Abstract
Introduction
The identification of the genetic risk factors that could discriminate non- thrombotic from thrombotic antiphospholipid antibodies (aPLA) carriers will improve prognosis of these patients. Several human studies have shown the presence of aPLAs associated with atherosclerotic plaque, which is a known risk factor for thrombosis. Hence, in order to determine the implication of atherosclerosis in the risk of developing thrombosis in aPLA positive patients, we performed a genetic association study with 3 candidate genes, APOH, LDLR and PCSK9.
Material & Methods
For genetic association study we analyzed 190 aPLA carriers -100 with non-thrombotic events and 90 with thrombotic events- and 557 healthy controls. Analyses were performed by χ2 test and were corrected by false discovery rate. To evaluate the functional implication of the newly established susceptibility loci, we performed expression analyses in 86 aPLA carrier individuals (43 with thrombotic manifestations and 43 without it) and in 45 healthy controls.
Results
Our results revealed significant associations after correction in SNPs located in LDLR gene with aPLA carriers and thrombotic aPLA carriers, when compared with healthy controls. The most significant association in LDLR gene was found between SNP rs129083082 and aPLA carriers in recessive model (adjusted P-value = 2.55 x 10−3; OR = 2.18; 95%CI = 1.49–3.21). Furthermore, our work detected significant allelic association after correction between thrombotic aPLA carriers and healthy controls in SNP rs562556 located in PCSK9 gene (adjusted P-value = 1.03 x 10−2; OR = 1.60; 95%CI = 1.24–2.06). Expression level study showed significantly decreased expression level of LDLR gene in aPLA carriers (P-value <0.0001; 95%CI 0.16–2.10; SE 0.38–1.27) in comparison to the control group.
Discussion
Our work has identified LDLR gene as a new susceptibility gene associated with the development of thrombosis in aPLA carriers, describing for the first time the deregulation of LDLR expression in individuals with aPLAs. Besides, thrombotic aPLA carriers also showed significant association with PCSK9 gene, a regulator of LDLR plasma levels. These results highlight the importance of atherosclerotic processes in the development of thrombosis in patients with aPLA.
Introduction
Antiphospholipid antibodies (aPLAs) are members of a heterogeneous family of immunoglobulins that recognize a variety of phospholipids or proteins that bind to phospholipids. The persistent presence of aPLAs can lead to the development of Antiphospholipid Syndrome (APS), a complex autoimmune disease characterized by venous and/or arterial thrombosis and/or pregnancy morbidity [1,2,3]. As a complex disease, APS is caused by a combination of genetic and environmental factors like some drugs or infections [4,5]. The genetic component involved in the development of APS is still largely unknown but, although there is no published data on familial aggregation, it may be as important as it is for other autoimmune diseases [6,7]. The main cause of death in APS patients is thrombosis, but albeit all APS individuals have aPLAs, only a fraction of APS patients have thrombotic manifestations and moreover, some aPLA carriers are asymptomatic with respect to APS and thrombosis [8,9]. Currently risk factors that discriminate non-thrombotic aPLA carriers from thrombotic aPLA carriers are still largely unknown. Therefore, the identification of the genetic risk factors involved in thrombotic phenotype will improve prognosis of these patients.
