Abstract
Human immunodeficiency virus type 1 (HIV-1) infection remains to be one of the major global health problems. It is thus necessary to identify novel therapeutic molecules to combat HIV-1. Natural antimicrobial peptides (AMPs) have been recognized as promising templates for developing topical microbicides. This review systematically discusses over 80 anti-HIV peptides annotated in the antimicrobial peptide database (http://aps.unmc.edu/AP). Such peptides have been discovered from bacteria, plants, and animals. Examples include gramicidin and bacteriocins from bacteria, cyclotides from plants, melittins and cecropins from insects, piscidins from fish, ascaphins, caerins, dermaseptins, esculentins, and maximins from amphibians, and cathelicidins and defensins from vertebrates. These peptides appear to work by different mechanisms and could block viral entry in multiple ways. As additional advantages, such anti-HIV peptides may possess other desired features such as antibacterial, antiparasital, spermicidal, and anticancer activity. With continued optimization of peptide stability, production, formulation and delivery methods, it is anticipated that some of these compounds may eventually become new anti-HIV drugs.
Keywords: antimicrobial peptides, cathelicidins, cecropins, cyclotides, database, defensins, HIV-1, host defense peptides, microbicides, peptide
1. INTRODUCTION
The first human immunodeficiency virus (HIV) case was documented in the 1980’s [1]. This virus has since caused many infections around the world. The United Nations estimates that there are now 34 million people living with HIV/AIDS (acquired immunodeficiency syndrome). This makes HIV/AIDS the fourth leading cause of death worldwide. As a consequence, it is important to develop novel therapeutic and preventative agents.
HIV type 1 (HIV-1), originated from chimpanzees Pan troglodytes troglodytes, was later transmitted to humans about one hundred years ago [2]. The virus infects not only T-lymphocytes, but also macrophages and dendritic cells [3, 4]. The life cycle of HIV-1 is complex, current knowledge can be found in review articles [5–7]. To facilitate my discussion, some major steps are depicted in Fig. 1. The HIV-1 infection starts from the attachment of viral glycoprotein gp120 to the CD4 receptor of T-lymphocytes. During this process, co-receptors also play an essential role. Depending on the type of co-receptors involved, the HIV-1 isolates are classified into three types: X4, R5, and combined X4R5 [8]. The X4 tropic virus uses the CXCR4 co-receptor, while the R5 strain utilizes the CCR5 co-receptor. Individuals with a 32-base deletion in the coding sequence of CCR5 are resistant to HIV-1 infection, underscoring the essential role of this co-receptor for viral infection [4]. For early-transmitted founder viruses, the R5 strain infects CD4+ lymphocytes but not macrophages. After fusion, HIV-1 releases its RNA into CD4 cells and “hijacks” the cells to produce more viruses. The viral RNA is necessarily transcribed into complementary DNA (cDNA) under the catalysis of reverse transcriptase. The DNA is then integrated into the host genome with the aid of viral integrase and reversely transcribed into mRNA for protein expression. After translation, protease processing, and assembly, new viral particles are released from the host cell and can infect other cells. Understanding the HIV-1 life cycle is important for developing anti-HIV-1 drugs.
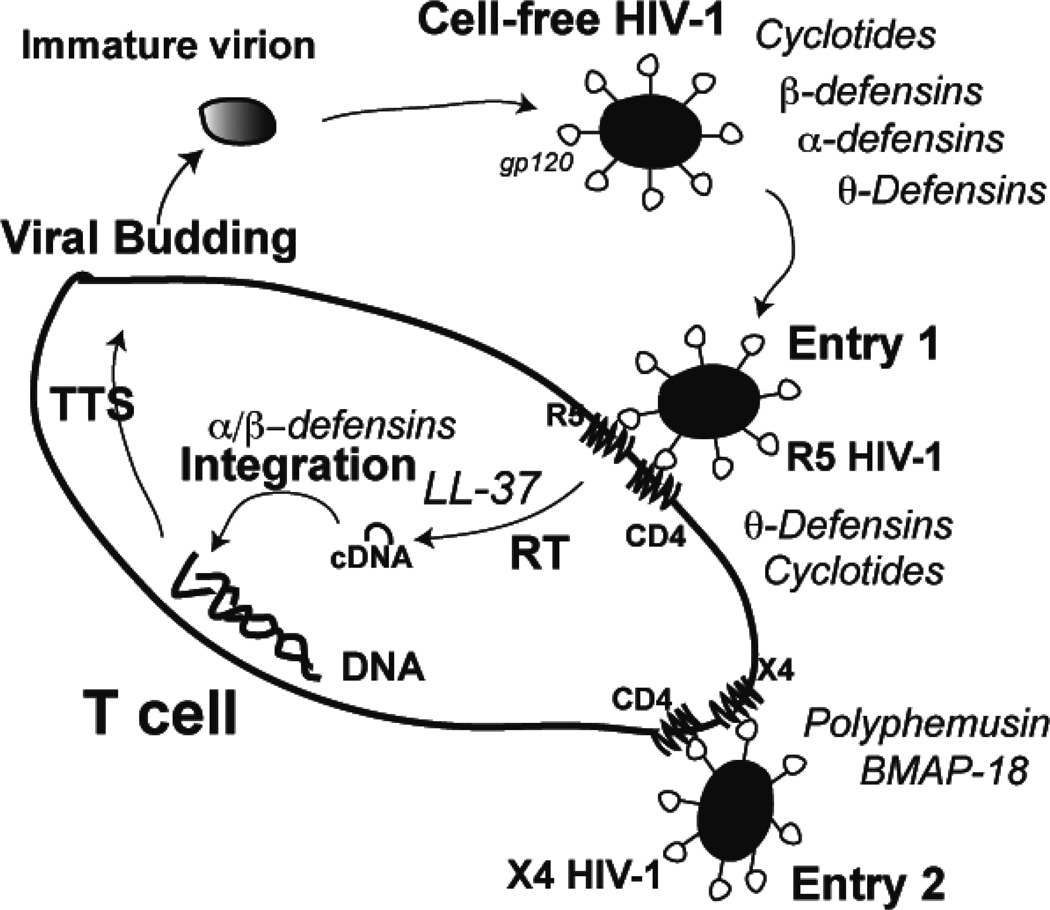
Fig. 1.
Natural AMPs could interfere with various stages of the HIV-1 life cycle. These include directly disruption of cell-free HIV-1 by defensins and cyclotides, blocking the entry of R5 HIV-1 by θ-defensins or the entry of X4 HIV-1 by polyphemusin analogs and BMAP-18, a likely LL-37 inhibition of viral reverse transcriptase required for reverse transcription (RT), interference by α or β-defensins after cDNA formation. The integrated viral genome is transcribed, translated, and assembled (TTS) into new virions, which leave the T cell via budding (see the text for references). This new construction is inspired by ref. [10].
Although substantial knowledge has been acquired about HIV-1 genetic sequences, immunological epitopes, drug resistance-associated mutations, and vaccine trials (http://www.hiv.lanl.gov/content/index), effective vaccines are not yet available, rendering it necessary to search for alternative therapeutic strategies such as anti-HIV microbicides [9, 10]. Natural compounds have been one of the major sources for developing modern medicine [11]. Although the emphasis was shifted to high-throughput screening (HTS) approaches in the 1990’s [12], there is continued interest in natural compounds, especially those with novel structural scaffolds. HIV-1 inhibitory activity of natural peptides might have been noticed in the early National Cancer Institute (NCI) anti-HIV-1 drug screening programs [13]. Recently, much research attention has been given to natural antimicrobial peptides (AMPs) due to the growing antibiotic resistance problem worldwide. These peptides are usually short (5–50 amino acids) and cationic (on average +3). As the essential defense molecules of innate immune systems of all life forms, cationic AMPs can effectively neutralize microbes. In in vitro experiments, AMPs were found to be active against various bacteria, viruses, fungi, and parasites [14–17]. In vivo, the deficiency of such molecules can cause various infectious diseases, whereas the expression of AMPs protects the host from infection, including viruses [18, 19]. These results underscore the protective role of natural AMPs against microbial infection.
AMPs vary in source, amino acid sequence, three-dimensional structure, and biological activity. This diversity makes it difficult to manage the AMP information manually. As a consequence, several databases have been established [20–28]. The antimicrobial peptide database (APD) (http://aps.unmc.edu/AP/main.html) [23, 24] has been widely utilized. Using the APD, users can readily obtain a set of peptides with defined activity (antibacterial, antiviral, antifungal, antiparasital, anti-cancer, insecticidal, spermicidal, and/or chemotactic). To aid our anti-HIV peptide project, we also annotated anti-HIV activity into the APD. When this manuscript was completed, more than 80 natural AMPs or their derivatives were registered as anti-HIV-1 peptides (Table 1). Most of these peptides are identified in plants and animals, including humans. This article describes natural anti-HIV-1 AMPs from bacteria, plants, and animals (Table 1) and highlights their mechanisms of action (Table 2). The article also summarizes database-derived methods for discovering and improving anti-HIV-1 peptides. Finally, the advantages and disadvantages in developing natural AMPs into anti-HIV-1 microbicides are discussed.
Table 1.
Known anti-HIV-1 antimicrobial peptides from different sources1.
| Source | no. of AMPs | no. of anti-HIV-1 AMPs |
|---|---|---|
| Bacteria | 171 | 5 |
| Plants | 289 | 26 |
| Animals | 1489 | 42 |
| Humans | 61 | 10 |
| Synthetic | 33 | 4 |
| Total | 2043 | 87 |
Obtained from the APD database at http://aps.unmc.edu/AP/main.html on September 12, 2012 [23, 24].
Table 2.
