Abstract
Native mass spectrometry (MS) has become an invaluable tool for the characterization of proteins and non-covalent protein complexes under near physiological solution conditions. Here we report the structural characterization of human hemoglobin (Hb), a 64 kDa oxygen-transporting protein complex, by high resolution native top-down mass spectrometry using electrospray ionization (ESI) and a 15-Tesla Fourier transform ion cyclotron resonance (FTICR) mass spectrometer. Native MS preserves the non-covalent interactions between the globin subunits, and electron capture dissociation (ECD) produces fragments directly from the intact Hb complex without dissociating the subunits. Using activated ion ECD, we observe the gradual unfolding process of the Hb complex in the gas phase. Without protein ion activation, the native Hb shows very limited ECD fragmentation from the N-termini, suggesting a tightly packed structure of the native complex and therefore low fragmentation efficiency. Precursor ion activation allows steady increase of N-terminal fragment ions, while the C-terminal fragments remain limited (38 c ions and 4 z ions on the α chain; 36 c ions and 2 z ions on the β chain). This ECD fragmentation pattern suggests that upon activation, the Hb complex starts to unfold from the N-termini of both subunits, whereas the C-terminal regions and therefore the potential regions involved in the subunit binding interactions remain intact. ECD-MS of the Hb dimer show similar fragmentation patterns as the Hb tetramer, providing further evidence for the hypothesized unfolding process of the Hb complex in the gas phase. Native top-down ECD-MS allows efficient probing of the Hb complex structure and the subunit binding interactions in the gas phase. It may provide a fast and effective means to probe the structure of novel protein complexes that are intractable to traditional structural characterization tools.
Keywords: Native mass spectrometry, electron capture dissociation, protein complex, gas phase structure, binding interface, hemoglobin
INTRODUCTION
Protein complexes are ubiquitous and important functional units in the cellular environment. Many biological processes require coordinated interactions of individual proteins to form higher order complex assemblies with distinct compositions and topology, which are crucial for the biological function. These macromolecular assemblies are widely present in nearly all organisms including Saccharomyces cerevisiae,1–4 Drosophila melanogaster,5, 6 Caenorhabditis elegans 7, 8 and Homo sapiens.9–11 It is estimated that each protein in yeast is involved in about eight interactions with other proteins, and the interaction network for human proteins is expected to be even larger.12 Many of these protein interactions and their perturbations are underlying causes for human diseases.13–18 Therefore, elucidating the structure of protein assemblies is an important aspect in understanding their biological functions and the molecular basis of disease.
Structural characterization of proteins is usually carried out by biophysical methods such as X-ray crystallography,19 nuclear magnetic resonance (NMR),20, 21 cryo-electron microscopy (cryoEM), etc.22, 23 However, these methods each have noticeable limitations. X-ray crystallography requires the growth of diffraction grade pure crystals to provide high resolution protein structural information; however, protein crystallization is challenging and requires significant efforts and optimization. It can be difficult to produce high quality mono-crystals for large heterogeneous protein complexes with high degree of conformational flexibility. NMR can provide structural information of proteins directly from the solution, but it has a practical size limit. In addition, these methods usually require relatively large quantities of sample and involve time-consuming procedures. Therefore, a fast and efficient method to probe the higher order structure of protein complexes is desirable.
Recently, mass spectrometry has emerged as an important tool for the study of native protein complexes and non-covalent interactions between biomolecules.24–27 This has benefited greatly from the development of soft ionization techniques such as electrospray ionization (ESI), which allows ionization and transfer of many types of large biomolecules and noncovalently-bound complexes into the gas phase, even for protein assemblies as large as megaDalton-sized viruses.28 The MS-based method has considerable advantages in specificity, sensitivity and speed, and consumes much less samples than biophysical methods. The stoichiometry of protein complexes can usually be confidently determined by the mass measurement. Additionally, when coupled with high resolution instruments, significant structural information can be obtained by direct fragmentation of protein complexes and interpretation of the product ions in a top-down MS format. For example, recent studies by Gross and coworkers showed that by using Fourier transform ion cyclotron resonance (FT-ICR) and electron capture dissociation (ECD), certain structural information, such as the flexible regions of several protein complexes and the non-covalent metal binding sites can be determined.29, 30 Given the growing evidence that tertiary and quaternary structures of proteins can be maintained in the gas phase on the time scale of typical MS experiments,24–31 high resolution native top-down MS may represent an especially valuable tool to probe the higher order structural investigations of protein complexes.
Hemoglobin (Hb) is the major respiratory protein in the red blood cells of vertebrates and mammalian organisms, and it plays a central role in binding and transporting oxygen to support essential energy and metabolic needs for life. Because of its critical physiological functions, hemoglobin has been extensively studied and its structural information has been well characterized over the past several decades.32 Mammalian Hbs are heterotetrameric protein complexes that are composed of two symmetric pairs of dimers consisting of α and β chains with 141 and 146 amino acid residues, respectively (MW α chain 15.1 kDa; β chain 15.9 kDa). The four globin chains are assembled together by non-covalent interactions such as hydrogen bonding, salt bridges and hydrophobic interactions. Additionally, each globin chain binds to a heme molecule in its native state. Although mass spectrometry has been used to study Hb in the past decades,33–40 sequence-specific structural characterization of intact Hb complex has not been reported until very recently.41–43 In this work, we used native human Hb complex as a test bed, given its well characterized structural information. We aim to explore the potential of high resolution native top-down MS as a structural characterization tool for probing the high order structural information of protein assemblies, such as subunit interactions and binding interfaces, and furthermore its application to address novel protein complexes.
EXPERIMENTAL SECTION
Samples and Sample Preparation
Chemicals were purchased from Sigma-Aldrich (St. Louis, MO) unless otherwise noted. All solutions were prepared in Milli-Q water (Millipore Corporation, Billerica, MA). Hb was prepared from blood samples collected from healthy human donors and the procedures were performed according to bioharzardous sample preparation protocols by the NIH. Briefly, human blood (30–40 mL) was collected with heparin anticoagulant from a healthy donor following appropriate institutional protocols. Red blood cells were collected by centrifugation at 700 x g for 10 min at 4 °C. The cells were washed three times with 0.9% NaCl and equilibrated with carbon monoxide (CO) for 5 min. The cells were then collected by centrifugation and lysed by resuspension in five volumes of water, followed by incubation on ice for 30 min. NaCl was added to a final concentration of 0.9%, resulting in aggregation of the membrane fractions into a gelatinous phase, which was removed by centrifugation for 15 min at 9500 x g. The supernatant containing Hb was supplemented with 1 mM EDTA and equilibrated with CO for 5 min. After adjusting the pH to 6.9, the hemolysate was applied to an SP Sepharose Fast Flow column (GE Healthcare) equilibrated with 10 mM NaH2PO4, 1 mM DTT, pH 6.9, and Hb was eluted with 10 mM Tris-HCl, pH 8.5. The fractions containing Hb were pooled, supplemented with 1 mM EDTA, and equilibrated with CO for 5 min. Subsequently, the sample was applied to a Q Sepharose Fast Flow column (GE Healthcare) equilibrated with 10 mM Tris-HCl, pH 8.5. Pure Hb was eluted with 30 mM NaH2PO4, pH 6.9, with a yield of 100 mg of protein/ml of blood. Hb concentrations were determined using Drabkin’s reagent (Sigma).