Candidate gene association studies and gene expression profiling have identified APS susceptibility genes involved in coagulation, inflammation and innate immune response [10,11,12,13,14,15,16,17,18,19]. However, and despite some experimental evidences connecting atherosclerosis and aPLA, none of these studies have focused their attention on genes related to atherosclerosis in aPLA carriers. It has been proposed that the development of thrombosis is induces by aPLAs through the propagation and amplification of hemostatic, inflammatory and pro-atherogenic responses in absence of physiological regulation [20,21]. Moreover, experimental models of atherosclerosis as well as human studies have described the presence of aPLAs in atherosclerotic plaques [22]. Among the most relevant predictors for arterial thrombosis and atherosclerotic cardiovascular diseases are those that target β2-glycoprotein I (β2GPI), a plasma protein encoded by the APOH gene [23,24]. Atherosclerotic plaques show high levels of β2GPI and oxidized low density lipoproteins (oxLDL), both targets of aPLAs, which can bind forming pro-atherogenic complexes [22,25]. These complexes are considered a risk factor to thrombosis and atherosclerosis in patients with an autoimmune background [26]. Low density lipoproteins (LDL) are removed from vessel by low density lipoprotein receptor (LDLR), encoded by LDLR gene, and their plasma levels are regulated by proprotein convertase subtilisin/kexin type 9 (PCSK9), a serine protease that promotes degradation of LDLR in liver [27]. Hence, the presence of genetic variants in APOH, LDLR and PCSK9 genes could promote pro-atherogenic responses modifying β2GPI and LDL plasma levels [12,28,29,30,31,32].
In this context, our work attempts to determine the implication of atherosclerosis in the risk of developing thrombosis in aPLA positive patients. For this purpose, we designed a candidate gene study with APOH, LDLR and PCSK9 genes, performing genetic association studies and gene expression analyses to compare individuals carrying aPLA with and without thrombosis, and healthy controls.
Materials and Methods
Samples
All subjects included in this study were Spanish Caucasian individuals. For the case group we collected individuals with persistently positive aPLA at medium-high titers from the Autoimmune Disease Research Unit of Hospital Universitario de Cruces (Barakaldo, Spain) during years 2008–2010. In the control group we included healthy individuals without family history of autoimmune diseases from the Basque Biobank for Research-OEHUN (Spain). The protocols for human subjects’ recruitment and study were approved by the Ethics Committee for Clinical Research of the Basque Country, and by the Ethics Committee for Research and Teaching of the University of the Basque Country (UPV/EHU). Samples and data from patients were provided by the Basque Biobank for Research-OEHUN (www.biobancovasco.org) and were processed following standard procedures with appropriate ethical approval. All subjects were informed about the study design and goals, and signed the informed consent.
The criteria for diagnostic of APS was based on the conclusions of the XI Antiphospholipid Antibodies International Congress held in Sydney in 2004 [2]. This criterion considers that a patient suffers the syndrome when displaying at least one clinical (vascular thrombosis and/or pathology of gestation) and one laboratory criteria: lupus anticoagulant (LA), anticardiolipin antibodies (aCL) and/or anti-β2GPI). The diagnostic criteria for systemic lupus erythematosus (SLE) was developed by the American College of Rheumatology in 1982 and revised in 1997. A patient that shows four or more criteria (facial erythema, discoid lupus, photosensitivity, oral ulcers, arthritis, serositis, renal impairment, altered CNS, blood disorder, immunological disorder, antinuclear antibodies) at some time during the clinical course of the disease is considered to suffer SLE. Antiphospholipid antibodies were considered positive when the patient shows aCL and/or LA. aCL must be present in the serum or plasma in medium or high titers (ie, > 40 GPLU / ml or MPLU / ml, or above the 99th percentile of the reference values of the laboratory; GPLU: IgG phospholipid units; MPLU: IgM phospholipid units) more than twice, with at least 12 weeks apart analysis. Detection was by ELISA standardized [33,34,35]. LA must be present in the plasma on two or more occasions at least 12 weeks apart analyzes. The detection was performed according to the indications of the International Society of Thrombosis and Haemostasis, scientific subcommittee antibody LA / phospholipid-dependent [36,37].
