Chimpanzee social activity is associated with diversity in the gut microbiome.
Keywords: Animals, chimpanzee, gut microbiota, animal behavior, social behavior, microbial communities
Abstract
Animal sociality facilitates the transmission of pathogenic microorganisms among hosts, but the extent to which sociality enables animals’ beneficial microbial associations is poorly understood. The question is critical because microbial communities, particularly those in the gut, are key regulators of host health. We show evidence that chimpanzee social interactions propagate microbial diversity in the gut microbiome both within and between host generations. Frequent social interaction promotes species richness within individual microbiomes as well as homogeneity among the gut community memberships of different chimpanzees. Sampling successive generations across multiple chimpanzee families suggests that infants inherited gut microorganisms primarily through social transmission. These results indicate that social behavior generates a pan-microbiome, preserving microbial diversity across evolutionary time scales and contributing to the evolution of host species–specific gut microbial communities.
Humans and other primates acquire their resident microorganisms from conspecifics through cohabitation and social interaction (1–3). Therefore, primate sociality may be essential to the long-term preservation of the diverse residents of the gut microbiome, many of which benefit hosts by training the immune system, metabolizing otherwise indigestible molecules, protecting against opportunistic infections, and synthesizing nutrients (4, 5). To evaluate the role of host social behavior in shaping gut microbial communities and in transmitting microbial diversity both within and between host generations, we coupled more than eight years (November 2000 to December 2008) of behavioral observation of Kasekela chimpanzees in Gombe, Tanzania, with deep sequencing of their gut microbiomes.
Across the study period, chimpanzees displayed consistent seasonal changes in social activity, allowing quantification of the influence of social behavior on the composition of chimpanzee gut microbiomes. During dry seasons, chimpanzees spend substantially more time alone or in small groups, whereas during wet seasons, chimpanzees forage together in larger groups (6). Accordingly, across the study period, we observed that chimpanzee sociability, calculated as the proportion of time individuals spent together on average [seasonal mean half-weight index of association (HWI)], was significantly higher each year during wet seasons (November through April) than during dry seasons (May through October) (fig. S1).
Between November 2000 and December 2008, 96 fecal samples were collected from 40 Kasekela individuals. In total, 14 individuals were sampled as infants (age 0 to 5 years, n = 27 samples), 2 as juveniles (age 5 to 8 years, n = 3 samples), 7 as adolescents (age 9 to 14 years, n = 9 samples), 18 as adults (age 16 to 34 years, n = 44 samples), and 6 as elderly (age >35 years, n = 13 samples). Samples were collected throughout wet and dry seasons and assigned to individual hosts by direct observation as well as mitochondrial D-loop and microsatellite genotyping (see table S1 for sample list). Libraries of 16S amplicons were prepared from genomic DNA extracted from fecal samples and sequenced to a mean depth of 103,040 reads per sample. Sequences were quality-filtered, clustered into phylotypes, and assigned to taxonomic groups in QIIME (Quantitative Insights into Microbial Ecology) (7). An Operational Taxonomic Unit (OTU) table containing the phylotype frequencies recovered from each sample is presented in table S2.
The gut microbiomes of socially interacting chimpanzees changed in parallel across the study period (Fig. 1A). Microbiomes sampled in the same season were, on average, more similar to one another than were microbiomes sampled in different seasons (t test; P = 0.0012; Fig. 1B). However, across the study period, microbial communities sampled during wet seasons did not differ consistently from those sampled during dry seasons in terms of either the relative abundances of bacterial phylotypes or community membership (fig. S2). Analysis of similarities (ANOSIM) of Bray-Curtis dissimilarities indicated that microbial communities recovered from samples collected after the midpoint of the study (around 1 May 2005) differed compositionally from those recovered from samples collected before the midpoint of the study (P = 0.0010). This difference was evident in the third principal coordinate of Bray-Curtis dissimilarities (Fig. 1A) but not in the first or second principal coordinates (fig. S3), and samples collected after the midpoint of the study did not differ in community membership, as measured by the Sorensen-Dice dissimilarity, from those collected before the midpoint of the study (fig. S3). Samples collected from different individuals in the same season were, on average, as similar to one another as were samples collected from the same individual in different seasons (fig. S4; P = 0.369). The parallel compositional changes in the gut microbiomes of Kasekela chimpanzees included the proliferation of several microbial species (table S3), the most abundant of which being a 97% bacterial OTU within Olsenella (fig. S5), a genus of Gram-positive, nonmotile, non–spore-forming Actinobacteria.
Fig. 1. Gut microbial communities of socially interacting chimpanzees change in parallel across host generations.
