Abstract
In silico prediction of unknown drug-target interactions (DTIs) has become a popular tool for drug repositioning and drug development. A key challenge in DTI prediction lies in integrating multiple types of data for accurate DTI prediction. Although recent studies have demonstrated that genomic, chemical and pharmacological data can provide reliable information for DTI prediction, it remains unclear whether functional information on proteins can also contribute to this task. Little work has been developed to combine such information with other data to identify new interactions between drugs and targets. In this paper, we introduce functional data into DTI prediction and construct biological space for targets using the functional similarity measure. We present a probabilistic graphical model, called conditional random field (CRF), to systematically integrate genomic, chemical, functional and pharmacological data plus the topology of DTI networks into a unified framework to predict missing DTIs. Tests on two benchmark datasets show that our method can achieve excellent prediction performance with the area under the precision-recall curve (AUPR) up to 94.9. These results demonstrate that our CRF model can successfully exploit heterogeneous data to capture the latent correlations of DTIs, and thus will be practically useful for drug repositioning.
Keywords: Drug-Target Interaction, Drug Repositioning, Conditional Random Field, Functional Similarity
1. Introduction
In recent years, drug repositioning or drug repurposing has become an increasingly popular trend in drug discovery.1–4 The main goal of drug repositioning is to reuse existing or abandoned drugs and identify their new therapeutic functions. Recent literature reveals that drugs often possess the so-called promiscuity property,5,6 that is, individual drugs can act on other off-target proteins in addition to the original target. This property provides a strong theoretical support for drug repositioning.
In silico prediction of drug-target interactions (DTIs) has been widely applied in drug repositioning, since it can significantly reduce time and cost of drug development. Molecular docking methods have been commonly used in predicting new DTIs if structure coordinates of both proteins and drugs are available.7–10 When three-dimensional (3D) structures of molecules are absent, we need to depend on other approaches to perform DTI prediction. The structure-free approaches can be roughly divided into two categories: ligand-based and network-based methods. Ligand-based methods exploit ligand similarity to identify new targets that can interact with a query drug.11,12 Although with some successful stories, ligand-based approaches have di±culty in identifying new interactions associated with novel binding scaffolds.13 Network-based methods14–20 detect the latent correlation features of DTIs to predict new interactions, and recently have become a popular tool for drug repositioning and drug development. A key challenge in network-based prediction approaches lies in integrating heterogeneous data for accurate DTI prediction. Traditional DTI prediction approaches often relate genomic and chemical data with DTI networks to perform new prediction.21 Recently, pharmacological data such as drug side-effets have also been taken into consideration,18,20,22–24 and the results suggest that incorporating more data into DTI prediction can further improve prediction accuracy. Most existing network-based approaches mainly rely on the sequence similarity to measure the closeness of two targets. The sequence similarity, however, is not necessarily sufficient enough to characterize the shared patterns of DTI profiles between two targets.
Functional similarity enables us to compare two proteins with respect to their molecular and biological functions.25 It is defined mainly based on Gene Ontology (GO) terms, which indicate the biological roles of gene products. This measure can identify functionally-related proteins regardless of homology, and hence provide additional information about the similarity of two targets aside from their genomic data. Based on functional similarity, we can construct biological space for proteins and analyze their DTI patterns from a different angle.
Although numerous approaches18,20,23,24,26 have been proposed to integrate genomic (i.e., protein sequences), chemical (i.e., chemical substructures of drugs) and pharmacological (i.e., drug side-effects) data for predicting unknown DTIs, functional information has not been well exploited in DTI prediction. To our knowledge, little work has been developed to systematically integrate functional information on proteins with the aforementioned data to predict missing interactions between drugs and targets. In this paper, we present a new approach to address the DTI prediction problem by systematically integrating large-scale chemical, pharmacological, genomic and functional data and DTI network information into a unified framework. Our method applies a probabilistic graphical model, called conditional random field (CRF), to encode the complicated network associated with drugs and targets, and predict new DTIs. We apply a stochastic gradient ascent approach plus the contrastive divergence (CD) algorithm27 to train our graphical model and capture the hidden correlations between drugs and targets. Tests on two benchmark datasets derived from multiple publicly-available databases show that our CRF model can effectively integrate multiple sources of information and achieve excellent prediction performance, with the area under the precision-recall curve (AUPR) up to 94.9. These results indicate that our approach can have potential applications in drug repositioning.
