Abstract
The Complex II family of enzymes, comprising the respiratory succinate dehydrogenases and fumarate reductases, catalyze reversible interconversion of succinate and fumarate. In contrast to the covalent flavin adenine dinucleotide (FAD) cofactor assembled in these enzymes, the soluble fumarate reductases (e.g. that from Shewanella frigidimarina) that assemble a noncovalent FAD cannot catalyze succinate oxidation but retain the ability to reduce fumarate. In this study, an SdhA-H45A variant that eliminates the site of the 8α-N3-histidyl covalent linkage between the protein and the FAD was examined. The variants SdhA-R286A/K/Y and -H242A/Y, that target residues thought to be important for substrate binding and catalysis were also studied. The variants SdhA-H45A and -R286A/K/Y resulted in assembly of a noncovalent FAD cofactor, which led to a significant decrease (−87 mV or more) in its reduction potential. The variant enzymes were studied by electron paramagnetic resonance spectroscopy following stand-alone reduction and potentiometric titrations. The “free” and “occupied” states of the active site were linked to the reduced and oxidized states of the FAD, respectively. Our data allows for a proposed model of succinate oxidation that is consistent with tunnel diode effects observed in the succinate dehydrogenase enzyme and a preference for fumarate reduction catalysis in fumarate reductase homologues that assemble a noncovalent FAD.
Succinate dehydrogenase (SdhCDAB), also known as mitochondrial Complex II and succinate:quinone oxidoreductase (SQR), is an essential Krebs cycle enzyme that couples succinate oxidation to ubiquinone (UQ) reduction. X-ray crystallographic structures of Sdh from Sus domesticus (1), Gallus gallus (2, 3) and Escherichia coli (4, 5), as well as those of the homologous fumarate reductase (Frd) from Wolinella succinogenes (6) and E. coli (7–9), show remarkable conservation of tertiary architecture in the Complex II family of enzymes. Succinate oxidation is initialized by a hydride transfer mechanism that donates 2 electrons to the covalent flavin adenine dinucleotide (FAD) cofactor in SdhA (10). The electrons are then singly funneled through three iron-sulfur clusters in SdhB ([2Fe-2S], [4Fe-4S] and [3Fe-4S]) towards the quinone-binding site where UQ is reduced to ubiquinol (UQH2). The E. coli, avian, and mammalian Sdh enzymes also contain a heme b moiety in the membrane anchor domain that is not required for catalysis (11, 12).
In E. coli, the paralogs Sdh and Frd play key roles in aerobic and anaerobic respiration, respectively. Although Sdh is optimized for succinate oxidation and Frd for fumarate reduction, the overexpressed enzymes can functionally replace each other in vivo (13, 14). In the E. coli paralogs, there is remarkable sequence conservation of residues surrounding the FAD, except for the presence of a conserved Gln (Q50) residue in the Sdh enzymes that is replaced by a conserved Glu (E49) residue in the Frd enzymes, suggesting that directionality of catalysis is partially governed by Coulombic effects (15). Other important determinants of functional directionality include the electrochemical profile of electron transfer through redox cofactors and the type of quinone species preferentially utilized (16). In addition, FAD reduction potentials (Em) may also impact the directionality of the reaction. In the soluble Shewanella frigidimarina fumarate reductase, the noncovalent FAD has an Em,7 value of −152 mV and the enzyme can only catalyze fumarate reduction (17, 18). In E. coli Sdh and Frd, the FAD cofactor is covalently bound to SdhA-H45 and FrdA-H44, respectively, via an 8α-N3-histidyl linkage (19, 20). Variants of E. coli Frd (FrdA-H44Y/C/S/R) and Saccharomyces cerevisiae Sdh (Sdh1-H90S) with non-covalently bound FAD lose succinate dehydrogenase activity but retain fumarate reductase activity (21, 22). In a FrdA-H44S variant, the Em,7 value of the non-covalent FAD was determined to be −134 mV using protein film voltammetry (23). The increased Em,7 values observed in enzymes having a covalently-attached FAD ((−55 mV in Frd and −79 mV in Sdh) (24, 25)) is believed to be a critical determinant of their ability to catalyze succinate oxidation as well as fumarate reduction (26). In vanillyl-alcohol oxidase, covalent attachment elevates the Em value of FAD by ~100 mV and increases the rate of catalysis by 1 order of magnitude (27). These observations support the hypothesis that covalent attachment is responsible for the high Em values reported for the FAD in both Sdh and Frd, which allows these enzymes to catalyze succinate oxidation.
The specific mechanism of covalent FAD attachment to SdhA remains elusive but is believed to be autocatalytic (28–30). Proper folding of the apoprotein and the presence of citric acid cycle intermediates appear to be prerequisites for covalent flavinylation (31). However, recent experiments show that recruitment of the FAD to the apoenzyme is chaperone-mediated. The Sdh5 protein in S. cerevisiae, and subsequently SdhE (previously named YgfY) in bacteria, was the first chaperone identified that is absolutely required for covalent FAD attachment to Sdh and Frd in vivo (32–34). In humans, individuals carrying germline mutations in the SDHAF2 gene (equivalent to SDH5 in yeast) exhibit a loss-of-function phenotype and have a tendency to develop paragangliomas or pheochromocytomas (33).
The dicarboxylate binding site is situated adjacent to the FAD molecule and can bind a range of substrates and inhibitors including succinate, fumarate, oxaloacetate (OAA), malonate, citrate, and 3-nitropropionate (3-NP). A compilation of X-ray crystallographic structures of Sdh with different inhibitors bound reveals a diversity of SdhA-R286 and SdhA-H242 side chain conformations, whereas those of other residues show little or no variability. A study of the W. succinogenes Frd enzyme showed that the positive charge of FrdA-R301 (equivalent to SdhA-R286) is important for catalysis and covalent FAD attachment (35). The E. coli FrdA-H232S variant (equivalent to SdhA-H242) is unable to oxidize succinate but retains the ability to reduce fumarate (36). Herein we examined substrate binding, catalysis, and the importance of covalent flavinylation by studying variants of SdhA-R286, SdhA-H242 and SdhA-H45 (Figure 1). Using redox potentiometry and electron paramagnetic resonance (EPR) spectroscopy, we report a definitive Em value for non-covalent FAD in Sdh. We also show that the EPR spectrum of the [2Fe-2S] cluster is sensitive to both inhibitor/substrate binding and to the redox state of the FAD.
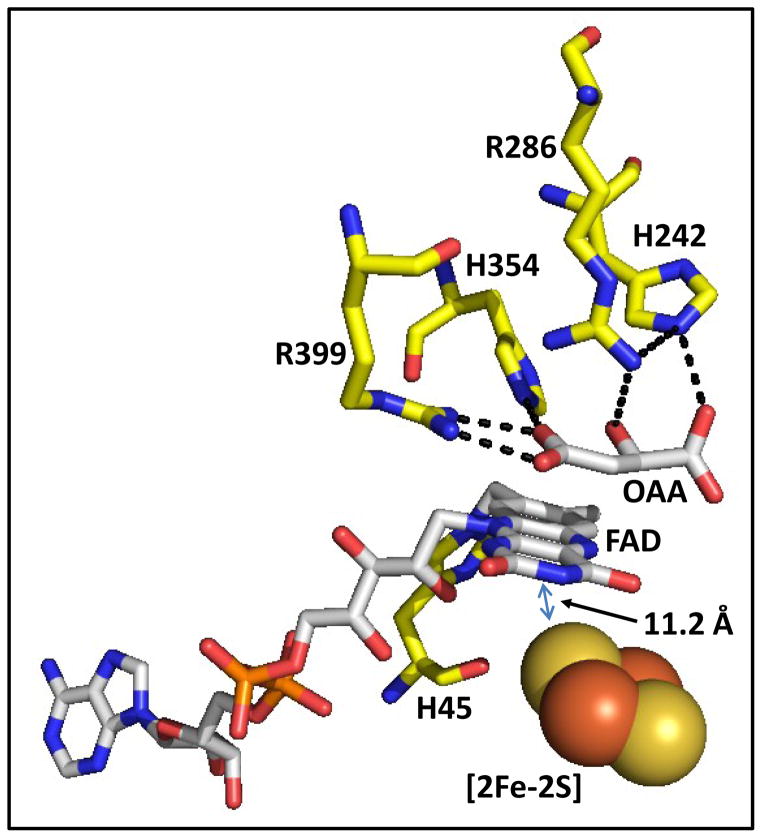
Figure 1. Overview of the dicarboxylic acid binding site in E. coli succinate dehydrogenase (PDB entry 2WDQ) (68).
Residues that H-bond to the co-crystallized inhibitor, oxaloacetate (OAA), are shown. SdhA-H45 is the site of covalent FAD attachment. The distance between the isoalloxazine ring and the [2Fe-2S] cluster located in SdhB subunit is also indicated.