Top-down FT-ICR Mass Spectrometry
Top-down MS of native human Hb A complexes was performed on an ultrahigh resolution 15-Tesla Bruker SolariX hybrid Qq-FTICR mass spectrometer. Native HbA samples (10 μM) were prepared in 20 mM ammonium acetate buffer (pH 6.8). The protein solutions were electrosprayed via nanoESI glass emitters (Proxeon/Thermo Scientific) at flow rates ranging from 20–50 nL/min. The electrospray source temperature was controlled by source heater and countercurrent nitrogen drying gas flow. The temperature was set between 180–200°C with a counter current gas flow of 2.5 L/min to provide optimal desolvation and heating of the native protein samples while maintaining the integrity of the non-covalent complex. MS experiments were performed in the broadband mode (m/z 600 – 8000) with the following settings: 1 M data size, capillary voltage 1000–1200 V, 0.5 sec source accumulation time, 1 sec ion accumulation time, 0.05 sec ion cooling time, 1 ms time of flight. The skimmer potential was tuned between 35–80 V to provide source activation of the Hb complex for sufficient desolvation.
Precursor ions of a single charge state were isolated by Q1 mass and a mass window size of 10 – 20 m/z. MS/MS experiments were performed with the following settings: 0.01 sec ECD pulse length, 1 V bias, 15 V ECD lens. ECD mass spectra were collected from data averaging of up to 800 scans with a transient length of 1.8 sec.
Data Analysis
MS/MS data were processed with DataAnalyis and Biotools (Bruker Daltonics). Briefly, monoisotopic masses ([M+H]+) were extracted by DataAnalysis software using a modified Thrash algorithm (SNAP ver 2.0, Bruker Daltonics) with the following settings: quality factor threshold 0.5; S/N threshold 2; maximum charge state, ≥ protein precursor charge state. Data were calibrated by internal product ions and further assigned using Biotools based on protein sequences determined by accurate mass measurements. A mass accuracy of 1.5 ppm was used for ECD product ion assignments. The assigned ions were manually confirmed to ensure the quality of the assignments.
RESULTS AND DISCUSSION
High Resolution Native MS of Human Hb
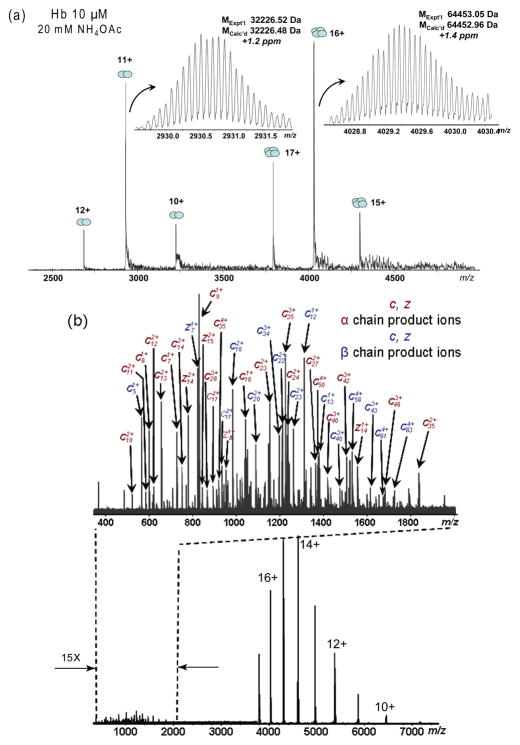
Native MS of human Hb electrosprayed from aqueous solutions (pH 6.8) showed the presence of two ion populations, corresponding to the Hb dimer (32 kDa, charge states 10+ to 12+) and tetramer (64 kDa, charge states 15+ to 17+), respectively (Figure 1(a)). This dimer/tetramer equilibrium is characteristic of the native Hb in the solution, indicating the preservation of the native Hb complex in the gas phase. The narrow charge state distributions of the dimer and tetramer ions suggest compact conformations of the native Hb in the gas phase.38, 39 High resolution, accurate mass measurements show the MW of 32,226.82 and 64,453.79 Da for the dimer and tetramer, respectively (from the most abundant isotopic masses; 10 ppm and 12 ppm, respectively, from its theoretical values; acquired with a resolving power of 175,000 at m/z 4029), which is consistent with the molecular formula of αhβh and (αhβh)2 (where α and β stand for the respective subunit globin chains, UniProtKB P69905 (HBA_HUMAN) and P68871 (HBB_HUMAN); h stands for the non-covalently bound heme, protoporphyrin IX (C34H32O4N4Fe)). Accurate mass measurements reveal that the Hb molecules are completely desolvated in the gas phase, and no apparent post-translational modifications are found. This is consistent with the Hb sample source from a healthy adult donor where the predominant form is Hemoglobin A (α2β2). The MS signal intensities indicate that the Hb complex are present at approximately 70% in the tetramer form and 30% in the dimer form, considering the ionization efficiency differences between the tetramer and dimer. This is consistent with the native MS results obtained on a quadrupole time-of-flight (QTOF) instrument in our laboratory42, 44 and those reported by other groups.33, 35, 40, 41
Figure 1.
Native top-down ECD MS of human hemoglobin (Hb). (a) ESI-FT-ICR mass spectrum of native human hemoglobin (10 μM Hb in 20 mM NH4OAc, pH 6.8); (b) ECD mass spectrum of Hb tetramers, showing extensive c/z•-product ions (m/z 400–2000) and charged-reduced precursors (m/z 3700–8000).
Activated Ion ECD Reveals the Hb Complex Unfolding in the Gas Phase
To obtain structural information on the Hb complex, top-down MS was performed on the Hb tetramer and dimer precursor ions. Collisionally activated dissociation (CAD) causes the Hb complex to simply dissociate into individual subunits and therefore cannot yield structurally useful information. This is observed in our experiments and was previously reported by other groups as well.36, 45 Alternatively, ECD represents a preferred method for the fragmentation of native protein complexes, owing to its ability to generate backbone fragmentations while preserving labile modifications46, 47 and non-covalent interactions.26, 27, 29, 30, 48 Figure 1(b) shows an ECD spectrum for the Hb tetramers (capillary voltage 1000–1300 V, source temperature 200 °C, funnel skimmer voltage 80 V). Precursor ions from all three charge states were selected for fragmentation to improve the product ion yields. Extensive fragmentations are observed from both subunit chains, while the Hb complex remains intact.