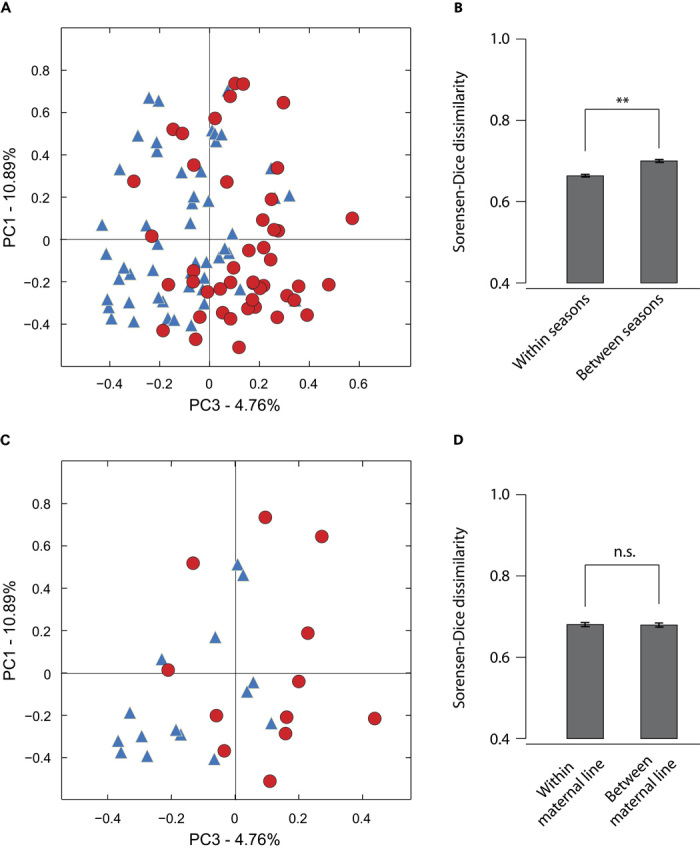
(A) Microbiome samples from individual chimpanzees plotted against the first and third principal coordinates of pairwise Bray-Curtis dissimilarities. Blue triangles represent samples collected before 1 May 2005, and red circles represent samples collected after 1 May 2005. (B) Mean Sorensen-Dice dissimilarity between microbiomes sampled in the same season (left) and between microbiomes sampled in different seasons (right). Error bars represent 95% confidence intervals (**P < 0.01). (C) Microbiome samples only from infants plotted against the first and third principal coordinates of pairwise Sorensen-Dice dissimilarities. Sample labels follow those in (A). (D) Mean Sorensen-Dice dissimilarity between gut microbiomes of the same maternal line (left) and between gut microbiomes of different maternal lines (right). Error bars represent 95% confidence intervals. n.s., not significant; α = 0.05.
Compositional changes in chimpanzee gut microbiomes that accrued over the study period were manifest in the next chimpanzee generation (Fig. 1C). We tested for vertical inheritance of gut microbial communities by asking whether hosts of the same maternal line shared more bacterial phylotypes on average than did unrelated hosts. For three of the four maternal lines analyzed, samples collected spanned two host generations, with the exception of the S family, for which three generations were sampled. For a schematic displaying the frequency at which each maternal line was sampled over the study period, see fig. S6. The inheritance of gut microbial communities across generations appeared to be primarily horizontal among socially-interacting hosts rather than vertical from parent to offspring: the community memberships of individual microbiomes were as similar to those of unrelated individuals as they were to those of kin (Fig. 1D). Although the gut microbiome’s initial inoculum typically comes from the mother (8), these results suggest that, over the course of a lifetime, hosts acquire most of their gut phylotypes through social interactions. The social inheritance of gut microbial communities may be essential to the preservation of microbial diversity over evolutionary time scales, as horizontal transfer of gut symbionts will eliminate bottleneck-induced extinctions that occur stochastically when the transmission of microorganisms is strictly from parent to offspring.
To further evaluate the degree to which social contact promotes cohesion among the gut microbiomes of interacting chimpanzees, we tested for a relationship between the compositional homogeneity of chimpanzee gut microbiomes during each season of sampling (as measured by the mean Sorensen-Dice dissimilarity between individual microbiomes) and the degree of sociability of chimpanzees during the season of sampling (as measured by the seasonal mean HWI as an indicator of general sociability). Chimpanzee gut microbiomes were more compositionally homogeneous during seasons in which hosts were more sociable than during seasons in which hosts were less sociable (Fig. 2). The mean Sorensen-Dice dissimilarity between individual gut microbiomes sampled during the same season showed an inverse relationship with the mean HWI of the season in which the microbiomes were sampled (P = 0.00020; R2 = 0.048), suggesting that the frequency of social contact is associated with the exchange of microorganisms among hosts. Such exchanges may occur through direct contact among interacting hosts, as has been observed in baboons (3), as well as through indirect transfer via feces deposited in the environment.
