme Fe in tissue surrounding microhemorrhage. The enhanced scattering signal that arises from resonance Raman spectroscopy has been used to map oxyhemoglobin and deoxyhemoglobin in blood vessels (43–45). In this investigation, a similar approach was used to map the distribution of hemoglobin at the site of microhemorrhage in ECM mice. Representative resonance Raman spectra from hemorrhaged tissue and from dried red blood cells are presented in fig. S9. On the basis of the appearance of the strong band at 1376 cm−1, oxyhemoglobin was likely to be the dominant form present (43–45). However, a small band is also observed at ~1358 cm−1, indicating some contribution from deoxyhemoglobin (43–45). It is assumed that during the chemical reaction characteristic of Fenton chemistry, the Fe ions will redox cycle and both ferric and ferrous forms will be present. Future studies may incorporate x-ray absorption spectroscopy speciation studies (57) to determine the relative proportions of ferric and ferrous Fe in heme and nonheme forms present in the brain parenchyma surrounding tissue hemorrhage.
CONCLUSIONS
The multimodal spectroscopic approach (FTIRM, PIXE, XFM, and resonance Raman spectroscopy) in combination with traditional histology, immunohistochemistry, and biochemical assays used in this study provided the unique capability of detecting an unprecedented level of biochemical and pathological information regarding disease mechanisms of CM. Specifically, the results provide the first direct evidence that elevated levels of anaerobic metabolites are not a uniform global condition during murine CM and are highly localized at the site of tissue microhemorrhage. In addition, this study shows the first strong, and direct, evidence for Fe-mediated protein oxidation following hemorrhage and hemoglobin breakdown during CM. These results are potentially valuable for future development of improved therapies for CM and indicate that anti-inflammatory therapy to prevent/minimize BBB damage and hemorrhage, development of greater efficacy of antioxidant delivery to the site of hemorrhage, and/or up-regulation of the native brain antioxidant capacity, such as neuroglobin (58), may result in improved patient outcome. The FTIR and resonance Raman spectroscopic techniques used in this study have a short penetration depth (typically <20 μm) and are not suitable for in vivo diagnostics or biochemical analysis in humans. However, continued development of micro–attenuated total reflection (micro-ATR) FTIR devices and fiber-optic Raman probes may allow, in the near future, biochemical information similar to that reported in this study to be detected in real time in vivo. This may provide significant benefit in identifying the extent of disease progression and the administration of an optimized therapy.
MATERIALS AND METHODS
This study used validated mouse models to generate ECM, experimental nonfatal mild malaria anemia without cerebral complications [experimental mild malaria anemia (EMMA)], experimental fatal malaria anemia without cerebral complications [experimental severe malaria anemia (ESMA)], and sham controls (7, 8, 29). Tissue harvesting and preparation of brain tissue were performed as in the Supplementary Materials, as described previously (30). The left hemisphere of the cerebellum was homogenized in phosphate-buffered saline (PBS) at 4°C, the supernatant was retained for biochemical assay of lactate (using the protocol described in the Supplementary Materials), and the pellet was retained for assay of host and parasite DNA content (using the protocol described in the Supplementary Materials). Four adjacent 10-μm-thick tissue sections were cut from the right hemisphere and mounted on a Si3N4 window for FTIRM/XFM analysis (27), on an aluminum slide (prepared in-house) for Raman spectroscopic analysis, and on standard glass microscope slides for routine H&E staining. Additional tissue sections were cut for further investigation of the elemental composition of blood vessels and scattering artifacts in FTIR spectra and for immunohistochemistry of plasma proteins (as described below).
FTIRM with a focal plane array detector was performed using equipment and protocols as described previously (29, 30), and described in detail below. In general, images were collected from an area of the cerebellum ranging from 170 μm × 170 μm to 680 μm × 680 μm, containing all three tissue layers (molecular layer, granular layer, and inner white matter). Following FTIRM, tissue sections were freeze-dried, and elemental mapping was performed on one set of tissue sections using PIXE and on the other set of tissue sections using XFM. Raman spectroscopic mapping was performed using 514-nm excitation, as described in detail in the Supplementary Materials. Following spectroscopic analysis, tissue sections were stained with H&E, in addition to the tissue section mounted on the glass microscope slide for routine histology.
The average molecular and elemental compositions were determined from regions of interest corresponding to hemorrhaged tissue, molecular layer, granular layer, and inner white matter, identified by a trained pathologist. A Student’s t test and 95% confidence limit were used to test for a significant difference between groups.
Animal models
All animals were 6-week-old female C57/B6 mice, given food and water ad libitum, and all experiments were approved by The University of Sydney Animal Ethics Committee and strictly followed Institutional Animal Care and Use Committee guidelines. ECM was induced in mice (n = 6) via intraperitoneal injection of diluted erythrocytes (200 μl) containing the malaria parasite Plasmodium berghei ANKA (59, 60). Experimental mild (noncerebral) malaria anemia and experimental severe (noncerebral) malaria anemia were induced in mice (n = 6 for both EMMA and ESMA) via intraperitoneal injection of diluted erythrocytes (200 μl) containing the malaria parasite P. berghei K173 (PBK) (60–62). In both cases, the 200-μl aliquot of blood contained a PRBC count of approximately 1 × 106. Sacrifice of mice infected with PBK at day 6 after inoculation equated to the mild anemia model, and sacrifice of mice at day 12 after infection equated to the severe anemia model (62, 63). Control (sham-infected) mice (n = 6) were injected with 200 μl of PBS via intraperitoneal injection.
Mice were examined for visual signs of illness at day 6 (ECM and EMMA) and day 12 (ESMA) after infection. EMMA mice displayed no visual signs of cerebral complications at day 6, and ESMA mice displayed no signs of cerebral complications at day 12 after infection. At day 12 after infection, ESMA mice displayed visual signs of severe malaria anemia (lethargy, hunched posture, and ruffled fur) (60–62). At day 6, ECM mice displayed signs of cerebral complications (impaired movement, impaired balance, and convulsions) (59, 61). Blood hematocrit and parasite counts were in the following ranges: sham mice, 40 to 50% and 0%, respectively; ECM and EMMA mice, 20 to 35% and 8 to 20%, respectively; ESMA mice, <15% and >50%, respectively [all were consistent with our previous reports (7, 60, 61)].
Tissue collection
Sham, ECM, and EMMA mice were sacrificed and brains were collected 6 days after infection, and ESMA mice were sacrificed 12 days after infection. Mice were anesthetized by inhalation of isoflurane vapor before sacrifice and were euthanized by cervical dislocation. Immediately after sacrifice, mouse brains were removed and the cerebellum was obtained and cut in half along the sagittal plane. The left hemisphere was immediately snap-frozen in liquid N2 and the right hemisphere was embedded in optimal cutting temperature medium and snap-frozen in a liquid nitrogen–cooled hexane slurry. In addition, the lungs of ECM mice were removed and snap-frozen in liquid nitrogen. All samples were stored at −80°C until required for analysis.