Properties and mechanisms of the three types of HIV-1 inhibitory defensins.
| Molecular shape | Non-circular | Circular | |
|---|---|---|---|
| Defensin type | α | β | θ |
| Representative | HNP-1 | HBD-2 | RTD-1 |
| Properties [23, 24] | |||
| APD ID | 176 | 524 | 445 |
| Year discovered | 1985 | 1997 | 1999 |
| Amino acids | 30 | 41 | 18 |
| Net charge | +3 | +7 | +5 |
| Hydrophobic content | 53% | 36% | 55% |
| 3 disulfide bonds C-C | 2–30; 4–19; 9–29 | 8–37; 15–30; 20–38 | 3–16; 5–14; 7–12 |
| 3D Structure | β (3 strands) | αβ (1 helix + 3 strands) | β (2 strands) |
| Mechanisms [99] | |||
| Cells used | PBMC | PBMC | PBMC |
| X4 EC50 (µM) | 2.3 | 6.9 | 3.5 |
| R5 EC50 (µM) | 2.6 | 5.8 | 3.6 |
| HIV-1 entry | No | No | Inhibition |
| After viral entry | Inhibition | Inhibition | No |
| Down-regulation of CXCR4 | Yes | Yes | Yes |
| Down-regulation of CCR5 or CD4 | No | No | No |
| Direct inactivation of R5 HIV-1 | Yes | Yes | No |
| Direct inactivation of X4 HIV-1 | Yes | Yes | Yes |
2. Bacterial AMPs with HIV-1 inhibitory activity
As of Sept 2012, there are 171 AMPs from bacteria (also called bacteriocins) in the APD. Among them, five bacteriocins are known to have anti-HIV-1 activity (Table 1). Most of bacteriocins are gene coded, but some are synthesized by multienzyme systems. Gramicidin is a classic example of non-ribosomally synthesized antibiotics. Isolated from a soil bacterium Bacillus brevisin (new name Brevibacillus brevis) in 1939, gramicidin is the first peptide antibiotic used clinically [29]. Gramicidin D has been approved by FDA for topical use. Both the N- and C-termini of gramicidin A are chemically modified (CHO at the N-terminus and NHCH2CH2OH at the C-terminus). In addition, amino acids 4, 6, 8, 10, and 12 are D-amino acids, allowing the formation of a special helical conformation in membranes. In lipid bilayer, two gramicidin molecules form a head-to-head dimer with the C-terminus exposed [30]. This pore allows ion passage. In addition to bactericidal activity, commercial gramicidin from Sigma, consisting of a mixture of 80% A, 6% B, and 14% C forms (for the amino acid sequence, visit the APD), showed inhibitory effects on HIV-1 infections [31]. In cervical explants, gramicidin (1 µg/ml) could protect the testing cells from infection by 71% [32]. The same peptide also possesses spermicidal effects. In the APD [24], maximin 3, magainin 2, dermaseptin-S1 and dermaseptin-S4 are also known to have both anti-HIV-1 and spermicidal activity. The combined contraceptive and anti-HIV-1 properties of these peptides are attractive for developing microbicides against sexual transmitted diseases (STIs). Whether gramicidin inhibits HIV-1 in vivo awaits to be tested.
Siamycin I, siamycin II, and RP 71955 are another three anti-HIV-1 peptides from the bacterial sources. These peptides are able to adopt a unique lasso structure. Here I use the crystal structure of BI-32169 (Fig. 2A) to illustrate this unique structure [33]. A ring structure is created due to a covalent bond between the NH of the N-terminal Gly1 and the side chain carboxylate of Asp9. Next, the tail (black in Fig. 2A) enters the ring structure. Finally, residue Cys19 at the C-terminal tail is locked by forming a disulfide bond with residue Cys6 in the ring structure. These anti-HIV peptides belong to class I lassos since they contain disulfide bonds [34]. The unique lasso scaffold makes these anti-HIV-1 compounds attractive due to their stability to proteases.
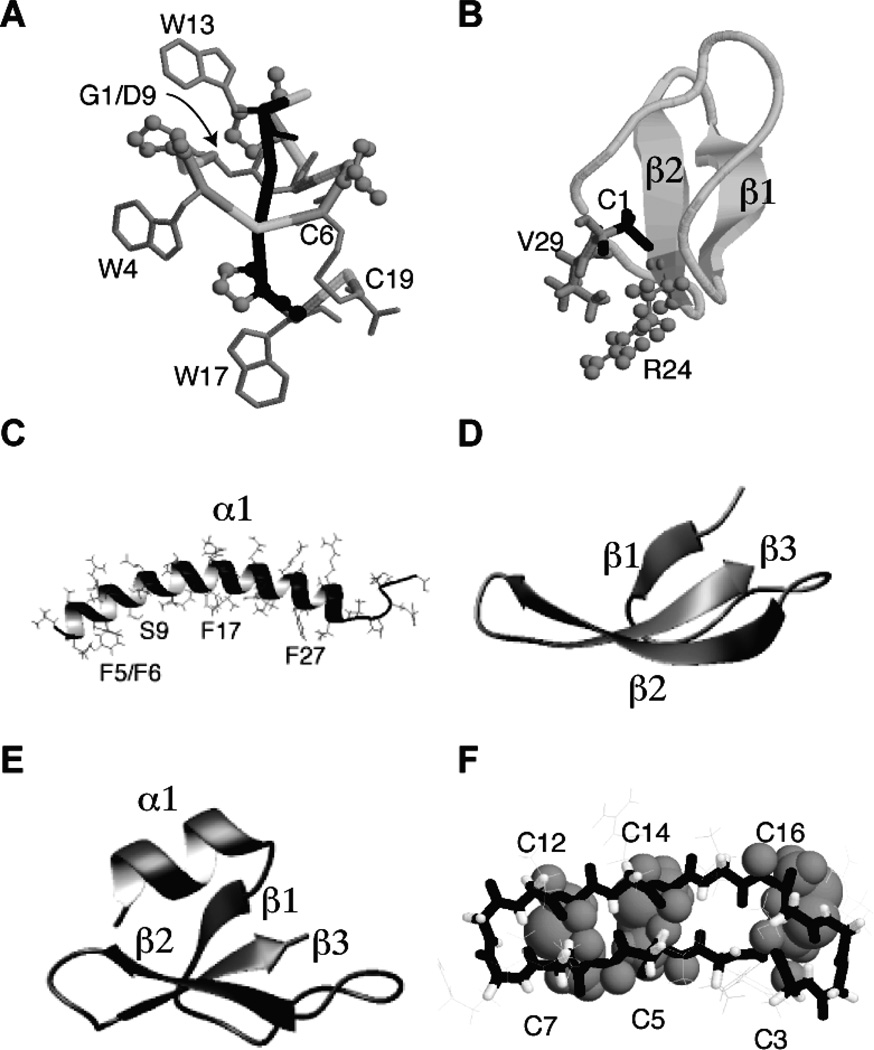
Fig. 2.
Three-dimensional structures of a bacterial lasso peptide (A), plant kalata B1 (B), human cathelicidin LL-37 (C), α-defensin 1 (HNP-1) (D), β-defensin-1 (HBD-1) (E), and rhesus theta defensin-1 (RTD-1) (F). Coordinates were obtained from the Protein Data Bank (PDB) (PDB IDs are 3NJW for the bacterial lasso BI-32169 [33]; 1JNU for membrane-bound kalata B1 [40]; 2K6O for membrane-bound LL-37 [78]; 3GNY for HNP-1 [90]; 1IJV for HBD-1 [133]; and 1HVZ for RTD-1 [134]. For clarity, only the three disulfide bonds in RTD-1 are displayed using space-filling model. A peptide bond forms between the N- and C-termini of kalata B1 and RTD-1. While the N-terminal amide of G1 and the carboxylic side chain of D9 form a macrolactam (pointed by an arrow in panel A), the C-terminal C19 forms a disulfide bond with C6. These structural scaffolds are remarkably stable to heat, chemicals, and proteases, making them ideal for peptide engineering through site-directed mutagenesis or peptide grafting (i.e. replacing an exposed loop of cyclotides with biologically active peptides) [135, 136]. For additional information on structural studies of natural AMPs, please refer to ref. [137].
3. Plant AMPs with HIV-1 inhibitory activity
In the APD [24], 289 AMPs are of plant sources. Among them, 26 are annotated as HIV-1 inhibitory peptides (Table 1). They are primarily cyclotides (in total 22) [35, 36]. The dominance of cyclotides may be attributed to dedicated screening efforts as well as growing interest in the circular structural scaffold of cyclotides (Fig. 2B). Structural features of cyclotides important for anti-HIV-1 activity have been appreciated. These include (1) the overall fold of these peptides; (2) the N and C-terminal cyclization via forming a peptide bond; and (3) regional hydrophobicity (e.g. a hydrophobic patch). One of the most potent cyclotides is also most hydrophobic, implying that this peptide targets the viral envelope [35]. In a series of peptides we recently designed, it is interesting to note that the most potent anti-HIV-1 peptide is also most hydrophobic based on HPLC retention time measurements [37].
As the first member in the family, kalata B1 is now used as a model peptide for cyclotide research (Fig. 2B). Isolated in 1973 [38], the circular structure of this peptide was not established until 1999 [39]. There is a peptide bond between Cys1 and Val29, probably enabling sidechain interactions between Val29 and the only cationic Arg24 (Fig. 2B) [40]. The cyclic structure is essential for anti-HIV-1 activity as an acyclic analog was inactive [41]. This observation indicates the importance of the fold of this small protein that enables hydrophobic side chains to cluster on the protein surface. This hydrophobic patch is proposed to be responsible for membrane binding. Interestingly, this patch preferentially binds to phosphatidylethanolamine (PE) and leads to pore formation (41–70 Å) in lipid bilayers [42].
Oligomerization on the membrane surface could be important for this process as well. Sando et al. [43] found that both the L and D-forms of kalata B1 have anti-HIV-1 activity, although not exactly identical. The peptide can act on viral membranes and disrupt HIV-1 particles. Because the HIV-1 envelope contains a high content of the PE lipid, specific recognition and peptide aggregation might be involved in extracting HIV-1 lipids, leading to collapse of the viral envelope.
4. Animal AMPs with HIV-1 inhibitory activity
Animals cover a broad range of species and constitute the major sources of AMPs registered in the APD (1489 out of a total of 2043) (Table 1). Because only a very small set (2%) of such AMPs was subjected to anti-HIV-1 assays, they are herein broadly grouped under the following subtitles: insects, amphibians, aquatic, and terrestrial animals.