Despite its advantages, ECD of native protein complexes has significant technical challenges. First, a native protein carries significantly less charges than its denatured forms, and the lack of charge can reduce the fragmentation efficiency severely. Denatured Hb monomers have an average charge state of greater than 20+, whereas the native Hb tetramer only carries 15 to 17 charges. Because the electron capture cross section scales with the square of the charge of the protein ions, the ECD efficiency of the Hb monomer is about twice as high as the Hb tetramer complex. Secondly, native protein complexes adopt tight globular conformations through strong inter- and intramolecular interactions, which may prevent the fragment ions from releasing and being detected.49 This effect is more significant for ECD of native protein complexes than for denatured proteins.
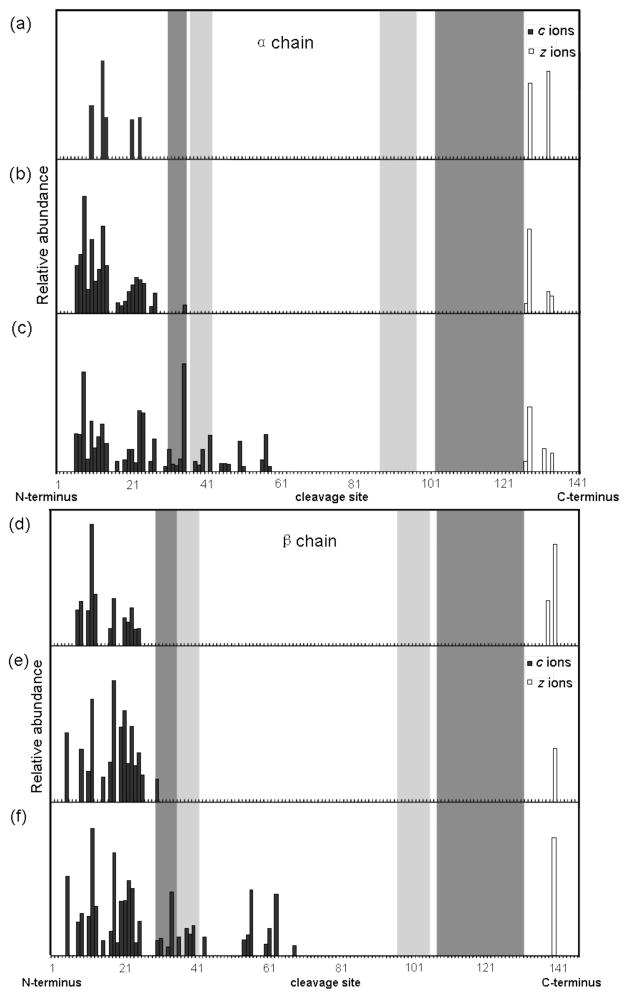
Figure 2(a) and (d) show the ECD product ions from the native Hb tetramer under normal ECD conditions. Without ion activation, only 5 c ions and 2 z• ions are observed from the α chain subunit, and 12 c ions and 2 z• ions are observed from the β chain subunit. Electron capture may induce more N-Cα bond cleavages on the subunit protein backbones, however these fragment intermediates and precursor complexes are held together by strong non-covalent interactions that prevent the fragment ions from being separated and observed. To overcome this problem, infrared irradiation of the protein ions prior to ECD in the ICR cell has been performed.49, 50 Additional ion activation disrupts the intramolecular non-covalent bonds within the gaseous protein ion’s secondary and tertiary structure to significantly increase the number of product ions observed from a protein. Here we activate the ions by increasing the ion source funnel skimmer (FS) potential, similar to the method used by Gross and coworkers.30 The increased FS potential induces collisions with the source gas to deposit internal energy to the protein ions. Figure 2(b) and (e) show the ECD fragmentation of the Hb tetramer under an elevated FS potential (60 V). Significantly more fragments ions are observed from both the α chain subunit (20 c ions, 4 z• ions) and the β chain subunit (15 c ions, 1 z• ions). A maximum of 36 c ions and 4 z• ions from the α chain, 22 c ions and 2 z• ions from β chain can be observed from ECD of the intact Hb complex, by increasing the FS energy to 80 V (Figure 2(c) and (f)). However, further increase of the FS potential causes the Hb complex to start dissociating and lose subunits.
Figure 2.
Top-down ECD analysis of human Hb complex tetramers. Sequence cleavage map of the α chain: (a) funnel skimmer (FS) potential 30 V; (b) 60 V; (c) 80 V. Sequence cleavage map of the β chain: (d) FS 30 V; (b) 60 V; (c) 80 V. Source temperature was set at 200 °C, ESI capillary voltage 1000–1300 V. Dark shaded bars denote the backbone sequences involved in the dimer interface interactions; light shade bars denote sequences involved in the tetramer interface interactions.
Comparison of the activated ion ECD data shows an interesting pattern from the intact Hb complex fragmentation. Despite increased product ion formation upon the Hb activation, the increased fragmentation is mainly observed from the N-terminal sequences of both subunit chains, whereas the C-terminal fragmentation remains very limited. This suggests that upon ion activation, the non-covalent interactions near the N-termini of the Hb complex are being disrupted and the Hb complex starts to unfold from the N-termini of both subunits. The unfolding process proceeds as the internal energy of the Hb complex increases and extends further into both subunits, while the C-terminal sequences of both subunits remain tightly associated by the intermolecular interactions that holds the Hb complex intact before its dissociation by higher energy collisional activation.