Tissue homogenization
Lung tissue and the left cerebellar hemisphere of control and ECM diseased mice were homogenized separately in 200 μl of PBS at 4°C. A 200-μl aliquot of each homogenate was centrifuged at 16,000g for 20 min at 4°C. The pellet was retained for parasite DNA analysis, and the supernatant fraction was retained for enzymatic lactate assays and bulk FTIR spectroscopic analyses.
Parasite DNA quantification
Parasite levels in brain and lung tissue were compared by calculating the ratio of a parasite gene to a mouse gene. The pellet from brain homogenates was resuspended in 200 μl of tissue lysis buffer (supplied with kit) and DNA was purified using the High Pure PCR Template Preparation Kit (Roche Applied Science). PCR amplification was performed using 1× ImmoMix (Bioline), 200 nM primer, and 1× SYBR Green I (Life Technologies) in a Rotor-Gene3000 (Corbett Research). The touchdown PCR amplification protocol comprised 35 cycles of 95°C for 15 s, 60°C (decreasing to 55°C over 6 cycles) for 15 s, and 72°C for 15 s. Purity of PCR products was assessed by melting curve analysis, and a standard curve derived from serial dilution of DNA from infected tissue was used to measure the levels of the host and parasite genes. P. berghei gene: carbamoyl phosphate synthetase II AF286897.1 (forward, TAAAACTGCTATTCAAACCGCC; reverse, GCTACCCCATTCTAGTGCGTACT). Mus musculus gene: tyrosine 3-monooxygenase activation protein, zeta polypeptide NM_001253807.1 (forward, TGTCACCAACCATTCCAACTTG; reverse, ACACTGAGTGGAGCCAGAAAGA).
LDH assay
Lactate concentrations in the supernatant fraction of cerebellum homogenates were determined using the Eton Bioscience enzymatic lactate assay kit. In general, lactate and NAD+ (nicotinamide adenine dinucleotide) in each sample were converted into pyruvate and NADH (reduced form of NAD+) by LDH. A tetrazolium salt was then reduced to formazan in an NADH-coupled enzyme reaction. The concentration of formazan (which is proportional to the lactate concentration) was quantified by spectrophotometric measurement at 490 nm.
Determination of FTIRM spectroscopic markers of lactate
Supernatant solution (100 μl) from control mice and ECM diseased mice was analyzed either without chemical modification or after the addition of 56 μg of lactate (C3H5O3Na, Sigma-Aldrich). An additional solution was prepared via the addition of 10 μl of LDH (Sigma-Aldrich) to 100 μl of the supernatant, and 1-μl aliquots were harvested for analysis at 1- and 5-min time points after LDH addition. FTIR spectroscopic analysis was performed on 1-μl aliquots of each solution.
Determination of FTIRM spectroscopic markers of protein oxidation
Spectroscopic markers of protein oxidation were analyzed from spectra collected from the supernatant solution of brain homogenates from control and ECM diseased mice and incubated without chemical modification or following incubation with 1 mM Fe(II) (FeSO4) or Fe(II) in the presence of 5 mM Desferal for 40 min. FTIR spectroscopic analysis was performed on 1-μl aliquots of each solution.
Bulk FTIR spectroscopic analyses
Bulk FTIR spectra were acquired from dried films prepared from the supernatant fractions of cerebellum homogenates. A 1-μl aliquot of each sample was transferred onto an infrared transparent silicon 96-well microtiter plate (each sample was analyzed in triplicate). Aliquots were spread over an area of approximately 50 mm2 and allowed to air dry for a period of 30 min to produce a dried film. FTIR analyses of the dried films were performed using a Bruker Tensor 27 FTIR spectrometer fitted with a high-throughput sampling accessory (HTS-XT), a thermal glowbar infrared source, and a mercury cadmium telluride detector. Spectra were collected over the range 400 to 4000 cm−1 at a resolution of 4 cm−1, with the co-addition of 256 scans per spectrum. A background spectrum was taken before each sample measurement using the same parameters as above.
Preparation of tissue sections: Complementary multimodal spectroscopic imaging
Four adjacent 10-μm-thick sections of cerebellum tissue were cut from the right-hand hemisphere of cryo-preserved cerebellum tissue from control, ECM, EMMA, and ESMA diseased mice. Tissue sections were cut at −14°C with a cryotome (Thermo Shandon). Two sections were melted onto Si3N4 membranes (Silson Ltd.) for FTIRM and XFM analyses. The dimensions of the Si3N4 windows are as follows: membrane size, 5 mm × 5 mm; membrane thickness, 500 nm; Si frame size, 10 mm × 10 mm; and frame thickness, 200 μm. Immediately after FTIR analysis of the sections mounted on Si3N4 windows, the sections were freeze-dried under vacuum at −80°C for 12 hours. The third section was mounted onto a thin sheet of Al (prepared in-house) for Raman spectroscopic analyses. The fourth section was mounted onto a standard glass microscope slide and fixed in 4% phosphate-buffered formalin (Sigma-Aldrich) for routine histological analysis.
Preparation of tissue sections for figs. S5 to S7
Three tissue sections were collected for preliminary studies: to confirm that compromised BBB was present in ECM diseased mice (section A), to determine the elemental distribution and morphology of cerebral blood vessels in elemental maps (section B), and for synchrotron radiation FTIRM investigation of the protein signature in ECM cerebellum tissue (section C). Two 7-μm-thick sections (A and C) and one 10-μm-thick cerebellum tissue section (B) were cut from each brain and mounted on a standard glass microscope slide (A), Si3N4 window (B), and low E Kevley slide (C). Tissue sections were freeze-dried as described previously. These tissue sections were cut at a position approximately 100 to 200 μm medial compared to the four adjacent sections described earlier.
FTIRM of cerebellum tissue sections with a focal place array detector
Images were acquired using a Bruker Vertex 80v FTIR spectrometer, coupled to a Hyperion 3000 microscope. The microscope is equipped with a liquid nitrogen–cooled 64 × 64 focal plane array detector. Opus software (version 6.5, Bruker) was used for instrument control and data collection. Spectra were acquired with the co-addition of 64 scans at 4 cm−1 resolution over the range 3600 to 900 cm−1. Images were collected in transmission mode using a 15× microscope objective and 2 × 2 pixel binning. The sampling area ranged from 170 μm × 170 μm (1024 spectra) to 680 μm × 680 μm (16,384 spectra). All data were collected from a region within the brain cerebellum containing the molecular, granular, and inner white matter layers. A background image (1024 spectra) was collected from a blank Si3N4 substrate before the collection of each sample image.
Synchrotron radiation FTIRM of cerebellum tissue sections (fig. S5)
Cerebellum tissue was mounted on a Kevley slide and analyzed in the mid-IR region. Data were collected over 4000 to 800 cm−1 with the co-addition of 128 scans on a Bruker Vertex 80v FTIR spectrometer with a 36× objective and a mercury cadmium telluride detector. Maps were collected with an aperture beam spot of 5 μm and a step size of 5 μm.