4.1. Insect AMPs
The discovery of cecropins by Hans Boman and colleagues in the 1980’s [44] is a milestone in the history of AMP research. The signal for expression of these peptides is transmitted via the toll pathway [45]. The toll appears to be a universal signaling pathway because it also exists in humans [46, 47]. The importance of the toll pathway has been recognized by the 2011 Nobel Prize.
The current APD collected 203 insect AMPs but only a few have been tested for anti-HIV-1 activity. Melittin and cecropin A are HIV-1 inhibitory [48]. Additional HIV-1 inhibitory peptides identified in our database screening [49] are ponericin L2 from ants [50], spinigerin from termites [51], and melectin from the Cleptoparasitic bees [52].
4.2. AMPs from amphibians
Since the discovery of magainins by Zasloff in 1987 [53], hundreds more AMPs have been documented in frogs. By the time this article was written, the current APD collected 842 AMPs from amphibians [24]. This accounts for ~40% of the peptide entries in our database. However, only 25 amphibian AMPs or their derivatives are known to have anti-HIV-1 activity. In spite of a small number, they account for ~60% of the anti-HIV-1 AMPs from the animal kingdom. Some amphibian AMPs are also capable of inhibiting other viruses such as channel catfish virus (CCV) and frog virus 3 (FV3) [54]. For amphibians, such a protective effect against FV3 is significant for frog survival.
It seems that maximin 3, isolated from skin secretions of the Chinese red belly toad Bombina maxima, is the first amphibian peptide to be tested for HIV-1 inhibitory activity (cell selectivity index of 7.6 at 0.77 µM) [55]. Lorin et al. [56] reported that dermaseptin S4 (DS4) inhibits both cell-free and cell-associated HIV-1 infection of T lymphocytes in vitro. Importantly, the peptide is effective against both R5 and X4 primary isolates, including a laboratory-adapted HIV-1 strain. Another 2005 paper reported HIV-1 inhibitory activity of several amphibian AMPs [57]. In particular, caerin 1.1, caerin 1.9, and maculentin 1.1 can stop HIV-1 infection in minutes, probably by blocking fusion between HIV-1 and host cells. Other peptides that displayed such an inhibitory effect are caerin 1.4, dahlein 5.6, dermaseptin S1, esculentin-1ARb, esculentin-2P, palustrin-3AR, and uperin 3.6. Both reports demonstrated that AMPs can inhibit viral transfer from dendritic cells to T cells.
Through our database screening project, additional anti-HIV-1 peptides have been identified from amphibians [21]. These new members are ascaphin-8, brevinin-2-related peptide, masculentin 1.3, ranatuerin 9, temporin-LTc, and peptide mutants of aurein 1.2, temporin-PTa, uperin 7.1, dermaseptin-S9, and maximin H5. Considering the large number (up to ~100) of natural AMPs in each of the 4000 types of frogs [58, 59] widely distributed on different continents [59–64], there is a great opportunity to identify lead compounds from amphibian AMPs for developing anti-HIV-1 microbicides.
4.3. AMPs from aquatic animals
Efforts have been made to identify anti-HIV-1 compounds from marine sources as well. A well-known example is cyanovirin-N, a protein from a blue green alga [65]. Aquatic animals also provide a good source for AMPs. For example, 64 fish AMPs have been registered into the APD [23, 24]. We showed that piscidin 1 possesses anti-HIV-1 activity [49]. In addition, polyphemusins and tachyplesins, isolated from horseshoe crabs, are HIV-1 inhibitory. T22 is a derivative of polyphemusin II where residues Phe5, Phe12 are converted to Tyr and residue Val7 to Lys. It has an excellent EC50 of 0.008 µg/ml (the concentration that inhibits viruses by 50%) and TC50 of 54 µg/ml (the concentration that causes 50% reduction in cell viability), leading to a cell selectivity index of 6750. T22 appears to inhibit viral fusion [66]. Since T22 binds to the envelope glycoprotein gp120 of HIV-1 and the cell receptor CD4 of T cells, it is established as a lectin [67]. In fact, the peptide binds to chemokine co-receptor CXCR4. T140, a derivative of T22, is the strongest inhibitor of the CXCR4 co-receptor. This peptide, however, does not inhibit R5-tropic HIV-1 [68]. Therefore, there is a need to search for novel anti-HIV-1 agents that inhibit the R5 viruses. This is clearly important considering the R5 viruses are the major strain transmitted via sexual intercourse [3].
4.4. AMPs from terrestrial animals
Cathelicidins, defensins, and histatins are major AMPs from humans [69]. Cathelicidins are a family of AMPs that share a common N-terminal “cathelin” domain in their precursor proteins [70]. However, the C-terminal antimicrobial regions of precursors can vary substantially in terms of sequence composition as well as 3D structure. Human cathelicidin LL-37 is a representative member of the helical family (Fig. 2C). Defensins discussed below all have three pairs of disulfide bonds. They are classified into three families based on their size and 3D structure. While human α-defensins only contain three β-strands (Fig. 2D), β-defensins are composed of one helical region and three β-strands (Fig. 2E). A third family of defensins is small and circular, thereby called β-defensins. The main regular structures are two antiparallel β-strands stabilized by three pairs of disulfide bonds (Fig. 2F).
Cathelicidins
Human cathelicidin LL-37 is a 37-residue antimicrobial peptide starting with a pair of leucines (LL). LL-37 and its precursor protein hCAP-18 were discovered by three laboratories in 1995 [71–73]. The therapeutic significance of this human AMP is now linked to Niels Finsen’s light therapy for tuberculosis. Light triggers the synthesis of vitamin D, which in turn induces the expression and release of human LL-37 that kills the bacterium [47, 74]. Indeed, the protective role of LL-37 against bacterial infection has been demonstrated in both clinical and animal models [19, 75]. In vitro, LL-37 is active against bacteria, fungi, and viruses. According to circular dichroism (CD), the peptide adopted a helical conformation at physiological pH, increased salt concentrations, or upon association with membranes [76, 77]. Detailed structural and dynamic analysis of human LL-37 by 3D triple-resonance solution-state NMR spectroscopy found a 30-residue long amphipahtic helix, while the C-terminal tail is unstructured and mobile (Fig. 2C) [78]. The long helix is demonstrated to directly interact with bacterial membranes. Interestingly, the hydrophobic surface of membrane-bound LL-37 is interrupted by a hydrophilic residue Ser9 that splits the hydrophobic surface into two domains [79]. These two membrane-binding domains (Fig. 2C) provide a structural basis for synergistic binding of LL-37 to lipopolysaccharides (LPS) observed earlier [80].
Several laboratories demonstrated the anti-HIV-1 activity of human cathelicidin LL-37 [81–83]. We reported an EC50 of 1.6 µM for human LL-37 [83]. It should be noted that the precursor protein of human cathelicidin can be cleaved in an alternative way. The protein in the semen is cleaved by gastricsin in vagina into ALL-38, which contains one additional alanine at the N-terminus compared to LL-37 [84]. It is safe to predict that ALL-38 is also inhibitory to HIV-1 and other pathogens so as to protect the sperm from infection. In addition, Yamasaki et al. detected fragments of LL-37 in human skin [85]. Because LL-23 and KR-20 are inactive against HIV-1 (Fig. 3), these two natural fragments may not be produced to protect humans from viral infection [83]. Rather, they have varying antimicrobial and immune modulating activity compared to LL-37 [85]. In particular, mutations of Ser9 of LL-23 led to peptides with distinct immune modulation abilities [79]. While LL-23 itself induces the production of chemokine MCP-1, LL-23V9 (i.e. Ser9 is changed to Val9) completely suppresses the release of this chemokine. This MCP-1 suppressing property of LL-23V9 might be used to develop anti-inflammatory agents [79].
Fig. 3.
Sequence and anti-HIV-1 activity of human cathelicidin LL-37 and its natural and artificial fragments. This figure was made to illustrate the active region based on partial data from Wang et al. [83].
Defensins
In the 1980’s, Lehrer and colleagues first isolated α-defensins from neutrophils [86]. Human α-defensins (HNP-1 to HNP-3) are essentially identical molecules that differ only by one residue at the N-terminus. These three defensins were initially found to have antiviral activity against herpes simplex virus (HSV) and influenza A virus (IAV) and later HIV-1 [87]. Like polyphemusin analogs [67], HNP-1 is also a lectin that binds to viral glycoprotein gp120 [88, 89] and such a binding is not affected by conversion of the L-form of this defensin to the D-form [88, 90]. High production of α-defensins1–3 by immature DCs appears as a host protective factor against progression of HIV-1 infection, suggesting potential diagnostic, therapeutic and preventive applications [91]. HNP-4 is more effective than HNP1–3 in protecting human PBMC from infection by X4 or R5 HIV-1 [92]. However, unlike HNP1–3, HNP-4 does not work primarily by binding to viral glycoproteins [88]. Thus, the exact mechanism how these defensins block HIV-1 is poorly understood. In fact, HNP-1 can inhibit HIV-1 entry in multiple ways, including disruption of the virus-receptor interactions, preventing its uptake, and interfering with the 6-helix bundle formation required for cell-virus fusion [139]. Another two members, HD-5 and HD-6, can also act on viruses. HD-5 binds to gp120 of HIV-1 in vitro, but not HD-6 [90]. They compete with HIV-1 entry inhibitors. Surprisingly, these defensins can concentrate on cell surfaces leading to increase in HIV-1 infectivity [93]. Such effects of HD5 and HD6 are undesired and could compromise the efficacy of other HIV-1 microbicides [94]. It is not yet clear whether the same process occurs in vivo.
