Abstract
The Notch signaling pathway plays a crucial role in skeletal development and homeostasis by regulating the proliferation and differentiation of osteoblasts and osteoclasts. However, the molecular mechanisms modulating the level and activity of Notch receptors in bone cells remain unknown. In this study, we uncovered that LNX2, an E3 ubiquitin ligase and Notch inhibitor Numb binding protein, was up-regulated during osteoclast differentiation. Knocking-down LNX2 expression in bone marrow macrophages by lentivirus-mediated short hairpin RNAs markedly inhibited osteoclast formation. Decreased LNX2 expression attenuated M-CSF-induced ERK and AKT activation and RANKL-stimulated activation of NF-kB and JNK pathways; therefore, accelerated osteoclast apoptosis. Additionally, loss of LNX2 led to an increased accumulation of Numb, which promoted the degradation of Notch and caused a reduction of the expression of the Notch downstream target gene, Hes1. We conclude that LNX2 regulates M-CSF/RANKL and the Notch signaling pathways during osteoclastogenesis.
Keywords: LNX2, osteoclast, bone resorption, RANKL, M-CSF, Notch, Numb
Introduction
Skeletal homeostasis in adults is maintained by a constant process called bone remodeling, in which old or damaged bone is removed by hematopoietic-derived osteoclasts followed by the deposit of new bone from mesenchymal-derived osteoblasts. Under physiological conditions, the amount of bone resorbed by osteoclasts is precisely replaced by new bone formation deposited by osteoblasts. Imbalance in these two processes leads to either loss of bone mass, in the case of osteoporosis, rheumatoid arthritis, and Paget’s disease of the bone, or to accumulation of disorganized bone mass, as in osteopetrosis and osteosclerosis [1, 2]. The generation, lifespan, and activity of osteoclasts and osteoblasts are regulated by systemic hormones as well as autocrine and paracrine cytokines and growth factors that are released from the bone matrix, secreted by neighboring cells in the bone microenvironment, or are anchored in the membrane of these cells [3]. These regulating factors are not only involved in the communications between osteoclasts and osteoblasts, but also play an important role in mediating cross talks between bone cells with vascular endothelial cells, neuronal cells, bone marrow stromal cells, adipocytes, and immune cells [4, 5].
Osteoclasts are derived from mononuclear precursors of the monocyte/macrophage lineage that eventually fuse to form multinucleated mature osteoclasts in a process controlled by two essential cytokines: the macrophage colony-stimulating factor (M-CSF) and the receptor activator of NF-κB ligand (RANKL) [6]. M-CSF stimulates the proliferation of macrophages and the survival of osteoclasts by activating ERK and the PI3K/AKT pathways. RANKL, on the other hand, induces the expression of osteoclast-specific genes and promotes the survival of osteoclasts by activating NF-κB, JNK, and intracellular calcium/NFATc1 pathways [7, 8]. Other soluble and membrane-bound cytokines and factors in bone marrow microenvironment exert their influence on osteoclast differentiation and function through modulating M-CSF, RANKL, and/or their downstream signaling pathways [9].
The Notch signaling pathway is evolutionarily conserved and plays a critical role in the determination of cell fate during embryonic development and postnatal tissue homeostasis. Notch signaling regulates proliferation, differentiation, and apoptosis in a cell-cell contact dependent manner [10]. Four Notch receptors (Notch1-4) and seven ligands (Delta-like 1, 3, 4; Jagged 1and 2; Delta/Notch-like epidermal growth factor-related receptor; and contactin/F3/NB-3) have been identified in mammals so far. Both Notch receptors and ligands are single-span transmembrane proteins localized at the cell surface. Interaction of Notch receptor-ligand triggers two sequential proteolytic cleavages: first by a metalloproteinase ADAM (a disintegrin and metalloproteinase) and the second by a γ-secretase complex. As a result, the Notch intracellular domain (NICD) is released from the plasma membrane and translocates to the nucleus where it binds to the transcription factor CSL (CBF1/RBPJκ, Su, and LAG1) and induces the expression of Notch target genes, including members of Hes (hairy and enhancer of split) and Hey (Hes related with YRPW motif) families [11].
Recent genetic studies in human and mice have reinforced an essential role of Notch signaling in bone development and homeostasis [12, 13]. Specifically, in vivo studies using mouse models with gain- and loss-of-function of Notch signaling in osteoblast lineage cells have revealed that Notch stimulates proliferation in osteoblast precursors; however, it inhibits their final differentiation to mature osteoblasts [14–16]. Nonetheless, in vitro studies by Nobta et al and Tezuka et al have suggested that Notch signaling promotes osteoblast differentiation and bone nodule formation. [17, 18]. The discrepancies between in vivo and in vitro work on the role of Notch signaling in osteoblasts may reflect the fact that the expression and interactions of Notch ligands and Notch receptors in bone marrow microenvironment are different from those in in vitro culture systems. In addition, deletion/activation of Notch signaling may exert different effects on osteoblast differentiation pathways when it occurs at early or late stages of osteoblast formation. These issues need further and future investigation. Deletion of Notch receptors in osteoclast precursor cells enhances osteoclast formation and bone resorption [19], suggesting an inhibitory role of Notch in osteoclastogenesis. On the other hand, activation of Notch signaling, especially Notch2, promotes osteoclast differentiation [20, 21]. These conflicting findings warrant that further characterization of the spatio-temporal and cell-context regulation of Notch signaling in bone cells is needed.
Numb, a membrane-associated adaptor protein critical for cell-fate determination, inhibits Notch signaling through the regulation of Notch endocytosis/recycling and the ubiquitin/proteasome degradation of NICD [22–25]. Numb binds to the Notch receptor and components of the Clathrin-dependent endocytic complexes [26–28]. Numb also helps to recruit several E3 ubiquitin ligases to the membrane-tethered Notch1 receptor and promote Notch1 degradation [29]. LNX (ligand of numb protein X) 1 and 2 are Ring finger and PDZ domain containing E3 ubiquitin ligases, sharing extensive homology with each other. LNX1 and LNX2 bind to Numb and foster its ubiquitylation, leading to proteasome-dependent degradation of Numb [30–32]. LNX proteins may enhance Notch signaling by lowering the level of Numb [33]. The expression of LNX and Numb and whether they regulate the Notch pathway in osteoclasts have not been examined and reported.
In this study, we provided evidence that LNX2 expression is up-regulated during osteoclast differentiation. We also showed that LNX2 promotes osteoclastogenesis at least in part, by modulating M-CSF and RANKL signaling pathways and by the Notch signaling through Numb.
MATERIALS AND METHODS
Antibodies and reagents
Antibodies were purchased from the following resources: mouse monoclonal anti-Cathepsin K (clone 182-12G5) (Millipore); rat anti-Notch2 (clone C651.6DbHN) (Developmental Studies Hybridoma Bank); rabbit polyclonal anti-ERK1/2, mouse monoclonal anti-phospho-ERK1/2 (Thr202/Tyr204), mouse monoclonal anti-Akt (pan) (clone 40D4), rabbit monoclonal anti-phosphor-Akt (Ser473) (clone 193H12), rabbit polyclonal anti-JNK, mouse monoclonal anti-phospho-JNK (Thr183/Tyr185) (clone G9), rabbit polyclonal anti-IKB-α, mouse monoclonal anti-phospho-IKB-α (Ser32/36) (clone 5A5), rabbit monoclonal anti-cleaved caspase-3 (catalog number 9664), rabbit polyclonal anti-PARP (catalog number 9542), rabbit monoclonal anti-Numb (C29G11) (catalog number 2756) (Cell Signaling Technology); mouse monoclonal anti-α-tubulin (clone DM1A) (Sigma-Aldrich); mouse monoclonal anti-beta actin (catalog number A00702) (GenScript).
Cell culture alpha-MEM and 10 × Penicillin-Streptomycin-L-Glutamine were purchased from Life Technologies and Sigma-Aldrich, respectively. Fetal bovine serum was purchased from Hyclone.