Streamline Raman spectroscopic mapping
Raman spectroscopic maps were acquired from 10-μm-thick tissue sections mounted on thin Al sheets. Raman spectroscopic maps were acquired following FTIR measurements using StreamLine mapping (Renishaw plc.) on a Renishaw Raman inVia Reflex spectrometer and controlled with Renishaw WiRE software (Renishaw, WiRE, version 3.2). The inVia Reflex spectrometer was equipped with an air-cooled charge-coupled device camera and a Leica DMLM microscope. Sample excitation was achieved using an Ar+ laser (Modu-Laser) emitting at 514.5 nm. Wave number calibration was achieved against the reference Si sample (Si band position, 520.5 ± 0.1 cm−1). Stokes Raman spectra were collected using a grating of 1200 lines per mm across the spectral range of 1900 to 650 cm−1. Spectra were collected with a 50× microscope objective, using 100% laser power, with an accumulation time of 10 s and an effective step size of 5 μm. The size of the maps collected ranged from 100 μm × 100 μm to 500 μm × 500 μm.
Elemental mapping with XFM at the Advanced Photon Source
Following FTIRM experiments, both sets of the 10-μm-thick tissue sections mounted on Si3N4 were immediately freeze-dried under vacuum conditions (12 hours at −80°C). The freeze-dried sections were stored on a desiccant, in the dark, at ambient conditions until required for elemental analyses with XFM or PIXE. Two-dimensional spatial maps of elements (P, S, Cl, K, Ca, Fe, Cu, and Zn) were determined for the 10-μm-thick freeze-dried tissue sections. All XFM measurements were performed at the 2-ID-E beamline at the Advanced Photon Source (APS), Argonne National Laboratory. X-ray fluorescence spectra were collected under a He atmosphere using monochromatic x-ray excitation (10 keV). The incident (excitation) beam was focused to 0.7 × 0.8 μm2 with a single zone plate and order sorting aperture device (63, 64). Fluorescence was detected 90° to the incident beam using a Si-drift detector (Vortex EM single-element SDD). Elemental maps of the entire cerebellum “fly scan maps” (65) (~3 × 3 mm2) were collected with 4-μm steps and a 40-ms dwell time before fine (high-resolution) elemental maps over an approximate area of interest of 200 × 200 μm2, with 0.7-μm steps and a 0.5-s dwell time.
Elemental mapping with XFM at the Australian Synchrotron
Freeze-dried tissue sections mounted on Si3N4 windows were analyzed at the XFM beamline at the Australian Synchrotron (66) to determine the elemental distribution of cerebral blood vessels. Two-dimensional spatial maps of elements (P, S, Cl, K, Ca, Fe, Cu, and Zn) were determined for the 10-μm-thick freeze-dried tissue sections. X-ray fluorescence spectra were collected in air using monochromatic x-ray excitation (10 keV). The incident (excitation) beam was focused to 2 × 2 μm2 full width at half maximum with a Kirkpatrick-Baez mirror pair. Fluorescence was detected 90° to the incident beam using a Si-drift detector (Vortex EM single-element SDD). Elemental maps were collected with 2-μm steps and a 4-s dwell time.
PIXE elemental analyses
All μ-PIXE analyses were performed using the Australian Nuclear Science and Technology Organization (ANSTO) High Energy Heavy Ion Microprobe (67–69). Analysis was performed with a 3-MeV proton beam and a 3- to 5-μm spot size. X-ray fluorescence spectra were collected over a 3 mm × 3 mm area using a high-purity Ge detector with an active area of 100 mm2. The detector was mounted at 90° to the incoming beam and the sample was mounted at 45° to the incoming beam. To reduce low-energy x-rays and to prevent scattered protons from entering the detector, a 100-μm-thick Mylar foil was placed in front of the detector. Elemental maps (P, S, Cl, K, Ca, Fe, Cu, and Zn) were collected from the same tissue regions analyzed using FTIR spectroscopic imaging. An obsidian standard (NBS 278) was used for calibration of the x-ray yield to allow quantitative measurements. The standard was mixed with 20% ultrapure carbon and pressed into a pellet to avoid charging.
H&E staining
The morphology and distribution of cerebral vessels, cell nuclei, collagen, and white matter were determined from H&E staining (70). Following staining, optical images were captured using a 4×, 10×, and 40× objective on an Olympus DP70 microscope controlled by DP Manager software (version 1.1.1.71, 2001 Olympus Optical).
Immunohistochemical staining of fibrinogen
The distribution of fibrinogen in the cerebellum tissue, as a marker of plasma proteins released into the brain due to compromised BBB, was studied with immunohistochemical staining following methods described previously (60, 71). Following staining, optical images were captured as described above.
Data processing of bulk FTIR spectra of dried films
Data analyses were performed using Opus Viewer 5.5 (Bruker Optic). Because of variation in the thickness of dried films, all spectra were preprocessed via vector normalization to the amide I band (1690 to 1610 cm−1). Second-derivative spectra were calculated from the normalized spectra using a nine-point Savitzky-Golay smoothing function.
Determination of regions of interest around cerebellar tissue layers
To determine the average molecular or elemental composition of cerebellar white matter, molecular layer, granular layer, and hemorrhaged vessels, overlap of FTIR functional group images or elemental maps with the adjacent H&E-stained section was performed and anatomical regions were defined. For FTIR functional group images, one further step—hierarchical cluster analysis—was performed, which was found to cluster spectra into the anatomical regions defined by overlay with the H&E image.
Fourier transform infrared microscopy
False-color functional group images were generated from inverted band intensities in second derivatives of the FTIR spectra using Cytospec version 1.2.04 software. The average spectrum from each tissue layer was calculated via a four-group Hierarchical Cluster Analysis (1700 to 1000 cm−1) and viewed using Opus Viewer 5.5.
Raman spectroscopic data analyses
Cosmic rays were removed from Raman spectra using a nearest-neighbor cosmic ray removal method (Renishaw WiRE 3 software tool). Noise-filtered Raman spectra were generated using a combination of direct classical least squares (Renishaw WiRE 3 component analysis) and alternating least-squares multivariate curve resolution (Renishaw’s Empty Modelling method). False-color functional group maps were then generated from the noise-filtered Raman spectra using Renishaw WiRE software (Renishaw, WiRE, version 3.2).
XFM data analyses
Quantitation (elemental area densities in micrograms per square centimeter) of P, S, Cl, K, Ca, Fe, Cu, and Zn was performed using the MAPS software package (version 1.6.5.03) by fitting the full fluorescence spectrum at every single point to modified Gaussians and comparing it with corresponding measurements on the thin-film standards NBS-1832 and NBS-1833 from the National Bureau of Standards/National Institute of Standards and Technology (72). The statistical significance of changes in the elemental content of treated cells compared to controls was assessed using Student’s t test and a 95% confidence limit.