Materials and Methods
Strains, Plasmids and Protein Preparation
E. coli strain TG1 (supE hsdΔ5 thi Δ (lac-proAB) F′ [traD36 proAB+ lacIq lacZΔM15]; GE Healthcare) was used for mutagenesis. E. coli strain DW35 (ΔfrdABCD, sdhC::kan) (37) transformed with pFAS plasmids (14) carrying mutant sdhCDAB constructs was used for protein expression and growth studies. Mutations were constructed using primers from Integrated DNA Technologies and the QuikChange protocol from Stratagene. All recombinant plasmids were verified by DNA sequencing. Variant enzymes, isolated as the major component of the cytoplasmic membrane fraction, were malonate-activated as previously described (11, 38). Additionally, the membranes were subsequently pelleted by centrifugation at 100,000 × g and resuspended in either 100 mM MOPS / 5 mM EDTA (pH 7) or 100 mM tricine / 5 mM EDTA (pH 8) to remove the malonate.
SDS-PAGE, Covalent Flavin Visualization and Flavin Estimation
Protein concentrations were estimated using a modified Lowry method (39) incorporating 1% (w/v) sodium dodecyl sulfate in the incubation mixture to solubilize membrane proteins (40). To analyze enzyme expression and covalent FAD attachment, 30 μg of protein was resolved on a 12 % SDS-PAGE gel followed by visualization using UV fluorescence and Coomassie Blue staining. The unstained gel was washed three times for 2 minutes in H2O followed by incubation in 10 % acetic acid at pH 3 before visualization by UV irradiation. The intensities of the SdhA bands from Coomassie Blue staining and UV fluorescence were quantified using ImageJ (41). Fluorometric quantitation of covalent flavin was also carried out in triplicate as previously described (42). To estimate the relative amounts of non-covalent FAD assembled, 10 μL of 55 % trichloroacetic acid was added to 100 μL of membrane preparation containing approximately 2 μg of protein. After incubation on ice for 15 minutes, the samples were centrifuged at 10,000 × g for 3 minutes and the supernatant fractions were collected. Fluorescence intensities at pH 7.0 and 3.3 were used to calculate the relative amounts of non-covalent FAD in each preparation (42).
Enzyme Assays
Succinate dependent reduction of MTT (2-(4,5-dimethyl-2-thiazolyl)-3,5-diphenyl-2H-tetrazolium bromide; ε = 17 mM−1 cm−1) was measured spectrophotometrically at 570 nm in the presence of 750 μM PMS (phenazine methosulfate) and 0.1 % Triton X-100 as previously described (43). Fumarate dependent oxidation of reduced BV (benzyl viologen, ε = 7.8 mM−1 cm−1) was monitored at 570 nm with excess sodium dithionite present at the beginning of the assay as previously described (44).
Redox Titrations and EPR Spectroscopy
Redox titrations were carried out anaerobically under argon at 25 °C on SdhCDAB-enriched membranes in 100 mM MOPS / 5 mM EDTA (pH 7). The following redox mediators were used at a concentration of 25 μM: 2,6-dichloroindophenol, toluylene blue, phenazine methosulfate, thionine, methylene blue, resorufin, indigotrisulfonate, indigocarmine, phenosafranine, and neutral red. EPR spectra were recorded using a Bruker Elexsys E500 spectrometer. For studies on the [2Fe-2S] cluster, the sample cavity was cooled to 40 K using an Oxford Instruments ESR900 flowing helium cryostat. For studies on the flavosemiquinone and ubisemiquinone radicals, a Bruker liquid nitrogen-evaporating cryostat (Bruker ER4111 VT variable temperature unit) was used to maintain the temperature at 150 K. In both cases, a microwave power of 20 mW, a microwave frequency of 9.38 GHz, and a modulation frequency of 100 kHz were used. Modulation amplitudes of 10 Gpp and 1 Gpp were used for spectra of the [2Fe-2S] cluster and flavosemiquinone, respectively. For preparation of EPR samples, dithionite was used at a final concentration of 2.5 mM, succinate and OAA. Incubations were carried at at 23 °C anaerobically under an argon atmosphere. Where appropriate, incubations with succinate and OAA were carried out for 4 minutes prior to dithionite addition for a subsequent 2 minutes.
Results
Expression and flavinylation of Sdh variants
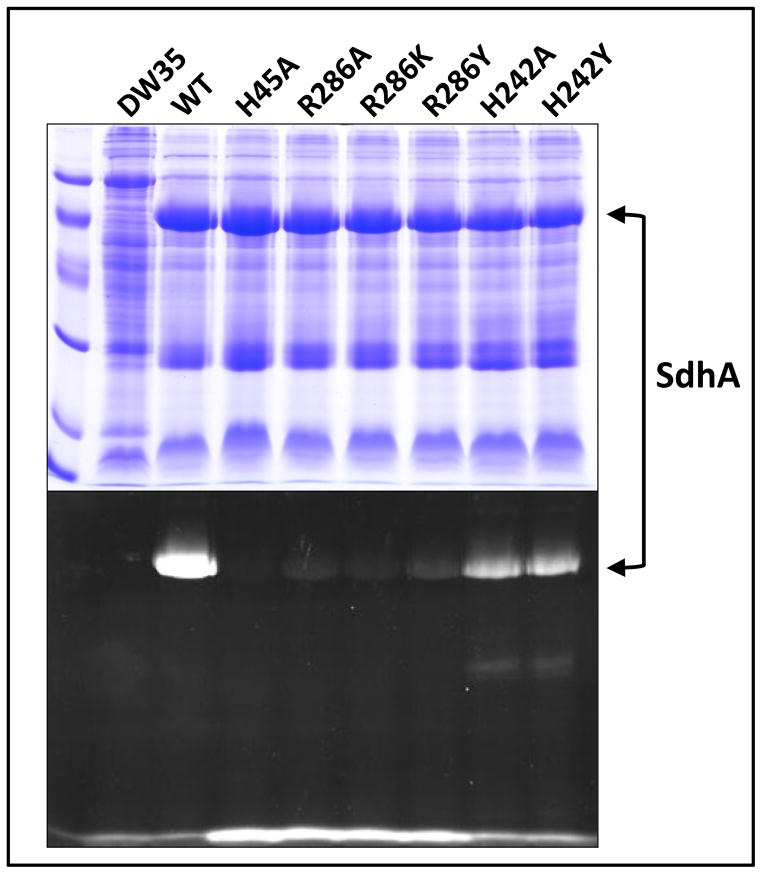
Using gel electrophoresis, we confirmed that all over-expressed variant enzymes assembled correctly to the cytoplasmic membrane (Figure 2A). The presence of covalently bound FAD was evaluated by UV trans-illumination of SDS-PAGE protein gels prior to Coomassie Blue staining (Figure 2B). Protein gels containing the wild-type enzyme show intensely fluorescence bands corresponding to the position of the SdhA subunit, whereas virtually no signal is observed for the SdhA-H45A variant, an enzyme predicted to lack covalent FAD based on studies of variants of E.coli Frd and S. cerevisiae Sdh (21, 22). Although the SdhA polypeptides in the SdhA-R286A/K/Y variants are clearly visible following staining with Coomassie Blue, their UV fluorescence intensities are negligible compared to that of the wild-type enzyme. The SdhA-H242A/Y variants both exhibit significant fluorescence at the position corresponding to SdhA, as well as a faint signal from a proteolytic fragment that migrates at a lower molecular weight. In the SdhA-H45A and SdhA-R286A/K/Y variant enzymes which do not assemble covalent FAD, intense fluorescence signals are observed at the dye front, which we interpret as arising from labile non-covalent FAD. Table 1 shows quantitation of the SdhA band observed in the Coomassie Blue stained gel presented in Figure 2, as well as relative quantitation of covalent and non-covalent FAD by fluorimetry.
Figure 2. Enzyme assembly and covalent FAD incorporation.
Proteins were separated on a 12 % SDS-PAGE gel and visualized by either Coomassie Blue staining (top) or UV excitation (bottom). Samples from left to right: low molecular weight standards, followed by enriched membrane preparations containing no Sdh (E. coli DW35), wild-type Sdh (WT), SdhA-H45A, -R286A, -R286K, -R286Y, -H242A and -H242Y enzymes.
Table 1.
Assembly and enzyme activity of variant enzymes.
| SdhA1 | Covalent FAD2 | Non-covalent FAD2 | Succinate:PMS/MTT3 | BV:Fumarate3 | |
|---|---|---|---|---|---|
| DW35 | 0 | 0 | 0 | 0.13 | 0.07 |
| SdhCDAB | 100 | 100 | 0 | 2.40 | 2.48 |
| H45A | 103 | 0 | 100 | 0.52 | 1.03 |
| R286A | 96 | 3 | 93 | 0.19 | 0.11 |
| R286K | 94 | 2 | 85 | 0.12 | 0.07 |
| R286Y | 84 | 4 | 77 | 0.10 | 0.07 |
| H242A | 84 | 32 | 18 | 0.13 | 1.27 |
| H242Y | 74 | 31 | 15 | 0.10 | 0.15 |
Relative amounts of SdhA Coomassie Blue staining were quantified from Figure 1 using ImageJ software.