Bone marrow macrophage and osteoclast cultures
Bone marrow macrophages (BMMs) were prepared as described previously [34]. Briefly, whole bone marrow was isolated from tibiae and femora of one or two 8–10 weeks C57/BL6J mice. Bone marrow cells were plated in α-10 medium (α-MEM, 10% heat-inactivated fetal bovine serum, 1 × Penicillin-Streptomycin-L-Glutamine solution) containing 1/10 volume of CMG 14–12 (conditioned medium supernatant containing recombinant M-CSF at 1µg/ml) [35] in petri-dishes. Cells were incubated at 37°C in 5% CO2, 95% air for 4–5 days. Fresh media and CMG 14–12 supernatant were replaced every the other day. Osteoclasts were generated after five days culture of BMMs with 1/100 volume of CMG 14–12 supernatant and 100 ng/ml of recombinant RANKL, generated as described before [36]. All animal procedures were approved by Institutional Animal Care and Use Committee at University of Arkansas for Medical Sciences.
Quantitative real time RT-PCR
BMMs were cultured in 6-well plates with M-CSF and/or RANKL for 5 days. Total RNA was purified using RNeasy mini kit (Qiagen) according to the manufacture’s protocol. First-strand cDNAs were synthesized from 0.5–1 µg of total RNA using the High Capacity cDNA Reverse Transcription kits (Life Technologies) following the manufacturer’s instructions. TaqMan quantitative real-time PCR was performed using the following Primers from Life Technologies: Lnx1 (Mm00495381_m1); Lnx2 (Mm00612746_m1); Acp5 (Mm00475698_m1); Ctsk (Mm00484039_m1); Hes1 (Mm01342805_m1). Samples were amplified using the StepOnePlus real-time PCR system (Life Technologies) with an initial denaturation at 95 °C for 10 min, followed by 40 cycles of 95 °C for 15 s and 60 °C for 1 min. The relative cDNA amount was calculated by normalizing to that of the mitochondrial gene Mrps2, which is steadily expressed in both BMMs and osteoclasts, using the ΔCt method [37].
Lentivirus mediated shRNA expression
The LKO.1 lentiviral vectors expressing shRNA sequence targeting mRNA of murine LNX2, TRCN0000040713/NM_080795.3-3299s1c1 and TRCN0000040715/NM_080795.3-933s1c1, were purchased from Sigma-Aldrich. 293-T cells were co-transfected with a LKO.1 gene transfer vector and virus packaging vectors, ΔH8.2 and VSVG by TransIT-LT1 transfection reagent (Mirus). Virus supernatants were collected after 48 hours transfection. BMMs were transduced with virus supernatant containing M-CSF and 20 µg/ml of Protamine (Sigma-Aldrich). Cells were then selected in α-10 medium containing M-CSF and 6 µg/ml puromycin (Sigma-Aldrich) for 3 days.
TRAP (tartrate-resistant acid phosphatase) staining
BMMs were cultured on 48-well tissue culture plate in α-10 medium with M-CSF and RANKL for 4–5 days. The cells were fixed with 4% paraformaldehyde/phosphate buffered saline (PBS) and TRAP was stained with NaK Tartrate and Napthol AS-BI phosphoric acid (Sigma-Aldrich) as described previously [38].
Cell proliferation and apoptosis Assays
BMMs were cultured with M-CSF for 3 days. BrdU was then added to the culture medium and incubated for 6 hours. BrdU incorporation rate was measured by the BrdU Cell Proliferation Assay Kit (Cell Signaling Technology) following the manufacturer's instructions. For the detection of osteoclast apoptosis, BMMs were cultured with M-CSF and RANKL for 3 days. Pre-osteoclasts were either untreated or serum and cytokine starved for 3 hours. Cell death was analyzed using cell death detection ELISA PLUS kit (Roche), which detects cytoplasmic histone-associated DNA fragmentation.
Immunoblotting
Cultured cells were washed with ice-cold PBS twice and lysed in 1 × RIPA buffer (Sigma) containing 1 mM DTT and Complete Mini EDTA-free protease inhibitor cocktail (Roche). After incubation on ice for 30 min, the cell lysates were clarified by centrifugation at 14,000 rpm for 15 min at 4 °C. Ten to thirty µg of total protein were subjected to 8% SDS-PAGE gels and transferred electrophoretically onto polyvinylidene difluoride membrane (EMD Millipore) by a semi-dry blotting system (Bio-Rad). The membrane was blocked in 5% fat-free milk/Tris-buffered saline for 1 hour and incubated with primary antibodies at 4°C overnight followed by secondary antibodies conjugated with horseradish peroxidase (Santa Cruz Biotechnology). After rinsing 3 times with Tris-buffered saline containing 0.1% Tween 20, the membrane was subjected to western blot analysis with enhanced chemiluminescent detection reagents (EMD Millipore). The densitometric measurements of each band in western blots of Figures 2 to 6 were performed by NIH ImageJ software. The data are presented in Supplementary Figure 1 as Ratios of densitometry intensity of specific bands to their respective ones in loading controls.
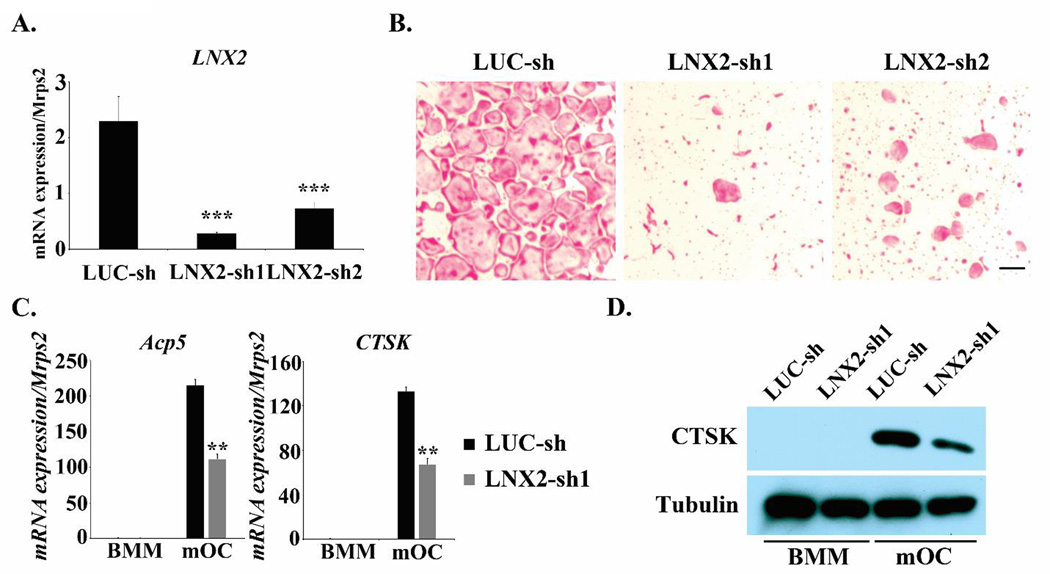
Fig. 2.
Knocking down of LNX2 expression markedly inhibits osteoclast formation. (A) BMMs were transduced with recombinant lenti-viruses expressing a control shRNA targeting fire fly luciferase (LUC-sh) or two LNX2-targeting shRNAs (LNX2-sh1 and LNX2-sh2), respectively. After selected with 6 µg/ml puromycin for 3 days, the cells were cultured with M-CSF plus RANKL for 5 days. Total RNAs were isolated from three independent cultures of each group. Quantitative real-time RT-PCR analysis of the expression of LNX2 was performed. Data were presented as mean ± S.D. *** p < 0.001 vs LUC-sh by one way ANOVA. (B) Lenti-viruses transduced BMMs were cultured with M-CSF and RANKL for 5 days. The cells were fixed and stained for TRAP. The scale bar = 20 µm. (C) The mRNA expression of osteoclast marker genes, TRAP (encoded by Acp5) and Cathepsin K (encoded by Ctsk), was measured by quantitative real-time PCR using TaqMan assay primers from Life Technologies., Data were presented as mean ± S.D, n = 3, ** p <0.01 vs LUC-sh by Student’s t-test. (D) The protein expression of Cathepsin K was detected by western blots. Tubulin served as a loading control. All experiments were repeated two to three times.