PIXE spectroscopic data analyses
Spectra from μ-PIXE maps were fitted and elemental concentrations [P, S, Cl, K, Ca, Fe, Cu, and Zn (in parts per million)] were determined using GeoPIXE software (67–69). The Bremsstrahlung background was fitted for a light element ratio of C10O5H2 and a density of 1.9.
Statistical analyses
Significant differences between band area and elemental concentration were determined using Student’s t test and a 95% confidence limit.
Supplementary Material
Acknowledgments
We thank J. Radford for her expert assistance and the Histopathology Laboratory at The University of Sydney for sample preparation. Funding: M.J.H. acknowledges funding from the Australian Government through an Australian Postgraduate Award scholarship, a John A. Lamberton scholarship from The University of Sydney, and an Australian Institute of Nuclear Science and Engineering (AINSE) postgraduate scholarship. We acknowledge funding support from the Australian Research Council (ARC) (PAL DP0208409, DP0664706, and DP0984722, including ARC Professorial Fellowships, GEG Discovery DP0774425, NHH Discovery DP0987074, and PAL and EAC LIEF LE0883036) and the National Health and Medical Research Council (NHH 512469, GEG 464893, and GEG 512101). We are also grateful for grants from AINSE (AINGRA08038 and ALNGRA10039) and the Australian Synchrotron for support of experiments at ANSTO, APS, and the Australian Synchrotron. Use of the APS, an Office of Science User Facility operated for the U.S. Department of Energy (DOE) Office of Science by Argonne National Laboratory, was supported by the U.S. DOE under contract no. DE-AC02-06CH11357. This research was undertaken in part on the infrared micro-spectroscopy and XFM beamlines at the Australian Synchrotron (Victoria, Australia). Author contributions: M.J.H. contributed 25% of the experimental research, drafted the manuscript, and was involved in all of the experiments outlined. J.B.A. contributed 5% of the experimental research and edited the manuscript. F.E.-A. contributed 7% of the experimental research and edited the manuscript. J.A.M. contributed 7% of the experimental research and edited the manuscript. *E.A.C. contributed 5% of the experimental research and edited the manuscript. H.J.B. contributed 6% of the experimental research and edited the manuscript. M.J.T. contributed 5% of the experimental research and edited the manuscript. D.P. contributed 5% of the experimental research and edited the manuscript. M.D.d.J. contributed 5% of the experimental research and edited the manuscript. R.S. contributed 5% of the experimental research and edited the manuscript. D.D.C. contributed 5% of the experimental research and edited the manuscript. S.V. contributed 5% of the experimental research and edited the manuscript. *G.E.G. contributed 5% of the experimental research and edited the manuscript. *N.H.H. contributed 5% of the experimental research and edited the manuscript. *P.A.L. contributed 5% of the experimental research and edited the manuscript. (*Coauthors who held a lead role as project supervisors and devised the project with M.J.H.) Competing interests: The authors declare that they have no competing interests. Data and materials availability: Requests for materials should be addressed to P.A.L. (peter.lay@sydney.edu.au). All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Additional data related to this paper may be requested from the authors.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/1/11/e1500911/DC1
Text
Fig. S1. Validation of second-derivative band intensity at 1127 cm−1 in the FTIR spectra as a marker for relative lactate concentration.
Fig. S2. Validation of second-derivative band intensity at 1627 cm−1 in the FTIR spectra as a marker for relative concentration of aggregated β sheet proteins and, therefore, protein oxidation.
Fig. S3. Correlation of FTIR maps of lipid distribution with H&E histology.
Fig. S4. HCA analysis of FTIR maps distinguishes four distinct regions: the molecular layer, granular layer, inner white matter, and hemorrhaged white matter.
Fig. S5. Principal components analysis of the average spectra for tissue layers determined from the HCA.
Fig. S6. Synchrotron radiation–based (SR) FTIR mapping of cerebellum tissue to identify Mie scattering and electric field standing wave spectral features.
Fig. S7. SR x-ray fluorescence elemental maps (2-μm step size) of healthy cerebellum blood vessels, showing location of Fe (white arrow) within the wall of the blood vessel.
Fig. S8. PIXE elemental maps of healthy and hemorrhaged cerebellum blood vessels.
Fig. S9. Resonance Raman spectra (514-nm excitation) collected from hemorrhaged tissue and dried red blood cells, showing characteristic enhanced intensity of hemoglobin bands.
REFERENCES AND NOTES
- 1.WHO World Malaria Report 2014 (World Health Organization, Geneva, 2014).
- 2.van Hensbroek M. B., Palmer A., Jaffar S., Schneider G., Kwiatkowski D., Residual neurologic sequelae after childhood cerebral malaria. J. Pediatr. 131, 125–129 (1997). [DOI] [PubMed] [Google Scholar]
- 3.Mackintosh C. L., Beeson J. G., Marsh K., Clinical features and pathogenesis of severe malaria. Trends Parasitol. 20, 597–603 (2004). [DOI] [PubMed] [Google Scholar]
- 4.Coltel N., Combes V., Hunt N. H., Grau G. E., Cerebral malaria—A neurovascular pathology with many riddles still to be solved. Curr. Neurovasc. Res. 1, 91–110 (2004). [DOI] [PubMed] [Google Scholar]
- 5.Warrel D. A., Veall N., Chanthavanich P., Karbwang J., White N. J., Looareesuwan S., Phillips R. E., Pongpaew P., Cerebral anaerobic glycolysis and reduced cerebral oxygen transport in human cerebral malaria. Lancet 332, 534–538 (1988). [DOI] [PubMed] [Google Scholar]
- 6.Penet M.-F., Viola A., Confort-Gouny S., Le Fur Y., Duhamel G., Kober F., Ibarrola D., Izquierdo M., Coltel N., Gharib B., Grau G. E., Cozzone P. J., Imaging experimental cerebral malaria in vivo: Significant role of ischemic brain edema. J. Neurosci. 25, 7352–7358 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rae C., McQuillan J. A., Parekh S. B., Bubb W. A., Weiser S., Balcar V. J., Hansen A. M., Ball H. J., Hunt N. H., Brain gene expression, metabolism, and bioenergetics: Interrelationships in murine models of cerebral and noncerebral malaria. FASEB J. 18, 499–510 (2004). [DOI] [PubMed] [Google Scholar]