Fig. 2. Chimpanzee sociability promotes cohesion among gut microbiomes.
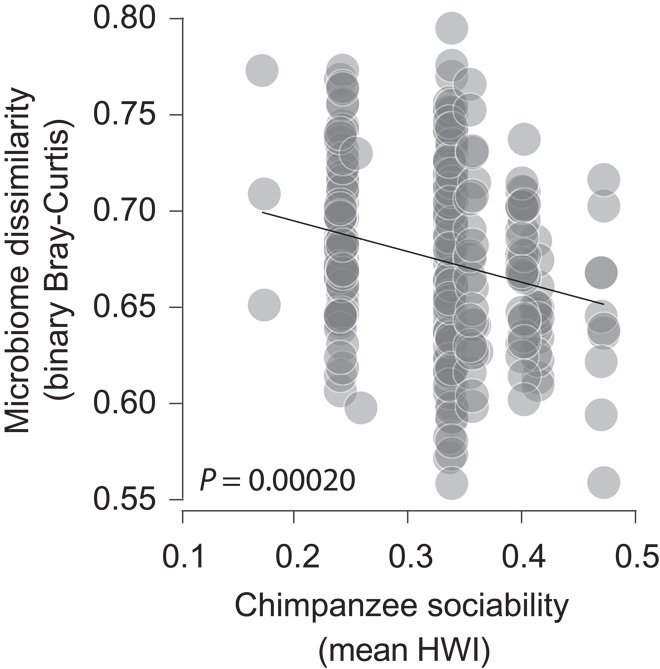
Differences in community memberships (Sorensen-Dice dissimilarities) of chimpanzee gut microbiomes sampled in the same season plotted against sociability of chimpanzees (mean HWI across all unique dyads of adult male or female chimpanzees) during the season of sampling. Points represent pairwise comparisons of microbiome community memberships.
A possible alternative explanation for the compositional homogeneity among chimpanzee gut microbiomes during seasons of high sociability is that chimpanzees consumed more similar diets during seasons of high sociability. We evaluated this hypothesis by testing for a positive association between dietary variability among chimpanzees and sociability as well as between dietary variability among chimpanzees and Bray-Curtis dissimilarity among microbiomes. Within-season dietary variation among chimpanzees, as measured by the variance in the proportion of food types across chimpanzee diets, was neither negatively associated with seasonal mean HWI nor positively associated with mean Bray-Curtis dissimilarity. In fact, variance across individuals in the proportion of fruit in the diet (table S4) was positively associated with HWI (P = 0.047; R2 = 0.17). Therefore, dietary convergence does not appear to explain the convergence among the chimpanzee-microbiome community memberships during periods of high sociability. This result indicates that although diet prominently influences the relative abundances of bacterial taxa in the gut microbiome (9), the gut microbiome’s community membership is populated primarily by dispersal from conspecific hosts.
Although we observed increases in the relative abundances of Olsenella and other phylotypes over time (fig. S5 and table S3), the degree of chimpanzee sociability (HWI) was not significantly associated with either the relative abundance or the presence/absence of any specific bacterial phylotypes (table S5). However, consistent with the exchange of microorganisms among socially interacting hosts, chimpanzee sociability was positively associated with species richness within individual juvenile, adolescent, and adult gut microbiomes (P = 0.0277; R2 = 0.076; Fig. 3A). Species richness within individual juvenile, adolescent, and adult gut microbiomes was also positively associated with dietary evenness (DE; P = 0.0265; R2 = 0.077; Fig. 3B), measured as Shannon’s evenness index of the proportion of fruit, leaves, and insects in the chimpanzee diet (table S6). We used likelihood ratio tests of generalized linear mixed-effects models to test whether sociability and DE each independently influences species richness within individual chimpanzee gut microbiomes (Supplementary Materials and Methods). This analysis revealed that species richness can be better predicted by a combination of sociability and DE than by DE alone (P = 0.00034) or sociability alone (P = 0.00099) (Fig. 3C), supporting the hypothesis that sociability and DE each independently promotes species richness within individual chimpanzee gut microbiomes. In many animals, microbial species richness in the microbiome provides protection against potential pathogens (10–12). In humans, low species richness in the gut microbiome has been associated with Clostridium difficile infection (13) and Crohn’s disease (14). Although social interactions can promote the spread of pathogens (1), our results suggest that sociality may also provide protection against disease by fostering commensal and mutualistic microbial diversity. However, further studies are necessary to test whether the fluctuations in microbial diversity we observed alter the protective effects of chimpanzee gut microbial communities.