Human β-defensins were first identified in 1995 [95]. Four β-defensins in humans are now well characterized, although 28 more are predicted in the genome [96]. These defensins have various biological functions, ranging from direct antimicrobial to immune modulation [87]. The relationships of these peptides with HIV-1 infection were also investigated. A single-nucleotide polymorphism in the HBD-1 gene was linked to HIV-1 infection in children [97]. Subsequent studies revealed that such nucleotide polymorphisms might play a role in mother-to-children HIV-1 transmission as well. Furthermore, HIV-1 infected patients appeared to have low mRNA levels for HBD-1 and HBD-2 in alveolar macrophages [98]. These results suggest the important protective role of these human defensins against HIV-1 infection. Indeed, HBD-2 and HBD-3 were demonstrated to have HIV-1 inhibitory effects in vitro at concentrations similar to those detected in vivo (~10 µM) [99], while HBD-1 (Fig. 2E) displayed a significantly lower potency or no activity [100, 101]. HBD-2 was found to inhibit the replication of both X4 and R5 HIV-1 viruses (Table 2) [99, 101]. In addition, HBD-2 and −3 could inactivate cell-free viruses [100, 101], while HBD-3 is an antagonist of the HIV-1 co-receptor CXCR4 [102]. Thus, HBD-2 and HBD-3 might play the major protective role in human innate immunity against HIV-1 infection. These results, if confirmed in vivo, could form the basis for developing topical virucides to stop HIV-1 transmission.
Circular θ-defensins (Fig. 2F) were first discovered in primate blood cells in 1999 [103]. The molecule is generated by tailoring and combining two copies of α-defensins. It possesses a fascinating ladder-like circular structure (Fig. 2F). This cyclic structure appears to be essential for antimicrobial activity, because the acyclic form of rhesus theta defensin-1 (RTD-1) is 3-fold less active against bacteria such as Staphylococcus aureus and Escherichia coli than the cyclic form [103]. One possibility is that the cyclic form is more effective in forming lipid-peptide domain than the acyclic form [104]. Another advantage is that the activities of these cyclic θ-defensins are not as sensitive to the effects of salts as acyclic α-defensins [103].
Note that θ-defensins are not expressed in humans due to premature stop codons [105]. It is not clear whether the lack of such molecules makes us more susceptible to HIV-1 infection. However, a synthetic peptide called retrocyclin-1, corresponding to the putative human θ-defensin, interfered with the early stage of viral infection of human CD4+ cells by both X4- and R5-tropic HIV-1, although it did not directly inactivate the virus [105]. Owen et al. [106] synthesized retrocyclin-1 entirely with D-amino acids. This D-form peptide (RC-112) is more potent against various HIV-1 subtypes than the L-form retrocyclin-1. The same group also obtained a more potent form RC-101 when one arginine of retrocyclin-1 was mutated to a lysine residue [107]. Interestingly, the silent human θ-defensin genes can be “reawakened” by using aminoglycosides that read-through the premature termination codon [108]. However, aminoglycosides are toxic to human cells. Furthermore, it is unclear whether other silent genes will be activated accidently, leading to unwanted outcomes (e.g. cancer). Therefore, the safety of aminoglycosides is questionable.
5. Mechanisms of action of natural AMPs
It is now evident that natural AMPs utilize a variety of mechanisms to inhibit HIV-1 replication. However, one should pay attention to peptide concentrations, virus types, and cell lines reported in the literature. When the concentrations of natural AMPs are too high, they may not have biological relevance. Also, certain cell lines and HIV-1 strains are more relevant than others. In the following, we focus on α, β, and θ defensins since they showed anti-HIV activity at concentrations similar to physiological ones. HNP-1, HBD-2, and RTD-1 were chosen as representatives because of their abundance in vivo [99]. As summarized in Table 2, these three defensins vary in peptide length, net charge, and hydrophobic content. However, they are all able to inactivate cell-free HIV-1 particles. While HNP-1 and HBD-2 inactivate both X4 and R5 viruses, RTD-1 only disrupts X4 HIV-1 (Table 2). Some AMPs are able to inhibit viral entry. This may be related to membrane-active properties of natural AMPs in general [16, 17, 109]. It is likely that 9-defensins cause lipid clustering in the HIV-1 envelope [104]. To date, a dozen of AMPs are known to exert lipid-clustering effects in model systems that mimic Gram-negative bacterial membranes, where phosphatidylglycerols (~30% PGs) and phosphatidyl-ethanolamines (~70% PEs) dominate [109]. Interestingly, cyclotides can directly interact with certain lipids such as PEs [43]. Some lantibiotics such as duramycins and cinnamycin are also known to target this lipid [110, 111]. This convergence in mechanism may be related to their similar amino acid compositions uncovered by previous bioinformatic analysis of lantibiotics and cyclotides [17]. Alternatively, 9-defensins bind to glycoproteins, thereby stoping viral entry [105, 112]. However, α and β-defensins appear to inhibit viral replication after HIV-1 entry (Table 2) [113] and Seidel et al. showed that the inhibition occurs after HIV-1 cDNA formation [99]. All these examples imply that defensins could inhibit HIV-1 in multiple mechanisms. In summary, the mechanisms of AMP action could be diverse and complex, making it more attractive to develop novel anti-HIV-1 microbicides to interfere at various stages of the HIV-1 life cycle (Fig. 1).
6. Methods for the discovery and improvement of anti-HIV-1 AMPs
In my laboratory, the first project completed by a graduate student was the construction of the APD started in 2002. The 2004 version of our database contained 525 AMPs [23]. Users quickly recognized its usefulness (~15,000 web hits per year). The peptide number increased to 1228 in the 2009 version [24]. This update further boosted the access of the APD (~100,000 web hits per year during 2010–2011). The current database possesses over 2000 peptide entries, primarily from natural sources. The increase in peptide entries makes the statistical results more reliable. These data enabled us to appreciate the significance of amino acid composition in terms of evolution, function, and structure of natural AMPs [140]. It also enriches our view on natural AMPs that can adopt a variety of structural scaffolds (Fig. 2). More importantly, it laid the basis for us to develop database approaches for identification of useful antimicrobials. In the following, I briefly describe our database-derived methods for discovering anti-HIV peptides.
6.1. Template optimization
To identify the best anti-HIV-1 region of LL-37, we synthesized a library of peptide segments. Like LL-37, these fragments are named by taking the single-lettered codes for the N-terminal two amino acids followed by peptide length. Because LL-23 and KR-20, the two terminally truncated natural fragments, are inactive against HIV-1 (Fig. 3), the anti-HIV region of LL-37 must be located in the central region. Indeed, SK-21, a synthetic fragment corresponding to residues 9–29 of LL-37, is HIV-1 inhibitory. However, the synthetic segment GI-20, corresponding to residues 13–32 (with the positions of IG swapped), displays the highest therapeutic index against HIV-1 (Fig. 3). We also tested the HIV-1 inhibitory activity of the major antimicrobial region (residues 17–32) of human LL-37 discovered by the NMR technique TOCSY-trim [114]. To best conserve peptide helicity, we added a glycine at the N-terminus and an amide group at the C-terminus. The peptide is hereinafter referred to as GF-17. Interestingly, GF-17 has an identical anti-HIV-1 activity (EC50) to GI-20. Thus, the additional residues at the N-terminus of GI-20 do not increase HIV-1 activity but reduce peptide cytotoxicity, thereby increasing therapeutic index (Fig. 3). In addition, FK-13 (a peptide corresponding to residues 17–29 of LL-37) [114] is identified as the smallest virucidal segment since KR-12, which is one residue shorter than FK-13, is only active against Escherichia coli but not HIV-1 [78]. These peptides are useful templates for developing anti-HIV-1 microbicides (Fig. 3). A subsequent in vitro assay suggests that the major antimicrobial region of LL-37 [114] inhibits the HIV-1 reverse transcriptase [115]. Interestingly, BMAP-18 [83], a fragment corresponding to the N-terminal 18 residues of bovine cathelicidin BAMP-27 [70], was found to be HIV-1 inhibitory with a therapeutic index of 22. Our ongoing studies suggest that this peptide inhibits HIV-1 entry (Buckheit et al., unpublished). It appears that cathelicidins, with various sequences and structures, may block HIV-1 replication at different stages of its life cycle.
6.2. Sequence shuffling
Another method to improve anti-HIV-1 activity of AMPs is sequence shuffling. Using aurein 1.2-F13W as a template (peptide sequence: GLFDIIKKIAESW) [116], we designed seven more peptides by re-arranging the 13 amino acid residues in the peptide but retaining the potential of each sequence to form an amphipathic helix. In addition, each peptide was designed by mimicking a natural peptide so that at least a group of N-terminal residues of the peptides were shared with a model peptide in the database. In total, eight peptides were obtained (for peptide sequences, please refer to the original paper). Compared to the original template, four peptides showed similar EC50 values. While two candidates became poorer in anti-HIV activity, another two peptides showed improved therapeutic indexes due to reduced cell toxicity. Hence, sequence shuffling provides a practical approach for optimizing an anti-HIV-1 peptide [49].
6.3. Database screening
As a pilot project, we also tested the idea of database screening [23, 24]. This approach will work if the library to be screened contains active compounds with desired activity. Thirty candidates were selected based on the following peptide properties: (1) length < 25 amino acid residues because longer peptides are more costly to synthesize; (2) net charge > 0, as anionic peptides tend to be inactive; (3) no cysteines, since peptides with multiple disulfide bonds are more costly to synthesize; (4) nontoxic to mammalian cells, since this is an undesired peptide property; (5) not synthetic, as we focus on natural peptides; and (6) anti-HIV-1 activity unknown, since we want to identify new candidates. The 30 AMPs originate from bacteria, spiders, insects, tunicates, fish, amphibians, and cattle. Eleven peptides have EC50 concentrations in the range of 0.63–7.1 µM. Four peptides show therapeutic indexes > 10. These four peptides include a temporin-PTa analog (EC50 0.63 µM), a mutant of temporin-LTc (EC50 0.83 µM), ponericin L2 (EC50 1.4 µM), and spinigerin (EC50 3.05 µM). This successful example demonstrates that the APD is a useful resource for identifying novel anti-HIV-1 peptides [49].
6.4. Database-aided de novo design
We also programmed a statistical interface for the APD [23]. In particular, amino acid composition profiles can be calculated for a single peptide or a group of AMPs with a common feature (e.g. peptide source, post-translational modification, structure, or activity). Fig. 4A shows the amino acid composition porfile of 797 AMPs from amphibians. It is clear that the amino acid use in natural AMPs is biased. In other words, some amino acids are more represented than others.