Given the well characterized structure of the Hb complex,51 it is of interest to correlate the ECD fragmentation patterns of the Hb complex with its structural elements such as the regions involved in the subunit interactions. Figure 2 highlights the four main subunit interfaces in the Hb tetramer. The light shaded bars indicate the tetramer interfaces between the two identical dimers α1β1 and α2β2, corresponding to Pro37-Tyr42, Ala88-Asn97 on the α chain, and Pro36-Phe41, His97-Leu105 on the β chain. The dark shaded bars indicate the dimer interfaces within the two dimers, corresponding to Arg31-Ser35, His103-Asp126 on the α chain, and Arg30-Tyr35, Asn108-Gln131 on the β chain. The quaternary structure of the Hb complex is formed by the tight association of α and β chain subunits into two pairs of identical dimers, and two dimers are further assembled into the Hb tetramer in a dynamic equilibrium. Therefore the non-covalent interactions involved in the dimer interfaces are stronger than those in the tetramer interfaces. Consistent with these structural features, when subjected to ECD without ion activation, only limited fragment ions are observed from residues outside the complex interfaces near N- and C-termini of both subunits, indicating an undisrupted near native state structure (Figure 2(a) and (d)). When subjected to ion activation, the N-terminal sequences that are not involved in subunit interactions start to unfold, giving rise to more fragment ions near the N-terminal ends of both subunits. Note that a few fragment ions start to be observed from the interface residues near the N-termini, suggesting these two shorter interfaces start to unravel as well, probably due to the more hydrophobic nature and therefore weaker interactions (Figure 2(b) and (e)). Higher activation energy causes the N-terminal interfaces to completely unravel and correspondingly extensive fragmentation from the unfolded N-terminal regions of both subunits results, whereas the C-terminal sequences of both subunits remain tightly locked by the stronger binding interfaces near the C-termini. As a result, only very few fragment ions are observed near the C-termini (Figure 2(c) and (f)).
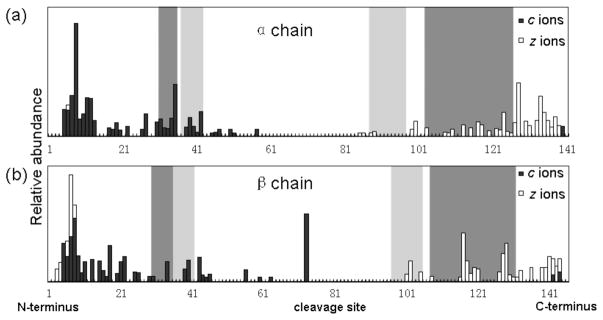
This hypothesis can be further confirmed by ECD fragmentation of denatured Hb monomers. Upon denaturing of the Hb complex, the Hb tetramer is dissociated into α and β subunit monomers and the binding interfaces are completely eliminated. As a result, extensive fragmentation is observed from both N- and C-termini of the monomers (Figure 3).
Figure 3.
Top-down ECD analysis of denatured Hb monomers in 50 % ACN, 0.1 % formic acid. (a) cleavage map of the α chain; (b) cleavage map of the β chain. Dark and light shaded bars denote the dimer and tetramer interfaces, respectively.
ECD of the Hb Dimer
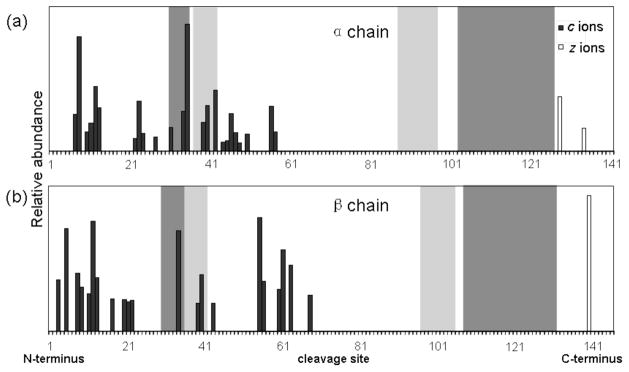
To obtain structural information for the Hb dimer, the dimer precursor ions are selected by the quadrupole and subjected to ECD fragmentation in the ICR cell. Figure 4 shows the ECD data of the Hb dimer with 80 V activation. The fragmentation pattern is similar to that of the Hb tetramer in that fragment ions are mainly produced from the N-terminal sequences of the subunits and C-terminal fragments are very limited. This is consistent with the fact that the Hb tetramer is formed from a dynamic equilibrium of two dimers; the Hb tetramer and dimer have very similar molecular shapes and identical dimer interfaces, except that the tetramer interfaces are not present in the dimer. Intuitively, one would expect more fragment ions from the dimer, given the exposed tetramer interface residues. However, the difference is not significant, possibly because of two factors. First, the reduced charge states of the dimer precursor ions, compared to those of the tetramer, reduce the electron capture efficiency of the dimer and therefore the fragmentation efficiency. Secondly, the tetramer interfaces are relatively smaller compared to the main dimer interface near the C-termini, and furthermore the tetramer interfaces are confined by the dimer interfaces along the subunit backbone. Therefore, upon activated ion ECD, the Hb dimer undergoes a similar unfolding process as the tetramer, giving rise to a generally similar fragmentation pattern that is characteristic of the native Hb structure. This is consistent with the general structural features of the Hb dimer according to the crystallographic structure.
Figure 4.
Top-down ECD analysis of human Hb complex dimers (FS potential 80 V). Backbone cleavage map of the α chain (a) and the β chain (b). Dark and light shaded bars denote the dimer and tetramer interfaces, respectively.
ECD has become a preferred method for characterizing labile post-translational modifications in top-down MS. However, mapping of protein non-covalent interactions directly by ECD is significantly more challenging. Protein complexes associated by weak non-covalent interactions often dissociate in the gas phase, before the protein primary structure is interrogated by ECD to provide the information concerning the binding sites or interface. For non-covalent protein complexes involving smaller ligands such as metals and peptides, the binding sites can be probed more readily by ECD as long as the protein-ligand complexes have sufficient stability in the gas phase.25, 48, 52, 53 Localizing protein-protein complexes binding interfaces by ECD requires a fine balance between maintaining the native structure of the protein complex and providing enough energy to facilitate fragmentation. In the Hb complex, ECD without protein ion activation only produces ions at the N- and C-termini of both subunits, suggesting the remaining part of the primary sequences are bound by non-covalent interactions and therefore intractable to fragmentation. Increasing the activation energy causes the interfaces formed by weaker interactions to dissociate and the Hb complex to unfold from the N-terminal sequences of both subunits, resulting in more fragment ions observed from the N-termini. At a high activation energy, the unfolding further extends and the fragmentation pattern provides a reference for the main interface of the Hb complex. From both the Hb tetramer and dimer ECD data, it can be deduced that the primary sequences between Gly59 and Asp126 on the α chain, Gly69 and Gln127 on the β chain are likely involved in the binding interfaces. This result is in general agreement with the characterized structure of the Hb complex by the crystallography results.32, 51
Correlation of ECD Fragmentation to the Hb Crystallographic Structure
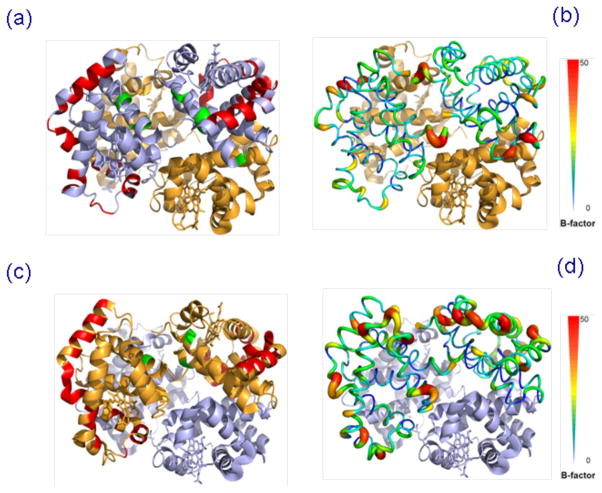
X-ray crystallography has remained a standard technique for protein structural characterization, and the complementary nature of MS as a structural biology tool has been well reviewed.54 Therefore, it is useful to draw a comparison with the crystallographic data to evaluate the potential utility of native protein ECD as a proxy technique for the high resolution structural method. Previously, Zhang et al reported correlations between the B-factors of several protein complexes and their native ECD MS data, and concluded that ECD fragmentation preferentially occurred on the flexible regions of protein crystal structures.29, 30 However, the authors did not provide the physical basis of such correlations. One explanation could be a possible correlation between the flexible regions and the surface residues of the protein structure, as the residues on the exterior of the protein structure are less restrained by the intra- and intermolecular bonds and therefore tend to have higher flexibility. However, this correlation is not apparent in the native Hb ECD data in our study. Figure 5 shows the ECD fragment ions highlighted on the Hb crystal structure, in comparison to the flexibility of the structure as indicated by the B-factor. It is apparent that some residues colored as high flexibility regions did not yield fragmentation, whereas certain regions with lower flexibility showed fragmentation. A recent work by Gross and coworkers on other proteins such as ubiquitin,55 as well as ECD of aldolase in our group56 showed poorer correlation with the B-factor. Therefore the correlations between ECD fragmentation and crystallographic structural flexibility do not seem to be converging as a general rule, but it is worth further investigation with more examples in the future to better define the correlation between solution phase and gas phase structures.