Amounts of covalent and non-covalent FAD (% relative to SdhCDAB and SdhA-H45A, respectively) were quantified by fluorimetry. Standard error: < 5 % of reported values.
Succinate:PMS/MTT and BV:fumarate activities are reported in μmol min−1 mg−1. Standard error: ±0.05 μmol min−1 mg−1.
As described above, the SdhA-H45A variant has no detectable covalent FAD, whereas the SdhA-R286 variants each contain less than 4 % covalent FAD. TCA precipitation results in the release of non-covalent FAD from the SdhA-H45A and SdhA-R286A/K/Y variants. In the SdhA-H242A and SdhA-H242Y variants, approximately 30 % of assembled enzyme contains covalently-bound FAD, while the remainder (approximately 15–18%) is assembled in the non-covalent form. The data also suggest that a significant amount of the SdhA-H242A and SdhA-H242Y variant enzymes are assembled with no FAD. Overall, these results suggest that SdhA-R286 plays an essential role in the mechanism of covalent flavinylation, and that SdhA-H242 plays a role in its efficiency and completeness.
Impact of flavinylation on enzyme catalysis
In vitro succinate:PMS/MTT and BV:fumarate assays were used to measure rates of succinate oxidation and fumarate reduction at the dicarboxylate binding site, respectively. All substitutions at SdhA-R286 and SdhA-H242, with the exception of the SdhA-H242A variant, essentially abolish succinate oxidase and fumarate reductase activities (Table 1). The SdhA-H242A variant enzyme retains ~50 % of BV:fumarate activity despite showing negligible succinate:PMS/MTT activity compared to the wild-type enzyme. This is similar to the results obtained for a FrdA-H232S variant (36). The SdhA-H45A variant with non-covalent FAD shows depressed rates of succinate/fumarate interconversion, with succinate oxidation affected (79 % decrease) to a larger degree than fumarate reduction (60 % decrease). This observation is consistent with flavinylation increasing the FAD Em,7 and rendering it more likely to participate in succinate oxidation.
Em,7 of non-covalent FAD and the [2Fe-2S] cluster in wild-type and variant enzymes
Although it is widely assumed that the Em value of a non-covalent FAD is lower than its covalent form, this has never been directly demonstrated in E. coli Sdh. We addressed this question using redox potentiometry followed by EPR spectroscopy. In the wild-type enzyme, the covalent FAD titrates with an Em,7 value of −100 mV (Table 2). In the SdhA-H45A variant, titration of the non-covalent FAD yields an Em,7 value of −187 mV, a decrease of 87 mV compared to the covalent FAD in the wild-type enzyme. Titrations of the non-covalent FAD cofactor in the SdhA-R286A/K/Y variants also yield Em,7 values that are significantly lower than that of the wild-type enzyme (Table 2). Specifically, the Em,7 values of the non-covalent FAD in SdhA-R286K and SdhA-R286Y is ~160 mV lower than that of the covalent FAD in wild-type Sdh. Titrations of the SdhA-H242A and SdhA-H242Y variants yield apparent FAD Em,7 values of −202 mV and −207 mV, respectively (Table 2). However, the intensities of the respective EPR signals in potentiometric titrations are much lower than those obtained for wild-type Sdh (Supplemental Figure 1), suggesting that the Em,7 being measured is that of the small amount of non-covalent FAD found in the SdhA-H242 variants, with the proportion of covalent flavosemiquinone in these enzymes likely being thermodynamically unstable, and thus essentially EPR invisible. Flavosemiquinone instability also likely explains the low signal intensities observed in potentiometric titrations of the SdhA-R286A variant. In order to gauge the quality of our redox titration data, the Em values of the ubisemiquinone species were examined as an internal control. In all the variant enzymes studied, the appearance of the ubisemiquinone species was aligned at an almost identical potential, indicating good reproducibility of conditions amongst the titrations (Supplemental Figure 1). Overall, our data demonstrate that in Sdh non-covalent FAD has a lower Em,7 than the covalently-bound form.
Table 2. Midpoint potentials of FAD, [2Fe-2S] cluster, and the occupied/free transition at pH 7.
200 μL samples poised at varying reduction potentials were frozen with liquid nitrogen-chilled ethanol and analyzed by EPR spectroscopy. The 2-electron FAD→FADH− transition gave rise to a transient radical species at g = 2.00 that was fitted according to Hastings et al. (67). The gz = 2.03 component was fitted to the Nernst equation to determine the midpoint potential of the [2Fe-2S] cluster. EFAD-FS1 was determined by plotting the height difference between the g = 1.92 “free” component and the g = 1.91 “occupied” component as in Figure 7. The error in Em values is approximately ± 10 mV.
| FAD Em,7 (mV) | [2Fe-2S] Em,7 (mV) | EFAD-FS1 (mV) | |
|---|---|---|---|
|
| |||
| WT Sdh | −100 | −15 | −125 |
| SdhA-H45A | −187 | −15 | −195 |
| SdhA-R286A | −209 | −18 | −255 |
| SdhA-R286K | −266 | −15 | −230 |
| SdhA-R286Y | −258 | +45 | -- |
| SdhA-H242A | −202 | −10 | ND |
| SdhA-H242Y | −207 | +40 | -- |
The [2Fe-2S] cluster lies in close proximity (~12 Å) to the isoalloxazine ring of the FAD. We therefore explored the possibility that variants of residues surrounding the isoalloxazine might elicit changes in [2Fe-2S] redox chemistry. In agreement with previous studies (45, 46), we obtained a [2Fe-2S] cluster Em,7 of approximately −15 mV in the wild-type enzyme. The majority of variants studied herein exhibited Em,7 values that do not differ significantly from that of the wild-type (Table 2). Exceptions to this are the Tyr substitutions of Arg286 and His242, which exhibit Em,7 values of +45 and +40 mV, respectively. These results indicate that in some cases, changes in the environment of the FAD can be propagated to the [2Fe-2S] cluster, suggesting that these two centres are electrostatically or conformationally linked in Sdh.
Communication between the FAD and the [2Fe-2S] cluster
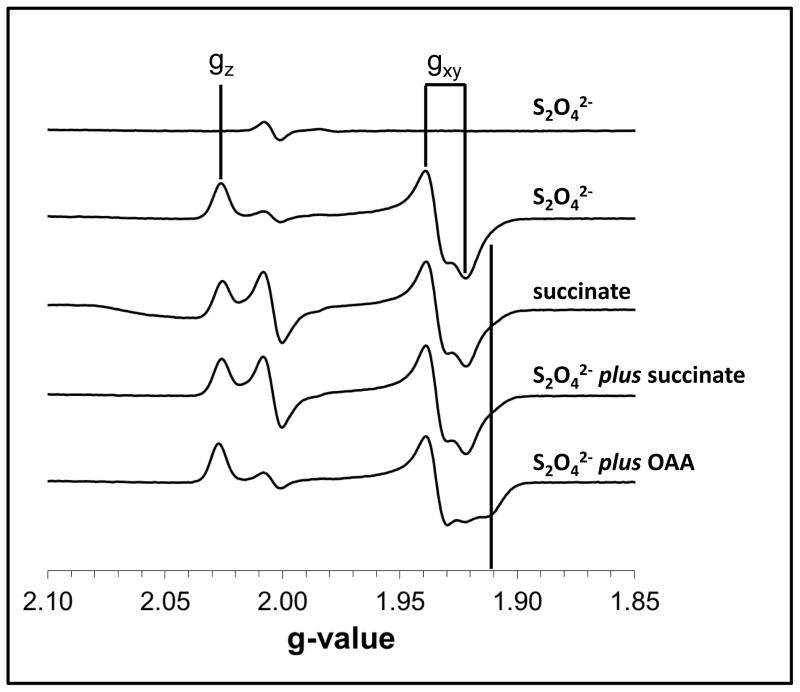
Given the proximity of the FAD isoalloxazine ring to the [2Fe-2S] cluster and the effects of the SdhA-R286Y and SdhA-H242Y substitutions on the Em of the latter, we hypothesized that substrate or inhibitor binding might elicit changes in the EPR lineshape of the cluster. Recently, a study of the Thermus thermophilus Sdh enzyme showed reduction of the enzyme by succinate yielded a reduced [2Fe-2S] EPR signal with higher rhombicity at the gxy feature (47). Figure 3 shows that when reduced with dithionite, the E. coli SdhCDAB [2Fe-2S] cluster EPR signal is comprised of a peak at gz = 2.03 and a multi-component peak-trough at gxy = 1.94 and 1.92. When the enzyme was solely reduced by succinate, or by dithionite in the presence of succinate, a small shoulder at g = 1.91 became apparent. When the enzyme was reduced with dithionite in the presence of OAA, the g = 1.91 feature became more prominent and, in fact, became a local trough, giving the gxy component a more rhombic feature that is reminiscent of the spectrum of succinate-reduced [2Fe-2S] cluster in the T. thermophilus enzyme.
Figure 3. The EPR line shape of the [2Fe-2S] cluster changes upon inhibitor binding.