- 8.Sanni L. A., Rae C., Maitland A., Stocker R., Hunt N. H., Is ischemia involved in the pathogenesis of murine cerebral malaria? Am. J. Pathol. 159, 1105–1112 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.White N. J., Looareesuwan S., Phillips R. E., Warrell D. A., Chanthavanich P., Pongpaew P., Pathophysiological and prognostic significance of cerebrospinal-fluid lactate in cerebral malaria. Lancet 325, 776–778 (1985). [DOI] [PubMed] [Google Scholar]
- 10.Molyneux M. E., Taylor T. E., Wirima J. J., Borgsteinj A., Clinical features and prognostic indicators in paediatric cerebral malaria: A study of 131 comatose Malawian children. Q. J. Med. 71, 441–459 (1989). [PubMed] [Google Scholar]
- 11.Coltel N., Combes V., Wassmer S. C., Chimini G., Grau G. E., Cell vesiculation and immunopathology: Implications in cerebral malaria. Microbes Infect. 8, 2305–2316 (2006). [DOI] [PubMed] [Google Scholar]
- 12.Hunt N. H., Golenser J., Chan-Ling T., Parekh S., Rae C., Potter S., Medana I. M., Miu J., Ball H. J., Immunopathogenesis of cerebral malaria. Int. J. Parasitol. 36, 569–582 (2006). [DOI] [PubMed] [Google Scholar]
- 13.Hunt N. H., Grau G. E., Cytokines: Accelerators and brakes in the pathogenesis of cerebral malaria. Trends Immunol. 24, 491–499 (2003). [DOI] [PubMed] [Google Scholar]
- 14.Cherubini A., Ruggiero C., Polidori M. C., Mecocci P., Potential markers of oxidative stress in stroke. Free Radic. Biol. Med. 39, 841–852 (2005). [DOI] [PubMed] [Google Scholar]
- 15.Clark I. A., Hunt N. H., Cowden W. B., Oxygen-derived free radicals in the pathogenesis of parasitic disease. Adv. Parasitol. 25, 1–44 (1986). [DOI] [PubMed] [Google Scholar]
- 16.Pamplona A., Hanscheid T., Epiphanio S., Mota M. M., Vigário A. M., Cerebral malaria and the hemolysis/methemoglobin/heme hypothesis: Shedding new light on an old disease. Int. J. Biochem. Cell Biol. 41, 711–716 (2009). [DOI] [PubMed] [Google Scholar]
- 17.Thumwood C. M., Hunt N. H., Cowden W. B., Clark I. A., Antioxidants can prevent cerebral malaria in Plasmodium berghei-infected mice. Br. J. Exp. Pathol. 70, 293–303 (1989). [PMC free article] [PubMed] [Google Scholar]
- 18.Sanni L. A., Fu S., Dean R. T., Bloomfield G., Stocker R., Chaudhri G., Dinauer M. C., Hunt N. H., Are reactive oxygen species involved in the pathogenesis of murine cerebral malaria. J. Infect. Dis. 179, 217–222 (1999). [DOI] [PubMed] [Google Scholar]
- 19.Parekh S. B., Bubb W. A., Hunt N. H., Rae C., Brain metabolic markers reflect susceptibility status in cytokine gene knockout mice with murine cerebral malaria. Int. J. Parasitol. 36, 1409–1418 (2006). [DOI] [PubMed] [Google Scholar]
- 20.Hanrieder J., Phan N. T. N., Kurczy M. E., Ewing A. G., Imaging mass spectrometry in neuroscience. ACS Chem. Neurosci. 4, 666–679 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Meguro R., Asano Y., Odagiri S., Li C., Iwatsuki H., Shoumura K., Nonheme-iron histochemistry for light and electron microscopy: A historical, theoretical and technical review. Arch. Histol. Cytol. 70, 1–19 (2007). [DOI] [PubMed] [Google Scholar]
- 22.Barelli H., Lebeau A., Vizzavona J., Delaere P., Chevallier N., Drouot C., Marambaud P., Ancolio K., Buxbaum J. D., Khorkova O., Heroux J., Sahasrabudhe S., Martinez J., Warter J. M., Mohr M., Checler F., Characterization of new polyclonal antibodies specific for 40 and 42 amino acid-long amyloid beta peptides: Their use to examine the cell biology of presenilins and the immunohistochemistry of sporadic Alzheimer’s disease and cerebral amyloid angiopathy cases. Mol. Med. 3, 695–707 (1997). [PMC free article] [PubMed] [Google Scholar]
- 23.Mirzaei H., Regnier F., Protein:protein aggregation induced by protein oxidation. J. Chromatogr. B 873, 8–14 (2008). [DOI] [PubMed] [Google Scholar]
- 24.Requena J. R., Chao C.-C., Levine R. L., Stadtman E. R., Glutamic and aminoadipic semialdehydes are the main carbonyl products of metal-catalyzed oxidation of proteins. Proc. Natl. Acad. Sci. U.S.A. 98, 69–74 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Penet M.-F., Kober F., Confort-Gouny S., Le Fur Y., Dalmasso C., Coltel N., Liprandi A., Gulian J.-M., Grau G. E., Cozzone P. J., Viola A., Magnetic resonance spectroscopy reveals an impaired brain metabolic profile in mice resistant to cerebral malaria infected with Plasmodium berghei ANKA. J. Biol. Chem. 282, 14505–14514 (2007). [DOI] [PubMed] [Google Scholar]
- 26.Aitken J. B., Carter E. A., Eastgate H., Hackett M. J., Harris H. H., Levina A., Lee Y.-C., Chen C.-I., Lai B., Vogt S., Lay P. A., Biomedical applications of X-ray absorption and vibrational spectroscopic microscopies in obtaining structural information from complex systems. Radiat. Phys. Chem. 79, 176–184 (2010). [Google Scholar]
- 27.Carter E. A., Rayner B. S., McLeod A. I., Wu L. E., Marshall C. P., Levina A., Aitken J. B., Witting P. K., Lai B., Cai Z., Vogt S., Lee Y.-C., Chen C.-I., Tobin M. J., Harris H. H., Lay P. A., Silicon nitride as a versatile growth substrate for microspectroscopic imaging and mapping of individual cells. Mol. BioSyst. 6, 1316–1322 (2010). [DOI] [PubMed] [Google Scholar]
- 28.Hackett M. J., Borondics F., Brown D., Hirschmugl C., Smith S. E., Paterson P. G., Nichol H., Pickering I. J., George G. N., Subcellular biochemical investigation of Purkinje neurons using synchrotron radiation Fourier transform infrared spectroscopic imaging with a focal plane array detector. ACS Chem. Neurosci. 4, 1071–1080 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hackett M. J., Lee J., El-Assaad F., McQuillan J. A., Carter E. A., Grau G. E., Hunt N. H., Lay P. A., FTIR imaging of brain tissue reveals crystalline creatine deposits are an ex vivo marker of localized ischemia during murine cerebral malaria: General implications for disease neurochemistry. ACS Chem. Neurosci. 3, 1017–1024 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hackett M. J., McQuillan J. A., El-Assaad F., Aitken J. B., Levina A., Cohen D. D., Siegele R., Carter E. A., Grau G. E., Hunt N. H., Lay P. A., Chemical alterations to murine brain tissue induced by formalin fixation: Implications for biospectroscopic imaging and mapping studies of disease pathogenesis. Analyst 136, 2941–2952 (2011). [DOI] [PubMed] [Google Scholar]