For this genetic association study one hundred and ninety aPLA carriers (aPLA+) and five hundred and sixty-two healthy controls were collected. To be considered in the aPLA+ case group, individuals had to exhibit medium-high aPLA levels on at least two occasions, analyzed twelve weeks apart. The aPLA+ group was subclassified as follows: non-thrombotic (aPLA+/th-, n = 100) and thrombotic (aPLA+/th+, n = 90). In the non-thrombotic group, we included patients exhibiting obstetric complications, patients with systemic lupus erythematosus and medium-high aPLA titers, and asymptomatic individuals with medium-high aPLA levels (S1 Table). The thrombotic group included patients with primary or secondary APS with one or more thrombotic manifestations. Four type of comparisons were carried out: aPLA+ vs. healthy controls; aPLA+/th- vs. healthy controls; aPLA+/th+ vs. healthy controls and aPLA+/th- vs. aPLA+/th+. The number and characteristics of individuals included in the study are shown in Table 1. For the gene expression study we analyzed eighty-six aPLA+ individuals, forty-three with thrombotic manifestations (aPLA+/th+), forty-three without thrombotic manifestations (aPLA+/th-), and forty-five healthy controls (Table 1).
Table 1. Characteristics of individuals included in the study.
| Analysis | Subgroup | N | Gender (%Females) | Age at inclusion (Years) |
|---|---|---|---|---|
| Genotyping | aPLA+ | 190 | 73.70% | 50.4±14.7 |
| aPLA+/th– | 100 | 85.00% | 50.3±15.1 | |
| aPLA+/th+ | 90 | 63.30% | 50.5±14.3 | |
| Controls | 557 | 52.90% | 43.2±10.4 | |
| Gene Expression | aPLA+ | 84 | 69.10% | 51.5±13.9 |
| aPLA+/th– | 41 | 85.40% | 51.0±13.6 | |
| aPLA+/th+ | 43 | 53.50% | 52.0±14.3 | |
| Controls | 45 | 82.20% | 45.8±13.1 |
DNA and RNA extraction
Genomic DNA was extracted from whole blood with Flexigen kit (QiagenInc, Valencia, California, USA) at the Basque Biobank for Research-OEHUN. DNA concentration was measured using a NanoDrop Spectrophotometer (NanoDrop Technologies, Inc, Wilmington, DE).
For RNA extraction, fresh peripheral blood mononuclear cells (PBMC) were isolated from heparinized venous blood by centrifugation on a Ficoll-Paque PLUS (GE Healthcare Bio-Sciences, Piscataway, NJ, USA). Total RNA from PBMCs was isolated with TRIzol Reagent (Invitrogen, Carlsbad, CA, USA) followed by RNeasy Mini Kit (Qiagen, Santa Clara, CA, USA). Purified total RNA was retrotranscribed to cDNA using High Capacity cDNA Reverse Transcription Kit (Applied Biosystems, Foster City, CA, USA), according to manufacturer’s instructions.
Genetic association analysis
A description of the candidate genes, including their physical location and biological functions is shown in Table 2. SNP selection was carried out by SNP Browser software (Applied Biosystems). We included SNPs that could affect function and/or transcription levels: non-synonymous SNPs and SNPs located in regulatory regions such as 5´UTR, 3´UTR, microRNA-binding sites, or transcription factor binding sites. We also included tagSNPs to cover the majority of all common variation in each gene. In total, a panel of 50 SNPs was selected (S2 Table). TaqMan® SNP Genotyping Assays (Applied Biosystems) was used for genotyping. This system uses two allele-specific MGB probes and two PCR primers to provide highly robust and accurate genotyping calls. Data were analyzed by Taqman Genotyper Software (Applied Biosystems).
Table 2. Candidate genes included in the study.