Spectra were of the following samples (as labeled in the figure): dithionite reduction of membranes prepared from DW35 cells lacking SdhCDAB; membranes enriched in wild-type SdhCDAB reduced with dithionite, succinate, dithionite plus succinate, and dithionite plus OAA. The g = 1.91 feature is marked for comparison.
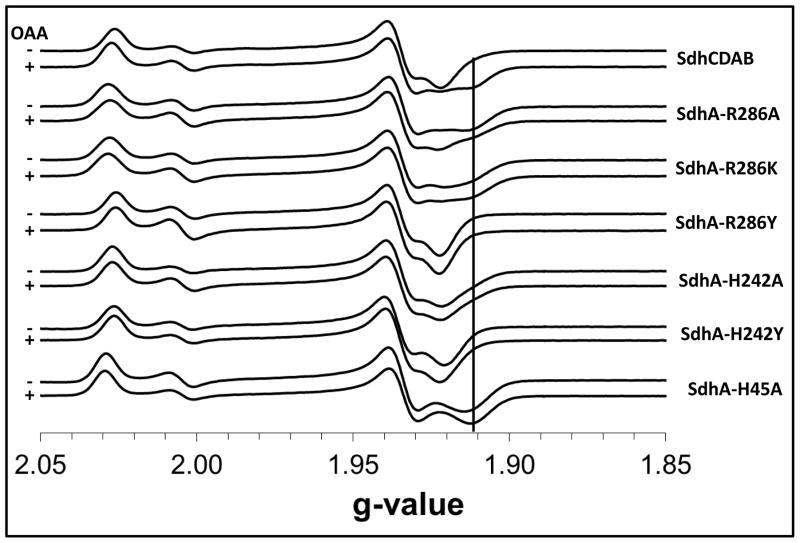
Figure 4 shows reduced EPR spectra of the [2Fe-2S] clusters in the variant enzymes studied herein. The SdhA-H45A variant gave rise to a noticeable change in the EPR lineshape of the [2Fe-2S] cluster and results in the disappearance of the g = 1.92 trough and its replacement by a trough at g = 1.91. Similarly, when the SdhA-H45A variant is reduced by succinate, or by dithionite in the presence of either succinate or OAA, a trough is observed at g = 1.91 and the g=1.92 trough is absent (Figure 4, Supplemental Figures 2 and 3). In the case of the SdhA-R286A and SdhA-R286K variants, dithionite reduction results in a spectrum with a significant g = 1.91 feature that appears to be unaffected by the presence of succinate and OAA (Figure 4 and Supplemental Figures 2 and 3). The [2Fe-2S] cluster in the SdhA-H242A variant behaves in a similar fashion, but the magnitude of the g = 1.91 feature is moderated such that a steady slope is observed under all reducing conditions. In stark contrast to this, the EPR lineshape of the reduced [2Fe-2S] cluster in the SdhA-R286Y and SdhA-H242Y variants shows absolutely no change under the different reduction conditions, and the appearance of the g = 1.91 feature could not be manifested using succinate or OAA (Figure 4, Supplemental Figures 3 and 4).
Figure 4. EPR line shapes of [2Fe-2S] clusters in variant enzymes.
Each pair of spectra were obtained by reducing the indicated enzymes with 2.5 mM dithionite in the absence (−, top) or presence (+, bottom) of 25 mM oxaloacetate (OAA). The g = 1.91 feature is marked for comparison.
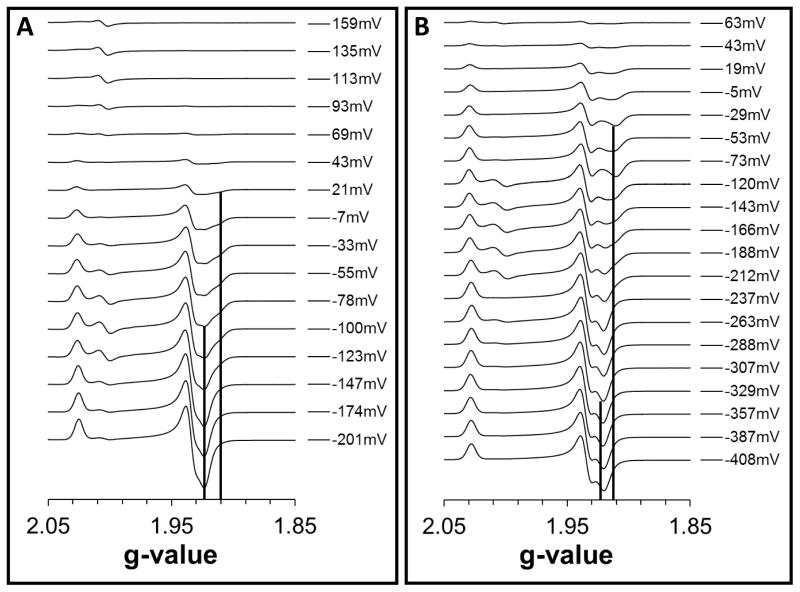
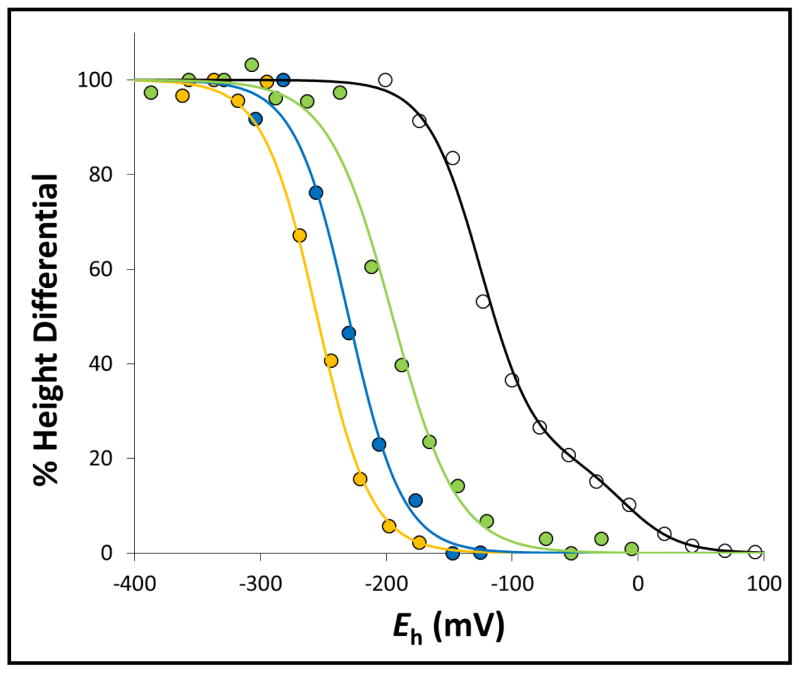
Surprisingly, the interplay between the native g = 1.92 trough and the inducible g = 1.91 trough is also present during redox titrations, wherein substrates and inhibitors are absent. As noted above, the g = 1.91 feature appears when the wild-type enzyme is reduced in the presence of substrate or inhibitor. In redox titrations of the wild-type enzyme, the magnitudes of the g = 1.92 and g = 1.91 features are potential-dependent (Figure 5A). The height differences between these two trough signals can be plotted against the ambient potential to yield a Nernstian relationship with a midpoint potential (“EFAD-FS1”) of −125 mV (Figure 6, Table 2). Interestingly, in the SdhA-H45A variant this value decreases to −195 mV (Figure 5B, Table 2). In the SdhA-R286A and SdhA-R286K variants, the EFAD-FS1 value decreases even further to −255 mV and −230 mV, respectively (Table 2). In redox titrations of the paralogous enzyme FrdABCD, the transition between g = 1.92 and g = 1.91 is not observed (Supplemental Figure 4).
Figure 5. Redox titrations of the [2Fe-2S] cluster.
Representative EPR spectra from redox titrations of the (A) wild-type SdhCDAB enzyme and (B) SdhA-H45A variant are shown. The positions of the g = 1.91 and g = 1.92 troughs are indicated by vertical lines.
Figure 6. Redox-dependent interplay between the g =1.91 and g = 1.92 components of the [2Fe-2S] cluster EPR spectrum.
The height differentials between the g = 1.91 and g = 1.92 components of the [2Fe-2S] cluster EPR signal as a function of ambient potential were plotted for wild-type SdhCDAB (black), SdhA-H45A (green), SdhA-R286A (yellow), and SdhA-R286K (blue) and fitted to the Nernst equation. For titration of the wild-type SdhCDAB enzyme, a second component corresponding to the reduction of the [2Fe-2S] cluster itself was also modeled.