Fig. 3. Chimpanzee sociability and DE each promotes species richness within individual gut microbiomes.
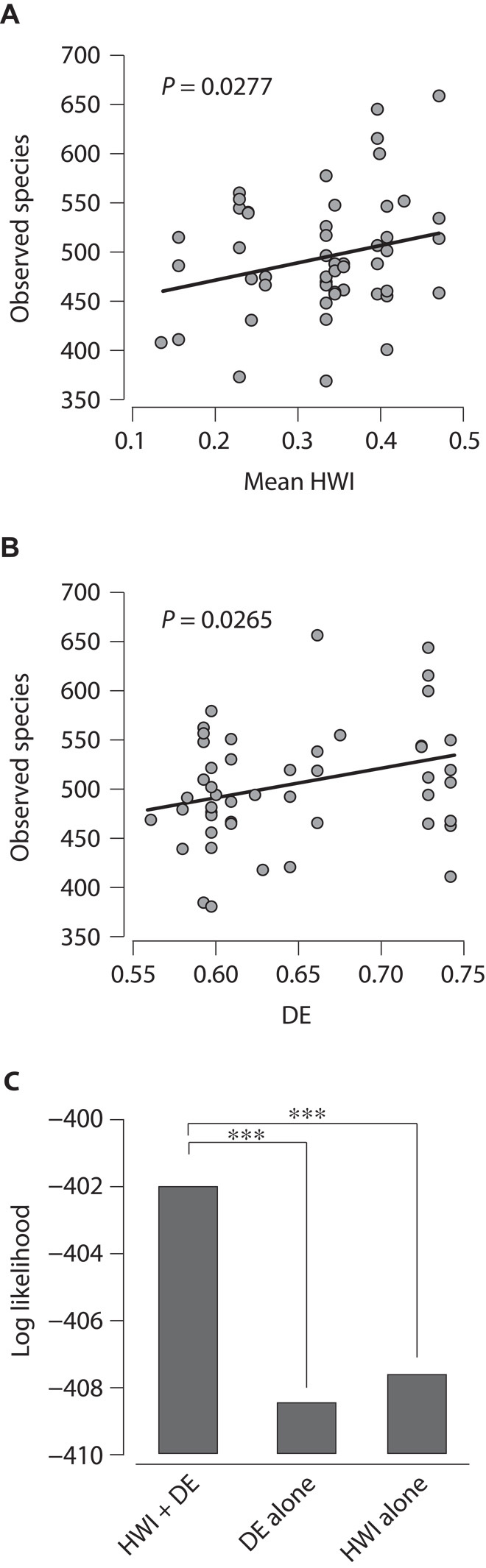
(A) Microbial species richness of individual gut microbiomes plotted against chimpanzee sociability during the season of sampling (mean HWI across all unique dyads of adult male or female chimpanzees). (B) Microbial species richness of individual gut microbiomes plotted against chimpanzee DE (Shannon’s evenness index of the proportions of dietary foodstuffs; table S6) during the season of sampling. In (A) and (B), the P value indicating whether the slope of the trend line differs from zero is shown. (C) Log likelihood of a model of species richness containing HWI and DE versus the log likelihoods of models of species richness containing HWI alone or DE alone. Significant differences were determined by the likelihood ratio test (***P < 0.001).
The Kasekela chimpanzees at Gombe cultivate a microbial metacommunity, in which chimpanzee gut microbiomes are connected to one another through the social interactions of their hosts. These results indicate that human social interactions may also generate microbial meta-communities comprised of individual microbiomes (15). The fact that the variability among hosts in microbiome community membership was negatively associated with host social activity (Fig. 2A), combined with the observation that microbial species richness per individual was positively associated with host social activity (Fig. 2B), fulfills long-standing predictions of metacommunity theory that increasing dispersal among community patches (that is, individual chimpanzees) should both homogenize the compositions of patches and foster the maintenance of species richness (16). Moreover, our results corroborate previous work that revealed strong associations between the degree of grooming contact between baboons and the degree of overlap between the species and genic compositions of baboon gut microbial communities (3). Together, observations in chimpanzees and baboons suggest that social transmission of gut microorganisms may be widespread across social animals.