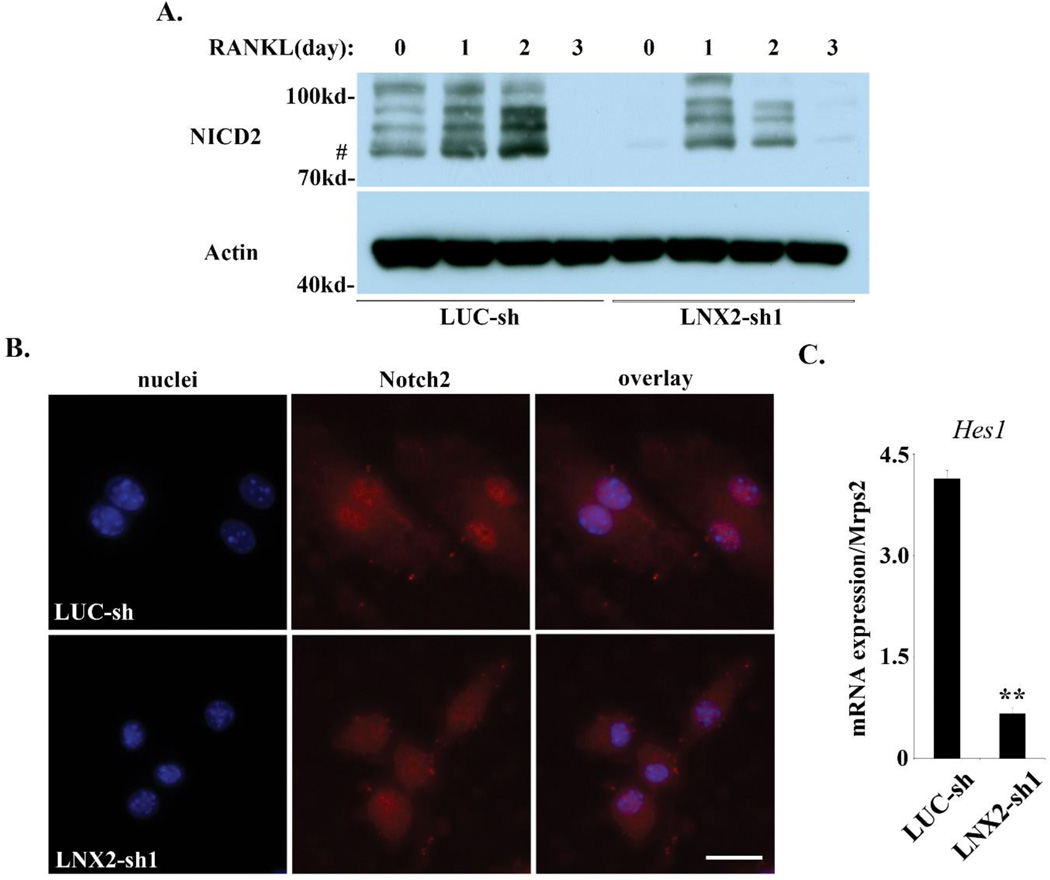
Fig. 6.
LNX2-deficiency results in decreased level and activity of Notch2. (A) BMMs were cultured with M-CSF alone or M-CSF and RANKL for three days. The level of Notch 2 intracellular domain (NICD2) was detected by western blots. Actin served as loading controls. # marks the expected 77kd molecular weight of NICD2. (B) Immunofluorescent staining of the nuclear localization of NICD2 in control and LNX2 knocking-down BMMs. A rat IgG control showed no labeling. The scale bar = 10 µm. (C) The mRNA expression of Notch target gene, Hes1, was examined by Quantitative real-time RT-PCR. ** p < 0.01 vs LUC-sh by Student’s t-test, n = 3. The data were representatives of two independent experiments.
Immunofluorescence
Immunofluorescence was performed using the protocol described previously [39]. In brief, cells grown on glass coverslips in a 24-well plate were fixed with 4% paraformaldehyde/PBS for 20 minutes, followed by permeabilizing and blocking in PBS/0.2% BSA/0.1% Triton-X100 for 30 minutes. The cells were then incubated with primary antibodies in PBS/0.2% BSA for 2 hours. Primary antibody labeling was visualized using fluorescent dye–conjugated secondary antibodies (Jackson ImmunoResearch Laboratories Inc.). The nucleus was stained with Hoechst 33258. Samples were mounted with 80% glycerol/PBS. Immunofluorescence-labeled cells were observed using a Carl Zeiss fluorescence microscope equipped with a charge-coupled-device camera.
Statistics
Data of 2-group comparisons were analyzed using a 2-tailed Student’s t test. For comparison of more than 2 groups, data were analyzed using one way Analysis of Variance (ANOVA) and the Bonferroni procedure was used for post-hoc comparison. For all graphs and in the text, data are represented as the mean ± standard deviation.
RESULTS
The expression of LNX1 and LNX2 is up-regulated during osteoclast differentiation
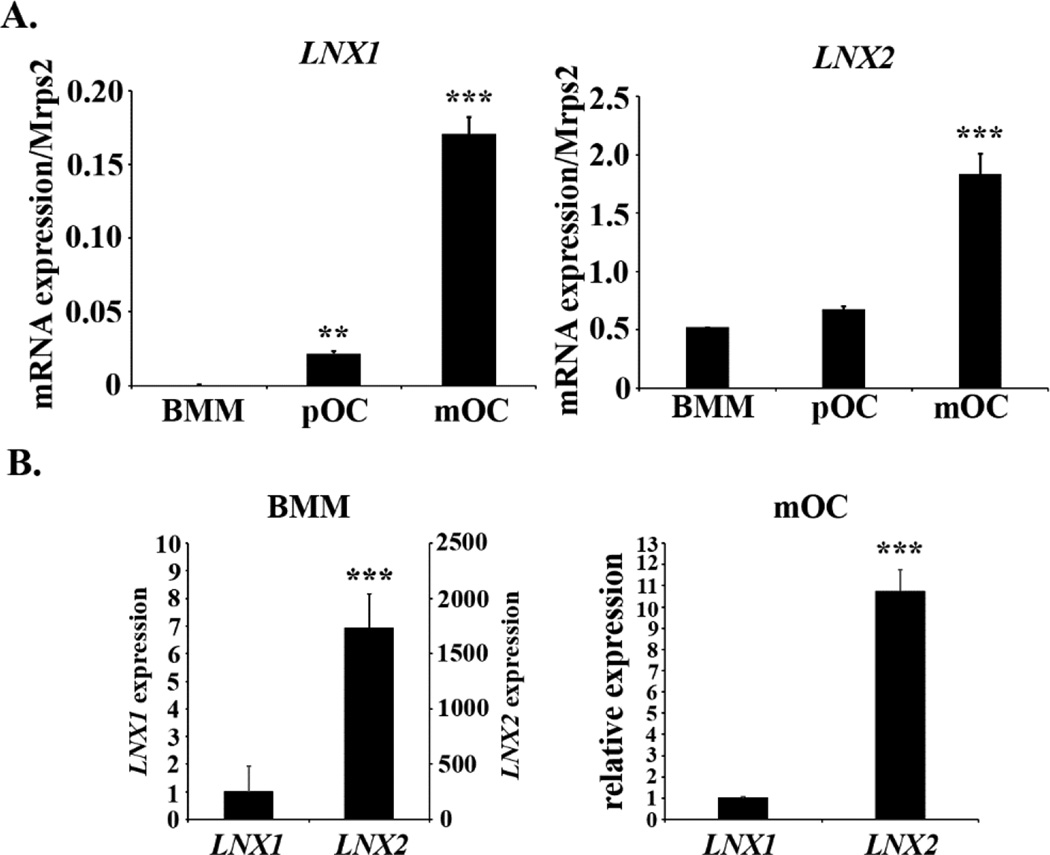
In search of novel molecules regulating osteoclast differentiation and/or function, we set out to examine the mRNA levels of LNX1 and LNX2, two highly homologous modulators of Numb and Notch, during the course of osteoclast formation in vitro using real-time PCR. Bone marrow macrophages were cultured with a combination of M-CSF and RANKL, two key cytokines required for osteoclast formation, to generate macrophages (BMM), pre-osteoclasts (pOC), and mature osteoclasts (mOC). As shown in Fig. 1A, both LNX mRNAs increased during osteoclast differentiation with the level of LNX2 reaching much higher level as compared to LNX1 (Fig. 1B), suggesting that LNX2 is the major LNX family member expressed in osteoclast lineage cells.