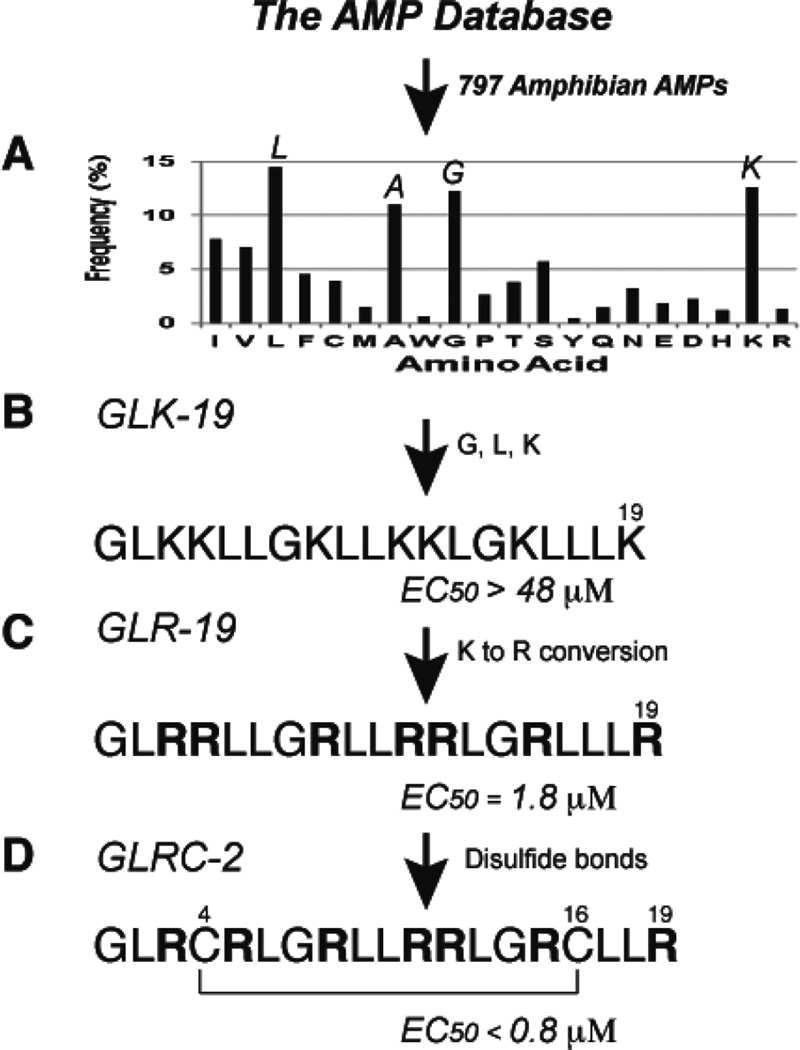
Fig. 4.
Database-guided de novo design of anti-HIV-1 peptides [138]. (A) The averaged amino acid composition profile of 797 amphibian AMPs calculated in the APD [23, 24]. The frequencies of L, A, G, and K are greater than 10%, thereby constituting a set of frequently occurring amino acids [24]. (B) Amino acids L, G, and K are utilized to design novel AMPs. One of them, GLK-19, is antibacterial [24] but not active against HIV-1 at 100 µg/ml. (C) GLR-19, an arginine version of GLK-19, is HIV-1 inhibitory [49]. (D) GLRC-2 is one of the best 19-residue GLRC peptides that showed anti-HIV-1 activity [37].
We define amino acids with >10% of the total amino acid composition as frequently occurring residues. Thus, A, G, L, and K are such residues in amphibian AMPs (Fig. 4A). Using G, L, and K, we designed a novel 19-residue peptide called GLK-19 (Fig. 4B) [24]. This peptide displays antibacterial activity against E. coli with a minimal inhibitory concentration (MIC) at 10 µM, which is similar to that of human LL-37 [79]. However, GLK-19 failed to inhibit HIV-1 with EC50 >48 µM (Fig. 4B).
Our database also enables us to investigate peptide sequence and activity relationships. We found that antiviral AMPs are rich in arginines [23]. To make use of this observation, we converted all the lysines in GLK-19 to arginines. Remarkably, GLR-19 became HIV-1 inhibitory with EC50 = 1.8 µM (Fig. 4C), underscoring the importance of arginines in blocking viral infection [24]. However, one should not generalize this observation since the peptide sequence also plays a role.
Because many antiviral peptides are rich in cysteines, we also introduced a disulfide bond at various positions of GLR-19 to generate a series of 19-residue polypeptides with varying loop sizes [37]. Since this series of peptides is composed of only amino acids G, L, R, and C, they are named as GLRC peptides. The peptide (GLRC-2) with a disulfide bond between Cys4 and Cys16 is found to be most potent with EC50 = 0.8 µM (Fig. 4D). The same peptide also shows inhibitory effects on HSV-2 (Herpes simplex virus type-2) as well as E. coli [37].
In summary, our major findings are as follows. First, it is possible to locate the most potent anti-HIV region in long peptides such as human LL-37 [83]. Second, our database screening suggests that natural AMPs are promising candidates for identifying novel anti-HIV peptides [49]. Third, the APD can be utilized to design novel anti-HIV peptides from the beginning [24, 37]. Finally, what we learned from bioinformatic analysis of natural AMPs can be used to enhance anti-HIV activity of peptides, irrespective of its polypeptide fold [37, 49]. All these examples indicate that the APD is a useful tool for identifying and engineering novel anti-HIV peptides. Importantly, our database approaches are not limited to the identification of HIV inhibitory peptides. For example, we have recently succeeded in screening our database or conducting ab initio design of novel peptides that are potent against community-associated methicillin-resistant Staphylococcus aureus (MRSA) USA300 [141, 142].
7. CONCLUDING REMARKS AND OUTLOOK
Nowadays more attention is paid to the development of peptide therapy [117]. The major advantages of peptides are small size, easy to optimize, and specific interactions. The specificity of peptides is one of the important advantages compared to small molecules that might not be sufficient to inhibit protein-protein interactions. The disadvantages of peptide therapies are possible toxicity, protease degradation and cost of production. Natural AMPs share the advantages and disadvantage of peptide therapies in general. Then why bother with AMPs? A plausible reason could be that the complex nature of many diseases, especially in the case of sexually transmitted diseases (STIs), requires comprehensive therapeutic strategies. Natural AMPs may hold the key to such complex problems. In addition to the diverse mechanisms of AMPs discussed above, the multiple beneficial effects of natural AMPs can be a plus. For example, simultaneous inhibition of both HIV-1 and HSV-2 is helpful since co-infection of these two viruses does occur [118]. AMPs are also known to have antibacterial activity. Thus, this beneficial effect may interfere with HIV-1 infection, although how bacteria may help viral infection is not yet well understood [119–121]. Also, patients, infected with HIV-1 and HSV-2, are more likely to infect Neisseria gonorrhoeae and Chlamydia trachomatis, two common bacterial causative agents of STIs [122]. The antibacterial effects of such anti-HIV-1 peptides could be essential, especially when such bacteria had developed resistance to traditional antibiotics. Needless to say, any peptide to be developed for this purpose must be selective enough so that it does not simultaneously destroy commensal bacteria (e.g. normal vaginal flora Lactobacillus). It is also known that HIV-1 patients are more susceptible to virus-caused cancer. Interestingly, some AMPs also kill cancer cells. Thus, the potential anti-cancer effect of anti-HIV-1 AMPs could reduce the likelihood of virus-caused cancer. In this case, AMPs could selectively act on cancer cells due to the exposure of phosphatidylserines (PS) while such anionic lipids are located in the inner membrane leaflet of normal human cells [123, 124]. As a third example, spermicidal effects of AMPs are beneficial to avoid unwanted pregnancies [125, 126]. It is these beneficial effects that make the development of AMP-based anti-HIV-1 peptides appealing.
AMPs exist widely in nature from bacteria, plants, invertebrates and vertebrates [14–17]. They possess diverse sequences, structural scaffolds, and functions (Fig. 2) [127–129]. Post-translational modification (PTM) plays an indispensible role in shaping this diversity (for a review, see ref [129]). At present, over 2000 natural AMPs have been manually registered into the APD through extensive literature search [23, 24], but only a small set of these peptides has been evaluated for anti-HIV activity [83]. This is probably due to the lack of efficient screening methods. Sometimes, it is not easy to obtain sufficient materials. In other cases, it remains to be a challenging task to synthesize compounds with a complex scaffold, especially those with extensive chemical modifications [129]. However, this situation will change with the development of high-throughput screening and peptide production methods that enable the evaluation of more peptides from natural sources collected in the APD or artificial combinatory libraries. Future evaluations should also consider the influence of cell and HIV types on anti-HIV activity of new compounds.
In order to obtain potent AMP candidates as leads for the development of topical microbicides, we have explored a variety of database strategies [23, 24]. These include anti-HIV-1 region mapping, peptide screening, sequence shuffling, de novo peptide design and improvement [37, 49, 83]. Through these studies, we learned that peptide length, sequence order, charge, amino acid composition, structure, and the folding of the peptide chain all play a role in determining the anti-HIV-1 activity of natural AMPs (Fig. 4). Such knowledge could be applied to next rounds of peptide engineering. The importance of peptide engineering has also been demonstrated by other laboratories. As a classic example, both therapeutic index and stability of polyphemusin II have been improved substantially [68]. More recently, extensive studies have been conducted for θ-defensins [103]. The circular β-hairpin structure of these peptides (Fig. 2F) is unique and makes them more potent and stable to proteases. In particular, these mini-defensins are also active against R5 HIV-1. One of the major problems is cost effective production. In the case of defensins, chemical synthesis may not be the final choice for production due to multiple disulfide bonds [17]. It is pleasing to see that progress has been made in recombinant production as well as formulation of these fascinating compounds [130–132]. These new advances bring such compounds closer to potential clinical applications. No doubt, the success with any of AMP-based topical microbicides will inspire the development of additional candidates that effectively inhibit various HIV-1 clades.
ACKNOWLEDGEMENTS
This study is supported by the grant R21AI082689 from the NIAID (National Institutes of Health, USA) to GW.