In summary, the following contributions are made in this paper: (1) Introduction of functional data into DTI prediction and construction of biological space for proteins using the functional similarity measure; (2) Development of a new machine learning approach that can systematically integrate heterogeneous data into a unified framework to predict unknown DTIs; and (3) Promising testing results on two benchmark datasets.
2. Methods
2.1. Conditional Random Field Framework
Conditional random field (CRF) is a probabilistic graphical model or a variant of Markov random field28–30 that was first proposed for object recognition and image segmentation.31 Now it has been widely used in many fields such as shallow parsing,32 named entity recognition,33 topic distillation,34 social recommendation35 and molecular structural modelling.36 We apply a binary CRF model34,35 to formulate our DTI prediction problem.
Let {di} ≤ i ≤ nd be the set of known drugs and {tj} 1 ≤ j ≤ nt, be the set of targets, where nd and nt represent the total numbers of drugs and targets respectively. We use X to denote observed data, including known DTIs and various similarity scores, such as sequence similarity scores for proteins and chemical similarity scores for drugs. In other words, X stands for a set of binary indicators representing known drug-target interactions, and positive variables representing observed similarity scores. For each drug di, we construct a CRF on an undirected graph G = (Vt, Et), where Vt = {tt} is the set of targets and each edge in Et represents the similarity between a pair of targets. Let vector Y = (y1, y2, ... ynt) denote the prediction, where each yj is a binary random variable representing the prediction of target tj, that is, yj = 1 if the predicted interaction between drug di and target tj is true, and yj = 0 otherwise. We call this model the target-based CRF. Similarly, for each target tj, we construct a CRF on an undirected graph G = (Vd, Ed), where Vd = {dd} is the set of drugs and each edge in Ed represents the similarity between a pair of drugs. We call the second model the drug-based CRF. For the convenience of description, next, we will mainly use the target-based model as an example to illustrate the learning and prediction procedures of our CRF model unless otherwise specified.
For each target-based CRF, we define a joint probability distribution conditioning on observation X. In the underlying graph, each node represents a target ti or its associated binary random variable yi, and each edge connecting two nodes represents the dependency between these two nodes. Hereinafter, we will slightly abuse the notation and use terms ‘node’ and ‘random variable’ interchangeably. The undirected graphical model possesses the so-called conditional independence property,37 which states that the conditional distribution of node yi is independent of all other nodes given its ‘neighbors’ (i.e., all other nodes that yi is connected to). By connecting similar proteins together, we indeed assume that the conditional state of a target depends only on the states of other proteins with high similarity. Details about how to construct edges between targets will be described in Section 3.1.
In a CRF model, the energy of a joint configuration Y given X can be defined as follows:
| (1) |
where f (yi|X) is a local node feature function defined based on the state of yi, g(yi, yj|X) is a relational edge feature function defined based on states of both yi and yj, and ai ≥ 0 and bij ≥ 0 are weight parameters that need to be learned from training data. In our DTI prediction framework, we let all target-based or drug-based CRFs share the same parameters ai and bij. Then the joint probability density function of Y given X can be defined as
| (2) |
where Z (X) = ΣY exp (−E(Y | X)) is the normalizing constant, also called partition function. We define functions f (·) and g (·) as followings:
| (3) |
| (4) |
where Hxi (yi) represents the observed feature of target ti, and Hxi,xj (yi − yj) represents the relational feature measure of yi and yj given observation X. In our framework, we let Hxi (yi) be the average number of observed drug interactions for target ti, and let Hxi,xj (yi − yj) be the difference between binary variables yi and yj. By defining the above two feature functions, we indeed add a penalization when (1) predictions for two connected nodes are different, and (2) the prediction of a given node deviates from its average state. Unlike in Ref. 35, which assumes that all nodes share the same parameter a and all edges share the same parameter b, here in our model all weight parameters ai, bij are set to be different values for individual nodes and edges. This parameter setting is more flexible to capture information from data and can avoid potential improper assumptions about weight parameters. Our test results (details are not shown in the paper) suggest that this new parameter setting can yield better performance than the original version35 which chooses a relatively rigid parameter setting.