| Gene Symbol | Gene Name | Gene ID | Cytoband | SNPs | Biological Functions | References |
|---|---|---|---|---|---|---|
| APOH | Apolipoprotein H | 350 | 17q23-qter | 14 | Blood coagulation | [11,21] |
| Fibrinolysis | ||||||
| Hemostasia | ||||||
| Apoptosis | ||||||
| Response to wounding | ||||||
| LDLR | LDL receptor | 3949 | 19p13.3 | 17 | Response to steroid hormone stimulus | [26–28] |
| Cholesterol homeostasis | ||||||
| LDL particle clearance | ||||||
| PCSK9 | proprotein convertase subtilisin/kexin type 9 | 255738 | 1p32.3 | 19 | Response to hormone stimulus | [23–25] |
| Sterol homeostasis | ||||||
| Apoptosis | ||||||
| Cellular response to stress | ||||||
| LDL particle clearance |
All data were quality-filtered according to the default parameters of Haploview software, v.4.3 [38]: SNPs with a call-rate lower than 90%, or those with deviations from Hardy-Weinberg equilibrium (HWE; p < 0.01) were excluded. After applying the quality control and excluding monomorphic SNPs, 38 SNPs remained for the association analysis. Allelic and genotypic association analyses were performed with PLINK software, v.1.07 [39], obtaining for each comparison the odds ratio (OR) with the 95% confidence interval (95%CI). Statistically significant results were those with p-values < 0.05 after false discovery rate (FDR) correction for multiple testing [40]. To determine statistical power of our analyses we used software CaTS (www.sph.umich.edu/csg/abecasis/CaTS/).
eQTLs analysis
We searched expression quantitative traits loci (eQTLs) in SNPs with significant P-value after correction in any of the subgroups studied. RegulomeDB (http://regulome.stanford.edu/) and Blood eQTL browser (http://genenetwork.nl/bloodeqtlbrowser/) were used to annotate SNPs with known and predicted regulatory elements.
Gene expression analysis
In the second stage, we evaluated the level of mRNA expression of those genes that showed significant genetic association with the presence of persistently high aPLA titers, and/or with a thrombotic phenotype. Forty-five healthy controls, forty-three thrombotic, and forty-three non-thrombotic aPLA carriers were analyzed by Real-time PCRusing TaqMan® Gene Expression Assay (Applied Biosystems). The reactions were run by triplicate on an ABI 7900HT Fast Real-Time PCR System, using standard cycling conditions. Threshold cycle (Ct) values in relative quantification mode were determined using 7900HT v.2.3 Sequence Detection Systems software (Applied Biosystems). The results were imported onto version 2.0 DataAssist™ software (Applied Biosystems), to determine the normalization factor of each candidate endogenous control genes. The YWHAZ gene, whose expression remained unchanged in all samples, was used as endogenous control. To detect statistically significant changes in gene expression between cases and controls, a relative expression software tool (REST) 2009 (Qiagen) was used [41].
Results
Table 3 summarizes the significant allelic associations detected by χ2 test after correction by false discovery rate (FDR). Two significant associations were detected; one located in low-density lipoprotein receptor gene (LDLR), and the other one in proprotein convertase subtilisin/kexin type 9 gene (PCSK9). SNP rs12983082 located in LDLR gene showed significant allelic association when comparing aPLA carriers with healthy controls (adjusted p-value = 0.0103; OR = 1.60; 95%CI = 1.24–2.06). The comparison of thrombotic aPLA carriers with healthy controls revealed significant allelic association in SNP rs562556 in PCSK9 gene (adjusted p-value = 0.0336; OR = 1.84; 95%CI = 1.28–2.66). Furthermore, genotypic association analyses also showed significant results in LDLR gene after correction (Table 4). These associations were evaluated under dominant, additive and recessive models. Thus, the comparison between aPLA carriers and healthy controls revealed significant associations with two SNPs located in LDLR gene, rs129083082 and rs1003723, and in both cases the best fitting model was recessive (rs12983082 adjusted P-value = 2.55 x 10−3; OR = 2.18; 95%CI = 1.49–3.21; rs1003723 adjusted P-value = 0.0106; OR = 1.96; 95%CI = 1.34–2.88). The statistical power of this analysis was 99% (MAF 0.59, OR 2.18, recessive model). Besides, SNP rs12983082 in LDLR gene also showed significant genotypic association when comparing thrombotic aPLA carriers and healthy controls (adjusted P-value = 0.0354; OR = 2.35; 95%CI = 1.42–3.91). Furthermore, with the aim of searching for the genetic factors that could contribute to the onset of thrombotic manifestations within aPLA+ individuals, we also compared thrombotic with non-thrombotic aPLA carriers. This comparison revealed significant allelic and genotypic association of SNP rs562556 located in PCSK9 before correction for multiple testing (Allelic level: P-value = 0.027; OR = 1.972; 95%CI = 1.06–2.80; Genotypic level: P-value = 0.032; OR = 1.92; 95%CI = 1.06–3.52). It could be a promising candidate SNP for differentiating thrombotic with non-thrombotic aPLA carriers to be analyzed in a larger sample.