Fig. 1.
The expression of LNX1 and LNX2 is up-regulated during osteoclast differentiation. Bone marrow macrophages (BMMs) were cultured with M-CSF alone or M-CSF plus RANKL for 2 and 4 days to generate macrophages (BMM), mononuclear pre-osteoclasts (p-OC) and mature osteoclasts (mOC), respectively. At these time points, more than 80% of cells were mononuclear pre-OCs and multinucleated mOCs in the respective cultures. Total RNAs were purified from three independent cultures of BMM, pOC, and mOC, respectively. Quantitative real-time RT-PCR analysis of the expression of LNX mRNA during osteoclast differentiation was performed using TaqMan assay primers from Life Technologies. The representative data from three experiments were presented as mean ± S.D. (A) data were analyzed using the ΔCt method. ** p < 0.01, *** p < 0.001 vs BMM by one-way ANOVA. (B) The relative levels of LNX1 and LNX2 cDNAs in BMM and mOC were analyzed using the delta ΔCt method. *** p < 0.001 vs LNX1 by a 2-tailed Student’s t test.
Knocking down LNX2 expression impairs osteoclast differentiation
To delineate the role of LNX2 in osteoclast biology we next knocked down LNX2 in macrophages by lentiviral transduction of shRNAs. To avoid potential unspecific and off-target effects of shRNA over-expression we selected two shRNAs (LNX2-sh1 and LNX2-sh2) which target different sites of murine LNX2 mRNA. A shRNA targeting firefly luciferase (Luc-sh) was used as a negative control. Transduced BMMs were then cultured with M-CSF and RANKL for 4 days and the mRNA level of LNX2 was measured by real-time PCR. Both LNX2 shRNAs significantly inhibited LNX2 expression (Fig. 2A) and led to reduced osteoclast formation as compared to Luc-sh transduced cells. This reduced osteoclast formation was evidenced by the decrease of multinucleated cells staining positively for TRAP, an osteoclast differentiation marker (Fig. 2B). Because LNX2-sh1 was more potent than LNX2-sh2 in inhibiting LNX2 expression and osteoclast formation, we subsequently used cells harboring this shRNA to examine the effects of LNX2 knock down on the mRNA and protein levels of osteoclast markers in macrophages and mature osteoclasts. As demonstrated in Fig. 2C–2D, the mRNA expression of TRAP (encoded by Acp5) and Cathepsin K (encoded by Ctsk), and the protein level of Cathepsin K, were significantly lower in LNX2 deficient osteoclasts, as compared to control cells. These results indicate that LNX2 plays a critical role in osteoclastogenesis.
Loss of LNX2 inhibits M-CSF and RANKL signalings and osteoclast
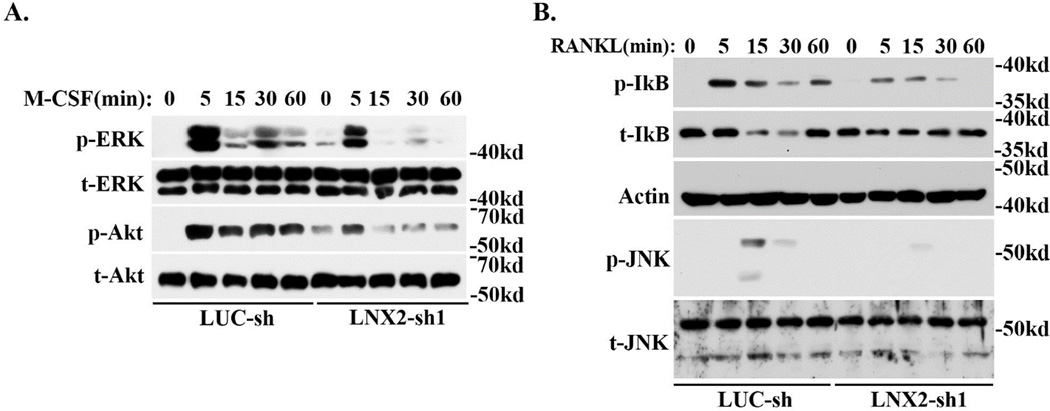
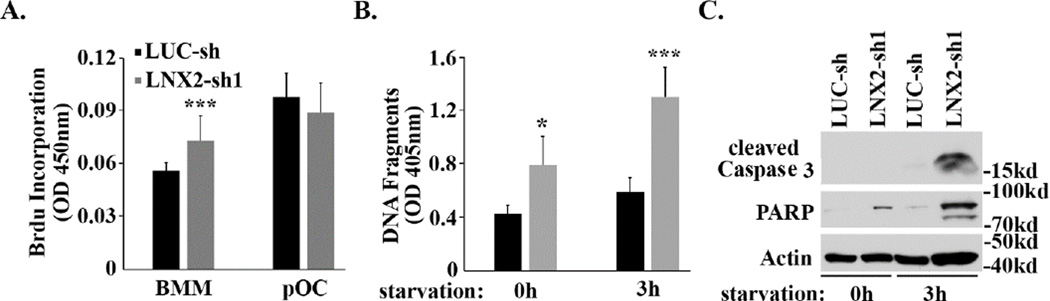
survival Since M-CSF and RANKL play a central role in osteoclast formation in vivo and in vitro, we next determined whether LNX2 regulates osteoclastogenesis through modulating the downstream signaling pathways of these two key cytokines. As shown in Fig. 3A, M-CSF-induced activation of ERK and AKT was blunted in LNX2 deficient macrophages as compared to control cells. Loss of LNX2 in osteoclast precursors also attenuated RANKL-stimulated activation of NF-kB and JNK, as demonstrated by decreased IkB phosphorylation/degradation and diminished JNK phosphorylation (Fig. 3B). ERK, NF-kB and JNK are critical for cell proliferation and survival of osteoclast precursors [40–44]. We then set out to resolve whether these cellular events were subject to LNX2 regulation. Loss of LNX2 slightly increased proliferation of macrophages but had little effect on pre-osteoclast proliferation, as measured by BrdU incorporation rate (Fig. 4A). The deficiency of LNX2 accelerated pre-osteoclast apoptosis under both basal and starvation conditions, as determined by cell death ELISA (Fig. 4B) and the levels of cleaved Caspase-3 and Poly (ADP-ribose) polymerase (PARP) (Fig. 4C). These data indicate that LNX2 plays an important role in survival of osteoclast precursors via modulating M-CSF and RANKL signaling pathways.
Fig. 3.
Reduction of LNX2 expression in macrophages inhibits M-CSF and RANKL signaling pathways. Lentivirus-transduced BMMs were serum-starved overnight and were then stimulated with 50ng/ml M-CSF (A) and 100ng/ml RANKL (B) for the indicated times. The activation of downstream signaling pathways was detected by western blots using specific antibodies. Total ERK and AKT in (A) and Actin and total JNK in (B) served as loading controls. The representative data from three experiments were presented.
Fig. 4.