Each primate species, including Homo sapiens, maintains a compositionally distinct gut microbiome that reflects the ecology and evolutionary history of its host (17–21). Viewing the gut microbial communities of a host population as a pan-microbiome cultivated by social interaction offers new insights into how the compositions of gut microbiomes might diverge over evolutionary time scales. The social transmission of microorganisms among hosts will create a greater diversity of microbial consortia on which selection can operate, potentially speeding up the rate of microbial adaptation (16). However, by reducing the efficiency at which selection can purge microbial associations that are detrimental to host fitness, social transmission will likely slow the rate at which gut microbial species evolve mutualistic effects (22). The frequent social transmission of microorganisms within host populations, coupled with the evolution and/or turnover of the microbial populations being transmitted, is sufficient to generate compositional differences between the gut microbial communities of host populations, even in the absence of evolutionary divergence between the host populations themselves. Microbiome divergence between isolated host populations may accrue in a manner analogous to genetic drift, in which the size and structure of host populations influence the rate at which host-associated microbial communities change over evolutionary time. Given the stark differences between human and chimpanzee social structures, our results indicate a need to thoroughly explore the relationship between social networks and microbiome composition in human societies.
The long-term monitoring of the gut microbiomes of Kasekela chimpanzees revealed a prominent role for social behavior in the preservation of commensal and mutualistic relationships between hosts and their resident microorganisms. Pan-microbiomes, such as the one cultivated by Kasekela chimpanzees in Gombe, are likely widespread across social animals, including humans, although the dynamics of the human pan-microbiome have not been investigated because of a lack of longitudinal monitoring of the microbiomes of human social groups. Our results suggest that changes in the human pan-microbiome occurring today as a result of modern life-styles (1, 21, 23, 24) could be inherited by future generations.
MATERIALS AND METHODS
Behavioral and dietary data collection
Data on social behavior and dietary composition were collected through all-day focal animal observation of identified individuals (25). Over the study period, 50 individuals were subjected to focal observation. During focal observations, arrivals and departures of other individuals were recorded continuously, as were food types consumed during each feeding bout. Note that the estimated proportions of each food type in chimpanzee diets are based on the observed time chimpanzees spent foraging on each food type rather than on direct measurements of the amount of food ingested.
For each individual, seasonal tendencies to associate with other chimpanzees were quantified using HWI, which estimates the proportion of time any two individuals spent together. To minimize sample bias and autocorrelation, HWI was calculated on the basis of the frequency of daily first encounters of an individual—either alone or in a group of individuals—with the focal individual rather than on total time in association, following previous work (26). HWI was calculated only for individuals seen >10 times in a given season and for those dyads for which both partners were alive at the beginning and end of the season. To quantify the composition of the diet of each focal individual in each season, the relative proportion of each food type was calculated as the ratio of time spent feeding on that food type and the total time spent feeding across all focal observations.
Sample collection and screening
Fecal samples were collected from chimpanzees under direct observation. The source individual for each sample was verified by genetic markers as described elsewhere (27). Samples were immediately suspended in RNAlater. Upon collection, samples were suspended in an equal volume of RNAlater and stored at −80°C. Fecal samples were tested for SIVcpz antibodies by Western blot analysis as reported previously by Keele et al. (28). The four samples that tested positive for SIVcpz came from a single infected individual that did not display an abnormal microbiome compositional profile, which can sometimes manifest in SIVcpz-positive chimpanzees (29).
Sample processing and sequencing
Genomic DNA was extracted from each sample by a bead-beating procedure (30). The V4 region of 16S recombinant DNA was amplified through polymerase chain reaction performed in triplicate using barcoded primers 515F and 806R following previously described protocols (21). Resulting amplicons were sequenced on the Illumina MiSeq platform.
Sequence quality control, taxonomy, and diversity estimates
Sequences were filtered in QIIME (7) using the split_libraries.py and default settings. High-quality reads were clustered into 97% OTUs (that is, phylotypes) via de novo uclust. Representative sequences were chosen from each OTU and assigned to taxonomic lineages with the RDP classifier using a confidence threshold of 80. To reduce spurious OTUs and to restrict our analyses to the long-term residents of the gut microbiome, OTUs present at a frequency of <0.0001 were removed from further analysis. Each sample was then subsampled to an even depth of 10,000 reads. Sorensen-Dice and Bray-Curtis dissimilarities for all pairwise comparisons of samples (β diversity) as well as observed microbial species per sample (α diversity) were calculated in QIIME.
Testing for parallel evolution of chimpanzee gut microbiomes
Principal coordinates of both weighted and Sorensen-Dice dissimilarities were calculated and PCoA plots were produced in QIIME. ANOSIM was implemented in QIIME to test whether microbiome samples from the second half of the study period differed compositionally from those of the first half and to test whether microbiome samples differed compositionally from year to year.