Table 3. Significant allelic associations detected.
| SNP# ID | Gene | Alleles (1/2) | Subgroup | N | MAFa | P-valueunadj | PFDRb | OR (95%CI) |
|---|---|---|---|---|---|---|---|---|
| rs12983082 | LDLR | C/T | Controls | 557 | 0.472 | |||
| aPLA+ | 190 | 0.588 | 2.71 x 10−4 | 0.0103 | 1.60 (1.24–2.06) | |||
| aPLA+/th+ | 90 | 0.597 | 3.85 x 10−3 | 0.0732 | 1.66 (1.17–2.35) | |||
| aPLA+/th- | 100 | 0.580 | 8.45 x 10−3 | 0.3212 | 1.55 (1.12–2.14) | |||
| aPLA+/th+ vs aPLA+/th-c | 90 | 0.597 | 0.7557 | 0.8906 | 1.07 (0.69–1.67) | |||
| rs562556 | PCSK9 | A/G | Controls | 557 | 0.189 | |||
| aPLA+ | 190 | 0.247 | 1.89 x 10−2 | 0.3263 | 1.40 (1.06–1.87) | |||
| aPLA+/th+ | 90 | 0.301 | 8.85 x 10−4 | 0.0336 | 1.84 (1.28–2.66) | |||
| aPLA+/th- | 100 | 0.200 | 0.7333 | 0.8607 | 1.07 (0.73–1.57) | |||
| aPLA+/th+ vs aPLA+/th-c | 90 | 0.301 | 2.72 x 10−2 | 0.8137 | 1.72 (1.06–2.80) |
a MAF, minor allele frequency.
b P-values adjusted using FDR [40].
c Comparison of aPLA+/th+ (N = 90) with aPLA+/th- (N = 100).
Table 4. Significant genotypic associations detected.
| N(%) | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| SNP# ID | Gene | Alleles (1/2) | Subgroup | N | Model | 1/1 | 1/2+2/2 | P-value unadja | PFDRb | OR (95%CI) |
| rs12983082 | LDLR | C/T | Controls | 557 | 103 (21.3) | 380 (78.7) | ||||
| aPLA+ | 190 | Recessive | 61 (37.2) | 103 (62.8) | 6.71 x 10−5 | 2.55 x 10−3 | 2.18 (1.49–3.21) | |||
| aPLA+/th+ | 90 | Recessive | 30 (38.9) | 47 (61.0) | 9.33 x 10−4 | 0.0354 | 2.35 (1.42–3.91) | |||
| aPLA+/th- | 100 | Recessive | 31 (35.6) | 56 (64.4) | 4.27 x 10−3 | 0.1271 | 2.04 (1.25–3.33) | |||
| aPLA+/th+ vs aPLA+/th-c | 90 | Recessive | 30 (38.9) | 47 (61.0) | 0.6599 | 1 | 1.15 (0.61–2.17) | |||
| rs1003723 | LDLR | T/C | Controls | 557 | 117 (22.6) | 400 (77.4) | ||||
| aPLA+ | 190 | Recessive | 58 (36.5) | 101 (63.5) | 5.56 x 10−4 | 0.0106 | 1.96 (1.34–2.88) | |||
| aPLA+/th+ | 90 | Recessive | 27 (36.5) | 47 (63.5) | 9.33 x 10−4 | 0.1610 | 1.96 (1.17–3.29) | |||
| aPLA+/th- | 100 | Recessive | 31 (36.5) | 54 (63.5) | 6.69 x 10−3 | 0.1271 | 1.96 (1.20–3.19) | |||
| aPLA+/th+ vs aPLA+/th-c | 90 | Recessive | n.d. | n.d. | ||||||
| rs562556 | PCSK9 | A/G | Controls | 557 | 189 (34.9) | 352 (65.1) | ||||
| aPLA+ | 190 | Dominant | 77 (43.3) | 101 (56.7) | 4.65 x 10−2 | 0.2947 | 1.42 (1.05–2.00) | |||
| aPLA+/th+ | 90 | Dominant | 43 (51.8) | 40 (48.2) | 3.46 x 10−3 | 0.1315 | 2.00 (1.26–3.19) | |||
| aPLA+/th- | 100 | Dominant | 34 (35.8) | 61 (64.2) | 0.8722 | 0.9969 | 1.04 (0.65–1.64) | |||
| aPLA+/th+ vs aPLA+/th-c | 90 | Dominant | 43 (51.8) | 40 (48.2) | 3.22 x 10−2 | 0.7766 | 1.92 (1.06–3.52) | |||
a Logistic regression.