Loss of LNX2 accelerates pre-osteoclast apoptosis. (A) BMMs were cultured with M-CSF alone (BMM) or M-CSF and RANKL (pOC) for two days. Cell proliferation rate was measured by BrdU incorporation ELISA. *** p < 0.001 vs BMM by one-way ANOVA. (B) and (C) BMMs were cultured with M-CSF and RANKL for two days to generate pre-osteoclasts. The cells were then either untreated (oh) or were serum and cytokine starved for 3 hours (3h). Apoptosis was assessed by (B) Cell Death Detection ELISA PLUS kit, which detects cytoplasmic histone-associated DNA fragmentation and (C) western blots using antibodies against cleaved Caspase-3 and PARP. Actin served as a loading control. Data in (A) and (B) were presented as mean ± S.D, n = 6, * p < 0.05, *** p <0.001 vs LUC-sh by one-way ANOVA. The experiments were repeated twice.
LNX2-deficient macrophages have increased Numb and decreased level and activity of Notch
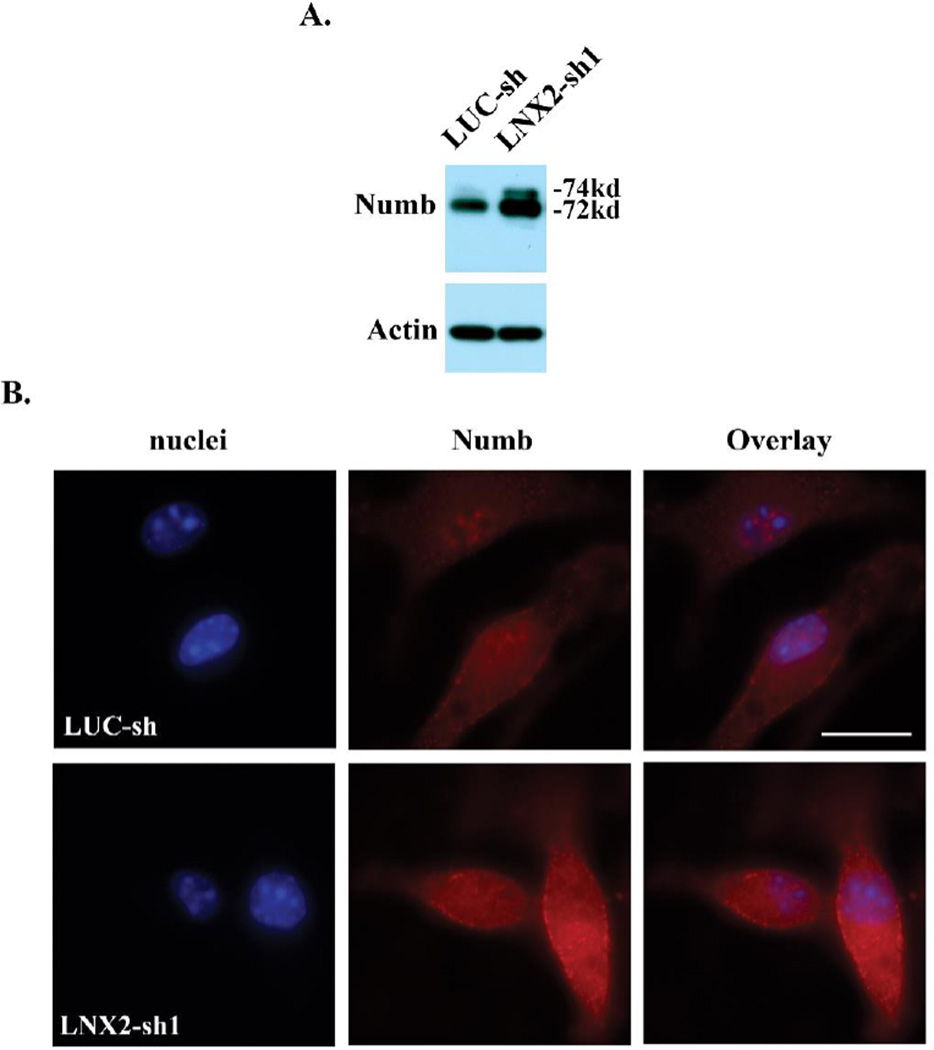
LNX proteins have been shown to bind Numb, leading to its ubiquitination and degradation [31]. We next tested whether LNX2 modulates Numb protein level in osteoclast precursor cells. Consistent with an inhibitory role of LNX proteins in Numb expression in other cell types [32], loss of LNX2 in macrophages resulted in an increased accumulation of both 74kd and 72kd isoforms of mammalian Numb protein, as demonstrated by western blots and immunofluorescence (Fig.5A and 5B).
Fig. 5.
Loss of LNX2 in BMMs increases the Numb protein level. (A) The protein expression of Numb in LUC-sh and LNX2-sh1 transduced BMMs was detected by western blots. Actin served as a loading control. (B) Immunofluorescent staining of Numb in BMMs. A rabbit IgG control showed no labeling. The scale bar = 10 µm. The data were representatives of three independent experiments.
Numb antagonizes the Notch signaling by (1) preventing the recycling of Notch receptor from endosomes back to the plasma membrane in Drosophila and mammalian cells [22, 23, 45]; and (2) simultaneously promoting trafficking of Notch through late endosomes/lysosomes for degradation. Thus, changes in Numb expression may alter the level and activity of Notch. To test if such is the case in osteoclast lineage cells, we examined the protein levels of the intracellular domains of Notch 2 (NICD2) which is the major Notch receptor in macrophages and osteoclasts [20, 46]. We also examined the expression of Notch downstream target gene, Hes1, in control and LNX2-deficient macrophages. Loss of LNX2 in osteoclast lineage cells dramatically decreased NICD2 level in both cytoplasm (Fig. 6A) and inhibited its accumulation in the nucleus (Fig. 6B). These changes were correlated with a reduction of Hes1 mRNA (Fig. 6C). These results indicate that LNX2 regulates the Notch signaling pathway in osteoclast precursor cells through Numb.
DISCUSSION
Although M-CSF and RANKL are two indispensable cytokines for osteoclastogenesis in vivo and in vitro [47–53], systemic hormones and other local cytokines and factors in bone marrow environment exert their fine-tuning regulations of osteoclasts through direct or indirect modulations of M-CSF and RANKL signaling pathways under physiological and pathological conditions [54, 55]. Recent genetic studies in human and mice have elucidated that Notch signaling is a critical regulator of cell fate decisions in osteoblast and osteoclast differentiation and cell-cell interactions during both skeletal development and homeostasis in adulthood [12, 13]. Thus, the identification of the molecular mechanisms that regulate this pathway in bone cells may help to develop novel therapeutics for the treatment of bone diseases. The proteolytic cleavages mediated by the ADAM family of enzymes and a γ-secretase complex have been well documented as key steps in Notch activation. Additionally, recent studies have revealed that the endocytic trafficking/recycling of the Notch receptors and their degradation through ubiquitin/proteasomes and lysosomes also play an important role in regulating the intracellular level and activity of Notch signaling cascade [56]. Numb and LNX proteins are evidently key molecules involved in these processes. However, their expression and function in bone cells have not been elucidated.
In this study, we have demonstrated that LNX2 is highly expressed and up-regulated during osteoclastogenesis in vitro. Nonetheless, LNX2 is crucial for osteoclast formation, as suppressing its expression in osteoclast precursors blunts osteoclastogenesis in vitro. Loss of LNX2 in osteoclast lineage cells accelerates apoptosis of osteoclast precursors, leading to decreased cell number/density and diminished formation of mature osteoclasts. Mechanistically, LNX2 deficiency in macrophages attenuates M-CSF-induced ERK and AKT activation and RANKL-stimulated activation of NF-kB and JNK pathways, which are indispensable for the survival of osteoclast lineage cells. In addition, down-regulation of LNX2 expression in macrophages leads to accumulation of Notch inhibitor, Numb, and decreased level and activity of Notch2. These results indicate that LNX2 is a critical molecule for osteoclastogenesis and a novel regulator of M-CSF/RANKL and Notch signalings in osteoclast lineage cells.