Testing for individual signatures in chimpanzee gut microbiomes
To test whether individual microbiomes maintained distinct compositions, a Student’s t test comparing the mean pairwise Bray-Curtis dissimilarity between samples collected from the same individual with the mean pairwise Bray-Curtis dissimilarity between samples collected from different individuals in the same season was performed.
Evaluating the inheritance of microbiome constituents
To test whether individual microbiomes resembled those of their kin more closely than they did those of unrelated individuals, a Student’s t test comparing the mean pairwise Bray-Curtis dissimilarity between samples collected from individuals of the same maternal line with the mean pairwise Bray-Curtis dissimilarity between samples collected from unrelated individuals was performed. This analysis considered four maternal lines: the F, G, S, and T families (table S1).
Identifying associations among chimpanzee sociability, diet, and microbiome composition
Regressions among sociability, diet composition, α diversity, and β diversity were performed in R. Diet composition was quantified in terms of the proportion of fruit, leaves, and insects. Multiple regressions were performed to test for associations between microbiome composition and the proportion of individual food types as well as DE, defined as Shannon’s evenness index of the proportions of food types listed in table S6. The regression of mean HWI and observed species as well as the regression of DE and observed species considered the microbiomes of juvenile, adolescent, and adult chimpanzees from years in which samples were collected during wet (15 November to 14 May) and dry (15 May to 14 November) seasons. Two samples with an α diversity >3 SDs from the mean were excluded from analysis. Significant associations were further evaluated through comparisons of mixed linear effects models.
Supplementary Material
Acknowledgments
We thank K. Hammond for aiding in the design of figures and the Jane Goodall Institute for supporting data collection. This research was conducted in compliance with Tanzanian regulations and had approval from Tanzania National Parks, the Tanzania Wildlife Research Institute, and the Tanzania Commission for Science and Technology. This research was approved by the Institutional Animal Care and Use Committee of Duke University (protocol A298-13-12). Funding: This work was supported by grants from the NIH (R01 AI58715 to B.H.H. and R01 GM101209 to H.O.), an NSF Graduate Research Fellowship (Award ID 2011119472) to A.H.M., an NSF Doctoral Dissertation Improvement Grant (Award ID 1407133) to H.O. and A.H.M., and NSF grant IOS-LTREB-1052693 to A.E.P. Author contributions: A.H.M. performed analyses and wrote the manuscript. S.F., M.L.W., and A.E.P. performed analyses. B.H.H. provided samples and reagents. H.O. provided samples and reagents and wrote the manuscript. Competing interests: The authors declare that they have no competing interests. Data and materials availability: Data needed to evaluate the conclusions in the paper are present in the paper, Supplementary Materials, and have been deposited to datadryad.org under accession SRX821575.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/2/1/e1500997/DC1
Materials and Methods
Fig. S1. Seasonal variation in chimpanzee sociability.
Fig. S2. Temporal shift in microbiome composition is not evident in PC2 of Bray-Curtis dissimilarities or PC1, PC2, or PC3 of Sorensen-Dice dissimilarities.
Fig. S3. Lack of differentiation between wet- and dry-season gut microbial communities.
Fig. S4. Testing for signatures of host individual in chimpanzee gut microbiomes.
Fig. S5. Proliferation of an Olsenella phylotype over time.
Fig. S6. Sampling of chimpanzee maternal lines.
Table S1. Fecal sample metadata.
Table S2. OTU table of chimpanzee gut microbiomes.
Table S3. Temporal shifts in bacterial frequencies.
Table S4. Among chimpanzee variance in fruit consumption across sampling seasons.
Table S5. Correlations between chimpanzee HWI and bacterial relative abundances.
Table S6. Consumption of food types by chimpanzees across seasons.