REFERENCES
- 1.Marx JL. Science. 1985;227:1449. doi: 10.1126/science.2983427. [DOI] [PubMed] [Google Scholar]
- 2.Gao F, Bailes E, Robertson DL, Chen Y, Rodenburg CM, Michael SF, Cummins LB, Arthur LO, Peeters M, Shaw GM, Sharp PM, Hahn BH. Nature. 1999;397:436. doi: 10.1038/17130. [DOI] [PubMed] [Google Scholar]
- 3.Rudnicka D, Schwartz O. Nat. Immunol. 2009;10:933. doi: 10.1038/ni0909-933. [DOI] [PubMed] [Google Scholar]
- 4.Liu R, Paxton WA, Choe S, Ceradini D, Martin SR, Horuk R, MacDonald ME, Stuhlmann H, Koup RA, Landau NR. Cell. 1996;86:367. doi: 10.1016/s0092-8674(00)80110-5. [DOI] [PubMed] [Google Scholar]
- 5.Greene WC, Peterlin BM. Nat. Med. 2002;8:673. doi: 10.1038/nm0702-673. [DOI] [PubMed] [Google Scholar]
- 6.Levy JA. Adv. Dent. Res. 2011;23:13. doi: 10.1177/0022034511398874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Friedrich BM, Dziuba N, Li G, Endsley MA, Murray JL, Ferguson MR. Virus Res. 2011;161:101. doi: 10.1016/j.virusres.2011.08.001. [DOI] [PubMed] [Google Scholar]
- 8.Berger EA, Doms RW, Fenyo EM, Korber BTM, Littman DR, Moore JP, Sattentau QJ, Schuitemaker H, Sodroski J, Weiss RA. Nature. 1998;391:240. doi: 10.1038/34571. [DOI] [PubMed] [Google Scholar]
- 9.Buckheit RW, Jr, Watson KM, Morrow KM, Ham AS. Antiviral Res. 2010;85:142. doi: 10.1016/j.antiviral.2009.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Turpin JA. Expert Opin. Investig. Drugs. 2002;11:1077. doi: 10.1517/13543784.11.8.1077. [DOI] [PubMed] [Google Scholar]
- 11.Koehn FE, Carter GT. Discov. Med. 2005;5:159. [PubMed] [Google Scholar]
- 12.Nisbet LJ, Moore M. Curr. Opin. Biotechnol. 1997;8:708. doi: 10.1016/s0958-1669(97)80124-3. [DOI] [PubMed] [Google Scholar]
- 13.Yang SS, Cragg GM, Newman DJ, Bader JP. J. Nat. Prod. 2001;64:265. doi: 10.1021/np0003995. [DOI] [PubMed] [Google Scholar]
- 14.Zasloff M. Nature. 2002;415:359. doi: 10.1038/415389a. [DOI] [PubMed] [Google Scholar]
- 15.Lehrer RI. Curr. Opin. Hematol. 2007;14:16. doi: 10.1097/00062752-200701000-00005. [DOI] [PubMed] [Google Scholar]
- 16.Mayer ML, Easton DM, Hancock REW. In: Antimicrobial Peptides: Discovery, Design and Novel Therapeutic Strategies. Wang G, editor. England: CABI; 2010. p. 195. [Google Scholar]
- 17.Wang G, Li X, Zasloff M. In: Antimicrobial Peptides: Discovery, Design and Novel Therapeutic Strategies. Wang G, editor. England: CABI; 2010. p. 1. [Google Scholar]
- 18.Wang Z, Lai Y, Bernard JJ, Macleod DT, Cogen AL, Moss B, Di Nardo A. J. Immunol. 2012;188:345. doi: 10.4049/jimmunol.1101703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee PHA, Ohtake T, Zaiou M, Murakami M, Rudisill JA, Lin KH, Gallo RL. Proc. Natl. Acad. Sci. USA. 2005;102:3750. doi: 10.1073/pnas.0500268102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tossi A. and associates. http://www.bbcm.univ.trieste.it/~tossi/amsdb.html.
- 21.Seshadri Sundararajan V, Gabere MN, Pretorius A, Adam S, Christoffels A, Lehväslaiho M, Archer JA, Bajic VB. Nucleic Acids Res. 2012;40:D1108. doi: 10.1093/nar/gkr1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Whitmore L, Wallace BA. Nucleic Acids Res. 2004;32:D593. doi: 10.1093/nar/gkh077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang Z, Wang G. Nucleic Acids Res. 2004;32:D590. doi: 10.1093/nar/gkh025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang G, Li X, Wang Z. Nucleic Acids Res. 37:D933. doi: 10.1093/nar/gkn823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gueguen Y, Garnier J, Robert L, Lefranc MP, Mougenot I, de Lorgeril J, Janech M, Gross PS, Warr GW, Cuthbertson B, Barracco MA, Bulet P, Aumelas A, Yang Y, Bo D, Xiang J, Tassanakajon A, Piquemal D, Bachère E. Dev. Comp. Immunol. 2006;30:283. doi: 10.1016/j.dci.2005.04.003. [DOI] [PubMed] [Google Scholar]
- 26.Seebah S, Suresh A, Zhou S, Choong YH, Chua H, Chuon D, Beuerman R, Verma C. Nucleic Acids Res. 2007;35:D265. doi: 10.1093/nar/gkl866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hammami R, Zouhir A, Ben Hamida J, Fliss I. BMC Microbiol. 2007;7:89. doi: 10.1186/1471-2180-7-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang CK, Kaas Q, Chiche L, Craik DJ. Nucleic Acids Res. 2008;36:D206. doi: 10.1093/nar/gkm953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dubos RJ. J. Exp. Med. 1939;70:1. doi: 10.1084/jem.70.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ketchem RR, Hu W, Cross TA. Science. 1993;261:1457. doi: 10.1126/science.7690158. [DOI] [PubMed] [Google Scholar]
- 31.Bourinbaiar AS, Krasinski K, Borkowsky W. Life Sci. 1994;54:PL5. doi: 10.1016/0024-3205(94)00579-6. [DOI] [PubMed] [Google Scholar]
- 32.Greenhead P, Hayes P, Watts PS, Laing KG, Griffin GE, Shattock RJ. J. Virol. 2000;74:5577. doi: 10.1128/jvi.74.12.5577-5586.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nar H, Schmid A, Puder C, Potterat O. ChemMedChem. 2010;5:1689. doi: 10.1002/cmdc.201000264. [DOI] [PubMed] [Google Scholar]
- 34.Knappe TA, Linne U, Robbel L, Marahiel MA. Chem. Biol. 2009;16:1290. doi: 10.1016/j.chembiol.2009.11.009. [DOI] [PubMed] [Google Scholar]
- 35.Ireland DC, Wang CK, Wilson JA, Gustafson KR, Craik DJ. Biopolymers. 2008;90:51. doi: 10.1002/bip.20886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gould A, Ji Y, Aboye TL, Camarero JA. Curr. Pharm. Des. 2011;17:4294. doi: 10.2174/138161211798999438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang G, Buckheit KW, Mishra B, Lushnikova T, Buckheit RW., Jr J. AIDS Clinical Res. 2011 [Google Scholar]
- 38.Gran L. Acta Pharmacol. Toxicol. (Copenh.) 1973;33:400. doi: 10.1111/j.1600-0773.1973.tb01541.x. [DOI] [PubMed] [Google Scholar]
- 39.Tam JP, Lu YA, Yang JL, Chiu KW. Proc. Natl. Acad. Sci. USA. 1999;96:8913. doi: 10.1073/pnas.96.16.8913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shenkarev ZO, Nadezhdin KD, Lyukmanova EN, Sobol VA, Skjeldal L, Arseniev AS. J. Inorg. Biochem. 2008;102:1246. doi: 10.1016/j.jinorgbio.2008.01.018. [DOI] [PubMed] [Google Scholar]
- 41.Cui T, Gao Y, Liew OW, Puah CM, Gutte B. J. Biotechnol. 2007;130:378. doi: 10.1016/j.jbiotec.2007.05.006. [DOI] [PubMed] [Google Scholar]
- 42.Huang YH, Colgrave ML, Daly NL, Keleshian A, Martinac B, Craik DJ. J. Biol. Chem. 2009;284:20699. doi: 10.1074/jbc.M109.003384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sando L, Henriques ST, Foley F, Simonsen SM, Daly NL, Hall KN, Gustafson KR, Aguilar MI, Craik DJ. Chembiochem. 2011;12:2456. doi: 10.1002/cbic.201100450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Steiner H, Hultmark D, Engström Å, Bennich H, Boman HG. Nature. 1981;292:246. doi: 10.1038/292246a0. [DOI] [PubMed] [Google Scholar]
- 45.Pan X, Zhou G, Wu J, Bian G, Lu P, Raikhel AS, Xi Z. Proc. Natl. Acad. Sci. USA. 2012;109:E23. doi: 10.1073/pnas.1116932108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Medzhitov R, Preston-Hurlburt P, Janeway CA., Jr Nature. 1997;388:394. doi: 10.1038/41131. [DOI] [PubMed] [Google Scholar]
- 47.Liu PT, Stenger S, Li H, Wenzel L, Tan BH, Krutzik SR, Ochoa MT, Schauber J, Wu K, Meinken C, Kamen DL, Wagner M, Bals R, Steinmeyer A, Zügel U, Gallo RL, Eisenberg D, Hewison M, Hollis BW, Adams JS, Bloom BR, Modlin RL. Science. 2006;311:1770. doi: 10.1126/science.1123933. [DOI] [PubMed] [Google Scholar]
- 48.Wachinger M, Klenschmidt A, Winder D, von Pechmann N, Ludvigsen A, Neumann M, Holle R, Salmons B, Erfle V, Brack-Werner R. J. Gen. Virol. 1998;79:731. doi: 10.1099/0022-1317-79-4-731. [DOI] [PubMed] [Google Scholar]