Figure 5.
Tetrameric Hb crystal structure (PDB ID: 1HHO) with (a) color highlights of ECD product ions on the α globin chain; (b) the B-factor putty representation of the α globin chain; (c) color highlights of ECD product ions on the β globin chain; (d) the B-factor putty representation of the β globin chain.
CONCLUSIONS
We performed a structural investigation of the human Hb complex by native top-down ECD MS. ECD showed the capability to generate fragment ions directly from the intact Hb complex without disrupting the intermolecular non-covalent interactions between globin subunits. By applying activated ion ECD fragmentation, the Hb complex structure can be gradually probed to yield information concerning the non-covalent binding interactions between the subunits. Specifically, protein ion activation disrupted the weaker binding interactions to induce the Hb complex structural unfolding from the N-termini of both subunits. This allowed the main binding interaction interfaces to be localized on the primary structures of the α and β subunits. Overall, our studies suggest that high resolution native top-down ECD MS may provide a fast and efficient tool to probe the higher order structure of protein complexes and may be applied to the structural investigation of novel protein complexes not amenable for conventional structural characterization methods.
Acknowledgments
Support from the US National Institutes of Health (R01 GM103479 and S10 RR028893 to J.A.L; R01 AI52217 to R.T.C) and the US Department of Energy (UCLA Institute of Genomics and Proteomics; DE-FC03-02ER63421) are acknowledged. We are grateful for additional helpful comments from Dr. Huilin Li and Piriya Wongkongkathep (UCLA).
References
- 1.Gavin AC, Aloy P, Grandi P, Krause R, Boesche M, Marzioch M, Rau C, Jensen LJ, Bastuck S, Dumpelfeld B, Edelmann A, Heurtier MA, Hoffman V, Hoefert C, Klein K, Hudak M, Michon AM, Schelder M, Schirle M, Remor M, Rudi T, Hooper S, Bauer A, Bouwmeester T, Casari G, Drewes G, Neubauer G, Rick JM, Kuster B, Bork P, Russell RB, Superti-Furga G. Proteome survey reveals modularity of the yeast cell machinery. Nature. 2006;440:631–636. doi: 10.1038/nature04532. [DOI] [PubMed] [Google Scholar]
- 2.Ho Y, Gruhler A, Heilbut A, Bader GD, Moore L, Adams SL, Millar A, Taylor P, Bennett K, Boutilier K, Yang LY, Wolting C, Donaldson I, Schandorff S, Shewnarane J, Vo M, Taggart J, Goudreault M, Muskat B, Alfarano C, Dewar D, Lin Z, Michalickova K, Willems AR, Sassi H, Nielsen PA, Rasmussen KJ, Andersen JR, Johansen LE, Hansen LH, Jespersen H, Podtelejnikov A, Nielsen E, Crawford J, Poulsen V, Sorensen BD, Matthiesen J, Hendrickson RC, Gleeson F, Pawson T, Moran MF, Durocher D, Mann M, Hogue CWV, Figeys D, Tyers M. Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature. 2002;415:180–183. doi: 10.1038/415180a. [DOI] [PubMed] [Google Scholar]
- 3.Krogan NJ, Cagney G, Yu HY, Zhong GQ, Guo XH, Ignatchenko A, Li J, Pu SY, Datta N, Tikuisis AP, Punna T, Peregrin-Alvarez JM, Shales M, Zhang X, Davey M, Robinson MD, Paccanaro A, Bray JE, Sheung A, Beattie B, Richards DP, Canadien V, Lalev A, Mena F, Wong P, Starostine A, Canete MM, Vlasblom J, Wu S, Orsi C, Collins SR, Chandran S, Haw R, Rilstone JJ, Gandi K, Thompson NJ, Musso G, St Onge P, Ghanny S, Lam MHY, Butland G, Altaf-Ui AM, Kanaya S, Shilatifard A, O’Shea E, Weissman JS, Ingles CJ, Hughes TR, Parkinson J, Gerstein M, Wodak SJ, Emili A, Greenblatt JF. Global landscape of protein complexes in the yeast Saccharomyces cerevisiae. Nature. 2006;440:637–643. doi: 10.1038/nature04670. [DOI] [PubMed] [Google Scholar]
- 4.Yu H, Braun P, Yildirim MA, Lemmens I, Venkatesan K, Sahalie J, Hirozane-Kishikawa T, Gebreab F, Li N, Simonis N, Hao T, Rual JF, Dricot A, Vazquez A, Murray RR, Simon C, Tardivo L, Tam S, Svrzikapa N, Fan C, de Smet AS, Motyl A, Hudson ME, Park J, Xin X, Cusick ME, Moore T, Boone C, Snyder M, Roth FP, Barabasi AL, Tavernier J, Hill DE, Vidal M. High-Quality Binary Protein Interaction Map of the Yeast Interactome Network. Science. 2008;322:104–110. doi: 10.1126/science.1158684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Formstecher E, Aresta S, Collura V, Hamburger A, Meil A, Trehin A, Reverdy C, Betin V, Maire S, Brun C, Jacq B, Arpin M, Bellaiche Y, Bellusci S, Benaroch P, Bornens M, Chanet R, Chavrier P, Delattre O, Doye V, Fehon R, Faye G, Galli T, Girault JA, Goud B, de Gunzburg J, Johannes L, Junier MP, Mirouse V, Mukherjee A, Papadopoulo D, Perez F, Plessis A, Rosse C, Saule S, Stoppa-Lyonnet D, Vincent A, White M, Legrain P, Wojcik J, Camonis J, Daviet L. Protein interaction mapping: A Drosophila case study. Genome Res. 2005;15:376–384. doi: 10.1101/gr.2659105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Giot L, Bader JS, Brouwer C, Chaudhuri A, Kuang B, Li Y, Hao YL, Ooi CE, Godwin B, Vitols E, Vijayadamodar G, Pochart P, Machineni H, Welsh M, Kong Y, Zerhusen B, Malcolm R, Varrone Z, Collis A, Minto M, Burgess S, McDaniel L, Stimpson E, Spriggs F, Williams J, Neurath K, Ioime N, Agee M, Voss E, Furtak K, Renzulli R, Aanensen N, Carrolla S, Bickelhaupt E, Lazovatsky Y, DaSilva A, Zhong J, Stanyon CA, Finley RL, White KP, Braverman M, Jarvie T, Gold S, Leach M, Knight J, Shimkets RA, McKenna MP, Chant J, Rothberg JM. A Protein Interaction Map of Drosophila melanogaster. Science. 2003;302:1727–1736. doi: 10.1126/science.1090289. [DOI] [PubMed] [Google Scholar]