2.2. Parameter Training
In the training process, we aim to learn parameters ai and bij from training data. We use stochastic gradient ascent38 as an optimization method to maximize the conditional log-likelihood of training data. To simplify the notation, we use vector θ to denote parameters (ai, bij), and function vector h to denote (f, g). Then the probability density function in Eq. (2) can be rewritten as
| (5) |
Thus we can derive the following conditional log-likelihood:
| (6) |
Since each component of θ is non-negative, we let . For simplicity, we use exp (θ′) to represent . Then we have
| (7) |
The gradient in Eq. (7) is
| (8) |
where Eθ(h(Y|X)) is the expectation of h (Y |X) and Y|X follows the distribution pθ defined in Eq. (5).
To apply the gradient ascent method, we need to deal with the expectation term in Eq. (8). It is algebraically intractable to directly calculate this expectation, and one possible solution is to employ some simulation techniques such as Markov Chain Monte Carlo (MCMC) to approximate its value. A Gibbs sampling method was used in Ref. 35 to sample a sequence of Y following the current distribution pθ and then approximate Eθ(h(Y|X)) by
| (9) |
where {ỹi}, 1 ≤ i ≤ L, is the sampled sequence, and L is the total number of sampling iterations. Sampling such sequence often proceeds as follows: We first randomly pick some initial value y0, and then sample each variable using the current value according to its conditional distribution. Normally, after some burn-in period, the distribution of yi can approximate distribution pθ.
Although Gibbs sampling is a popular method to approximate the expectation, it su®ers from heavy computational cost, which is impractical in our case. Here we apply another sampling algorithm, called contrastive divergence (CD), which was first proposed in Ref. 27. The CD algorithm has been successfully used to train restricted Boltzmann machines39 and it can be easily implemented. The basic idea of the CD algorithm is to substitute Eθ(h (Y|X)) in Eq. (8) by EpT (h(Y|X)), where pT represents the distribution of data transformed by T cycles of Gibbs sampling.27 In practice, T is often chosen to be one. Although the CD algorithm may lead to biased estimates, the bias is small in general.40 In practice, the CD algorithm can provide an effcient method to approximate the log-likelihood function.27,39,40
2.3. Predicting New Drug-Target Interactions
To predict unknown drug-target interactions for a query drug given observation X, we compute the conditional probability distribution p (yk|y−k, X) for each target tk, where y−k denote the all other targets except tk. For i ≠ k, yi = 1 if target yi is known to interact with the query drug, and yi = 0 otherwise. We then calculate the conditional expectation of yk as the prediction score of the interaction between target yk and the query drug.
3. Results 3.1. Constructing Conditional Random Field
In our CRF model, an edge connecting two nodes indicates the relational dependency between them, and we assume that two connected nodes should share high similarity. One natural approach for constructing edges in the underlying graph is to connect two nodes if their similarity score is above a threshold. By choosing different threshold values we should be able to tune the number of edges in the graph. This construction method, which we call the threshold-based approach, could yield an unbalanced graph in which some nodes may have much fewer neighbors than others. This situation would make it difficult for inferring the states of those neighbor-free nodes. To avoid this problem, we used another approach to construct the underlying graph. For each node ti, let Ni be the set of top K nodes that have the highest similarity scores with ti, and we connect two nodes ti and tj if ti ∈ Nj or tj ∈ Ni. We refer to this new approach as the degree-based approach, which ensures that the degree of each node in the underlying graph is at least K and roughly balanced, and thus can prevent the existence of ‘isolated’ nodes. In practice, we should not choose a large value of K in order to train our CRF model efficiently on a large-scale dataset. Our sensitivity analysis shows that our results did not vary much for different K values (Supplementary Material S2). We can also combine the above two approaches to get an integration-based approach for constructing edges, that is, we connected two nodes mainly based on a similarity score threshold but also added more connections to a node if its degree is less than K. The comparison results show that different construction approaches did not influence much on prediction performance when choosing K ≥ 2 (Supplementary Material S3). In the following analysis, the underlying graph of our CRF model was constructed mainly based on the degree-based approach, unless otherwise specified. We chose K = 4 when a single similarity measure was used and K = 2 when multiple similarity measures were used. This parameter was fixed throughout all our tests.