- 31.Hackett M. J., Siegele R., El-Assaad F., McQuillan J. A., Aitken J. B., Carter E. A., Grau G. E., Hunt N. H., Cohen D., Lay P. A., Investigation of the mouse cerebellum using STIM and μ-PIXE spectrometric and FTIR spectroscopic mapping and imaging. Nucl. Instrum. Methods Phys. Res. Sect. B 269, 2260–2263 (2011). [Google Scholar]
- 32.Hackett M. J., Smith S. E., Paterson P. G., Nichol H., Pickering I. J., George G. N., X-ray absorption spectroscopy at the sulfur K-edge: A new tool to investigate the biochemical mechanisms of neurodegeneration. ACS Chem. Neurosci. 3, 178–185 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Silasi G., Klahr A. C., Hackett M. J., Auriat A. M., Nichol H., Colbourne F., Prolonged therapeutic hypothermia does not adversely impact neuroplasticity after global ischemia in rats. J. Cereb. Blood Flow Metab. 32, 1525–1534 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Miller L. M., Wang Q., Telivala T. P., Smith R. J., Lanzirotti A., Miklossy J., Synchrotron-based infrared and X-ray imaging shows focalized accumulation of Cu and Zn co-localized with β-amyloid deposits in Alzheimer’s disease. J. Struct. Biol. 155, 30–37 (2006). [DOI] [PubMed] [Google Scholar]
- 35.Szczerbowska-Boruchowska M., Dumas P., Kastyak M. Z., Chwiej J., Lankosz M., Adamek D., Krygowska-Wajs A., Biomolecular investigation of human substantia nigra in Parkinson’s disease by synchrotron radiation Fourier transform infrared microspectroscopy. Arch. Biochem. Biophys. 459, 241–248 (2007). [DOI] [PubMed] [Google Scholar]
- 36.Heraud P., Caine S., Campanale N., Karnezis T., McNaughton D., Wood B. R., Tobin M. J., Bernard C. C. A., Early detection of the chemical changes occurring during the induction and prevention of autoimmune-mediated demyelination detected by FT-IR imaging. Neuroimage 49, 1180–1189 (2010). [DOI] [PubMed] [Google Scholar]
- 37.Kneipp J., Miller L. M., Joncic M., Kittel M., Lasch P., Beekes M., Naumann D., In situ identification of protein structural changes in prion-infected tissue. Biochim. Biophys. Acta 1639, 152–158 (2003). [DOI] [PubMed] [Google Scholar]
- 38.Chwiej J., Dulinska J., Janeczko K., Dumas P., Eichert D., Dudala J., Setkowicz Z., Synchrotron FTIR micro-spectroscopy study of the rat hippocampal formation after pilocarpine-evoked seizures. J. Chem. Neuroanat. 40, 140–147 (2010). [DOI] [PubMed] [Google Scholar]
- 39.Petibois C., Déléris G., Chemical mapping of tumor progression by FT-IR imaging: Towards molecular histopathology. Trends Biotechnol. 24, 455–462 (2006). [DOI] [PubMed] [Google Scholar]
- 40.Chwiej J., Adamek D., Szczerbowska-Boruchowska M., Krygowska-Wajs A., Wojcik S., Falkenberg G., Manka A., Lankosz M., Investigations of differences in iron oxidation state inside single neurons from substantia nigra of Parkinson’s disease and control patients using the micro-XANES technique. J. Biol. Inorg. Chem. 12, 204–211 (2007). [DOI] [PubMed] [Google Scholar]
- 41.Pushie M. J., Pickering I. J., Martin G. R., Tsutsui S., Jirik F. R., George G. N., Prion protein expression level alters regional copper, iron and zinc content in the mouse brain. Metallomics 3, 206–214 (2011). [DOI] [PubMed] [Google Scholar]
- 42.Chwiej J., Winiarski W., Ciarach M., Janeczko K., Lankosz M., Rickers K., Setkowicz Z., The role of trace elements in the pathogenesis and progress of pilocarpine-induced epileptic seizures. J. Biol. Inorg. Chem. 13, 1267–1274 (2008). [DOI] [PubMed] [Google Scholar]
- 43.Ogawa M., Harada Y., Yamaoka Y., Fujita K., Yaku H., Takamatsu T., Label-free biochemical imaging of heart tissue with high-speed spontaneous Raman microscopy. Biochem. Biophys. Res. Commun. 382, 370–374 (2009). [DOI] [PubMed] [Google Scholar]
- 44.Torres Filho I. P., Terner J., Pittman R. N., Somera L. G. III, Ward K. R., Hemoglobin oxygen saturation measurements using resonance Raman intravital microscopy. Am. J. Physiol. Heart Circ. Physiol. 289, H488–H495 (2005). [DOI] [PubMed] [Google Scholar]
- 45.Ward K. R., Barbee R. W., Reynolds P. S., Torres Filho I. P., Tiba M. H., Torres L., Pittman R. N., Terner J., Oxygenation monitoring of tissue vasculature by resonance Raman spectroscopy. Anal. Chem. 79, 1514–1518 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Grau G. E., de Kossodo S., Cerebral malaria: Mediators, mechanical obstruction or more? Parasitol. Today 10, 408–409 (1994). [DOI] [PubMed] [Google Scholar]
- 47.N. H. Hunt, C. M. Thumwood, I. A. Clark, W. Cowden, Are reactive oxygen species involved in cerebral malaria?, in Free Radicals: Chemistry, Pathology and Medicine, C. Rice-Evans, T. Dormandy, Eds. (Richelieu Press, London, 1988), pp. 405–414. [Google Scholar]
- 48.Thumwood C. M., Hunt N. H., Clark I. A., Cowden W. B., Breakdown of the blood-brain barrier in murine cerebral malaria. Parasitology 96, 579–589 (1988). [DOI] [PubMed] [Google Scholar]
- 49.Hempel C., Combes V., Hunt N. H., Kurtzhals J. A. L., Grau G. E. R., CNS hypoxia is more pronounced in murine cerebral than noncerebral malaria and is reversed by erythropoietin. Am. J. Pathol. 179, 1939–1950 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lackner P., Beer R., Heussler V., Goebel G., Rudzki D., Helbok R., Tannich E., Schmutzhard E., Behavioural and histopathological alterations in mice with cerebral malaria. Neuropathol. Appl. Neurobiol. 32, 177–188 (2006). [DOI] [PubMed] [Google Scholar]
- 51.Hoffman M., Weinberg J. B., Tumor necrosis factor-α induces increased hydrogen peroxide production and Fc receptor expression, but not increased Ia antigen expression by peritoneal macrophages. J. Leukoc. Biol. 42, 704–707 (1987). [DOI] [PubMed] [Google Scholar]