Discussion
In the Complex II family of enzymes, the covalent attachment of a FAD moiety via an 8α-N3-histidyl linkage is absolutely conserved. How is the FAD cofactor recruited to the apoenzyme and how does flavinylation proceed thereafter? In the case of SdhCDAB, it was recently discovered that the SdhE chaperone directly interacts with the SdhA subunit to mediate flavinylation (32, 48), and the absence of the equivalent SdhAF2 chaperone in humans results in paraganglioma tumorigenesis (33). Following insertion of FAD into its binding pocket and repositioning of the capping domain, covalent attachment is thought to be an autocatalytic process, a common theme that is conserved in other flavoenzymes (49–51). In this study, we show the SdhA-R286 residue to be absolutely essential for flavinylation. In a prior study on the W. succinogenes fumarate reductase, covalent attachment of FAD was abolished in a FrdA-R301E variant (equivalent to residue SdhA-R286) but was retained in a FrdA-R301K variant (35). Preliminary studies on a FrdA-R287K variant suggested covalent attachment of FAD was retained (unpublished data, Maklashina et al.). Interestingly, all three substitutions of SdhA-R286 studied herein resulted in assembly of non-covalent FAD, including the Lys variant. It has been proposed that the guanidinium side chain of the conserved SdhA-R286 residue can: (i) H-bond to and stabilize the substrate in the dicarboxylate binding site, and (ii) act as a proton donor for fumarate reduction and a proton acceptor during succinate oxidation (9, 15, 35, 52). In comparison, the imidazole ring of SdhA-H242 plays a more limited role by H-bonding to the substrate and is not believed to be involved in proton shuttling during catalysis. The observation that the SdhA-H242Y variant enzyme does not turnover but can still incorporate a covalent flavin suggests that substrate binding (see below for discussion on substrate binding), but not enzyme turnover, is necessary for flavinylation. A study on the Bacillus subtilis Sdh enzyme has also shown that residues not involved in catalysis or substrate binding can hinder FAD recruitment and flavinylation (53).
Removal of the covalent linkage via substitution of the His residue results in elimination of succinate dehydrogenase activity and partial retention of fumarate reductase activity (21, 22). It is widely believed that covalent attachment increases the Em value of the FAD cofactor by ~100 mV to facilitate electron transfer between its isoalloxazine ring and succinate/fumarate (16, 21, 54). This assertion is based on two key observations. First, mutational studies on other flavoenzymes such as vanillyl-alcohol oxidase (27) and cholesterol oxidase (55) demonstrated that removal of the covalent linkage resulted in a ~100 mV decrease in the Em value of the FAD. Second, in the homologous soluble fumarate reductase from S. frigidimarina, the FAD is assembled in the non-covalent form and has a Em value that is approximately 100 mV lower than that of the covalent FAD in E. coli Frd (56). In this study, we show definitively using redox potentiometry on the SdhCDAB system that non-covalent FAD in a SdhA-H45A variant does indeed have a decreased midpoint potential (Em,7 = −187 mV) versus the wild-type SdhCDAB enzyme (Em,7 = −100 mV). We also showed the Em value of the covalent FAD can be lowered via SdhA-H242A and SdhA-H242Y mutations. In agreement with similar studies, the SdhA-H45A and SdhA-H242A variants show that decreasing the Em value of the FAD resulted in loss of succinate oxidase and fumarate reductase activities, with the former affected to a greater degree. When we examined whether the [2Fe-2S] cluster can be reduced by succinate, we noted that there was enough succinate oxidase activity to elicit significant reduction (Supplemental Figure 2). In the case of the SdhA-H242Y variant, the lack of succinate-dependent [2Fe-2S] cluster reduction may be partly explained by its increased Em value in comparison with the wild-type (+40mV versus −15mV).
Our observations that substrates and inhibitors can alter the EPR lineshape of the reduced [2Fe-2S] cluster enabled us to explore its communication with the substrate binding site. A recent study on the T. thermophilus Sdh showed that specific reduction of the enzyme by succinate caused the gxy component of the [2Fe-2S] cluster signal to become more rhombic (47). Using the variant enzymes herein, we extended the study to include OAA. As detailed in the results section, a feature at g = 1.91 became apparent when succinate was used as the sole reductant, or was used in conjunction with dithionite to reduce the enzyme. This trough feature became even more apparent when the enzyme was pre-incubated with a strong inhibitor such as OAA (Ki = 70 nM) (15) (Figure 3). In agreement with the T. thermophilus study, we also believe that the EPR lineshape changes are suggestive of two distinct conformations of the enzyme: an “occupied” state with substrate/inhibitor bound (g = 1.91 trough) and a “free” state without substrate/inhibitor bound (g = 1.92 trough) that are in rapid equilibrium. This is consistent with the hypothesis that structural changes occur, especially of the hinge region and the capping domain, to allow opening and closing of the active site during catalysis (5, 9, 26, 57).
Reduction of the SdhA-R286A, SdhA-R286K and SdhA-H242A variants using dithionite alone elicited the appearance of the g = 1.91 feature, suggesting that these variant enzymes are at least partially present in a state that mimics the occupied state of the wild-type enzyme. Additionally, the lack of change observed in their EPR spectra when succinate and OAA were added before reduction by dithionite also indicates that these variant enzymes are in the occupied state, and that they are tightly locked in this conformation. Interestingly, both Tyr substitutions of SdhA-R286 and SdhA-H242 resulted in the absence of the g = 1.91 feature under all conditions. One possible explanation to account for this is that the Tyr side chain forms a strong H-bond with SdhA-E255, preventing entry of the substrate into the active site and locking it in the free state. But, given that substrate binding is absolutely necessary for meaningful flavinylation and that ~31% of the assembled SdhA-H242Y variant contained covalently-attached FAD (Table 1), it is possible that only this proportion of enzyme was in a substrate-binding competent comformation during maturation. Indeed, OAA binding was observed, but to a lesser extent, in a FrdA-H232S variant (equivalent to SdhA-H242) (36). The more likely explanation is that the Tyr side chain raises the pKa of the gxy components of the [2Fe-2S] cluster EPR signal. In T. thermophilus Sdh, the axial signal representing the free state occurs when pH < pKa and the rhombic signal representing the occupied state occurs when pH > pKa (47). If the pKa was indeed raised by Tyr substitutions of SdhA-R286 and SdhA-H242, then the equilibrium at any given pH would be shifted toward the free state, the form that we exclusively observed for these two Tyr variants. By analogy, since only the g = 1.91 trough was observed for the SdhA-H45A variant, the non-covalent FAD must be decreasing the pKa of the “free” ⬄ “occupied” transition such that the “occupied” form becomes dominant at pH 7 in our experiments. Thus, one important role of protein flavinylation is to maintain the pKa of the free/occupied transition near physiological pH to allow enzyme turnover in vivo.
The free/occupied transition became even more intriguing when we examined the EPR spectra from redox titrations. In the wild-type SdhCDAB enzyme, a hint of the g = 1.91 feature was noticeable at potentials above approximately −150 mV. When the ambient potential was further decreased, this feature slowly disappeared, concurrent with an increase in signal intensity of the trough at g = 1.92. The height difference between the g = 1.91 and g=1.92 troughs can be fitted to a Nernstian function with a midpoint potential of −125 mV (Figure 6 and Table 2). Thus, the switch between the occupied (g = 1.91) and free (g = 1.92) states for the wild-type SdhCDAB enzyme occurs at −125 mV, which we have termed “EFAD-FS1”. In the SdhA-H45A variant wherein a non-covalent FAD is assembled, the EFAD-FS1 value was decreased to −195 mV, which approximately equals the decrease observed for the Em,7 value of a non-covalent FAD versus its covalent form. Similarly, in the SdhA-R286A and SdhA-R286K variants, the EFAD-FS1 values were decreased further, also consistent with the decreases observed for their non-covalent FAD moieties. Altogether, the EPR data suggests that the EFAD-FS1 value, which reports the transition between the occupied and free states of the active site, is dependent on the redox state of the FAD cofactor.