Cell number/density, regulated by proliferation and survival of osteoclast precursors, is critical for osteoclastogenesis in cultures. In this study, we found that loss of LNX2 in osteoclast precursors induced apoptosis in pre-osteoclasts with slightly increase in proliferation of macrophages (Fig 4). Meanwhile, we did not observe increased cell death of mature osteoclasts in day-4 (Fig 2B) and day-5 (data not shown) of control and LNX2 knocking-down cultures. Thus, the declined number of mature osteoclasts in LNX2-deficient cultures was likely due to increased apoptosis of pre-osteoclasts, leading to decreased number/density of mono-nuclear osteoclast precursors. Of note, however, we cannot exclude whether LNX2 regulates mature osteoclast survival/death. The time-lapse live cell imaging has been proved to be a very useful technique to uncover detailed cellular events, including apoptosis during osteoclast differentiation [57]. It will be interesting to employ this technique in the future, to identify at which stages the LNX2-deficient cells undergo apoptosis in the course of osteoclast differentiation.
LNX1 and LNX2 were originally identified as Numb binding proteins [30, 32] and have been since shown to regulate Numb protein level by targeting it for ubiquitin-dependent degradation [25]. Thus, LNX2 may regulate Notch2 level via Numb. On the other hand, how LNX2 modulates M-CSF and RANKL signaling pathways remain unidentified. Both LNX1 and LNX2 interact with several cell surface receptors and promote their ubiquitylation and endocytosis [58–60]. It is likely that LNX2 may interact with c-fms and RANK, receptors for M-CSF and RANKL, respectively, and regulates their kinetics between the plasma membrane and the intracellular endosomes, thereby influences receptor signalings. Another possibility is that LNX2, functioning as an E3 ubiquitin ligase, regulates the stability of signaling molecules involved in pathways downstream of M-CSF and RANKL. Identification of novel LNX2-interracting proteins in osteoclast lineage cells, as it being done for LNX1 in other cell types [61, 62], will solve this issue. In addition, generation and characterization of skeletal phenotypes of LNX2-deficient mice, which are not available at the moment, will help to define whether LNX2 regulates osteoclastogenesis in vivo.
While the work from Yamada et al [63] and Bai et al [19] suggests that Notch signaling is inhibitory for osteoclastogenesis, recent in vitro and in vivo studies [20, 21, 64, 65] elucidate that Notch receptors, especially Notch2, are required for osteoclast formation through optimizing RANKL-induced NF-kB activation. Our recently published [46] and the present work supports a positive role of Notch signaling in osteoclastogenesis. The explanations for discrepancy about the opposite functions of Notch signaling in osteoclasts remain unclear. One possibility is that the contradictory findings may result from the experimental differences between in vivo situations and in vitro culture conditions.
Supplementary Material
Acknowledgments
The authors would like to thank Drs. Stavros C Manolagas, Charles A Obrien, and Ms. GibAnn Berryhill for the critics of the manuscript prior submission. Erin Hogan is thanked for her support in microscopes. The work was supported by NIH grants AR062012 and P01 AG13918.
Footnotes
Conflict of Interest
Jian Zhou, Toshifumi Fujiwara, Shiqiao Ye,, Xiaolin Li, Haibo Zhao state that they have no conflicts of interest.
REFERENCES
- 1.Rachner TD, Khosla S, Hofbauer LC. Osteoporosis: now and the future. Lancet. 2011;377:1276–1287. doi: 10.1016/S0140-6736(10)62349-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sobacchi C, Schulz A, Coxon FP, Villa A, Helfrich MH. Osteopetrosis: genetics, treatment and new insights into osteoclast function. Nature reviews. Endocrinology. 2013 doi: 10.1038/nrendo.2013.137. [DOI] [PubMed] [Google Scholar]
- 3.Henriksen K, Karsdal MA, John Martin T. Osteoclast-Derived Coupling Factors in Bone Remodeling. Calcified tissue international. 2013 doi: 10.1007/s00223-013-9741-7. [DOI] [PubMed] [Google Scholar]
- 4.Henriksen K, Neutzsky-Wulff AV, Bonewald LF, Karsdal MA. Local communication on and within bone controls bone remodeling. Bone. 2009;44:1026–1033. doi: 10.1016/j.bone.2009.03.671. [DOI] [PubMed] [Google Scholar]
- 5.Sims NA, Gooi JH. Bone remodeling: Multiple cellular interactions required for coupling of bone formation and resorption. Seminars in cell & developmental biology. 2008;19:444–451. doi: 10.1016/j.semcdb.2008.07.016. [DOI] [PubMed] [Google Scholar]
- 6.Boyle WJ, Simonet WS, Lacey DL. Osteoclast differentiation and activation. Nature. 2003;423:337–342. doi: 10.1038/nature01658. [DOI] [PubMed] [Google Scholar]
- 7.Nakashima T, Hayashi M, Takayanagi H. New insights into osteoclastogenic signaling mechanisms. Trends in endocrinology and metabolism: TEM. 2012;23:582–590. doi: 10.1016/j.tem.2012.05.005. [DOI] [PubMed] [Google Scholar]
- 8.Teitelbaum SL, Ross FP. Genetic regulation of osteoclast development and function. Nature reviews. Genetics. 2003;4:638–649. doi: 10.1038/nrg1122. [DOI] [PubMed] [Google Scholar]
- 9.Negishi-Koga T, Takayanagi H. Bone cell communication factors and Semaphorins. BoneKEy reports. 2012;1:183. doi: 10.1038/bonekey.2012.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kopan R, Ilagan MX. The canonical Notch signaling pathway: unfolding the activation mechanism. Cell. 2009;137:216–233. doi: 10.1016/j.cell.2009.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hori K, Sen A, Artavanis-Tsakonas S. Notch signaling at a glance. Journal of cell science. 2013;126:2135–2140. doi: 10.1242/jcs.127308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Engin F, Lee B. NOTCHing the bone: insights into multi-functionality. Bone. 2010;46:274–280. doi: 10.1016/j.bone.2009.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Regan J, Long F. Notch signaling and bone remodeling. Current osteoporosis reports. 2013;11:126–129. doi: 10.1007/s11914-013-0145-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Canalis E, Parker K, Feng JQ, Zanotti S. Osteoblast lineage-specific effects of notch activation in the skeleton. Endocrinology. 2013;154:623–634. doi: 10.1210/en.2012-1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Engin F, Yao Z, Yang T, Zhou G, Bertin T, Jiang MM, Chen Y, Wang L, Zheng H, Sutton RE, Boyce BF, Lee B. Dimorphic effects of Notch signaling in bone homeostasis. Nature medicine. 2008;14:299–305. doi: 10.1038/nm1712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hilton MJ, Tu X, Wu X, Bai S, Zhao H, Kobayashi T, Kronenberg HM, Teitelbaum SL, Ross FP, Kopan R, Long F. Notch signaling maintains bone marrow mesenchymal progenitors by suppressing osteoblast differentiation. Nature medicine. 2008;14:306–314. doi: 10.1038/nm1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tezuka K, Yasuda M, Watanabe N, Morimura N, Kuroda K, Miyatani S, Hozumi N. Stimulation of osteoblastic cell differentiation by Notch. Journal of bone and mineral research : the official journal of the American Society for Bone and Mineral Research. 2002;17:231–239. doi: 10.1359/jbmr.2002.17.2.231. [DOI] [PubMed] [Google Scholar]
