TO THE EDITOR
Telomeres are tandem repeats of the non-coding DNA structures at the end of human chromosomes that protect the coding DNA and the integrity of the genome (Blackburn, 1991). The ability to sustain telomere length confers unlimited proliferative capacity to cancer cells. In a majority of cancers, telomere length is maintained by the activity of the enzyme telomerase (Kim et al., 1994), whose catalytic subunit is encoded by the telomerase reverse transcriptase (TERT) gene. However, until recently, the underlying mechanisms for telomerase activation in cancer cells were largely unknown.
Recurrent transcription activating mutations of the TERT promoter were first described in melanoma and subsequently in other tumor types (Horn et al., 2013; Vinagre et al., 2013). These mutations upregulate TERT expression by recruiting the multimeric GA-binding protein transcription factor that specifically binds to the mutant promoter (Bell et al., 2015). In addition to the mutations, DNA methylation of the TERT promoter is likely to play a role in TERT expression (Guilleret and Benhattar, 2004). Recently, it was shown that a region of the TERT promoter upstream of the transcription start site is methylated in malignant telomerase-expressing pediatric brain tumors but not in telomerase-negative normal brain tissues or low-grade tumors (Castelo-Branco et al., 2013).
We previously showed that an aggressive form of pediatric melanoma developing within giant congenital nevi (GCN) retains the wild-type TERT promoter (Lu et al., 2015). To determine whether epigenetic modifications may play a role in telomerase expression in this melanoma subtype, we analyzed the DNA methylation profile of a CpG-rich region of the TERT promoter, shown previously to be differentially methylated between normal and malignant tissues (Castelo-Branco et al., 2013), in 13 melanomas (3 arising in GCN; 7 conventional; and 3 spitzoid) and 10 benign or borderline melanocytic tumors (1 GCN; 3 GCN with nodular proliferation; and 6 borderline spitzoid melanocytic neoplasms) from 23 pediatric and adult patients. The human investigations were performed after approval by the local institutional review boards. Written, informed patient consent was waived because the research involved no more than minimal risk to the subjects. The status of the TERT promoter, BRAF and NRAS mutations, and kinase fusions was available for the spitzoid tumors and a subset of melanoma samples from our prior studies (Lee et al., 2015; Lu et al., 2015).
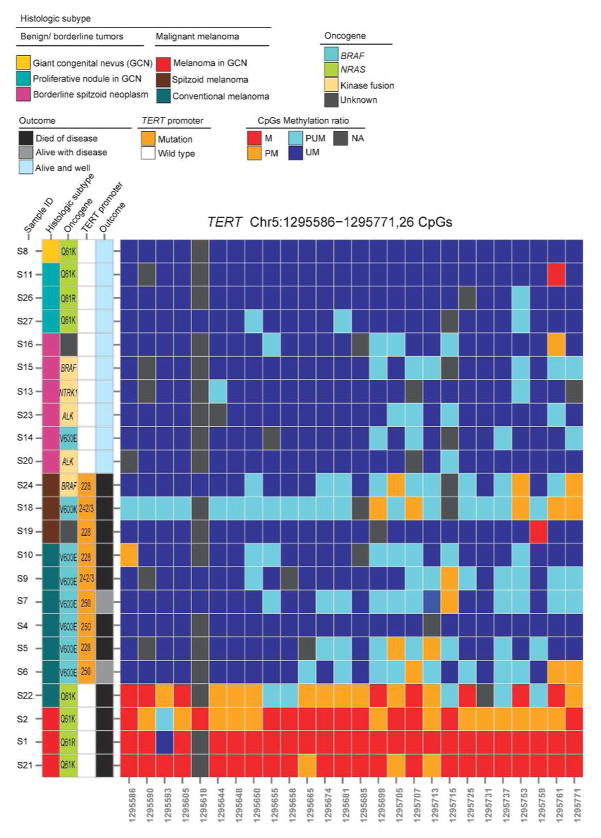
Supplementary Table S1 and Figure 1 summarize the demographic and outcome data and the primary driver oncogene for the 23 study subjects. PCR–Sanger sequencing identified a hotspot TERT promoter mutation [4 C228T (chr5:1,295,228); 3 C250T (chr5: 1,295,250), and 2 CC242/243TT (chr5: 1,295,242-3)] in 9 of 13 melanoma samples (6/7 conventional; 3/3 spitzoid) but not in the 10 benign or borderline melanocytic neoplasms (Figure 1). The DNA methylation status of a region of the TERT promoter, from 482 bp to 667 bp upstream of the ATG start site [chr5:1295586–1295771 (GRCh37/hg19)] (Supplementary Figure S1 online), encompassing 26 CpG sites, was assayed by next-generation bisulfite sequencing (Methods, Supplementary Material online). For each CpG site, the methylation ratio (Beta-value) was measured in the range of 0 to 1 (Supplementary Table S2 online). The methylation status was defined as follows: >0.7, methylated (Figure 1, red); 0.5–0.7, partially methylated (orange); 0.3 to <0.5, partially unmethylated (cyan); and <0.3, unmethylated (blue). Supplementary Table S3 shows the total number of methylated Cs and unmethylated Cs in the sequenced region for each sample. Remarkably, almost all 26 CpG sites in the sequenced region were highly methylated in the 3 melanomas arising in GCN (S1, S2, S21), and the one conventional melanoma bearing wild-type TERT promoter (S22), whereas the CpG sites remained predominantly unmethylated in the 9 mutant TERT promoter melanomas and the 10 benign or borderline melanocytic neoplasms (Figure 1).
