To The Editor
Over the last decade, large-scale, primarily ENU based, chemical mutagenesis projects (Clark et al, 2004) inspired the development of high-throughput, broad coverage means to analyze mutant mouse phenotypes. The dark skin mutations, GnaqMhdadsk1 and GnaqMhdadsk10, arose in just such an ENU mutagenesis screen. These dark skin mutations are due to missense mutations in the Gnaq gene (Van Raamsdonk et al, 2004; Fitch et al, 2003). A third dominant mutation, GnaqM1J, arose an ENU mutagenesis screen using the C57BL/6J mouse strain at The Jackson Laboratory. We report here the phenotype and molecular defect of this new allelic mutation.
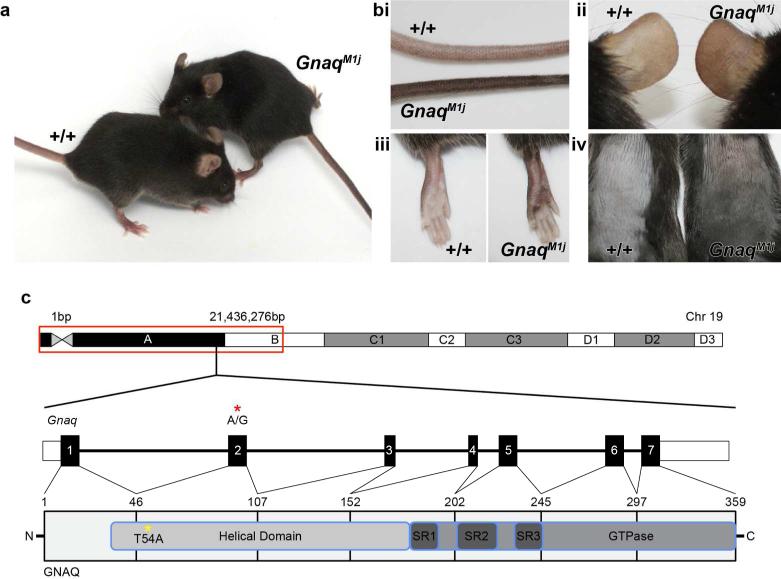
The GnaqM1J deviant mice presented with abnormally pigmented tails inherited in a dominant manner (Figure 1 A). Further observations indicated pigmentary changes throughout the skin of deviant mice (Figure 1 Bi-iv). GnaqM1J mice exhibited unusually dark pigmented tails (Figure 1 Bi), ears (Figure 1 Bii), lower legs and foot pads, (Figure 1 Biii) and dorsal skin (Figure 1 Biv) as compared to wild-type litter mates.
Figure 1.
Mutant mice (Left, A) compared with wildtype controls (Right, C57BL/6J +/+) had unusually darkly pigmented tails (i), ears (ii), lower legs (iii), foot pads (iii) and dorsal skin (iv). (C) SNP mapping identified a 21Mb interval on proximal Chr 19 that includes the causative variant. Gnaq, located within the mapping interval, consists of 7 exons. Targeted exome sequencing identified an A to G substitution in exon 2 of this gene (red asterisk; GAGCA\TCCTT), which results in a T54A change in the helical domain of GNAQ (yellow asterisk).