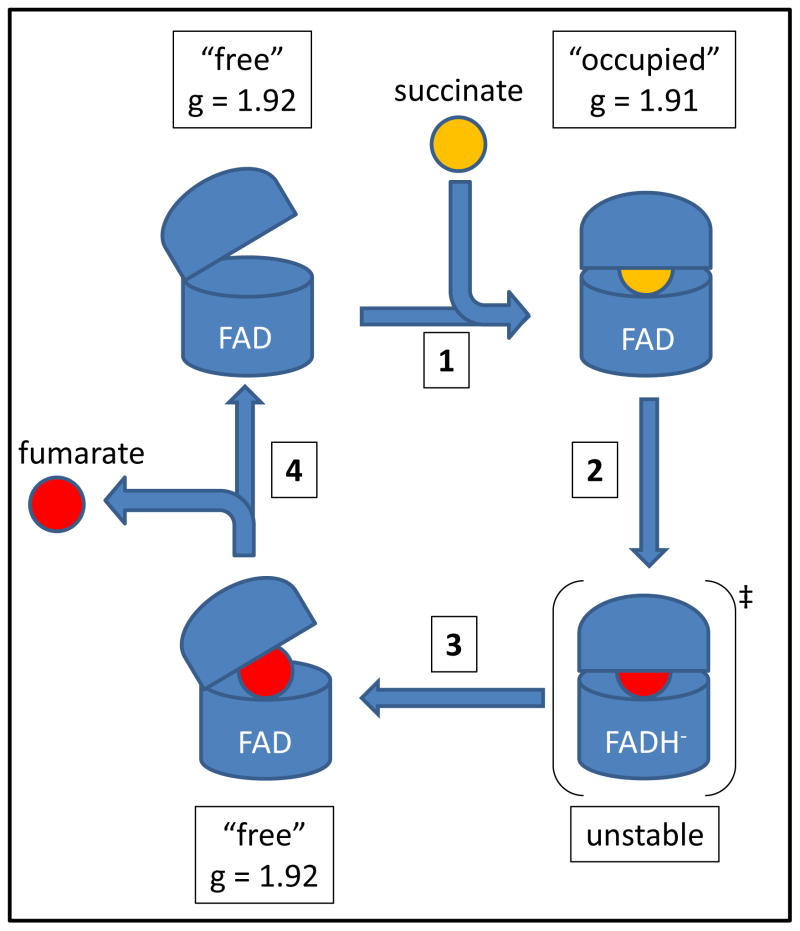
Based on this newfound knowledge, we constructed a simple model to visualize succinate oxidation at the FAD binding domain (Figure 7). First, the oxidized enzyme assumes a conformation where the dicarboxylic acid binding site is “free” (g = 1.92). Next, succinate binds and induces the “occupied” conformation (g = 1.91). Succinate is then oxidized to fumarate and 2 electrons are transferred to the FAD via a hydride transfer. The combination of the “occupied” form and a reduced FADH− molecule represents a high energy transition state that can drive conformational change back to the “free” form, resulting in product (fumarate) release. Alternatively, oxidation of FADH− by the [Fe-S] clusters in SdhB may also induce conformational change to the “free” form. In E. coli Sdh, movements of the capping and FAD binding domains are important for activating the C2-C3 bond of fumarate/succinate to attain the transition state (5, 9). In X-ray crystallographic studies of W. succinogenes fumarate reductase, the capping domain has been captured at different positions relative to the FAD binding domain, hinting at structural changes that occur during enzyme turnover (35, 58). In spite of this, the isoalloxazine ring always assumes a planar conformation in all available X-ray crystal structures of Complex II enzymes, indicating that the reduced FADH− form is highly unstable. In studies of FAD in solution and in other flavoeznymes, reduction of the isoalloxazine ring (involving the N5-C4a-C10a-N1 atoms) results in a bent structure in solution or leads to conformational change of the host protein (59–63). A previous study had shown that the biophysical properties of the FAD were altered depending on the redox state of the [2Fe-2S] cluster (25). The EPR data on the [2Fe-2S] cluster presented in this study provides complementary data that strongly suggests a conformational shift between the free and occupied states occurs when the redox state of the FAD is altered.
Figure 7. Proposed model for succinate oxidation and fumarate release.
(1) Succinate binds to the enzyme to induce conformational change from the “free” to the “occupied” form. (2) Succinate is oxidized, and FAD is reduced to FADH−. (3) The high energy “occupied”/FADH− transition state drives the conformational change to the “free” form. (4) Fumarate is released from the “free” form, and electrons are passed to SdhB.
The proposed mechanism of succinate oxidation, similar to the one proposed in 1992 (64), has important ramifications for enzyme catalysis. First, another succinate molecule cannot bind until the FADH− cofactor has distributed its electrons to the iron-sulfur clusters in SdhB. However, this should not limit the rate of catalysis since electron transfer is orders of magnitude faster than the rates of fumarate release and succinate binding. Second, the gating mechanism presented by the redox state of the FAD cofactor implies that the reversible reactions of succinate oxidation and fumarate reduction do not occur as mirror images of each other. In the voltammetry studies done on the SdhAB dimer, the soluble enzyme exhibits a “tunnel diode” behavior in the presence of fumarate (the rate of fumarate reduction actually decreases with increasing driving force) (64–66). Further, the studies showed that the dissociation constant for the substrate-FrdAB complex when FAD is reduced is highest compared to those of its semiquinone and quinone forms (66). Although the data presented herein do not directly explain the tunnel diode mechanism, there is undeniably a strong connection: both the EFAD-FS1 transition and the Eswitch potential at which the tunnel diode effect is turned on closely match the Em of the FAD.
The model presented in Figure 7 incorporates these findings and aligns with the tunnel diode behavior; that is, when the flavin is reduced (i.e., higher driving force), the equilibrium is shifted towards the “free” state, preventing fumarate binding and its subsequent reduction. In variant enzymes (and soluble Sh. frigidimarina fumarate reductase) that assemble a noncovalent FAD, two factors come into play. First, the Em value of the FAD cofactor is decreased, resulting in lower occupancy of the FAD by electrons, which makes fumarate binding and reduction favorable. Second, although the “free” state with oxidized FAD also makes succinate binding more favorable, the succinate/fumarate couple of +30 mV is energetically incapable of driving FAD reduction (−185 mV or lower for noncovalent FAD). This explains why fumarate reductase activity, but not succinate dehydrogenase activity, is preferentially retained when the Em value of the FAD is decreased.
Summary
Through a series of EPR experiments, we have provided, for the first time, a definitive midpoint potential of the non-covalent FAD cofactor in the SdhCDAB enzyme. Noncovalent FAD does indeed have a decreased Em,7 value that is decreased (ΔEm,7 = −87 mV) compared to its covalent counterpart in the wild-type enzyme. Substitutions made at the SdhA-R286 position resulted in enzymes that assembled noncovalent FADs and were incapable of catalysis. EPR spectroscopy also suggested the existence of two conformational states, “free” and “occupied”, that are tightly coupled to the redox state of the FAD and reduction of the [2Fe-2S] cluster. A model for succinate oxidation was also presented that takes into account the tunnel diode behavior of Sdh and the preference for fumarate reduction in variant enzymes that lack covalent flavin.
Supplementary Material
Acknowledgments
Funding: This work was supported by the Canadian Institutes of Health Research (grant MDP98735 to JHW), the Department of Veterans Affairs Merit grant BX001077 to GC, and National Institutes of Health grant GM61606 to GC.
Abbreviations
- FAD
flavin adenine dinucleotide
- Frd
fumarate reductase
- OAA
oxaloacetate
- Sdh
succinate dehydrogenase
- SdhCDAB, succinate
ubiquinone oxidoreductase (bacterial, e.g. E. coli)
- UQ
ubiquinone
- UQH2
ubiquinol
Footnotes
Supporting information is available online at http://pubs.acs.org
References
- 1.Sun F, Huo X, Zhai Y, Wang A, Xu J, Su D, Bartlam M, Rao Z. Crystal structure of mitochondrial respiratory membrane protein complex II. Cell. 2005;121:1043–1057. doi: 10.1016/j.cell.2005.05.025. [DOI] [PubMed] [Google Scholar]
- 2.Huang LS, Shen JT, Wang AC, Berry EA. Crystallographic studies of the binding of ligands to the dicarboxylate site of Complex II, and the identity of the ligand in the “oxaloacetate-inhibited” state. Biochim Biophys Acta. 2006;1757:1073–1083. doi: 10.1016/j.bbabio.2006.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Huang LS, Sun G, Cobessi D, Wang AC, Shen JT, Tung EY, Anderson VE, Berry EA. 3-nitropropionic acid is a suicide inhibitor of mitochondrial respiration that, upon oxidation by complex II, forms a covalent adduct with a catalytic base arginine in the active site of the enzyme. J Biol Chem. 2006;281:5965–5972. doi: 10.1074/jbc.M511270200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yankovskaya V, Horsefield R, Tornroth S, Luna-Chavez C, Miyoshi H, Leger C, Byrne B, Cecchini G, Iwata S. Architecture of succinate dehydrogenase and reactive oxygen species generation. Science. 2003;299:700–704. doi: 10.1126/science.1079605. [DOI] [PubMed] [Google Scholar]
- 5.Tomasiak TM, Maklashina E, Cecchini G, Iverson TM. A threonine on the active site loop controls transition state formation in Escherichia coli respiratory complex II. J Biol Chem. 2008;283:15460–15468. doi: 10.1074/jbc.M801372200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lancaster CR. Wolinella succinogenes quinol:fumarate reductase-2.2-A resolution crystal structure and the E-pathway hypothesis of coupled transmembrane proton and electron transfer. Biochim Biophys Acta. 2002;1565:215–231. doi: 10.1016/s0005-2736(02)00571-0. [DOI] [PubMed] [Google Scholar]