We tested the following six different approaches in our conditional random field framework:
Genomic approach (GEN): The target-based CRF was constructed using the sequence similarity measure.
Functional approach (FUN): The target-based CRF was constructed using the functional similarity measure.
Integrated Genomic-Functional approach (IGF): The target-based CRF was constructed using the sequence and functional similarity measures simultaneously. In other words, two nodes were connected if they satisfied the sequence or functional similarity criterion.
Chemical approach (CHEM): The drug-based CRF was constructed using the chemical similarity measure.
Pharmacological approach (PHAR): The drug-based CRF was constructed using the sidee®ect similarity measure.
Integrated Chemical-Pharmacological approach (ICP): The drug-based CRF was constructed using the chemical and side-effect similarity measures simultaneously. In other words, an edge was constructed if it was valid under the chemical or side-effect similarity measure criterion.
In addition, we investigated the combination of two independent predictions from target-based and drug-based CRFs respectively. For any given drug-target pair, let Sd denote the prediction score using the drug-based CRF model and St denote the prediction score using the target-based CRF model. Then our final score for this query drug-target pair is
| (10) |
In the current version of our program, we fixed α = 0.5. By fine-tuning the parameter ®, we may achieve better results than our current tests. Our final approach integrated chemical, pharmacological, genomic and functional data simultaneously:
Full Integration approach (FI): The final prediction was the simple linear combination of both integrated chemical- pharmacological (ICP) and integrated genomic-functional (IGF) approaches using Eq. (10).
Our program was implemented in Matlab (2010 b) based on the UGM package developed by Mark Schmidt (http://www.di.ens.fr/~mschmidt/Software/UGM.html). UGM is a Matlab toolbox that implements various tasks in discrete undirected graphical models with pairwise potentials. We used the default parameters of functions in the UGM package throughout all our tests.
3.2. Datasets
To demonstrate the predictive power of our approach, we first tested it on a dataset derived from the KEGG database41,42 which contains experimentally-verified drug-target interactions. We call this dataset the first dataset. All drugs in the first dataset have molecular weight more than 100. In order to obtain pharmacological information we only included those drugs that also have side-e®ect records in the SIDER database.43 As a consequence, in total 875 drugs and 249 proteins with 2596 drug-target interactions were obtained in the first dataset.
To compare with other existing approaches, we tested our algorithm on another dataset that has been published in Ref. 24, where all drugs have records in SIDER, JAPIC and AERS. JAPIC and AERS are two public databases about drug side-effects. More details about these two databases can be found in Ref. 24. The data we tested here is slightly different from the original data which contains 359 drugs and 226 proteins with 1188 drug-target interactions. We excluded six proteins that do not have any GO annotation and two drugs that have no interaction with the remaining proteins. Thus the new dataset includes 357 drugs and 220 proteins with 1174 drug-target interactions. We call this new dataset the second dataset. Descriptive statistics about the first and second datasets are provided in Supplementary Material S1.
Chemical similarities between drugs were calculated using the graph kernel approach,44 where chemical structure information of drugs was taken from the KEGG database. Side-effect similarities between drugs were calculated using the same method as in Ref. 24, where pharmacological information was obtained from the SIDER database. Sequence similarities between proteins were computed using local alignment kernel approach.45 Functional similarities between proteins were calculated using online software FunSimMat,46,47 in which functional similarity scores were derived from GO terms annotated with biological process and molecular function. In both datasets that we have tested, most pairs of proteins or drugs were dissimilar. In the first dataset, less than 3% of all drug pairs had chemical similarity score greater than 0.85 (all similarity scores were normalized to 1), and less than 1% of all protein pairs had sequence similarity score greater than 0.85. In the second dataset, less than 2% of all drug pairs had chemical similarity score greater than 0.85, and less than 1% of all protein pairs had sequence similarity score greater than 0.85.
3.3. Performance Evaluation
We used the Receiver Operator Characteristic (ROC) curve and the Precision-Recall (PR) curve to evaluate the performance of our algorithm. In addition, we also computed the AUC (area under ROC curve) and AUPR (area under PR curve) scores. In our performance evaluation, true positives were those correctly predicted interactions, while false positives were those predicted interactions that were not present in the tested dataset. For highly-unbalanced data, the PR curve is usually considered to be a better criterion to assess the prediction performance, since it can punish more false positive examples.16,19,48 Thus our analysis mainly focused on AUPR, although in many cases AUC and AUPR were positively correlated. Our tests were performed mainly using a 10-fold cross-validation procedure. In this procedure, all DTIs were randomly partitioned into 10 equal size subsamples. Each subsample was in turn used as validation data to test our algorithm, and the remaining nine subsamples were used as training data.