- 49.Wang G, Watson KM, Peterkofsky A, Buckheit RW., Jr Antimicrob. Agents Chemother. 2010;54:1343. doi: 10.1128/AAC.01448-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Orivel J, Redeker V, Le Caer JP, Krier F, Revol-Junelles AM, Longeon A, Chaffotte A, Dejean A, Rossier J. J. Biol. Chem. 2001;276:17823. doi: 10.1074/jbc.M100216200. [DOI] [PubMed] [Google Scholar]
- 51.Lamberty M, Zachary D, Lanot R, Bordereau C, Robert A, Hoffmann JA, Bulet P. J. Biol. Chem. 2001;276:4085. doi: 10.1074/jbc.M002998200. [DOI] [PubMed] [Google Scholar]
- 52.Cerovský V, Hovorka O, Cvacka J, Voburka Z, Bednárová L, Borovicková L, Slaninová J, Fucík V. Chembiochem. 2008;9:2815. doi: 10.1002/cbic.200800476. [DOI] [PubMed] [Google Scholar]
- 53.Zasloff M. Proc. Natl. Acad. Sci. USA. 1987;84:5449. doi: 10.1073/pnas.84.15.5449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chinchar VG, Bryan L, Silphadaung U, Noga E, Wade D, Rollins-Smith L. Virology. 2004;323:268. doi: 10.1016/j.virol.2004.02.029. [DOI] [PubMed] [Google Scholar]
- 55.Lai R, Zheng YT, Shen JH, Liu GJ, Liu H, Lee WH, Tang SZ, Zhang Y. Peptides. 2002;23:427. doi: 10.1016/s0196-9781(01)00641-6. [DOI] [PubMed] [Google Scholar]
- 56.Lorin C, Saidi H, Belaid A, Zairi A, Baleux F, Hocini H, Bélec L, Hani K, Tangy F. Virology. 2005;334:264. doi: 10.1016/j.virol.2005.02.002. [DOI] [PubMed] [Google Scholar]
- 57.VanCompernolle SE, Taylor RJ, Oswald-Richter K, Jiang J, Youree BE, Bowie JH, Tyler MJ, Conlon JM, Wade D, Aiken C, Dermody TS, KewalRamani VN, Rollins-Smith LA, Unutmaz D. J. Virol. 2005;79:11598. doi: 10.1128/JVI.79.18.11598-11606.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ma Y, Liu C, Liu X, Wu J, Yang H, Wang Y, Li J, Yu H, Lai R. Genomics. 2010;95:66. doi: 10.1016/j.ygeno.2009.09.004. [DOI] [PubMed] [Google Scholar]
- 59.Yang X, Lee WH, Zhang Y. J. Proteome Res. 2012;11:306. doi: 10.1021/pr200782u. [DOI] [PubMed] [Google Scholar]
- 60.Azevedo Calderon A, Silva Ade A, Ciancaglini P, Stábeli RG. Amino Acids. 2011;40:29. doi: 10.1007/s00726-010-0622-3. [DOI] [PubMed] [Google Scholar]
- 61.Conlon JM. Cell. Mol. Life Sci. 2011;68:2303. doi: 10.1007/s00018-011-0720-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jackway RJ, Pukala TL, Donnellan SC, Sherman PJ, Tyler MJ, Bowie JH. Peptides. 2011;32:161. doi: 10.1016/j.peptides.2010.09.019. [DOI] [PubMed] [Google Scholar]
- 63.Nicolas P, El Amri C. Biochim. Biophys. Acta. 2009;1788:1537. doi: 10.1016/j.bbamem.2008.09.006. [DOI] [PubMed] [Google Scholar]
- 64.Simmaco M, Kreil G, Barra D. Biochim. Biophys. Acta. 1788:1551. doi: 10.1016/j.bbamem.2009.01.004. [DOI] [PubMed] [Google Scholar]
- 65.Tziveleka LA, Vagias C, Roussis V. Curr. Top. Med. Chem. 2003;3:1512. doi: 10.2174/1568026033451790. [DOI] [PubMed] [Google Scholar]
- 66.Nakashima H, Masuda M, Murakami T, Koyanagi Y, Matsumoto A, Fujii N, Yamamoto N. Antimicrob. Agents Chemother. 1992;36:1249. doi: 10.1128/aac.36.6.1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tamamura H, Otaka A, Murakami T, Ishihara T, Ibuka T, Waki M, Matsumoto A, Yamamoto N, Fujii N. Biochem. Biophys. Res. Commun. 1996;219:555. doi: 10.1006/bbrc.1996.0272. [DOI] [PubMed] [Google Scholar]
- 68.Murakami T, Zhang TY, Koyanagi Y, Tanaka Y, Kim J, Suzuki Y, Minoguchi S, Tamamura H, Waki M, Matsumoto A, Fujii N, Shida H, Hoxie JA, Peiper SC, Yamamoto N. J. Virol. 1999;73:7489. doi: 10.1128/jvi.73.9.7489-7496.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.De Smet K, Contreras R. Biotechnol Lett. 2005;27:1337. doi: 10.1007/s10529-005-0936-5. [DOI] [PubMed] [Google Scholar]
- 70.Zanetti M. Curr. Issues Mol. Biol. 2005;7:179. [PubMed] [Google Scholar]
- 71.Agerberth B, Gunne H, Odeberg J, Kogner P, Boman HG, Gudmundsson GH. Proc. Natl. Acad. Sci. USA. 1995;92:195. doi: 10.1073/pnas.92.1.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Cowland JB, Johnsen AH, Borregaard N. FEBS Lett. 1995;368:173. doi: 10.1016/0014-5793(95)00634-l. [DOI] [PubMed] [Google Scholar]
- 73.Larrick JW, Hirata M, Balint RF, Lee J, Zhong J, Wright SC. Infect. Immun. 1995;63:1291. doi: 10.1128/iai.63.4.1291-1297.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.White JH, Bitton AJ. In: Antimicrobial peptides: discovery, design and novel therapeutic strategies. Wang G, editor. Vol. 181 England: CABI; 2010. [Google Scholar]
- 75.Putsep K, Carlsson G, Boman HG, Andersson M. Lancet. 2002;360:1144. doi: 10.1016/S0140-6736(02)11201-3. [DOI] [PubMed] [Google Scholar]
- 76.Johansson J, Gudmundsson GH, Rottenberg ME, Berndt KD, Agerberth B. J. Biol. Chem. 1998;273:3718. doi: 10.1074/jbc.273.6.3718. [DOI] [PubMed] [Google Scholar]
- 77.Oren Z, Lerman JC, Gudmundsson GH, Agerberth B, Shai Y. Biochem. J. 1999;341:501. [PMC free article] [PubMed] [Google Scholar]
- 78.Wang G. J. Biol. Chem. 2008;283:32637. doi: 10.1074/jbc.M805533200. [DOI] [PubMed] [Google Scholar]
- 79.Wang G, Elliott M, Cogen AL, Ezell EL, Gallo RL, Hancock RE. Biochemistry. 2012;51:653. doi: 10.1021/bi2016266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Turner J, Cho Y, Dinh N-N, Waring AJ, Lehrer RI. Antimicrob. Agents Chemother. 1998;42:2206. doi: 10.1128/aac.42.9.2206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Steinstraesser L, Tippler B, Mertens J, Lamme E, Homann HH, Lehnhardt M, Wildner O, Steinau HU, Uberla K. Retrovirology. 2005;2:2. doi: 10.1186/1742-4690-2-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Bergman P, Walter-Jallow L, Broliden K, Agerberth B, Söderlund J. Curr. HIV Res. 2007;5:410. doi: 10.2174/157016207781023947. [DOI] [PubMed] [Google Scholar]
- 83.Wang G, Watson KM, Buckheit RW., Jr Antimicrob. Agents Chemother. 2008;52:3438. doi: 10.1128/AAC.00452-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Sørensen OE, Gram L, Johnsen AH, Andersson E, Bangsbøll S, Tjabringa GS, Hiemstra PS, Malm J, Egesten A, Borregaard N. J. Biol. Chem. 2003;278:28540. doi: 10.1074/jbc.M301608200. [DOI] [PubMed] [Google Scholar]
- 85.Yamasaki K, Schauber J, Coda A, Lin H, Dorschner RA, Schechter NM, Bonnart C, Descargues P, Hovnanian A, Gallo RL. FASEB J. 2006;20:2068. doi: 10.1096/fj.06-6075com. [DOI] [PubMed] [Google Scholar]
- 86.Selsted ME, Harwig SS, Ganz T, Schilling JW, Lehrer RI. J. Clin. Invest. 1985;76:1436. doi: 10.1172/JCI112121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Lehrer RI, Lu W. Immun. Rev. 2012;245:84. doi: 10.1111/j.1600-065X.2011.01082.x. [DOI] [PubMed] [Google Scholar]
- 88.Wang W, Owen SM, Rudolph DL, Cole AM, Hong T, Waring AJ, Lal RB, Lehrer RI. J. Immunol. 2004;173:515. doi: 10.4049/jimmunol.173.1.515. [DOI] [PubMed] [Google Scholar]
- 89.Furci L, Sironi F, Tolazzi M, Vassena L, Lusso P. Blood. 2007;109:2928. doi: 10.1182/blood-2006-05-024489. [DOI] [PubMed] [Google Scholar]
- 90.Wei G, de Leeuw E, Pazgier M, Yuan W, Zou G, Wang J, Ericksen B, Lu WY, Lehrer RI, Lu W. J. Biol. Chem. 2009;284:29180. doi: 10.1074/jbc.M109.018085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Rodríguez-García M, Climent N, Oliva H, Casanova V, Franco R, Leon A, Gatell JM, García F, Gallart T. PLoS One. 2010;5:e9436. doi: 10.1371/journal.pone.0009436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Wu Z, Cocchi F, Gentles D, Ericksen B, Lubkowski J, Devico A, Lehrer RI, Lu W. FEBS Lett. 2005;579:162. doi: 10.1016/j.febslet.2004.11.062. [DOI] [PubMed] [Google Scholar]
- 93.Rapista A, Ding J, Benito B, Lo YT, Neiditch MB, Lu W, Chang TL. Retroviology. 2011;8:45. doi: 10.1186/1742-4690-8-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Ding J, Rapista A, Teleshova N, Lu W, Klotman ME, Chang TL. J. Innate Immun. 2011;3:208. doi: 10.1159/000322355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Bensch KW, Raida M, Mägert HJ, Schulz-Knappe P, Forssmann WG. FEBS Lett. 1995;368:331. doi: 10.1016/0014-5793(95)00687-5. [DOI] [PubMed] [Google Scholar]