- 7.Li SM, Armstrong CM, Bertin N, Ge H, Milstein S, Boxem M, Vidalain PO, Han JDJ, Chesneau A, Hao T, Goldberg DS, Li N, Martinez M, Rual JF, Lamesch P, Xu L, Tewari M, Wong SL, Zhang LV, Berriz GF, Jacotot L, Vaglio P, Reboul J, Hirozane-Kishikawa T, Li QR, Gabel HW, Elewa A, Baumgartner B, Rose DJ, Yu HY, Bosak S, Sequerra R, Fraser A, Mango SE, Saxton WM, Strome S, van den Heuvel S, Piano F, Vandenhaute J, Sardet C, Gerstein M, Doucette-Stamm L, Gunsalus KC, Harper JW, Cusick ME, Roth FP, Hill DE, Vidal M. A Map of the Interactome Network of the Metazoan C. elegans. Science. 2004;303:540–543. doi: 10.1126/science.1091403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tewari M, Hu PJ, Ahn JS, Ayivi-Guedehoussou N, Vidalain PO, Li SM, Milstein S, Armstrong CM, Boxem M, Butler MD, Busiguina S, Rual JF, Ibarrola N, Chaklos ST, Bertin N, Vaglio P, Edgley ML, King KV, Albert PS, Vandenhaute J, Pandey A, Riddle DL, Ruvkun G, Vidal M. Systematic Interactome Mapping and Genetic Perturbation Analysis of a C. elegans TGF-β Signaling Network. Molecular Cell. 2004;13:469–482. doi: 10.1016/S1097-2765(04)00033-4. [DOI] [PubMed] [Google Scholar]
- 9.Stelzl U, Worm U, Lalowski M, Haenig C, Brembeck FH, Goehler H, Stroedicke M, Zenkner M, Schoenherr A, Koeppen S, Timm J, Mintzlaff S, Abraham C, Bock N, Kietzmann S, Goedde A, Toksoz E, Droege A, Krobitsch S, Korn B, Birchmeier W, Lehrach H, Wanker EE. A Human Protein-Protein Interaction Network: A Resource for Annotating the Proteome. Cell. 2005;122:957–968. doi: 10.1016/j.cell.2005.08.029. [DOI] [PubMed] [Google Scholar]
- 10.Rual JF, Venkatesan K, Hao T, Hirozane-Kishikawa T, Dricot A, Li N, Berriz GF, Gibbons FD, Dreze M, Ayivi-Guedehoussou N, Klitgord N, Simon C, Boxem M, Milstein S, Rosenberg J, Goldberg DS, Zhang LV, Wong SL, Franklin G, Li SM, Albala JS, Lim JH, Fraughton C, Llamosas E, Cevik S, Bex C, Lamesch P, Sikorski RS, Vandenhaute J, Zoghbi HY, Smolyar A, Bosak S, Sequerra R, Doucette-Stamm L, Cusick ME, Hill DE, Roth FP, Vidal M. Towards a proteome-scale map of the human protein–protein interaction network. Nature. 2005;437:1173–1178. doi: 10.1038/nature04209. [DOI] [PubMed] [Google Scholar]
- 11.Gandhi TKB, Zhong J, Mathivanan S, Karthick L, Chandrika KN, Mohan SS, Sharma S, Pinkert S, Nagaraju S, Periaswamy B, Mishra G, Nandakumar K, Shen BY, Deshpande N, Nayak R, Sarker M, Boeke JD, Parmigiani G, Schultz J, Bader JS, Pandey A. Analysis of the human protein interactome and comparison with yeast, worm and fly interaction datasets. Nature Genetics. 2006;38:285–293. doi: 10.1038/ng1747. [DOI] [PubMed] [Google Scholar]
- 12.Schreiber G. The Nitty-Gritty of Protein Interactions. Structure. 2005;13:1737–1738. doi: 10.1016/j.str.2005.11.001. [DOI] [PubMed] [Google Scholar]
- 13.Goehler H, Lalowski M, Stelzl U, Waelter S, Stroedicke M, Worm U, Droege A, Lindenberg KS, Knoblich M, Haenig C, Herbst M, Suopanki J, Scherzinger E, Abraham C, Bauer B, Hasenbank R, Fritzsche A, Ludewig AH, Buessow K, Coleman SH, Gutekunst CA, Landwehrmeyer BG, Lehrach H, Wanker EE. Protein Interaction Network Links GIT1, an Enhancer of Huntingtin Aggregation, to Huntington’s Disease,” M. olecular Cell. 2004;18:853–865. doi: 10.1016/j.molcel.2005.06.031. [DOI] [PubMed] [Google Scholar]
- 14.Lim J, Hao T, Shaw C, Patel AJ, Szabo G, Rual JF, Fisk CJ, Li N, Smolyar A, Hill DE, Barabasi AL, Vidal M, Zoghbi HY. A Protein–Protein Interaction Network for Human Inherited Ataxias and Disorders of Purkinje Cell Degeneration. Cell. 2006;125:801–814. doi: 10.1016/j.cell.2006.03.032. [DOI] [PubMed] [Google Scholar]
- 15.Boutell JM, Thomas P, Neal JW, Weston VJ, Duce J, Harper PS, Jones AL. Aberrant Interactions of Transcriptional Repressor Proteins with the Huntington’s Disease Gene Product, Huntingtin. Human Molecular Genetics. 1999;8:1647–1655. doi: 10.1093/hmg/8.9.1647. [DOI] [PubMed] [Google Scholar]
- 16.Li SH, Li XJ. Huntingtin–protein interactions and the pathogenesis of Huntington’s disease. Trends in Genetics. 2004;20:146–154. doi: 10.1016/j.tig.2004.01.008. [DOI] [PubMed] [Google Scholar]
- 17.Ryan DP, Matthews JM. Protein–protein interactions in human disease. Curr Opin Struct Biol. 2005;15:441–446. doi: 10.1016/j.sbi.2005.06.001. [DOI] [PubMed] [Google Scholar]