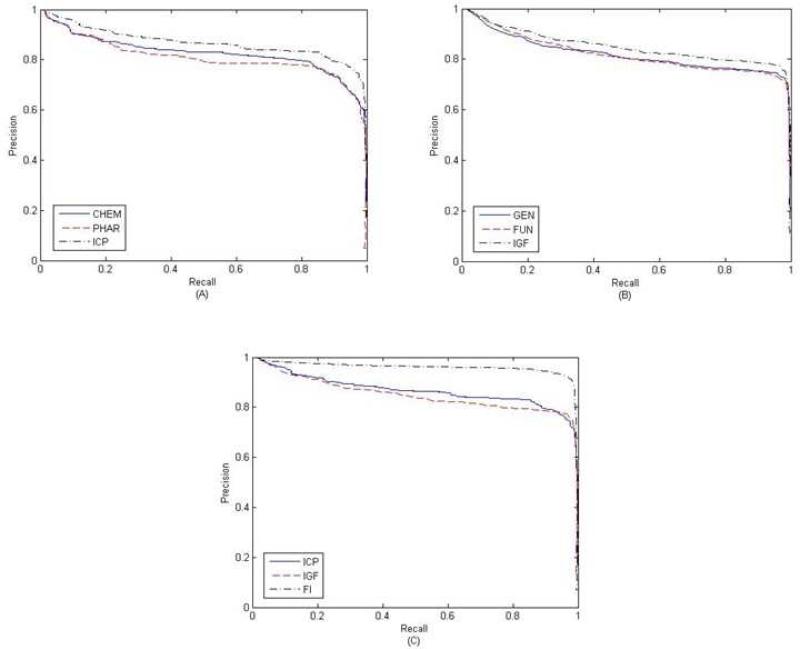
Table 1 summarizes the test results on the first dataset using the 10-fold cross-validation procedure. Under the target-based CRF framework, integrating both genomic and functional data achieved better performance than other two approaches, with the AUPR score improved by > 3%. When both chemical and pharmacological data were integrated into the drug-based CRF framework, the results outperformed each single-similarity based approach with the AUPR score improved by > 4%. When integrating all available information, the FI approach achieved the best performance with AUPR > 94. Figure 1 shows the AUPR curves for different approaches tested on the first dataset. These results demonstrate that incorporating additional information about drugs and proteins can further improve prediction accuracy. To check the robustness of our model, we also performed a 5-fold cross-validation test, and only observed a slight decrease in AUC and AUPR values (Supplementary Material S4).
Table 1.
Prediction results on the first dataset using 10-fold cross-validation. Both AUC and AUPR scores are normalized to 100. The best result is shown in bold.
| Approach | Evaluation Criterion | ||
|---|---|---|---|
| AUC | AUPR | ||
| Target-based CRF | GEN | 97.3 | 80.7 |
| FUN | 97.7 | 80.9 | |
| IGF | 98.0 | 83.9 | |
| Drug-based CRF | CHEM | 96.0 | 81.5 |
| PHAR | 96.6 | 79.9 | |
| ICP | 98.1 | 85.9 | |
| Full Integration Approach (FI) | 99.2 | 94.9 | |
Fig. 1.
PR curves for different approaches on the first dataset. (A) PR curves for drug-based CRFs. (B) PR curves for target-based CRFs. (C) PR curves for the FI approach.