- 96.Scheetz T, Bartlett JA, Walters JD, Schutte BC, Casavant TL, McCray PB., Jr Immunol. Rev. 2002;190:137. doi: 10.1034/j.1600-065x.2002.19010.x. [DOI] [PubMed] [Google Scholar]
- 97.Braida L, Boniotto M, Pontillo A, Tovo PA, Amoroso A, Crovella S. AIDS. 2004;18:1598. doi: 10.1097/01.aids.0000131363.82951.fb. [DOI] [PubMed] [Google Scholar]
- 98.Alp S, Skrygan M, Schlottmann R, Kreuter A, Otte JM, Schmidt WE, Brockmeyer NH, Bastian A. Eur. J. Med. Res. 2005;10:1. [PubMed] [Google Scholar]
- 99.Seidel A, Ye Y, de Armas LR, Soto M, Yarosh W, Marcsisin RA, Tran D, Selsted ME, Camerini D. PLoS One. 2010;5:e9737. doi: 10.1371/journal.pone.0009737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Quiñones-Mateu ME, Lederman MM, Feng Z, Chakraborty B, Weber J, Rangel HR, Marotta ML, Mirza M, Jiang B, Kiser P, Medvik K, Sieg SF, Weinberg A. AIDS. 2003;17:F39. doi: 10.1097/00002030-200311070-00001. [DOI] [PubMed] [Google Scholar]
- 101.Sun L, Finnegan CM, Kish-Catalone T, Blumenthal R, Garzino-Demo P, La Terra Maggiore GM, Berrone S, Kleinman C, Wu Z, Abdelwahab S, Lu W, Garzino-Demo A. J. Virol. 2005;79:14318. doi: 10.1128/JVI.79.22.14318-14329.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Feng Z, Dubyak GR, Lederman MM, Weinberg A. J. Immunol. 2006;177:782. doi: 10.4049/jimmunol.177.2.782. [DOI] [PubMed] [Google Scholar]
- 103.Tang YQ, Yuan J, Osapay G, Osapay K, Tran D, Miller CJ, Ouellette AJ, Selsted ME. Science. 1999;286:498. doi: 10.1126/science.286.5439.498. [DOI] [PubMed] [Google Scholar]
- 104.Abuja PM, Zenz A, Trabi M, Craik DJ, Lohner K. FEBS Lett. 2004;566:301. doi: 10.1016/j.febslet.2004.03.112. [DOI] [PubMed] [Google Scholar]
- 105.Cole AM, Hong T, Boo LM, Nguyen T, Zhao C, Bristol G, Zack JA, Waring AJ, Yang OO, Lehrer RI. Proc. Natl. Acad. Sci. USA. 2002;99:1813. doi: 10.1073/pnas.052706399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Owen SM, Rudolph D, Wang W, Cole AM, Sherman MA, Waring AJ, Lehrer RI, Lal RB. J. Peptide Res. 2004;63:469. doi: 10.1111/j.1399-3011.2004.00155.x. [DOI] [PubMed] [Google Scholar]
- 107.Owen SM, Rudolph D, Wang W, Cole AM, Waring AJ, Lal RB, Lehrer RI. AIDS Res. Hum. Retroviruses. 2004;20:1157. doi: 10.1089/aid.2004.20.1157. [DOI] [PubMed] [Google Scholar]
- 108.Venkataraman N, Cole AL, Ruchala P, Waring AJ, Lehrer RI, Stuchlik O, Pohl J, Cole AM. PLoS Biol. 2009;7:e95. doi: 10.1371/journal.pbio.1000095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Epand RM, Epand RF. J. Pept. Sci. 2011;17:298. doi: 10.1002/psc.1319. [DOI] [PubMed] [Google Scholar]
- 110.Clejan S, Guffanti AA, Cohen MA, Krulwich TA. J. Bacteriol. 1989;171:1744. doi: 10.1128/jb.171.3.1744-1746.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Zhao M. Amino Acids. 2011;41:1071. doi: 10.1007/s00726-009-0386-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Cole AM. Protein Pept. Lett. 2005;12:41. doi: 10.2174/0929866053406101. [DOI] [PubMed] [Google Scholar]
- 113.Chang TL, Francois F, Mosoian A, Klotman ME. J. Virol. 2003;77:6777. doi: 10.1128/JVI.77.12.6777-6784.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Li X, Li Y, Han H, Miller DW, Wang G. J. Am. Chem. Soc. 2006;128:5776. doi: 10.1021/ja0584875. [DOI] [PubMed] [Google Scholar]
- 115.Wong JH, Legowska A, Rolka K, Ng TB, Hui M, Cho CH. Peptides. 2011;32:1117. doi: 10.1016/j.peptides.2011.08.018. [DOI] [PubMed] [Google Scholar]
- 116.Li X, Li Y, Peterkofsky A, Wang G. Biochim. Biophys. Acta. 2006;1758:1203. doi: 10.1016/j.bbamem.2006.03.032. [DOI] [PubMed] [Google Scholar]
- 117.Huther A, Dietrich U. AIDS Rev. 2007;9:208. [PubMed] [Google Scholar]
- 118.Barnabas RV, Celum C. Curr. HIV Res. 2012;10:228. doi: 10.2174/157016212800618156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Broxmeyer L, Cantwell A. Med. Hypotheses. 2008;71:741. doi: 10.1016/j.mehy.2008.06.012. [DOI] [PubMed] [Google Scholar]
- 120.Rieg G, Butler DM, Smith DM, Daar ES. Int. J. STD AIDS. 2010;21:207. doi: 10.1258/ijsa.2009.009331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Nagot N, Ouedraogo A, Defer MC, Vallo R, Mayaud P, Van de Perre P. Sex Transm. Infect. 2007;83:365. doi: 10.1136/sti.2007.024794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Venkatesh KK, van der Straten A, Mayer KH, Blanchard K, Ramjee G, Lurie MN, Chipato T, Padian NS, de Bruyn G. Sex Transm. Dis. 2011;38:562. doi: 10.1097/OLQ.0b013e31820a8c2c. [DOI] [PubMed] [Google Scholar]
- 123.Wu WK, Wang G, Coffelt SB, Betancourt AM, Lee CW, Fan D, Wu K, Yu J, Sung JJ, Cho CH. Int. J. Cancer. 2010;127:1741. doi: 10.1002/ijc.25489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Schröder-Borm H, Bakalova R, Andrä J. FEBS Lett. 2005;579:6128. doi: 10.1016/j.febslet.2005.09.084. [DOI] [PubMed] [Google Scholar]
- 125.Silkin L, Hamza S, Kaufman S, Cobb SL, Vederas JC. Bioorg. Med. Chem. Lett. 2008;18:3103. doi: 10.1016/j.bmcl.2007.11.024. [DOI] [PubMed] [Google Scholar]
- 126.Reddy KV, Aranha C, Gupta SM, Yedery RD. Reproduction. 2004;128:117. doi: 10.1530/rep.1.00028. [DOI] [PubMed] [Google Scholar]
- 127.Epand RM, Vogel HJ. Biochim. Biophys. Acta. 1999;1462:11. doi: 10.1016/s0005-2736(99)00198-4. [DOI] [PubMed] [Google Scholar]
- 128.Tossi A, Sandri L. Curr. Pharm. Des. 2002;8:743. doi: 10.2174/1381612023395475. [DOI] [PubMed] [Google Scholar]
- 129.Wang G. Curr. Biotechnol. 2012;1:72. doi: 10.2174/2211550111201010072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Sassi AB, Cost MR, Cole AL, Cole AM, Patton DL, Gupta P, Rohan LC. Antimicrob. Agents Chemother. 2011;55:2282. doi: 10.1128/AAC.01190-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Lee S-B, Li B, Jin S, Daniell H. Plant Biotechnol. J. 2011;9:100. doi: 10.1111/j.1467-7652.2010.00538.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Gould A, Li Y, Majumder S, Garcia AE, Carlsson P, Shekhtman A, Camarero JA. Mol. Biosyst. 2012;8:1359. doi: 10.1039/c2mb05451e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Hoover DM, Chertov O, Lubkowski J. J. Biol. Chem. 2001;276:39021. doi: 10.1074/jbc.M103830200. [DOI] [PubMed] [Google Scholar]
- 134.Trabi M, Schirra HJ, Craik DJ. Biochemistry. 2001;40:4211. doi: 10.1021/bi002028t. [DOI] [PubMed] [Google Scholar]
- 135.Knappe TA, Manzenrieder F, Mas-Moruno C, Linne U, Sasse F, Kessler H, Xie X, Marahiel MA. Angew. Chem. Int. Ed. Engl. 2011;50:8714. doi: 10.1002/anie.201102190. [DOI] [PubMed] [Google Scholar]
- 136.Wong CT, Rowlands DK, Wong CH, Lo TW, Nguyen GK, Li HY, Tam JP. Angew. Chem. Int. Ed. Engl. 2012;51:5620. doi: 10.1002/anie.201200984. [DOI] [PubMed] [Google Scholar]
- 137.Wang G. In: Antimicrobial Peptides: Discovery, Design and Novel Therapeutic Strategies. Wang G, editor. England: CABI; 2010. p. 141. [Google Scholar]
- 138.Wang G. In: Antimicrobial Peptides: Discovery, Design and Novel Therapeutic Strategies. Wang G, editor. England: CABI; 2010. p. 72. [Google Scholar]
- 139.Demirkhanyan LH, Marin M, Padilla-Parra S, Zhan C, Miyauchi K, Jean-Baptiste M, Novitskiy G, Lu W, Melikyan GB. J. Biol. Chem. 2012;287:28821. doi: 10.1074/jbc.M112.375949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Mishra B, Wang G. Frontier Immunol. 2012;3:221. doi: 10.3389/fimmu.2012.00221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Menousek J, Mishra B, Hanke ML, Heim CE, Kielian T, Wang G. Int. J. Antimicrob. Agents. 2012;39:402. doi: 10.1016/j.ijantimicag.2012.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Mishra B, Wang G. J. Am. Chem. Soc. 2012;134:12426. doi: 10.1021/ja305644e. [DOI] [PMC free article] [PubMed] [Google Scholar]