- 52.Nathan C. F., Murray H. W., Wiebe M. E., Rubin B. Y., Identification of interferon-γ as the lymphokine that activates human macrophage oxidative metabolism and antimicrobial activity. J. Exp. Med. 158, 670–689 (1983). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Huang F.-P., Xi G., Keep R. F., Hua Y., Nemoianu A., Hoff J. T., Brain edema after experimental intracerebral hemorrhage: Role of hemoglobin degradation products. J. Neurosurg. 96, 287–293 (2002). [DOI] [PubMed] [Google Scholar]
- 54.Wu J., Hua Y., Keep R. F., Schallert T., Hoff J. T., Xi G., Oxidative brain injury from extravasated erythrocytes after intracerebral hemorrhage. Brain Res. 953, 45–52 (2002). [DOI] [PubMed] [Google Scholar]
- 55.Gordeuk V., Thuma P., Brittenham G., McLaren C., Parry D., Backenstose A., Biemba G., Msiska R., Holmes L., McKinley E., Vargas L., Gilkeson R., Poltera A. A., Effect of iron chelation therapy on recovery from deep coma in children with cerebral malaria. N. Engl. J. Med. 327, 1473–1477 (1992). [DOI] [PubMed] [Google Scholar]
- 56.Thuma P. E., Mabeza G. F., Biemba G., Bhat G. J., McLaren C. E., Moyo V. M., Zulu S., Khumalo H., Mabeza P., M’Hango A., Parry D., Poltera A. A., Brittenham G. M., Gordeuk V. R., Effect of iron chelation therapy on mortality in Zambian children with cerebral malaria. Trans. R. Soc. Trop. Med. Hyg. 92, 214–218 (1998). [DOI] [PubMed] [Google Scholar]
- 57.Aitken J. B., Levina A., Lay P. A., Studies on the biotransformations and biodistributions of metal-containing drugs using X-ray absorption spectroscopy. Curr. Top. Med. Chem. 11, 553–571 (2011). [DOI] [PubMed] [Google Scholar]
- 58.Duong T. T. H., Witting P. K., Antao S. T., Parry S. N., Kennerson M., Lai B., Vogt S., Lay P. A., Harris H. H., Multiple protective activities of neuroglobin in cultured neuronal cells exposed to hypoxia re-oxygenation injury. J. Neurochem. 108, 1143–1154 (2009). [DOI] [PubMed] [Google Scholar]
- 59.Ma N., Hunt N. H., Madigan M. C., Chan-Ling T., Correlation between enhanced vascular permeability, up-regulation of cellular adhesion molecules and monocyte adhesion to the endothelium in the retina during the development of fatal murine cerebral malaria. Am. J. Pathol. 149, 1745–1762 (1996). [PMC free article] [PubMed] [Google Scholar]
- 60.Miu J., Mitchell A. J., Müller M., Carter S. L., Manders P. M., McQuillan J. A., Saunders B. M., Ball H. J., Lu B., Campbell I. L., Hunt N. H., Chemokine gene expression during fatal murine cerebral malaria and protection due to CXCR3 deficiency. J. Immunol. 180, 1217–1230 (2008). [DOI] [PubMed] [Google Scholar]
- 61.Neill A. L., Hunt N. H., Pathology of fatal and resolving Plasmodium berghei cerebral malaria in mice. Parasitology 105, 165–175 (1992). [DOI] [PubMed] [Google Scholar]
- 62.Mitchell A. J., Hansen A. M., Hee L., Ball H. J., Potter S. M., Walker J. C., Hunt N. H., Early cytokine production is associated with protection from murine cerebral malaria. Infect. Immun. 73, 5645–5653 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Harris H. H., Vogt S., Eastgate H., Lay P. A., A link between copper and dental caries in human teeth identified by X-ray fluorescence elemental mapping. J. Biol. Inorg. Chem. 13, 303–306 (2008). [DOI] [PubMed] [Google Scholar]
- 64.Yun W., Lai B., Cai Z., Maser J., Legnini D., Gluskin E., Chen Z., Krasnoperova A. A., Vladimirsky Y., Cerrina F., Di Fabrizio E., Gentili M., Nanometer focusing of hard x rays by phase zone plates. Rev. Sci. Instrum. 70, 2238 (1999). [Google Scholar]
- 65.Vogt S., Feser M., Legnini D., Kirz J., Maser J., Fast differential phase-contrast imaging and total fluorescence yield mapping in a hard X-ray fluorescence microprobe. AIP Conf. Proc. 705, 1348 (2004). [Google Scholar]
- 66.Paterson D., de Jonge M. D., Howard D. L., Lewis W., McKinlay J., Starritt A., Kusel M., Ryan C. G., Kirkham R., Moorhead G., Siddons D. P., The X-ray fluorescence microscopy beamline at the Australian Synchrotron. AIP Conf. Proc. 1365, 219–222 (2011). [Google Scholar]
- 67.Ryan C. G., van Achterbergh E., Jamieson D. N., Advances in Dynamic Analysis PIXE imaging: Correction for spatial variation of pile-up components. Nucl. Instrum. Methods Phys. Res. Sect. B 231, 162–169 (2005). [Google Scholar]
- 68.Ryan C. G., van Achterbergh E., Yeats C. J., Drieberg S. L., Mark G., McInnes B. M., Win T. T., Cripps G., Suter G. F., Quantitative, high sensitivity, high resolution, nuclear microprobe imaging of fluids, melts and minerals. Nucl. Instrum. Methods Phys. Res. Sect. B 188, 18–27 (2002). [Google Scholar]
- 69.Siegele R., Cohen D. D., Dytlewski N., The ANSTO high energy heavy ion microprobe. Nucl. Instrum. Methods Phys. Res. Sect. B 158, 31–38 (1999). [Google Scholar]
- 70.Harris H., On the rapid conversion of haematoxylin into haematein in staining reactions. J. Appl. Microsc. 3, 777–780 (1900). [Google Scholar]
- 71.Potter S., Chan-Ling T., Ball H. J., Mansour H., Mitchell A., Maluish L., Hunt N. H., Perforin mediated apoptosis of cerebral microvascular endothelial cells during experimental cerebral malaria. Int. J. Parasitol. 36, 485–496 (2006). [DOI] [PubMed] [Google Scholar]
- 72.Vogt S., MAPS: A set of software tools for analysis and visualization of 3D X-ray fluorescence data sets. J. Phys. IV 104, 635–638 (2003). [Google Scholar]
- 73.Combes V., De Souza J. B., Rénia L., Hunt N. H., Grau G. E., Cerebral malaria: Which parasite? Which model? Drug Discov. Today Dis. Models 2, 141–147 (2005). [Google Scholar]
- 74.Kneipp J., Lasch P., Baldauf E., Beekes M., Naumann D., Detection of pathological molecular alterations in scrapie-infected hamster brain by Fourier transform infrared (FT-IR) spectroscopy. Biochim. Biophys. Acta 1501, 189–199 (2000). [DOI] [PubMed] [Google Scholar]
- 75.Petibois C., Gionnet K., Gonçalves M., Perromat A., Moenner M., Déléris G., Analytical performances of FT-IR spectrometry and imaging for concentration measurements within biological fluids, cells and tissues. Analyst 131, 640–647 (2006). [DOI] [PubMed] [Google Scholar]