3.4. Comparison Results
To compare with other existing approaches, we tested our algorithm on the second dataset, i.e., the benchmark dataset published in Ref. 24. Here we mainly compared our approach with the pairwise kernel regression (PKR) method proposed in Ref. 24, which claimed that PKR outperformed many other state-of-the-art methods on the same data. As in Ref. 24, we also tested seven different approaches, including AERS-freq-based pharmacogenomic approach (AERS-freq), AERS-bit-based pharmacogenomic approach (AERS-bit), SIDER-based pharmacogenomic approach (SIDER), JAPIC-based pharmacogenomic approach (JAPIC), chemogenomic approach (CHEM), integrated pharmacogenomic approach (INTEG-P) and integrated pharmaco-chemogenomic approach (INTEG-PC). These different methods, as suggested by their names, are defined mainly based on input data, and more details about them can be found in Ref. 24 or Supplementary Material S5 of this paper. In addition, we tested an additional approach that combines chemical, side-effect, sequence and functional data together. This approach was not included in Ref. 24 and we referred to it as ‘INTEG-ALL’. Table 2 shows the comparison results between our conditional random field (CRF) model and the pairwise kernel regression (PKR) model.
Table 2.
The comparison results between our CRF and PKR methods. The second dataset was tested in our CRF model using 3-fold cross-validation. The results for PKR were taken from Ref. 24 in which pair-wise cross-validation corresponds to our 3-fold cross-validation test here. Note that the INTEG-ALL approach was absent in Ref. 24. The best score is shown in bold.
| Approach | AUPR | |
|---|---|---|
| CRF | PKR | |
| AERS-freq | 85.7 | 80.6 |
| AERS-bit | 85.4 | 81.3 |
| SIDER | 87.3 | 76.8 |
| JAPIC | 91.2 | 87.7 |
| CHEM | 87.7 | 79.7 |
| INTEG-P | 90.7 | 87.4 |
| INTEG-PC | 90.4 | 88.5 |
| INTEG-ALL | 91.5 | \ |
As shown in table 2, our method outperformed the PKR model over all different tests. In particular, our approach can improve the AUPR score by up to 10.5 when only SIDER-based information was used. Furthermore, the results produced by CRF were not as sensitive to different input data as those produced by PKR. For example, the AUPR score of PKR based on JAPIC was about 10% larger than that based on SIDER, whereas the test of our algorithm on SIDER-based data can still yield decent performance. These comparison results indicate that our method is more robust to input data than PKR, and may have a better capacity to handle noise in data.a
4. Conclusion
In this article, we introduced functional data into DTI prediction and developed a probabilistic graphical model to predict new drug-target interactions using known drug-target interactions and various similarity scores for both drugs and targets. Our model can integrate chemical, pharmacological, genomic and functional data systematically, and predict new DTI interactions with high accuracy. We demonstrated that incorporating functional information of targets can further improve prediction performance.
Currently, our algorithm uses a simple linear combination of independent predictions from drug-based and target-based CRFs respectively. In the future, we will extend our model into a more sophisticated framework that can better integrate both drug-based and target-based CRF models. In addition, we will incorporate other data such as drug-drug interaction (DDI) and protein-protein interaction (PPI) information into DTI prediction. We hope that by incorporating these additional information our model can reveal mechanism of drug action to a greater extent. Currently we only evaluated our approach based on benchmark data. We will explore the practical applications of our prediction algorithm, e.g., identifying novel drug-target interactions for drug repositioning.
Supplementary Material
Acknowledgements
This work was supported in part by the National Basic Research Program of China Grant 2011CBA00300, 2011CBA00301, the National Natural Science Foundation of China Grant 61033001, 61061130540. We thank the anonymous reviewers for their helpful comments.
Footnotes
Supplementary Material is available at http://iiis.tsinghua.edu.cn/~compbio/papers/psb2014/psb2014_sm.pdf.
Although our dataset were slightly differently from the original data tested in the PKR model (six proteins and two drugs were excluded from the original dataset), the tiny difference between two datasets should not change the conclusions that we draw here.