- 18.Xia WM, Zhang JM, Perez R, Koo EH, Selkoe DJ. Interaction between amyloid precursor protein and presenilins in mammalian cells: Implications for the pathogenesis of Alzheimer disease. Proc Natl Acad Sci USA. 1997;94:8208–8213. doi: 10.1073/pnas.94.15.8208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ilari A, Savino C. Protein Structure Determination by X-Ray Crystallography Bioinformatics: Data, Sequence Analysis and Evolution. In: Keith JM, editor. Meth Mol Biol. Vol. 452. Human Press; Totowa, NJ: 2008. pp. 63–87. [DOI] [PubMed] [Google Scholar]
- 20.Wuthrich K. Protein structure determination in solution by NMR spectroscopy. J Biol Chem. 1990;265:22059–22062. [PubMed] [Google Scholar]
- 21.Wuthrich K. The way to NMR structures of proteins. Nature Struct Mol Biol. 2001;8:923–925. doi: 10.1038/nsb1101-923. [DOI] [PubMed] [Google Scholar]
- 22.Heymann JB, Conway JF, Steven AC. Molecular dynamics of protein complexes from four-dimensional cryo-electron microscopy. J Struct Biol. 2004;147:291–301. doi: 10.1016/j.jsb.2004.02.006. [DOI] [PubMed] [Google Scholar]
- 23.Zhou ZH. Atomic resolution cryo electron microscopy of macromolecular complexes. In: Pt B, Ludtke SJ, Prasad BVV, editors. Advances in Protein Chemistry and Structural Biology, Vol 82: Recent Advances in Electron Cryomicroscopy. Elsevier; 2011. pp. 1–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Loo JA. Studying noncovalent protein complexes by electrospray ionization mass spectrometry. Mass Spectrom Rev. 1997;16:1–23. doi: 10.1002/(SICI)1098-2787(1997)16:1<1::AID-MAS1>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 25.Loo JA. Electrospray ionization mass spectrometry: a technology for studying noncovalent macromolecular complexes. Int J Mass Spectrom. 2000;200:175–186. doi: 10.1016/S1387-3806(00)00298-0. [DOI] [Google Scholar]
- 26.Xie Y, Zhang J, Yin S, Loo JA. Top-Down ESI-ECD-FT-ICR Mass Spectrometry Localizes Noncovalent Protein-Ligand Binding Sites. J Am Chem Soc. 2006;128:14432–14433. doi: 10.1016/S1387-3806(00)00298-0. [DOI] [PubMed] [Google Scholar]
- 27.Yin S, Loo JA. Elucidating the Site of Protein-ATP Binding by Top-Down Mass Spectrometry. J Am Soc Mass Spectrom. 2010;21:899–907. doi: 10.1016/j.jasms.2010.01.002. [DOI] [PubMed] [Google Scholar]
- 28.Snijder J, Rose RJ, Veesler D, Johnson JE, Heck AJR. Studying 18 MDa Virus Assemblies with Native Mass Spectrometry. Angew Chem Int Ed. 2013;52:4020–4023. doi: 10.1002/anie.201210197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang H, Cui W, Wen J, Blankenship RE, Gross ML. Native Electrospray and Electron-Capture Dissociation in FTICR Mass Spectrometry Provide Top-Down Sequencing of a Protein Component in an Intact Protein Assembly. J Am Soc Mass Spectrom. 2010;21:1966–1968. doi: 10.1016/j.jasms.2010.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang H, Cui W, Wen J, Blankenship RE, Gross ML. Native Electrospray and Electron-Capture Dissociation FTICR Mass Spectrometry for Top-Down Studies of Protein Assemblies. Anal Chem. 2011;83:5598–5606. doi: 10.1021/ac200695d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Breuker K, Brüeschweiler S, Tollinger M. Electrostatic Stabilization of a Native Protein Structure in the Gas Phase. Angew Chem Int Ed. 2011;50:873–877. doi: 10.1002/anie.201005112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Perutz MF. Stereochemistry of Cooperative Effects in Haemoglobin: Haem–Haem Interaction and the Problem of Allostery. Nature. 1970;228:726–739. doi: 10.1038/228726a0. [DOI] [PubMed] [Google Scholar]
- 33.Kuprowski MC, Boys BL, Konermann L. Analysis of Protein Mixtures by Electrospray Mass Spectrometry: Effects of Conformation and Desolvation Behavior on the Signal Intensities of Hemoglobin Subunits. J Am Soc Mass Spectrom. 2007;18:1279–1285. doi: 10.1016/j.jasms.2007.04.002. [DOI] [PubMed] [Google Scholar]
- 34.Liu J, Konermann L. Protein–Protein Binding Affinities in Solution Determined by Electrospray Mass Spectrometry. J Am Soc Mass Spectrom. 2011;22:408–417. doi: 10.1007/s13361-010-0052-1. [DOI] [PubMed] [Google Scholar]
- 35.Griffith WP, Kaltashov IA. Highly Asymmetric Interactions between Globin Chains during Hemoglobin Assembly Revealed by Electrospray Ionization Mass Spectrometry,” B. iochemistry. 2003;42:10024–10033. doi: 10.1021/bi034035y. [DOI] [PubMed] [Google Scholar]
- 36.Versluis C, Heck AJR. Gas-phase dissociation of hemoglobin. Int J Mass Spectrom. 2001;210:637–649. doi: 10.1016/S1387-3806(01)00428-6. [DOI] [Google Scholar]
- 37.Griffith WP, Kaltashov IA. Mass Spectrometry in the Study of Hemoglobin: from Covalent Structure to Higher Order Assembly. Curr Org Chem. 2006;10:535–553. doi: 10.2174/138527206776055303. [DOI] [Google Scholar]
- 38.Katta V, Chait BT. Observation of the heme-globin complex in native myoglobin by electrospray-ionization mass spectrometry. J Am Chem Soc. 1991;113:8534–8535. doi: 10.1021/ja00022a058. [DOI] [Google Scholar]