Figure 1.
Association of the mutational status and the methylation profile of the TERT promoter with disease characteristics and outcome data for 23 patients with melanocytic tumors. The 26 CpG sites were aberrantly methylated in wild-type TERT promoter melanomas (the last 4 rows) but were predominantly unmethylated in low-grade or benign melanocytic tumors (atypical spitzoid neoplasms and GCN with proliferative nodules) and in mutant TERT promoter melanomas. Methylation panel color code: M, methylated; PM, partially methylated; PUM, partially unmethylated; UM, unmethylated; NA, not available.
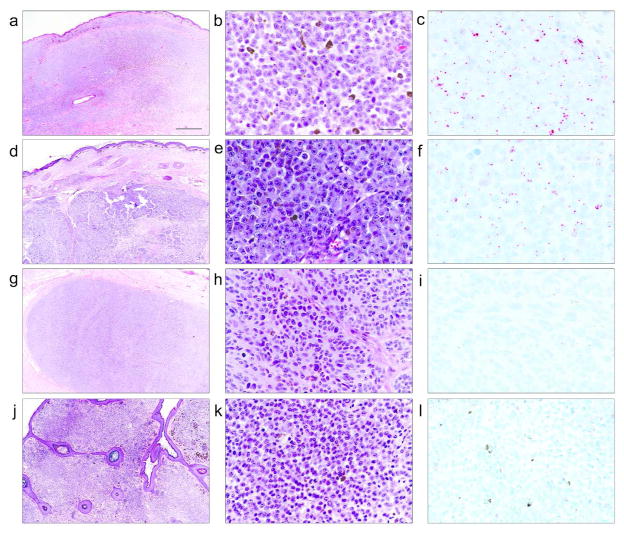
Next, we evaluated the association of TERT promoter CpG methylation with telomerase expression by TERT mRNA in situ hybridization (ISH) and by gene expression analysis (Methods, Supplementary Material online). TERT mRNA ISH revealed distinct, intracellular punctate signals in melanomas arising in GCN (Figure 2c and 2f) but not in the proliferative nodules in GCN (Figures 2i and 2l). The TERT promoter methylation level was calculated as the log2 ratio of the total number of methylated Cs versus the total number of unmethylated Cs in the sequenced region [logit (B-value)]. The TERT expression level was measured by using RNA sequencing data available for a subset of samples. An association analysis revealed a strong correlation between TERT promoter methylation and TERT expression level (P = 0.0422, adjusted r2 = 0.5145; Supplementary Figure S2 online).
Figure 2.
Photomicrographs (H&E–stained) and TERT mRNA in situ hybridization (ISH) for 2 melanomas in GCN (2 top panels) and 2 proliferative nodules in GCN (2 bottom panels). mRNA ISH shows numerous high-resolution red intracellular punctate signals in malignant melanocytes (c and f) and no signals above the background level in melanocytes of proliferative nodules (i and l). Scale bars= 1000 μm (a) and 50 μm (b).
Our data demonstrate that epigenetic modification through TERT promoter CpG methylation is an alternative pathway for TERT reactivation in melanoma. Although epigenetic remodeling by promoter methylation is generally considered a signature of gene silencing, TERT expression is paradoxically increased by promoter methylation (Guilleret and Benhattar, 2004). Although the exact mechanism underlying CpG DNA methylation in TERT upregulation is not known, one possible mechanism is by inhibiting transcriptional repressors such as CTCF (Renaud et al., 2007), SIN3A, or MAZ (Xu et al., 2013) from binding to the target DNA-binding sites in the region (Figure S1). Also, even when the promoter is largely methylated, a small region of the core promoter upstream of the transcription start site remains unmethylated to allow for the continued transcriptional activity of TERT (Renaud et al., 2007; Zinn et al., 2007).
Individuals with GCN are at increased risk for developing melanoma (Figures 2a-b and 2d-e) that occurs most frequently in the first decade of life (Bittencourt et al., 2000). A much more frequent change in these nevi than melanoma is the development of clonal proliferations often in the form of nodules (Figures 2g-h and 2j-k), which may suggest or mimic melanoma on clinical or histological grounds (Yelamos et al., 2014). The differential pattern for TERT promoter methylation and telomerase expression between melanomas in GCN and proliferative nodules demonstrated in our study is consistent with the benign clinical course of proliferative nodules compared with the invariably aggressive behavior of melanoma arising in GCN. Further studies in a larger number of patients are needed to determine the potential diagnostic value of TERT promoter methylation assays for ambiguous proliferative lesions within GCN.