- 18.Nobta M, Tsukazaki T, Shibata Y, Xin C, Moriishi T, Sakano S, Shindo H, Yamaguchi A. Critical regulation of bone morphogenetic protein-induced osteoblastic differentiation by Delta1/Jagged1-activated Notch1 signaling. The Journal of biological chemistry. 2005;280:15842–15848. doi: 10.1074/jbc.M412891200. [DOI] [PubMed] [Google Scholar]
- 19.Bai S, Kopan R, Zou W, Hilton MJ, Ong CT, Long F, Ross FP, Teitelbaum SL. NOTCH1 regulates osteoclastogenesis directly in osteoclast precursors and indirectly via osteoblast lineage cells. The Journal of biological chemistry. 2008;283:6509–6518. doi: 10.1074/jbc.M707000200. [DOI] [PubMed] [Google Scholar]
- 20.Fukushima H, Nakao A, Okamoto F, Shin M, Kajiya H, Sakano S, Bigas A, Jimi E, Okabe K. The association of Notch2 and NF-kappaB accelerates RANKL-induced osteoclastogenesis. Molecular and cellular biology. 2008;28:6402–6412. doi: 10.1128/MCB.00299-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Simpson MA, Irving MD, Asilmaz E, Gray MJ, Dafou D, Elmslie FV, Mansour S, Holder SE, Brain CE, Burton BK, Kim KH, Pauli RM, Aftimos S, Stewart H, Kim CA, Holder-Espinasse M, Robertson SP, Drake WM, Trembath RC. Mutations in NOTCH2 cause Hajdu-Cheney syndrome, a disorder of severe and progressive bone loss. Nature genetics. 2011;43:303–305. doi: 10.1038/ng.779. [DOI] [PubMed] [Google Scholar]
- 22.Cotton M, Benhra N, Le Borgne R. Numb inhibits the recycling of Sanpodo in Drosophila sensory organ precursor. Current biology : CB. 2013;23:581–587. doi: 10.1016/j.cub.2013.02.020. [DOI] [PubMed] [Google Scholar]
- 23.Couturier L, Mazouni K, Schweisguth F. Numb localizes at endosomes and controls the endosomal sorting of notch after asymmetric division in Drosophila. Current biology : CB. 2013;23:588–593. doi: 10.1016/j.cub.2013.03.002. [DOI] [PubMed] [Google Scholar]
- 24.Giebel B, Wodarz A. Notch signaling: numb makes the difference. Current biology : CB. 2012;22:R133–R135. doi: 10.1016/j.cub.2012.01.006. [DOI] [PubMed] [Google Scholar]
- 25.McGill MA, McGlade CJ. Mammalian numb proteins promote Notch1 receptor ubiquitination and degradation of the Notch1 intracellular domain. The Journal of biological chemistry. 2003;278:23196–23203. doi: 10.1074/jbc.M302827200. [DOI] [PubMed] [Google Scholar]
- 26.Berdnik D, Torok T, Gonzalez-Gaitan M, Knoblich JA. The endocytic protein alpha-Adaptin is required for numb-mediated asymmetric cell division in Drosophila. Developmental cell. 2002;3:221–231. doi: 10.1016/s1534-5807(02)00215-0. [DOI] [PubMed] [Google Scholar]
- 27.Guo M, Jan LY, Jan YN. Control of daughter cell fates during asymmetric division: interaction of Numb and Notch. Neuron. 1996;17:27–41. doi: 10.1016/s0896-6273(00)80278-0. [DOI] [PubMed] [Google Scholar]
- 28.Song Y, Lu B. Interaction of Notch signaling modulator Numb with alpha-Adaptin regulates endocytosis of Notch pathway components and cell fate determination of neural stem cells. The Journal of biological chemistry. 2012;287:17716–17728. doi: 10.1074/jbc.M112.360719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gulino A, Di Marcotullio L, Screpanti I. The multiple functions of Numb. Experimental cell research. 2010;316:900–906. doi: 10.1016/j.yexcr.2009.11.017. [DOI] [PubMed] [Google Scholar]
- 30.Dho SE, Jacob S, Wolting CD, French MB, Rohrschneider LR, McGlade CJ. The mammalian numb phosphotyrosine-binding domain. Characterization of binding specificity and identification of a novel PDZ domain-containing numb binding protein, LNX. The Journal of biological chemistry. 1998;273:9179–9187. doi: 10.1074/jbc.273.15.9179. [DOI] [PubMed] [Google Scholar]
- 31.Nie J, McGill MA, Dermer M, Dho SE, Wolting CD, McGlade CJ. LNX functions as a RING type E3 ubiquitin ligase that targets the cell fate determinant Numb for ubiquitin-dependent degradation. The EMBO journal. 2002;21:93–102. doi: 10.1093/emboj/21.1.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rice DS, Northcutt GM, Kurschner C. The Lnx family proteins function as molecular scaffolds for Numb family proteins. Molecular and cellular neurosciences. 2001;18:525–540. doi: 10.1006/mcne.2001.1024. [DOI] [PubMed] [Google Scholar]
- 33.Lai EC. Protein degradation: four E3s for the notch pathway. Current biology : CB. 2002;12:R74–R78. doi: 10.1016/s0960-9822(01)00679-0. [DOI] [PubMed] [Google Scholar]
- 34.Ye S, Fowler TW, Pavlos NJ, Ng PY, Liang K, Feng Y, Zheng M, Kurten R, Manolagas SC, Zhao H. LIS1 regulates osteoclast formation and function through its interactions with dynein/dynactin and Plekhm1. PloS one. 2011;6:e27285. doi: 10.1371/journal.pone.0027285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Takeshita S, Kaji K, Kudo A. Identification and characterization of the new osteoclast progenitor with macrophage phenotypes being able to differentiate into mature osteoclasts. Journal of bone and mineral research : the official journal of the American Society for Bone and Mineral Research. 2000;15:1477–1488. doi: 10.1359/jbmr.2000.15.8.1477. [DOI] [PubMed] [Google Scholar]
- 36.Feng X, Novack DV, Faccio R, Ory DS, Aya K, Boyer MI, McHugh KP, Ross FP, Teitelbaum SL. A Glanzmann's mutation in beta 3 integrin specifically impairs osteoclast function. The Journal of clinical investigation. 2001;107:1137–1144. doi: 10.1172/JCI12040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nature protocols. 2008;3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- 38.Jilka RL, Hangoc G, Girasole G, Passeri G, Williams DC, Abrams JS, Boyce B, Broxmeyer H, Manolagas SC. Increased osteoclast development after estrogen loss: mediation by interleukin-6. Science (New York, N.Y.) 1992;257:88–91. doi: 10.1126/science.1621100. [DOI] [PubMed] [Google Scholar]
- 39.Zhao H, Laitala-Leinonen T, Parikka V, Vaananen HK. Downregulation of small GTPase Rab7 impairs osteoclast polarization and bone resorption. The Journal of biological chemistry. 2001;276:39295–39302. doi: 10.1074/jbc.M010999200. [DOI] [PubMed] [Google Scholar]
- 40.David JP, Sabapathy K, Hoffmann O, Idarraga MH, Wagner EF. JNK1 modulates osteoclastogenesis through both c-Jun phosphorylation-dependent and -independent mechanisms. Journal of cell science. 2002;115:4317–4325. doi: 10.1242/jcs.00082. [DOI] [PubMed] [Google Scholar]
- 41.Franzoso G, Carlson L, Xing L, Poljak L, Shores EW, Brown KD, Leonardi A, Tran T, Boyce BF, Siebenlist U. Requirement for NF-kappaB in osteoclast and B-cell development. Genes & development. 1997;11:3482–3496. doi: 10.1101/gad.11.24.3482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gingery A, Bradley E, Shaw A, Oursler MJ. Phosphatidylinositol 3-kinase coordinately activates the MEK/ERK and AKT/NFkappaB pathways to maintain osteoclast survival. Journal of cellular biochemistry. 2003;89:165–179. doi: 10.1002/jcb.10503. [DOI] [PubMed] [Google Scholar]