b P-values adjusted using FDR [40] Significant P-values in bold.
c Comparison of aPLA+/th+ (N = 90) with aPLA+/th- (N = 100).
* Allele combinations under recessive model. In Dominant model (rs562556), 1/1 genotype includes homozygotes for allele 2 and 1/2+2/2 column includes the heterozygote plus allele 1 homozygote. n.d, no data.
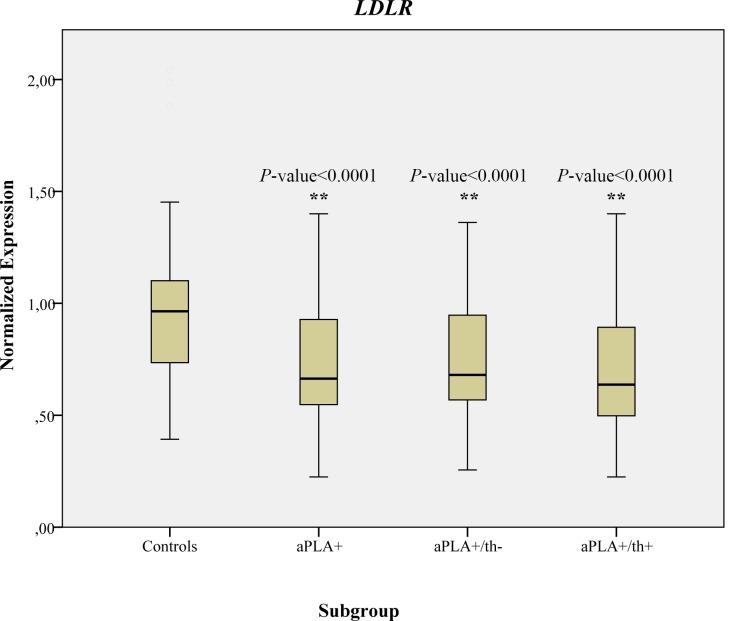
Finally, to evaluate if these SNPs could affect gene expression levels, we searched for data in eQTLs databases, RegulomeDB (http://regulome.stanford.edu/) [42] and Blood eQTL Browser (http://genenetwork.nl/bloodeqtlbrowser/) [43]. Thereby, we found significant putative functional effects related with SNP rs12983082 located in LDLR gene. RegulomeDB scored this SNP as 3a (being 1 the highest score), which means that it is likely to affect transcription factor binding, any motif and DNase peak. Interestingly, the expression of LDLR gene was significantly decreased in aPLA carriers (P-value < 0.0001; 95%CI 0.16–2.10; SE 0.38–1.27) in comparison to control group, regardless of thrombotic phenotype, aPLA+/th- (P-value < 0.0001; 95%CI 0.14–2.14; SE 0.34–1.27) and aPLA+/th+ (P-value < 0.0001; 95%CI 0.17–2.08; SE 0.40–1.27) (Fig 1). PCSK9 was excluded from further analyses given their undetectable level of expression at the mRNA level in PBMCs derived from all samples.