- 7.Iverson TM, Luna-Chavez C, Cecchini G, Rees DC. Structure of the Escherichia coli fumarate reductase respiratory complex. Science. 1999;284:1961–1966. doi: 10.1126/science.284.5422.1961. [DOI] [PubMed] [Google Scholar]
- 8.Iverson TM, Luna-Chavez C, Croal LR, Cecchini G, Rees DC. Crystallographic studies of the Escherichia coli quinol-fumarate reductase with inhibitors bound to the quinol-binding site. J Biol Chem. 2002;277:16124–16130. doi: 10.1074/jbc.M200815200. [DOI] [PubMed] [Google Scholar]
- 9.Tomasiak TM, Archuleta TL, Andrell J, Luna-Chavez C, Davis TA, Sarwar M, Ham AJ, McDonald WH, Yankovskaya V, Stern HA, Johnston JN, Maklashina E, Cecchini G, Iverson TM. Geometric restraint drives on- and off-pathway catalysis by the Escherichia coli menaquinol:fumarate reductase. J Biol Chem. 2011;286:3047–3056. doi: 10.1074/jbc.M110.192849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vik SB, Hatefi Y. Possible occurrence and role of an essential histidyl residue in succinate dehydrogenase. Proc Natl Acad Sci USA. 1981;78:6749–6753. doi: 10.1073/pnas.78.11.6749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tran QM, Rothery RA, Maklashina E, Cecchini G, Weiner JH. Escherichia coli succinate dehydrogenase variant lacking the heme b. Proc Natl Acad Sci. 2007;104:18007–18012. doi: 10.1073/pnas.0707732104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Oyedotun KS, Sit CS, Lemire BD. The Saccharomyces cerevisiae succinate dehydrogenase does not require heme for ubiquinone reduction. Biochim Biophys Acta. 2007;1767:1436–1445. doi: 10.1016/j.bbabio.2007.09.008. [DOI] [PubMed] [Google Scholar]
- 13.Guest JR. Partial replacement of succinate dehydrogenase function by phage- and plasmid-specified fumarate reductase in Escherichia coli. J Gen Microbiol. 1981;122:171–179. doi: 10.1099/00221287-122-2-171. [DOI] [PubMed] [Google Scholar]
- 14.Maklashina E, Berthold DA, Cecchini G. Anaerobic expression of Escherichia coli succinate dehydrogenase: functional replacement of fumarate reductase in the respiratory chain during anaerobic growth. J Bacteriol. 1998;180:5989–5996. doi: 10.1128/jb.180.22.5989-5996.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Maklashina E, Iverson TM, Sher Y, Kotlyar V, Andrell J, Mirza O, Hudson JM, Armstrong FA, Rothery RA, Weiner JH, Cecchini G. Fumarate reductase and succinate oxidase activity of Escherichia coli complex II homologs are perturbed differently by mutation of the flavin binding domain. J Biol Chem. 2006;281:11357–11365. doi: 10.1074/jbc.M512544200. [DOI] [PubMed] [Google Scholar]
- 16.Maklashina E, Cecchini G, Dikanov SA. Defining a direction: electron transfer and catalysis in Escherichia coli complex II enzymes. Biochim Biophys Acta. 2013;1827:668–678. doi: 10.1016/j.bbabio.2013.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Turner KL, Doherty MK, Heering HA, Armstrong FA, Reid GA, Chapman SK. Redox properties of flavocytochrome c3 from Shewanella frigidimarina NCIMB400. Biochemistry. 1999;38:3302–3309. doi: 10.1021/bi9826308. [DOI] [PubMed] [Google Scholar]
- 18.Dobbin PS, Butt JN, Powell AK, Reid GA, Richardson DJ. Characterization of a flavocytochrome that is induced during the anaerobic respiration of Fe3+ by Shewanella frigidimarina NCIMB400. Biochem J. 1999;342( Pt 2):439–448. [PMC free article] [PubMed] [Google Scholar]
- 19.Walker WH, Singer TP. Identification of the covalently bound flavin of succinate dehydrogenase as 8-alpha-(histidyl) flavin adenine dinucleotide. J Biol Chem. 1970;245:4224–4225. [PubMed] [Google Scholar]
- 20.Weiner JH, Dickie P. Fumarate reductase of Escherichia coli. Elucidation of the covalent-flavin component. J Biol Chem. 1979;254:8590–8593. [PubMed] [Google Scholar]
- 21.Blaut M, Whittaker K, Valdovinos A, Ackrell BA, Gunsalus RP, Cecchini G. Fumarate reductase mutants of Escherichia coli that lack covalently bound flavin. J Biol Chem. 1989;264:13599–13604. [PubMed] [Google Scholar]
- 22.Robinson KM, Rothery RA, Weiner JH, Lemire BD. The covalent attachment of FAD to the flavoprotein of Saccharomyces cerevisiae succinate dehydrogenase is not necessary for import and assembly into mitochondria. Eur J Biochem. 1994;222:983–990. doi: 10.1111/j.1432-1033.1994.tb18949.x. [DOI] [PubMed] [Google Scholar]
- 23.Heffron K. PhD thesis. University of Oxford; 2001. Studies of the redox and catalytic properties of the anaerobic respiratory enzymes of Escherichia coli. [Google Scholar]
- 24.Ackrell BA, Cochran B, Cecchini G. Interactions of oxaloacetate with Escherichia coli fumarate reductase. Arch Biochem Biophys. 1989;268:26–34. doi: 10.1016/0003-9861(89)90561-4. [DOI] [PubMed] [Google Scholar]
- 25.Ohnishi T, King TE, Salerno JC, Blum H, Bowyer JR, Maida T. Thermodynamic and electron paramagnetic resonance characterization of flavin in succinate dehydrogenase. J Biol Chem. 1981;256:5577–5582. [PubMed] [Google Scholar]
- 26.Cecchini G, Schroder I, Gunsalus RP, Maklashina E. Succinate dehydrogenase and fumarate reductase from Escherichia coli. Biochim Biophys Acta. 2002;1553:140–157. doi: 10.1016/s0005-2728(01)00238-9. [DOI] [PubMed] [Google Scholar]
- 27.Fraaije MW, van den Heuvel RH, van Berkel WJ, Mattevi A. Covalent flavinylation is essential for efficient redox catalysis in vanillyl-alcohol oxidase. J Biol Chem. 1999;274:35514–35520. doi: 10.1074/jbc.274.50.35514. [DOI] [PubMed] [Google Scholar]
- 28.Mewies M, McIntire WS, Scrutton NS. Covalent attachment of flavin adenine dinucleotide (FAD) and flavin mononucleotide (FMN) to enzymes: the current state of affairs. Protein Sci. 1998;7:7–20. doi: 10.1002/pro.5560070102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cecchini G. Function and structure of complex II of the respiratory chain. Annu Rev Biochem. 2003;72:77–109. doi: 10.1146/annurev.biochem.72.121801.161700. [DOI] [PubMed] [Google Scholar]
- 30.Kim HJ, Winge DR. Emerging concepts in the flavinylation of succinate dehydrogenase. Biochim Biophys Acta. 2013;1827:627–636. doi: 10.1016/j.bbabio.2013.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Robinson KM, Lemire BD. Covalent attachment of FAD to the yeast succinate dehydrogenase flavoprotein requires import into mitochondria, presequence removal, and folding. J Biol Chem. 1996;271:4055–4060. doi: 10.1074/jbc.271.8.4055. [DOI] [PubMed] [Google Scholar]
- 32.McNeil MB, Clulow JS, Wilf NM, Salmond GP, Fineran PC. SdhE is a conserved protein required for flavinylation of succinate dehydrogenase in bacteria. J Biol Chem. 2012;287:18418–18428. doi: 10.1074/jbc.M111.293803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hao HX, Khalimonchuk O, Schraders M, Dephoure N, Bayley JP, Kunst H, Devilee P, Cremers CW, Schiffman JD, Bentz BG, Gygi SP, Winge DR, Kremer H, Rutter J. SDH5, a gene required for flavination of succinate dehydrogenase, is mutated in paraganglioma. Science. 2009;325:1139–1142. doi: 10.1126/science.1175689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McNeil MB, Hampton HG, Hards KJ, Watson BN, Cook GM, Fineran PC. The succinate dehydrogenase assembly factor, SdhE, is required for the flavinylation and activation of fumarate reductase in bacteria. FEBS Lett. 2014;588:414–421. doi: 10.1016/j.febslet.2013.12.019. [DOI] [PubMed] [Google Scholar]
- 35.Lancaster CR, Gross R, Simon J. A third crystal form of Wolinella succinogenes quinol:fumarate reductase reveals domain closure at the site of fumarate reduction. Eur J Biochem. 2001;268:1820–1827. [PubMed] [Google Scholar]
- 36.Schroder I, Gunsalus RP, Ackrell BA, Cochran B, Cecchini G. Identification of active site residues of Escherichia coli fumarate reductase by site-directed mutagenesis. J Biol Chem. 1991;266:13572–13579. [PubMed] [Google Scholar]