In our cohort, TERT promoter hypermethylation or promoter mutations occurred in all melanoma samples but in none of the the benign or low-grade melanocytic lesions, suggesting that a panel incorporating TERT promoter methylation and mutation assays may help discriminate between benign/borderline and overtly malignant melanocytic neoplasms. Future studies need to assess the potential use of these assays for diagnostic or prognostic purposes in the clinic.
In summary, we demonstrate that in subsets of malignant melanoma, TERT is upregulated epigenetically by a methylation-dependent mechanism. These findings have potential therapeutic implications. TERT promoter CpG hypermethylation is a reversible phenomenon. Treatment with DNA demethylating agents reduced TERT expression and telomerase activity in telomerase-positive cell lines (Guilleret and Benhattar, 2003; Renaud et al., 2007). Together, these findings provide a rationale for developing a therapeutic strategy in preclinical studies through epigenetic modifications at TERT promoter regulatory sites in melanomas with the CpG island methylator phenotype.
Supplementary Material
Acknowledgments
Funding: This research was supported in part by the National Cancer Institute of the National Institutes of Health under Award Number P30CA021765 and by ALSAC.
Footnotes
Competing Interests: The authors declare that they have no competing interests.
References
- Bell RJ, Rube HT, Kreig A, et al. Cancer. The transcription factor GABP selectively binds and activates the mutant TERT promoter in cancer. Science. 2015;348:1036–9. doi: 10.1126/science.aab0015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bittencourt FV, Marghoob AA, Kopf AW, et al. Large congenital melanocytic nevi and the risk for development of malignant melanoma and neurocutaneous melanocytosis. Pediatrics. 2000;106:736–41. doi: 10.1542/peds.106.4.736. [DOI] [PubMed] [Google Scholar]
- Blackburn EH. Structure and function of telomeres. Nature. 1991;350:569–73. doi: 10.1038/350569a0. [DOI] [PubMed] [Google Scholar]
- Castelo-Branco P, Choufani S, Mack S, et al. Methylation of the TERT promoter and risk stratification of childhood brain tumours: an integrative genomic and molecular study. Lancet Oncol. 2013;14:534–42. doi: 10.1016/S1470-2045(13)70110-4. [DOI] [PubMed] [Google Scholar]
- Guilleret I, Benhattar J. Demethylation of the human telomerase catalytic subunit (hTERT) gene promoter reduced hTERT expression and telomerase activity and shortened telomeres. Exp Cell Res. 2003;289:326–34. doi: 10.1016/s0014-4827(03)00281-7. [DOI] [PubMed] [Google Scholar]
- Guilleret I, Benhattar J. Unusual distribution of DNA methylation within the hTERT CpG island in tissues and cell lines. Biochem Biophys Res Commun. 2004;325:1037–43. doi: 10.1016/j.bbrc.2004.10.137. [DOI] [PubMed] [Google Scholar]
- Horn S, Figl A, Rachakonda PS, et al. TERT promoter mutations in familial and sporadic melanoma. Science. 2013;339:959–61. doi: 10.1126/science.1230062. [DOI] [PubMed] [Google Scholar]
- Kim NW, Piatyszek MA, Prowse KR, et al. Specific association of human telomerase activity with immortal cells and cancer. Science. 1994;266:2011–5. doi: 10.1126/science.7605428. [DOI] [PubMed] [Google Scholar]
- Lee S, Barnhill RL, Dummer R, et al. TERT Promoter Mutations Are Predictive of Aggressive Clinical Behavior in Patients with Spitzoid Melanocytic Neoplasms. Sci Rep. 2015;5:11200. doi: 10.1038/srep11200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu C, Zhang J, Nagahawatte P, et al. The genomic landscape of childhood and adolescent melanoma. J Invest Dermatol. 2015;135:816–23. doi: 10.1038/jid.2014.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renaud S, Loukinov D, Abdullaev Z, et al. Dual role of DNA methylation inside and outside of CTCF-binding regions in the transcriptional regulation of the telomerase hTERT gene. Nucleic Acids Res. 2007;35:1245–56. doi: 10.1093/nar/gkl1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinagre J, Almeida A, Populo H, et al. Frequency of TERT promoter mutations in human cancers. Nature communications. 2013;4:2185. doi: 10.1038/ncomms3185. [DOI] [PubMed] [Google Scholar]
- Xu M, Katzenellenbogen RA, Grandori C, et al. An unbiased in vivo screen reveals multiple transcription factors that control HPV E6-regulated hTERT in keratinocytes. Virology. 2013;446:17–24. doi: 10.1016/j.virol.2013.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yelamos O, Arva NC, Obregon R, et al. A Comparative Study of Proliferative Nodules and Lethal Melanomas in Congenital Nevi From Children. Am J Surg Pathol. 2014 doi: 10.1097/PAS.0000000000000351. [DOI] [PubMed] [Google Scholar]
- Zinn RL, Pruitt K, Eguchi S, et al. hTERT is expressed in cancer cell lines despite promoter DNA methylation by preservation of unmethylated DNA and active chromatin around the transcription start site. Cancer Res. 2007;67:194–201. doi: 10.1158/0008-5472.CAN-06-3396. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.