- 43.Iotsova V, Caamano J, Loy J, Yang Y, Lewin A, Bravo R. Osteopetrosis in mice lacking NF-kappaB1 and NF-kappaB2. Nature medicine. 1997;3:1285–1289. doi: 10.1038/nm1197-1285. [DOI] [PubMed] [Google Scholar]
- 44.Miyazaki T, Katagiri H, Kanegae Y, Takayanagi H, Sawada Y, Yamamoto A, Pando MP, Asano T, Verma IM, Oda H, Nakamura K, Tanaka S. Reciprocal role of ERK and NF-kappaB pathways in survival and activation of osteoclasts. The Journal of cell biology. 2000;148:333–342. doi: 10.1083/jcb.148.2.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.McGill MA, Dho SE, Weinmaster G, McGlade CJ. Numb regulates post-endocytic trafficking and degradation of Notch1. The Journal of biological chemistry. 2009;284:26427–26438. doi: 10.1074/jbc.M109.014845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhou J, Fujiwara T, Ye S, Li X, Zhao H. Downregulation of Notch modulators, tetraspanin 5 and 10, inhibits osteoclastogenesis in vitro. Calcified tissue international. 2014;95:209–217. doi: 10.1007/s00223-014-9883-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dougall WC, Glaccum M, Charrier K, Rohrbach K, Brasel K, De Smedt T, Daro E, Smith J, Tometsko ME, Maliszewski CR, Armstrong A, Shen V, Bain S, Cosman D, Anderson D, Morrissey PJ, Peschon JJ, Schuh J. RANK is essential for osteoclast and lymph node development. Genes & development. 1999;13:2412–2424. doi: 10.1101/gad.13.18.2412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fuller K, Wong B, Fox S, Choi Y, Chambers TJ. TRANCE is necessary and sufficient for osteoblast-mediated activation of bone resorption in osteoclasts. The Journal of experimental medicine. 1998;188:997–1001. doi: 10.1084/jem.188.5.997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hsu H, Lacey DL, Dunstan CR, Solovyev I, Colombero A, Timms E, Tan HL, Elliott G, Kelley MJ, Sarosi I, Wang L, Xia XZ, Elliott R, Chiu L, Black T, Scully S, Capparelli C, Morony S, Shimamoto G, Bass MB, Boyle WJ. Tumor necrosis factor receptor family member RANK mediates osteoclast differentiation and activation induced by osteoprotegerin ligand. Proceedings of the National Academy of Sciences of the United States of America. 1999;96:3540–3545. doi: 10.1073/pnas.96.7.3540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lacey DL, Timms E, Tan HL, Kelley MJ, Dunstan CR, Burgess T, Elliott R, Colombero A, Elliott G, Scully S, Hsu H, Sullivan J, Hawkins N, Davy E, Capparelli C, Eli A, Qian YX, Kaufman S, Sarosi I, Shalhoub V, Senaldi G, Guo J, Delaney J, Boyle WJ. Osteoprotegerin ligand is a cytokine that regulates osteoclast differentiation and activation. Cell. 1998;93:165–176. doi: 10.1016/s0092-8674(00)81569-x. [DOI] [PubMed] [Google Scholar]
- 51.Wiktor-Jedrzejczak W, Bartocci A, Ferrante AW, Jr, Ahmed-Ansari A, Sell KW, Pollard JW, Stanley ER. Total absence of colony-stimulating factor 1 in the macrophage-deficient osteopetrotic (op/op) mouse. Proceedings of the National Academy of Sciences of the United States of America. 1990;87:4828–4832. doi: 10.1073/pnas.87.12.4828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yasuda H, Shima N, Nakagawa N, Yamaguchi K, Kinosaki M, Mochizuki S, Tomoyasu A, Yano K, Goto M, Murakami A, Tsuda E, Morinaga T, Higashio K, Udagawa N, Takahashi N, Suda T. Osteoclast differentiation factor is a ligand for osteoprotegerin/osteoclastogenesis-inhibitory factor and is identical to TRANCE/RANKL. Proceedings of the National Academy of Sciences of the United States of America. 1998;95:3597–3602. doi: 10.1073/pnas.95.7.3597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yoshida H, Hayashi S, Kunisada T, Ogawa M, Nishikawa S, Okamura H, Sudo T, Shultz LD, Nishikawa S. The murine mutation osteopetrosis is in the coding region of the macrophage colony stimulating factor gene. Nature. 1990;345:442–444. doi: 10.1038/345442a0. [DOI] [PubMed] [Google Scholar]
- 54.Manolagas SC. Birth and death of bone cells: basic regulatory mechanisms and implications for the pathogenesis and treatment of osteoporosis. Endocrine reviews. 2000;21:115–137. doi: 10.1210/edrv.21.2.0395. [DOI] [PubMed] [Google Scholar]
- 55.Roodman GD. Regulation of osteoclast differentiation. Annals of the New York Academy of Sciences. 2006;1068:100–109. doi: 10.1196/annals.1346.013. [DOI] [PubMed] [Google Scholar]
- 56.Kandachar V, Roegiers F. Endocytosis and control of Notch signaling. Current opinion in cell biology. 2012;24:534–540. doi: 10.1016/j.ceb.2012.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jansen ID, Vermeer JA, Bloemen V, Stap J, Everts V. Osteoclast fusion and fission. Calcified tissue international. 2012;90:515–522. doi: 10.1007/s00223-012-9600-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.D'Agostino M, Tornillo G, Caporaso MG, Barone MV, Ghigo E, Bonatti S, Mottola G. Ligand of Numb proteins LNX1p80 and LNX2 interact with the human glycoprotein CD8alpha and promote its ubiquitylation and endocytosis. Journal of cell science. 2011;124:3545–3556. doi: 10.1242/jcs.081224. [DOI] [PubMed] [Google Scholar]
- 59.Kansaku A, Hirabayashi S, Mori H, Fujiwara N, Kawata A, Ikeda M, Rokukawa C, Kurihara H, Hata Y. Ligand-of-Numb protein X is an endocytic scaffold for junctional adhesion molecule 4. Oncogene. 2006;25:5071–5084. doi: 10.1038/sj.onc.1209468. [DOI] [PubMed] [Google Scholar]
- 60.Takahashi S, Iwamoto N, Sasaki H, Ohashi M, Oda Y, Tsukita S, Furuse M. The E3 ubiquitin ligase LNX1p80 promotes the removal of claudins from tight junctions in MDCK cells. Journal of cell science. 2009;122:985–994. doi: 10.1242/jcs.040055. [DOI] [PubMed] [Google Scholar]
- 61.Guo Z, Song E, Ma S, Wang X, Gao S, Shao C, Hu S, Jia L, Tian R, Xu T, Gao Y. Proteomics strategy to identify substrates of LNX, a PDZ domain-containing E3 ubiquitin ligase. Journal of proteome research. 2012;11:4847–4862. doi: 10.1021/pr300674c. [DOI] [PubMed] [Google Scholar]
- 62.Wolting CD, Griffiths EK, Sarao R, Prevost BC, Wybenga-Groot LE, McGlade CJ. Biochemical and computational analysis of LNX1 interacting proteins. PloS one. 2011;6:e26248. doi: 10.1371/journal.pone.0026248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yamada T, Yamazaki H, Yamane T, Yoshino M, Okuyama H, Tsuneto M, Kurino T, Hayashi S, Sakano S. Regulation of osteoclast development by Notch signaling directed to osteoclast precursors and through stromal cells. Blood. 2003;101:2227–2234. doi: 10.1182/blood-2002-06-1740. [DOI] [PubMed] [Google Scholar]
- 64.Choi YH, Ann EJ, Yoon JH, Mo JS, Kim MY, Park HS. Calcium/calmodulin-dependent protein kinase IV (CaMKIV) enhances osteoclast differentiation via the up-regulation of Notch1 protein stability. Biochimica et biophysica acta. 2013;1833:69–79. doi: 10.1016/j.bbamcr.2012.10.018. [DOI] [PubMed] [Google Scholar]
- 65.Duan L, de Vos P, Fan M, Ren Y. Notch is activated in RANKL-induced osteoclast differentiation and resorption. Frontiers in bioscience : a journal and virtual library. 2008;13:7064–7071. doi: 10.2741/3210. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.