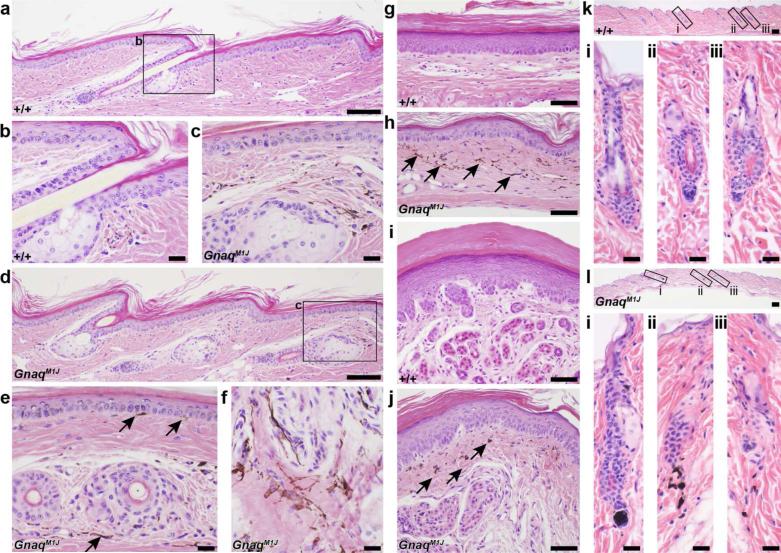
To define histological lesions, a complete set of tissues were harvested from 3 males and 3 females, mutant and control, 12 week old mice and processed routinely. Paraffin sections were stained with hematoxylin and eosin (H&E), and examined by an experienced board certified veterinary anatomic pathologist (JPS) (Silva and Sundberg, 2012). Histological analysis of tail skin taken from wild type C57BL/6J mice demonstrated low levels of interfollicular epidermal pigment and dermal hyper pigmentation (Figure 2 A,B). By contrast, GnaqM1J mutant mice tail sections exhibited high levels of dermal hyper pigmentation and interfollicular epidermal pigmentation in similar regions of the tail (Figure 2 C,D). This was more evident in cross sections of GnaqM1J tail skin (Figure 2E, arrows), especially in oblique sections of the dermis (Figure 2 F). In addition, sections through ventral leg skin (Figure 2G,H) and foot pads with eccrine glands (Figure 2I,J) revealed dermal hyper pigmentation, but no interfollicular pigment in the epidermis. Also, wild-type dorsal skin sections and hair follicles (Figure 2Ki-iii) exhibited no pigment evident in the dermal papilla or residual pigment in the dermis below the hair follicle in the fibrous track of the former catagen follicle, while dorsal skin sections from mutant mouse (Figure 2Li-iii) exhibit abnormal pigmentation in the dermal papilla region extending into the underlying dermis in the fibrous streak remaining at the end of catagen. Immunohistochemistry was also performed whereby paraffin sections from mutant and control animals were stained with antibodies specific to the following antigens: Smooth Muscle Actin, CD31, LYVE1, and VEGFA. The immunohistochemical stains were used to examine vascular abnormalities in these mice, yet no differences were observed between mutant and control mice (data not shown).
Figure 2.
Normal tail skin (A,B) had small amounts of interfollicular epidermal pigmentation and dermal pigmentation compared to the same location in the mutant mice (C,D). This was more evident in cross sections of the tail (E) and oblique sections in the dermis (F). Similarly, ventral leg skin (G,H) and foot pads with eccrine glands (I,J) had dermal hyper-pigmentation in mutant mice (H,J) but not in the controls (G,I). Epidermal pigmentation was not evident in these regions. Arrows indicate areas of pigmentation and enlarged areas of the low magnification tail skin images. GNAQ wildtype mice have no pigment evident in the dermal papilla or residual pigment in the dermis below the hair follicle in the fibrous track of the former catagen follicle (Ki-iii), while mutant animals exhibit abnormal pigmentation in the dermal papilla region extending into the underlying dermis in the fibrous streak remaining at the end of catagen (Li-iii).
SNP mapping was performed to isolate the genomic region containing the mutant allele by backcrossing the original C57BL6/J deviant mouse to the inbred mouse strain FVB/NJ. Mapping studies revealed linkage to proximal mouse chromosome 19 near SNP marker 21,436,276 bp (GRCm38) with a logarithm of odds (LOD) score equal to 5.3. Targeted exome sequencing of Gnaq identified a single base pair variant resulting in an A to G missense mutation in exon 2 at 16,219,611 bp (Figure 1 C). This variant is predicted to result in a Threonine to Alanine change at amino acid 54 (T54A). GNAQ consists of an alpha- helical domain, three flexible switch domains and a GTPase domain (Kumar et al, 2009). The T54A resides in the N-terminus of the alpha-helical domain. The “Sorting Tolerant From Intolerant” (SIFT) algorithm identified the T54A variant as a deleterious mutation in the helical domain of GNAQ (SIFT score 0.01), which may affect proper domain functioning. (The Uniprot Consortium, 2014) (Figure 1 C). Previous ENU induced mutations GnaqMhdadsk1 and GnaqMhdadsk10, affected the C-terminus of the alpha-helical domain and GTPase-domain, respectively (Van Raamsdonk et al 2004, Fitch et al, 2003). All three ENU-induced mutations, affecting different areas of the GNAQ protein, resulted in similar, but not identical, pigmentary changes to the skin of affected mice. Also, the differences in phenotype between the previously described alleles (GnaqMhdadsk1 and GnaqMhdadsk10) and GnaqM1J, may be due to genetic background of the mouse strain, whereby the previous alleles were found on the C3HeB/FeJ strain, with a wild-type agouti locus (A/A), and the new allele was found on the C57BL/6J background, with a mutant agouti allele (a/a) producing only black pigment.