- 39.Konermann L, Douglas DJ. Unfolding of proteins monitored by electrospray ionization mass spectrometry: a comparison of positive and negative ion modes. J Am Soc Mass Spectrom. 1998;9:1248–1254. doi: 10.1016/S1044-0305(98)00103-2. [DOI] [PubMed] [Google Scholar]
- 40.Boys BL, Konermann L. Folding and Assembly of Hemoglobin Monitored by Electrospray Mass Spectrometry Using an On-line Dialysis System. J Am Soc Mass Spectrom. 2007;18:8–16. doi: 10.1016/j.jasms.2006.08.013. [DOI] [PubMed] [Google Scholar]
- 41.Cui W, Zhang H, Gross ML. Fourier Transform Ion Cyclotron Resonance Mass Spectrometry of Intact Heterogeneous Non-Covalent Protein Complexes. Proceedings of the 59th ASMS Conference on Mass Spectrometry and Allied Topics; Denver, Colorado. June 5–9, 2011. [Google Scholar]
- 42.Zhang J, Malmirchegini GR, Clubb RT, Loo JA. Deciphering the Mechanism and Dynamics of Hemoglobin Capture by Staphylococcus aureus Cell-surface Protein, IsdH, by Native MS. Proceedings of the 60th ASMS Conference on Mass Spectrometry and Allied Topics; Vancouver, Canada. May 20–24, 2012. [Google Scholar]
- 43.Lermyte F, Williams JP, Brown JM, Martin EM, Sobott F. Extensive Charge Reduction and Dissociation of Intact Protein Complexes Following Electron Transfer on a Quadrupole-Ion Mobility-Time-of-Flight MS. J Am Soc Mass Spectrom. 2015 doi: 10.1007/s13361-015-1124-z. in press. [DOI] [PubMed] [Google Scholar]
- 44.Spirig T, Malmirchegini GR, Zhang J, Robson SA, Sjodt M, Liu M, Kumar KK, Dickson CF, Gell DA, Lei B, Loo JA, Clubb RT. Staphylococcus aureus Uses a Novel Multidomain Receptor to Break Apart Human Hemoglobin and Steal Its Heme. J Biol Chem. 2013;288:1065–1078. doi: 10.1074/jbc.M112.419119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Benesch JLP. Collisional Activation of Protein Complexes: Picking Up the Pieces. J Am Soc Mass Spectrom. 2009;20:341–348. doi: 10.1016/j.jasms.2008.11.014. [DOI] [PubMed] [Google Scholar]
- 46.Zhang J, Dong X, Hacker TA, Ge Y. Deciphering Modifications in Swine Cardiac Troponin I by Top-Down High-Resolution Tandem Mass Spectrometry. J Am Soc Mass Spectrom. 2010;21:940–948. doi: 10.1016/j.jasms.2010.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang J, Guy MJ, Norman HS, Chen YC, Xu Q, Dong X, Guner H, Wang S, Kohmoto T, Young KH, Moss RL, Ge Y. Top-Down Quantitative Proteomics Identified Phosphorylation of Cardiac Troponin I as a Candidate Biomarker for Chronic Heart Failure. J Proteome Res. 2011;10:4054–4065. doi: 10.1021/pr200258m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Clarke DJ, Murray E, Hupp T, Mackay CL, Langridge-Smith PRR. Mapping a Noncovalent Protein–Peptide Interface by Top-Down FTICR Mass Spectrometry Using Electron Capture Dissociation. J Am Soc Mass Spectrom. 2011;22:1432–1440. doi: 10.1007/s13361-011-0155-3. [DOI] [PubMed] [Google Scholar]
- 49.Horn DM, Ge Y, McLafferty FW. Activated Ion Electron Capture Dissociation for Mass Spectral Sequencing of Larger (42 kDa) Proteins. Anal Chem. 2000;72:4778–4784. doi: 10.1021/ac000494i. [DOI] [PubMed] [Google Scholar]
- 50.Hakansson K, Chalmers MJ, Quinn JP, McFarland MA, Hendrickson CL, Marshall AG. Combined Electron Capture and Infrared Multiphoton Dissociation for Multistage MS/MS in a Fourier Transform Ion Cyclotron Resonance Mass Spectrometer. Anal Chem. 2003;75:3256–3262. doi: 10.1021/ac030015q. [DOI] [PubMed] [Google Scholar]
- 51.Perutz MF, Rossmann MG, Cullis AF, Muirhead H, Will G, North ACT. Structure of Haemoglobin: A Three-Dimensional Fourier Synthesis at 5.5-Å. Resolution, Obtained by X-Ray Analysis. Nature. 1960;185:416–422. doi: 10.1038/185416a0. [DOI] [PubMed] [Google Scholar]
- 52.Yin S, Loo JA. Top-Down Mass Spectrometry of Supercharged Native Protein-Ligand Complexes. Int J Mass Spectrom. 2011;300:118–122. doi: 10.1016/j.ijms.2010.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li H, Wongkongkathep P, Van Orden SL, Ogorzalek Loo RR, Loo JA. Revealing Ligand Binding Sites and Quantifying Subunit Variants of Non-Covalent Protein Complexes in a Single Native Top-Down FTICR MS Experiment. J Am Soc Mass Spectrom. 2014;25:2060–2068. doi: 10.1007/s13361-014-0928-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sharon M, Robinson CV. The Role of Mass Spectrometry in Structure Elucidation of Dynamic Protein Complexes. Annu Rev Biochem. 2007;76:167–93. doi: 10.1146/annurev.biochem.76.061005.090816. [DOI] [PubMed] [Google Scholar]
- 55.Zhang H, Cui W, Gross ML. Native electrospray ionization and electron-capture dissociation for comparison of protein structure in solution and the gas phase. Int J Mass Spectrom. 2013;354–355:288–291. doi: 10.1016/j.ijms.2013.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Li H, Wolff JJ, Van Orden SL, Loo JA. Native Top-Down Electrospray Ionization-Mass Spectrometry of 158 kDa Protein Complex by High-Resolution Fourier Transform Ion Cyclotron Resonance Mass Spectrometry. Anal Chem. 2014;86:317–320. doi: 10.1021/ac4033214. [DOI] [PMC free article] [PubMed] [Google Scholar]