REFERENCES AND NOTES
- 1.Yatsunenko T., Rey F. E., Manary M. J., Trehan I., Dominguez-Bello M. G., Contreras M., Magris M., Hidalgo G., Baldassano R. N., Anokhin A. P., Heath A. C., Warner B., Reeder J., Kuczynski J., Caporaso J. G., Lozupone C. A., Lauber C., Clemente J. C., Knights D., Knight R., Gordon J. I., Human gut microbiome viewed across age and geography. Nature 486, 222–227 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Song S. J., Lauber C., Costello E. K., Lozupone C. A., Humphrey G., Berg-Lyons D., Caporaso J. G., Knights D., Clemente J. C., Nakielny S., Gordon J. I., Fierer N., Knight R., Cohabiting family members share microbiota with one another and with their dogs. eLife 2, e00458 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tung J., Barreiro L. B., Burns M. B., Grenier J. C., Lynch J., Grieneisen L. E., Altmann J., Alberts S. C., Blekhman R., Archie E. A., Social networks predict gut microbiome composition in wild baboons. eLife 4, e05224 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kau A. L., Ahern P. P., Griffin N. W., Goodman A. L., Gordon J. I., Human nutrition, the gut microbiome and the immune system. Nature 474, 327–336 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Khosravi A., Mazmanian S. K., Disruption of the gut microbiome as a risk factor for microbial infections. Curr. Opin. Microbiol. 16, 221–227 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.van Lawick-Goodall J., The behaviour of free-living chimpanzees in the Gombe Stream Reserve. Anim. Behav. Monogr. 1, 161–311 (1968). [Google Scholar]
- 7.Caporaso J. G., Kuczynski J., Stombaugh J., Bittinger K., Bushman F. D., Costello E. K., Fierer N., Peña A. G., Goodrich J. K., Gordon J. I., Huttley G. A., Kelley S. T., Knights D., Koenig J. E., Ley R. E., Lozupone C. A., McDonald D., Muegge B. D., Pirrung M., Reeder J., Sevinsky J. R., Turnbaugh P. J., Walters W. A., Widmann J., Yatsunenko T., Zaneveld J., Knight R., QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7, 335–336 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Grölund M.-M., Lehtonen O.-P., Eerola E., Kero P., Fecal microflora in healthy infants born by different methods of delivery: Permanent changes in intestinal flora after cesarean delivery. J. Pediatr. Gastroenterol. Nutr. 28, 19–25 (1999). [DOI] [PubMed] [Google Scholar]
- 9.David L. A., Maurice C. F., Carmody R. N., Gootenberg D. B., Button J. E., Wolfe B. E., Ling A. V., Devlin A. S., Varma Y., Fischbach M. A., Biddinger S. B., Dutton R. J., Turnbaugh P. J., Diet rapidly and reproducibly alters the human gut microbiome. Nature 505, 559–563 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dillon R. J., Vennard C. T., Buckling A., Charnley A. K., Diversity of locust gut bacteria protects against pathogen invasion. Ecol. Lett. 8, 1291–1298 (2005). [Google Scholar]
- 11.Koch H., Schmid-Hempel P., Socially transmitted gut microbiota protect bumble bees against an intestinal parasite. Proc. Natl. Acad. Sci. U.S.A. 108, 19288–19292 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lawley T. D., Clare S., Walker A. W., Stares M. D., Connor T. R., Raisen C., Goulding D., Rad R., Schreiber F., Brandt C., Deakin L. J., Pickard D. J., Duncan S. H., Flint H. J., Clark T. G., Parkhill J., Dougan G., Targeted restoration of the intestinal microbiota with a simple, defined bacteriotherapy resolves relapsing Clostridium difficile disease in mice. PLOS Pathog. 8, e1002995 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chang J. Y., Antonopoulos D. A., Kalra A., Tonelli A., Khalife W. T., Schmidt T. M., Young V. B., Decreased diversity of the fecal microbiome in recurrent Clostridium difficile-associated diarrhea. J. Infect. Dis. 197, 435–438 (2008). [DOI] [PubMed] [Google Scholar]
- 14.Manichanh C., Rigottier-Gois L., Bonnaud E., Gloux K., Pelletier E., Frangeul L., Nalin R., Jarrin C., Chardon P., Marteau P., Roca J., Dore J., Reduced diversity of faecal microbiota in Crohn’s disease revealed by a metagenomic approach. Gut 55, 205–211 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.The Human Microbiome Project Consortium , Structure, function and diversity of the healthy human microbiome. Nature 486, 207–214 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wilson D. S., Complex interactions in metacommunities, with implications for biodiversity and higher levels of selection. Ecology 73, 1984–2000 (1992). [Google Scholar]