- 37.Westenberg DJ, Gunsalus RP, Ackrell BA, Sices H, Cecchini G. Escherichia coli fumarate reductase frdC and frdD mutants. Identification of amino acid residues involved in catalytic activity with quinones. J Biol Chem. 1993;268:815–822. [PubMed] [Google Scholar]
- 38.Cheng VW, Tran QM, Boroumand N, Rothery RA, Maklashina E, Cecchini G, Weiner JH. A conserved lysine residue controls iron-sulfur cluster redox chemistry in Escherichia coli fumarate reductase. Biochim Biophys Acta. 2013;1827:1141–1147. doi: 10.1016/j.bbabio.2013.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lowry OH, Rosebrough NJ, Farr AL, Randall RJ. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951;193:265–275. [PubMed] [Google Scholar]
- 40.Markwell MA, Haas SM, Bieber LL, Tolbert NE. A modification of the Lowry procedure to simplify protein determination in membrane and lipoprotein samples. Anal Biochem. 1978;87:206–210. doi: 10.1016/0003-2697(78)90586-9. [DOI] [PubMed] [Google Scholar]
- 41.Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Singer TP, Edmondson DE. Structure, properties, and determination of covalently bound flavins. Methods Enzymol. 1980;66:253–264. doi: 10.1016/0076-6879(80)66466-0. [DOI] [PubMed] [Google Scholar]
- 43.Kita K, Vibat CR, Meinhardt S, Guest JR, Gennis RB. One-step purification from Escherichia coli of complex II (succinate: ubiquinone oxidoreductase) associated with succinate-reducible cytochrome b556. J Biol Chem. 1989;264:2672–2677. [PubMed] [Google Scholar]
- 44.Dickie P, Weiner JH. Purification and characterization of membrane-bound fumarate reductase from anaerobically grown Escherichia coli. Can J Biochem. 1979;57:813–821. doi: 10.1139/o79-101. [DOI] [PubMed] [Google Scholar]
- 45.Condon C, Cammack R, Patil DS, Owen P. The succinate dehydrogenase of Escherichia coli Immunochemical resolution and biophysical characterization of a 4-subunit enzyme complex. J Biol Chem. 1985;260:9427–9434. [PubMed] [Google Scholar]
- 46.Cheng VW, Ma E, Zhao Z, Rothery RA, Weiner JH. The iron-sulfur clusters in Escherichia coli succinate dehydrogenase direct electron flow. J Biol Chem. 2006;281:27662–27668. doi: 10.1074/jbc.M604900200. [DOI] [PubMed] [Google Scholar]
- 47.Kolaj-Robin O, Noor MR, O’Kane SR, Baymann F, Soulimane T. Atypical features of Thermus thermophilus succinate:quinone reductase. PLoS One. 2013;8:e53559. doi: 10.1371/journal.pone.0053559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.McNeil MB, Fineran PC. The Conserved RGxxE Motif of the Bacterial FAD Assembly Factor SdhE Is Required for Succinate Dehydrogenase Flavinylation and Activity. Biochemistry. 2013;52:7628–7640. doi: 10.1021/bi401006a. [DOI] [PubMed] [Google Scholar]
- 49.Brandsch R, Bichler V. Autoflavinylation of apo6-hydroxy-D-nicotine oxidase. J Biol Chem. 1991;266:19056–19062. [PubMed] [Google Scholar]
- 50.Edmondson DE, Newton-Vinson P. The covalent FAD of monoamine oxidase: structural and functional role and mechanism of the flavinylation reaction. Antioxid Redox Signal. 2001;3:789–806. doi: 10.1089/15230860152664984. [DOI] [PubMed] [Google Scholar]
- 51.Jin J, Mazon H, van den Heuvel RH, Heck AJ, Janssen DB, Fraaije MW. Covalent flavinylation of vanillyl-alcohol oxidase is an autocatalytic process. FEBS J. 2008;275:5191–5200. doi: 10.1111/j.1742-4658.2008.06649.x. [DOI] [PubMed] [Google Scholar]
- 52.Pankhurst KL, Mowat CG, Rothery EL, Hudson JM, Jones AK, Miles CS, Walkinshaw MD, Armstrong FA, Reid GA, Chapman SK. A proton delivery pathway in the soluble fumarate reductase from Shewanella frigidimarina. J Biol Chem. 2006;281:20589–20597. doi: 10.1074/jbc.M603077200. [DOI] [PubMed] [Google Scholar]
- 53.Maguire JJ, Magnusson K, Hederstedt L. Bacillus subtilis mutant succinate dehydrogenase lacking covalently bound flavin: identification of the primary defect and studies on the iron-sulfur clusters in mutated and wild-type enzyme. Biochemistry. 1986;25:5202–5208. doi: 10.1021/bi00366a033. [DOI] [PubMed] [Google Scholar]
- 54.Heuts DP, Scrutton NS, McIntire WS, Fraaije MW. What’s in a covalent bond? On the role and formation of covalently bound flavin cofactors. FEBS J. 2009;276:3405–3427. doi: 10.1111/j.1742-4658.2009.07053.x. [DOI] [PubMed] [Google Scholar]
- 55.Motteran L, Pilone MS, Molla G, Ghisla S, Pollegioni L. Cholesterol oxidase from Brevibacterium sterolicum. The relationship between covalent flavinylation and redox properties. J Biol Chem. 2001;276:18024–18030. doi: 10.1074/jbc.M010953200. [DOI] [PubMed] [Google Scholar]
- 56.Jeuken LJ, Jones AK, Chapman SK, Cecchini G, Armstrong FA. Electron-transfer mechanisms through biological redox chains in multicenter enzymes. J Am Chem Soc. 2002;124:5702–5713. doi: 10.1021/ja012638w. [DOI] [PubMed] [Google Scholar]
- 57.Iverson TM, Luna-Chavez C, Schroder I, Cecchini G, Rees DC. Analyzing your complexes: structure of the quinol-fumarate reductase respiratory complex. Curr Opin Struct Biol. 2000;10:448–455. doi: 10.1016/s0959-440x(00)00113-5. [DOI] [PubMed] [Google Scholar]
- 58.Lancaster CR, Kroger A, Auer M, Michel H. Structure of fumarate reductase from Wolinella succinogenes at 2.2 A resolution. Nature. 1999;402:377–385. doi: 10.1038/46483. [DOI] [PubMed] [Google Scholar]
- 59.Xia C, Hamdane D, Shen AL, Choi V, Kasper CB, Pearl NM, Zhang H, Im SC, Waskell L, Kim JJ. Conformational changes of NADPH-cytochrome P450 oxidoreductase are essential for catalysis and cofactor binding. J Biol Chem. 2011;286:16246–16260. doi: 10.1074/jbc.M111.230532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhu W, Becker DF. Flavin redox state triggers conformational changes in the PutA protein from Escherichia coli. Biochemistry. 2003;42:5469–5477. doi: 10.1021/bi0272196. [DOI] [PubMed] [Google Scholar]
- 61.Wille G, Ritter M, Friedemann R, Mantele W, Hubner G. Redox-triggered FTIR difference spectra of FAD in aqueous solution and bound to flavoproteins. Biochemistry. 2003;42:14814–14821. doi: 10.1021/bi035219f. [DOI] [PubMed] [Google Scholar]
- 62.Kao YT, Saxena C, He TF, Guo L, Wang L, Sancar A, Zhong D. Ultrafast dynamics of flavins in five redox states. J Am Chem Soc. 2008;130:13132–13139. doi: 10.1021/ja8045469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ryan KS, Chakraborty S, Howard-Jones AR, Walsh CT, Ballou DP, Drennan CL. The FAD cofactor of RebC shifts to an IN conformation upon flavin reduction. Biochemistry. 2008;47:13506–13513. doi: 10.1021/bi801229w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sucheta A, Ackrell BA, Cochran B, Armstrong FA. Diode-like behaviour of a mitochondrial electron-transport enzyme. Nature. 1992;356:361–362. doi: 10.1038/356361a0. [DOI] [PubMed] [Google Scholar]
- 65.Pershad HR, Hirst J, Cochran B, Ackrell BA, Armstrong FA. Voltammetric studies of bidirectional catalytic electron transport in Escherichia coli succinate dehydrogenase: comparison with the enzyme from beef heart mitochondria. Biochim Biophys Acta. 1999;1412:262–272. doi: 10.1016/s0005-2728(99)00066-3. [DOI] [PubMed] [Google Scholar]
- 66.Leger C, Heffron K, Pershad HR, Maklashina E, Luna-Chavez C, Cecchini G, Ackrell BA, Armstrong FA. Enzyme electrokinetics: energetics of succinate oxidation by fumarate reductase and succinate dehydrogenase. Biochemistry. 2001;40:11234–11245. doi: 10.1021/bi010889b. [DOI] [PubMed] [Google Scholar]
- 67.Hastings SF, Kaysser TM, Jiang F, Salerno JC, Gennis RB, Ingledew WJ. Identification of a stable semiquinone intermediate in the purified and membrane bound ubiquinol oxidase-cytochrome bd from Escherichia coli. Eur J Biochem. 1998;255:317–323. doi: 10.1046/j.1432-1327.1998.2550317.x. [DOI] [PubMed] [Google Scholar]
- 68.Ruprecht J, Yankovskaya V, Maklashina E, Iwata S, Cecchini G. Structure of Escherichia coli succinate:quinone oxidoreductase with an occupied and empty quinone-binding site. J Biol Chem. 2009;284:29836–29846. doi: 10.1074/jbc.M109.010058. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.