References
- 1.Dudley JT, Deshpande T, Butte AJ. Brief Bioinform. 2011 Jul;12:303. doi: 10.1093/bib/bbr013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dudley JT, Sirota M, Shenoy M, Pai RK, Roedder S, Chiang AP, Morgan AA, Sarwal MM, Pasricha PJ, Butte AJ. Sci Transl Med. 2011 Aug;3:96ra76. doi: 10.1126/scitranslmed.3002648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lussier YA, Chen JL. Sci Transl Med. 2011 Aug;3:96ps35. doi: 10.1126/scitranslmed.3001512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Xie L, Xie L, Kinnings SL, Bourne PE. Annu Rev Pharmacol Toxicol. 2012;52:361. doi: 10.1146/annurev-pharmtox-010611-134630. [DOI] [PubMed] [Google Scholar]
- 5.Ekins S, Williams AJ, Krasowski MD, Freundlich JS. Drug discovery today. 2011;16:298. doi: 10.1016/j.drudis.2011.02.016. [DOI] [PubMed] [Google Scholar]
- 6.Blatt J, Corey SJ. Drug discovery today. 2012;18:4. doi: 10.1016/j.drudis.2012.07.009. [DOI] [PubMed] [Google Scholar]
- 7.Cheng AC, Coleman RG, Smyth KT, Cao Q, Soulard P, Caffrey DR, Salzberg AC, Huang ES. Nature biotechnology. 2007;25:71. doi: 10.1038/nbt1273. [DOI] [PubMed] [Google Scholar]
- 8.Morris GM, Huey R, Lindstrom W, Sanner MF, Belew RK, Goodsell DS, Olson AJ. Journal of computational chemistry. 2009;30:2785. doi: 10.1002/jcc.21256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kinnings SL, Liu N, Tonge PJ, Jackson RM, Xie L, Bourne PE. J Chem Inf Model. 2011;51:408. doi: 10.1021/ci100369f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Donald BR. Algorithms in structural molecular biology. The MIT Press; 2011. [Google Scholar]
- 11.Keiser MJ, Roth BL, Armbruster BN, Ernsberger P, Irwin JJ, Shoichet BK. Nature biotechnology. 2007;25:197. doi: 10.1038/nbt1284. [DOI] [PubMed] [Google Scholar]
- 12.Keiser MJ, Setola V, Irwin JJ, Laggner C, Abbas AI, Hufeisen SJ, Jensen NH, Kuijer MB, Matos RC, Tran TB, et al. Nature. 2009;462:175. doi: 10.1038/nature08506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yabuuchi H, Niijima S, Takematsu H, Ida T, Hirokawa T, Hara T, Ogawa T, Minowa Y, Tsujimoto G, Okuno Y. Mol Syst Biol. 2011 Mar;7:472. doi: 10.1038/msb.2011.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cheng F, Liu C, Jiang J, Lu W, Li W, Liu G, Zhou W, Huang J, Tang Y. PLoS Computational Biology. 2012;8:e1002503. doi: 10.1371/journal.pcbi.1002503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen X, Liu M-X, Yan G-Y. Molecular BioSystems. 2012;8:1970. doi: 10.1039/c2mb00002d. [DOI] [PubMed] [Google Scholar]
- 16.van Laarhoven T, Nabuurs SB, Marchiori E. Bioinformatics. 2011;27:3036. doi: 10.1093/bioinformatics/btr500. [DOI] [PubMed] [Google Scholar]
- 17.Mei J-P, Kwoh C-K, Yang P, Li X-L, Zheng J. Bioinformatics. 2013;29:238. doi: 10.1093/bioinformatics/bts670. [DOI] [PubMed] [Google Scholar]
- 18.Shi Y, Zhang X, Liao X, Lin G, Schuurmans D. Pac Symp Biocomput. 2013;18:41. [PubMed] [Google Scholar]
- 19.Wang Y, Zeng J. Bioinformatics. 2013;29:i126. doi: 10.1093/bioinformatics/btt234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang W, Yang S, Li J. Pac Symp Biocomput. 2013;18:53. [PMC free article] [PubMed] [Google Scholar]
- 21.Yamanishi Y, Araki M, Gutteridge A, Honda W, Kanehisa M. Bioinformatics. 2008;24:i232. doi: 10.1093/bioinformatics/btn162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Campillos M, Kuhn M, Gavin A-C, Jensen LJ, Bork P. Science. 2008;321:263. doi: 10.1126/science.1158140. [DOI] [PubMed] [Google Scholar]
- 23.Yamanishi Y, Kotera M, Kanehisa M, Goto S. Bioinformatics. 2010;26:i246. doi: 10.1093/bioinformatics/btq176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Takarabe M, Kotera M, Nishimura Y, Goto S, Yamanishi Y. Bioinformatics. 2012;28:i611. doi: 10.1093/bioinformatics/bts413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schlicker A, Domingues FS, Rahnenführer J, Lengauer T. BMC bioinformatics. 2006;7:302. doi: 10.1186/1471-2105-7-302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhao S, Li S. PloS one. 2010;5:e11764. doi: 10.1371/journal.pone.0011764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hinton GE. Neural computation. 2002;14:1771. doi: 10.1162/089976602760128018. [DOI] [PubMed] [Google Scholar]