- 17.Ley R. E., Hamady M., Lozupone C., Turnbaugh P. J., Ramey R. R., Bircher J. S., Schlegel M. L., Tucker T. A., Schrenzel M. D., Knight R., Gordon J. I., Evolution of mammals and their gut microbes. Science 320, 1647–1651 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yildirim S., Yeoman C. J., Sipos M., Torralba M., Wilson B. A., Goldberg T. L., Stumpf R. M., Leigh S. R., White B. A., Nelson K. E., Characterization of the fecal microbiome from non-human wild primates reveals species specific microbial communities. PLOS One 5, e13963 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ochman H., Worobey M., Kuo C.-H., Ndjango J.-B. N., Peeters M., Hahn B. H., Hugenholtz P., Evolutionary relationships of wild hominids recapitulated by gut microbial communities. PLOS Biol. 8, e1000546 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moeller A. H., Peeters M., Ndjango J.-B., Li Y., Hahn B. H., Ochman H., Sympatric chimpanzees and gorillas harbor convergent gut microbial communities. Genome Res. 23, 1715–1720 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Moeller A. H., Li Y., Ngole E. M., Ahuka-Mundeke S., Lonsdorf E. V., Pusey A. E., Peeters M., Hahn B. H., Ochman H., Rapid changes in the gut microbiome during human evolution. Proc. Natl. Acad. Sci. U.S.A. 111, 16431–16435 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ewald P. W., Transmission modes and evolution of the parasitism-mutualism continuum. Ann. N. Y. Acad. Sci. 503, 295–306 (1987). [DOI] [PubMed] [Google Scholar]
- 23.Clemente J. C., Pehrsson E. C., Blaser M. J., Sandhu K., Gao Z., Wang B., Magris M., Hidalgo G., Contreras M., Noya-Alarcón Ó., Lander O., McDonald J., Cox M., Walter J., Oh P. L., Ruiz J. F., Rodriguez S., Shen N., Song S. J., Metcalf J., Knight R., Dantas G., Dominguez-Bello M. G., The microbiome of uncontacted Amerindians. Sci. Adv. 1, e1500183 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Blaser M. J., Falkow S., What are the consequences of the disappearing human microbiota? Nat. Rev. Microbiol. 7, 887–894 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.J. Goodall, The Chimpanzees of Gombe: Patterns of Behavior (Belknap Press of Harvard Univ. Press, Cambridge, MA, 1986). [Google Scholar]
- 26.Foerster S., McLellan K., Schroepfer-Walker K., Murray C. M., Krupenye C., Gilby I. C., Pusey A. E., Social bonds in the dispersing sex: Partner preferences among adult female chimpanzees. Anim. Behav. 105, 139–152 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Degnan P. H., Pusey A. E., Lonsdorf E. V., Goodall J., Wroblewski E. E., Wilsong M. L., Rudicell R. S., Hahn B. H., Ochman H., Factors associated with the diversification of the gut microbial communities within chimpanzees from Gombe National Park. Proc. Natl. Acad. Sci. U.S.A. 109, 13034–13039 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Keele B. F., Jones J. H., Terio K. A., Estes J. D., Rudicell R. S., Wilson M. L., Li Y., Learn G. H., Beasley T. M., Schumacher-Stankey J., Wroblewski E., Mosser A., Raphael J., Kamenya S., Lonsdorf E. V., Travis D. A., Mlengeya T., Kinsel M. J., Else J. G., Silvestri G., Goodall J., Sharp P. M., Shaw G. M., Pusey A. E., Hahn B. H., Increased mortality and AIDS-like immunopathology in wild chimpanzees infected with SIVcpz. Nature 460, 515–519 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Moeller A. H., Shilts M., Li Y., Rudicell R. S., Lonsdorf E. V., Pusey A. E., Wilson M. L., Hahn B. H., Ochman H., SIV-induced instability of the chimpanzee gut microbiome. Cell Host Microbe 14, 340–345 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Goodman A. L., Kallstrom G., Faith J. J., Reyes A., Moore A., Dantas G., Gordon J. I., Extensive personal human gut microbiota culture collections characterized and manipulated in gnotobiotic mice. Proc. Natl. Acad. Sci. U.S.A. 108, 6252–6257 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/2/1/e1500997/DC1
Materials and Methods
Fig. S1. Seasonal variation in chimpanzee sociability.
Fig. S2. Temporal shift in microbiome composition is not evident in PC2 of Bray-Curtis dissimilarities or PC1, PC2, or PC3 of Sorensen-Dice dissimilarities.
Fig. S3. Lack of differentiation between wet- and dry-season gut microbial communities.
Fig. S4. Testing for signatures of host individual in chimpanzee gut microbiomes.
Fig. S5. Proliferation of an Olsenella phylotype over time.
Fig. S6. Sampling of chimpanzee maternal lines.
Table S1. Fecal sample metadata.
Table S2. OTU table of chimpanzee gut microbiomes.
Table S3. Temporal shifts in bacterial frequencies.
Table S4. Among chimpanzee variance in fruit consumption across sampling seasons.
Table S5. Correlations between chimpanzee HWI and bacterial relative abundances.
Table S6. Consumption of food types by chimpanzees across seasons.