- 76.Petibois C., Drogat B., Bikfalvi A., Déléris G., Moenner M., Histological mapping of biochemical changes in solid tumors by FT-IR spectral imaging. FEBS Lett. 581, 5469–5474 (2007). [DOI] [PubMed] [Google Scholar]
- 77.Petibois C., Melin A.-M., Perromat A., Cazorla G., Déléris G., Glucose and lactate concentration determination on single microsamples by Fourier-transform infrared spectroscopy. J. Lab. Clin. Med. 135, 210–215 (2000). [DOI] [PubMed] [Google Scholar]
- 78.Liu K.-Z., Mantsch H. H., Simultaneous quantitation from infrared spectra of glucose concentrations, lactate concentrations, and lecithin/sphingomyelin ratios in amniotic fluid. Am. J. Obstet. Gynecol. 180, 696–702 (1999). [DOI] [PubMed] [Google Scholar]
- 79.Gómez-Fernández J. C., Villalain J., The use of FT-IR for quantitative studies of the apparent pKa of lipid carboxyl groups and the dehydration degree of the phosphate group of phospholipids. Chem. Phys. Lipids 96, 41–52 (1998). [DOI] [PubMed] [Google Scholar]
- 80.Kretlow A., Wang Q., Kneipp J., Lasch P., Beekes M., Miller L., Naumann D., FTIR-microspectroscopy of prion-infected nervous tissue. Biochim. Biophys. Acta 1758, 948–959 (2006). [DOI] [PubMed] [Google Scholar]
- 81.Wang Q., Kretlow A., Beekes M., Naumann D., Miller L., In situ characterization of prion protein structure and metal accumulation in scrapie-infected cells by synchrotron infrared and X-ray imaging. Vib. Spectrosc. 38, 61–69 (2005). [Google Scholar]
- 82.Leskovjan A. C., Lanzirotti A., Miller L. M., Amyloid plaques in PSAPP mice bind less metal than plaques in human Alzheimer’s disease. Neuroimage 47, 1215–1220 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Miller L. M., Bourassa M. W., Smith R. J., FTIR spectroscopic imaging of protein aggregation in living cells. Biochim. Biophys. Acta 1828, 2339–2346 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kuzyk A., Kastyak M., Agrawal V., Gallant M., Sivakumar G., Rak M., Del Bigio M. R., Westaway D., Julian R., Gough K. M., Association among amyloid plaque, lipid, and creatine in hippocampus of TgCRND8 mouse model for Alzheimer disease. J. Biol. Chem. 285, 31202–31207 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kastyak-Ibrahim M. Z., Nasse M. J., Rak M., Hirschmugl C., Del Bigio M. R., Albensi B. C., Gough K. M., Biochemical label-free tissue imaging with subcellular-resolution synchrotron FTIR with focal plane array detector. Neuroimage 60, 376–383 (2012). [DOI] [PubMed] [Google Scholar]
- 86.Gough K. M., Tzadu L., Kastyak M. Z., Kuzyk A. C., Julian R. L., Theoretical and experimental considerations for interpretation of amide I bands in tissue. Vib. Spectrosc. 53, 71–76 (2010). [Google Scholar]
- 87.Leskovjan A. C., Kretlow A., Miller L. M., Fourier transform infrared imaging showing reduced unsaturated lipid content in the hippocampus of a mouse model of Alzheimer’s disease. Anal. Chem. 82, 2711–2716 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Bassan P., Byrne H. J., Lee J., Bonnier F., Clarke C., Dumas P., Gazi E., Brown M. D., Clarke N. W., Gardner P., Reflection contributions to the dispersion artefact in FTIR spectra of single biological cells. Analyst 134, 1171–1175 (2009). [DOI] [PubMed] [Google Scholar]
- 89.Bassan P., Kohler A., Martens H., Lee J., Byrne H. J., Dumas P., Gazi E., Brown M., Clarke N., Gardner P., Resonant Mie scattering (RMieS) correction of infrared spectra from highly scattering biological samples. Analyst 135, 268–277 (2010). [DOI] [PubMed] [Google Scholar]
- 90.Bassan P., Sachdeva A., Kohler A., Hughes C., Henderson A., Boyle J., Shanks J. H., Brown M., Clarke N. W., Gardner P., FTIR microscopy of biological cells and tissue: Data analysis using resonant Mie scattering (RMieS) EMSC algorithm. Analyst 137, 1370–1377 (2012). [DOI] [PubMed] [Google Scholar]
- 91.Bassan P., Lee J., Sachdeva A., Pissardini J., Dorling K. M., Fletcher J. S., Henderson A., Gardner P., The inherent problem of transflection-mode infrared spectroscopic microscopy and the ramifications for biomedical single point and imaging applications. Analyst 138, 144–157 (2013). [DOI] [PubMed] [Google Scholar]
- 92.Filik J., Frogley M. D., Pijanka J. K., Wehbe K., Cinque G., Electric field standing wave artefacts in FTIR micro-spectroscopy of biological materials. Analyst 137, 853–861 (2012). [DOI] [PubMed] [Google Scholar]
- 93.Popescu B. F. G., Robinson C. A., Rajput A., Rajput A. H., Harder S. L., Nichol H., Iron, copper and zinc distribution of the cerebellum. Cerebellum 8, 74–79 (2009). [DOI] [PubMed] [Google Scholar]
- 94.Popescu B. F. G., Robinson C. A., Chapman L. D., Nichol H., Synchrotron X-ray fluorescence reveals abnormal metal distributions in brain and spinal cord in spinocerebellar ataxia: A case report. Cerebellum 8, 340–351 (2009). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/1/11/e1500911/DC1
Text
Fig. S1. Validation of second-derivative band intensity at 1127 cm−1 in the FTIR spectra as a marker for relative lactate concentration.
Fig. S2. Validation of second-derivative band intensity at 1627 cm−1 in the FTIR spectra as a marker for relative concentration of aggregated β sheet proteins and, therefore, protein oxidation.
Fig. S3. Correlation of FTIR maps of lipid distribution with H&E histology.
Fig. S4. HCA analysis of FTIR maps distinguishes four distinct regions: the molecular layer, granular layer, inner white matter, and hemorrhaged white matter.
Fig. S5. Principal components analysis of the average spectra for tissue layers determined from the HCA.
Fig. S6. Synchrotron radiation–based (SR) FTIR mapping of cerebellum tissue to identify Mie scattering and electric field standing wave spectral features.
Fig. S7. SR x-ray fluorescence elemental maps (2-μm step size) of healthy cerebellum blood vessels, showing location of Fe (white arrow) within the wall of the blood vessel.
Fig. S8. PIXE elemental maps of healthy and hemorrhaged cerebellum blood vessels.
Fig. S9. Resonance Raman spectra (514-nm excitation) collected from hemorrhaged tissue and dried red blood cells, showing characteristic enhanced intensity of hemoglobin bands.