A multitude of signaling pathways operate in the skin, whose coordination is essential for proper skin development and differentiation (Supplementary Figure 1 and 2). Yet, the molecular mechanisms governing these processes are still largely unknown. We report here a new, publicly available, ENU induced mutant (GnaqM1J) that results in similar, but more extensive, skin hyperpigmentation defects as the GnaqMhdadsk1 and GnaqMhdadsk10 alleles (Fitch et al, 2003).
Early in mouse development migrating melanoblasts localize to the dermis and epidermis (Fitch et al, 2003). Previously described mutations in Gnaq (GnaqMhdadsk1 and GnaqMhdadsk10) cause increases in the number of differentiated melanoblasts resulting in increased pigmentation in the dermis of mutant mice (Fitch et al, 2003). Although GnaqM1J is similar to the previous mutations, in that increased dermal pigmentation is observed in adult mice, unlike the previous mutants, it results in both dermal and epidermal hyperpigmentation in tail skin. In light of these mutations, we hypothesize that the genes highlighted in Supplementary Figures 1 and 2 work in a coordinate manner to direct development, localization, and differentiation of skin melanoblasts.
Studies using hairless mice with various degrees of cutaneous pigmentation suggested that pigment may play a role in barrier function (Man et al, 2014). As such, mutations in genes regulating skin pigmentation may seriously affect the function of the skin. Gnaq can be tied to barrier formations by way of its interactions with other barrier genes, like Cav1 (Supplementary Figure 2).
Mutations affecting skin color are infrequent in the mice, as most affect hair color (Dadras et al, 2014; Sundberg and Silva, 2012). Having single gene mutations on the inbred and very commonly used C57BL/6J background provides a new tool for refined evaluation of pigment in the skin of mice.
Supplementary Material
Acknowledgements
The authors thank Jesse Hammer for his technical assistance in preparing the figures.
Research reported in this publication was supported by the National Institutes of Health (R01AR063781 to JPS; CHP,LD, HF supported by P40 OD010972). The Jackson Laboratory Shared Scientific Services were supported in part by a Basic Cancer Center Core Grant from the National Cancer Institute (CA34196). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Abbreviations
- Gnaq
guanine nucleotide binding protein alpha q polypeptide
- M1J
Mutation 1 Jackson
- ENU
N-ethyl-N-nitrosourea
- bp
base pairs
- SNP
single nucleotide polymorphism
- SIFT
Sorting Tolerant From Intolerant algorithm
- GTP
guanosine 5’-triphosphate
- LOD
logarithm of odds
Footnotes
Conflicts of Interest: The authors have no conflicts of interest to report.
References
- Clark AT, Goldowitz D, Takahashi JS, et al. Implementing large-scale ENU mutagenesis screens in North America. Genetica. 2004;122:51–64. doi: 10.1007/s10709-004-1436-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dadras S, Silva KA, King LE, et al. Transplantable malignant melanoma in LT.B6 congenic mice resembling pigmented epitheliod melanocytoma in humans. J Invest Dermatol. 2014;134:1772–1775. doi: 10.1038/jid.2014.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitch KR, McGowan KA, van Raamsdonk CD, et al. Genetics of dark skin in mice. Genes Dev. 2003;17(2):214–228. doi: 10.1101/gad.1023703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar P, Henikoff S, Ng PC. Predicting the effects of coding non-synonymouse variant on protein function using the SIFT algorithm. Nat Protoc. 2009;4(7):1073–81. doi: 10.1038/nprot.2009.86. [DOI] [PubMed] [Google Scholar]
- Man MQ, Lin TK, Santiago JL, et al. Basis for enhanced barrier function of pigmented skin. J. Invest. Dermatol. 2014;134(9):2399–407. doi: 10.1038/jid.2014.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva K, Sundberg J. In: Necropsy Methods. Hedrich H, editor. AcademicPress; London: 2012. pp. 779–806. [Google Scholar]
- Sundberg JP, Silva KA. What color is the skin of a mouse? Vet Pathol Jan. 2012;49(1):142–5. doi: 10.1177/0300985811417244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The UniProt Consortium. Activities at the Universal Protein Resource (UniProt) Nucleic Acids Res. 2014;42(D1):D191–D198. doi: 10.1093/nar/gkt1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Raamsdonk CD, Fitch KR, Fuchs H, et al. Effects of G-protein mutations on skin color. Nat Genet. 2004;36(9):961–8. doi: 10.1038/ng1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.