- 28.Kollar D, Friedman N. Probabilistic graphical models: principles and techniques. The MIT Press; 2009. [Google Scholar]
- 29.Zeng J, Zhou P, Donald BR. A markov random field framework for protein side-chain resonance assignment, in Research in Computational Molecular Biology. 2010 [Google Scholar]
- 30.Zeng J, Zhou P, Donald BR. Journal of biomolecular NMR. 2011;50:371. doi: 10.1007/s10858-011-9522-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lafferty J, McCallum A, Pereira FC. Conditional random fields: Probabilistic models for segmenting and labeling sequence data. Proceedings of the 18th International Conference on Machine Learning (ICML-2001) 2001 [Google Scholar]
- 32.Sha F, Pereira F. Shallow parsing with conditional random fields. Proceedings of the 2003 Conference of the North American Chapter of the Association for Computational Linguistics on Human Language Technology-Volume. 2003;1 [Google Scholar]
- 33.Settles B. Biomedical named entity recognition using conditional random fields and rich feature sets; Proceedings of the International Joint Workshop on Natural Language Processing in Biomedicine and its Applications; 2004. [Google Scholar]
- 34.Qin T, Liu T-Y, Zhang X-D, Wang D-S, Li H. tech. rep., Technical Report MSR-TR-2008-156. Microsoft Corporation; 2008. Global ranking of documents using continuous conditional random fields. [Google Scholar]
- 35.Xin X, King I, Deng H, Lyu MR. A social recommendation framework based on multi-scale continuous conditional random fields. Proceedings of the 18th ACM conference on Information and knowledge management. 2009 [Google Scholar]
- 36.Wang Z, Xu J. Bioinformatics. 2011 Jul;27:i102. doi: 10.1093/bioinformatics/btr232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bishop CM, et al. Pattern recognition and machine learning. Springer; New York: 2006. [Google Scholar]
- 38.Bertsekas DP, Nedić A, Ozdaglar AE. Convex analysis and optimization. Athena Scientific; Belmont: 2003. [Google Scholar]
- 39.Salakhutdinov R, Mnih A, Hinton G. Restricted boltzmann machines for collaborative filtering. Proceedings of the 24th international conference on Machine learning. 2007 [Google Scholar]
- 40.Carreira-Perpignan GEHMA. On contrastive divergence learning. Artificial Intelligence and Statistics. 2005 [Google Scholar]
- 41.Kanehisa M, Goto S, Hattori M, Aoki-Kinoshita KF, Itoh M, Kawashima S, Katayama T, Araki M, Hirakawa M. Nucleic acids research. 2006;34:D354. doi: 10.1093/nar/gkj102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kanehisa M, Araki M, Goto S, Hattori M, Hirakawa M, Itoh M, Katayama T, Kawashima S, Okuda S, Tokimatsu T, et al. Nucleic acids research. 2008;36:D480. doi: 10.1093/nar/gkm882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kuhn M, Campillos M, Letunic I, Jensen LJ, Bork P. Molecular systems biology. 2010;6 doi: 10.1038/msb.2009.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mahé P, Ueda N, Akutsu T, Perret J-L, Vert J-P. Journal of chemical information and modeling. 2005;45:939. doi: 10.1021/ci050039t. [DOI] [PubMed] [Google Scholar]
- 45.Saigo H, Vert J-P, Ueda N, Akutsu T. Bioinformatics. 2004;20:1682. doi: 10.1093/bioinformatics/bth141. [DOI] [PubMed] [Google Scholar]
- 46.Schlicker A, Albrecht M. Nucleic acids research. 2008;36:D434. doi: 10.1093/nar/gkm806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schlicker A, Albrecht M. Nucleic acids research. 2010;38:D244. doi: 10.1093/nar/gkp979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bleakley K, Yamanishi Y. Bioinformatics. 2009;25:2397. doi: 10.1093/bioinformatics/btp433. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.