Fig 1. Normalized gene expression of LDLR gene in healthy controls, aPLA+, aPLA+/th- and aPLA+/th+ individuals.
Two asterisks denote statistically significant differences between case and controls (P-value < 0.0001).
Discussion
High aPLA titers are commonly recognized as a thrombosis risk factor, whereby a fraction of antibody carriers will develop the full anti-phospholipid syndrome. Identification of the risk factors that trigger thrombotic manifestations in aPLA carriers should help prevent the development of these symptoms in APS. We searched for susceptibility genes involved in atherogenic processes by a combination of genetic association study and gene expression analyses in aPLA+ individuals with and without thrombosis. Our results have revealed that two genes involved in cholesterol metabolism [44], LDLR and PCSK9, are associated with the presence of aPLA and with the susceptibility of developing thrombosis among individuals with high aPLA titers.
Low density lipoprotein receptor (LDLR) removes from the vessels low density lipoproteins (LDL), which contain cholesterol esters, phospholipids, triglycerides and apolipoproteins [44], and its deficiencies have been previously related with development of hypercholesterolemia and atherosclerotic lesions [45,46]. The reduced LDLR mRNA levels that were observed in the present study in aPLA+ carriers suggest a link between LDLR expression and aPLA production in an autoimmunity-prone background. It has been described that loss of murine ldlr expression in the context of a pro-autoimmune background leads to immune hyperactivity, characterized by increased activation of B and CD4+ T cells, and to an elevated production of aPLAs against cardiolipin and oxidized LDL [47]. Besides, atherosclerotic plaques in humans commonly exhibit high levels of aPLAs, β2GPI and oxidized LDL (oxLDL), all of them aPLAs targets [22,25]. Whether low LDLR expression levels in PBMCs are associated with thrombotic manifestations in patients with aPLAs remains to be clarified.
Our results also detected a significant association between thrombotic aPLA carriers and PCSK9, a gene that encodes a regulator of LDLR plasma levels. Besides, this gene is the only one that showed significant associations before correction in the comparison between thrombotic and non-thrombotic aPLA carriers. All these observations suggest that PCSK9 could play a role in the thrombotic phenotype. Unfortunately, PCSK9 is not expressed at detectable levels in PBMCs and further studies will be necessary to uncover their contribution to thrombosis in patients with aPLAs.
In contrast, we did not find significant genetic association between aPLA carriers and APOH, which has been previously studied in this kind of patients due to the hypothesis that some structural defects in β2GPI could be related with the stimulation of autoantibodies production [22]. Yasuda et al. (2005) detected significant associations, but as in the case of our work, those results could not be replicated in independent cohorts [48,49].
It should be stressed that in the present study we included individuals with arterial and venous thrombosis in thrombotic risk group (aPLA+/thr+), and that the arterial thrombosis was the most common (60%). This fact may explain, in part, the strong association found with atherosclerosis-related genes, but this also suggests that OR (odds ratio) levels of significance could be even higher if the study was focused only on arterial thrombosis in patients with aPLA+. In any case, these results support the widely proposed hypothesis of the implication of atherosclerosis in cardiovascular risk for several autoimmune diseases [26,50].
In summary, our results suggest that LDLR gene and PCSK9 gene might play a role in the production of aPLA and in the development of thrombotic APS, although further validation studies will be needed in order to confirm these findings in larger independent cohorts. Moreover, for the translation of these results to the clinical practice it would be useful the study of specific subgroups as the aPLA high risk profile (presence of lupus anticoagulant (LA) and/or persistent aCL at moderate/high titers), the type/number of thrombotic events (arterial/venous) or the exclusive analysis of primary APS in cohort with sufficient sample size.
Supporting Information
(DOC)
(DOC)
Acknowledgments
We wish to acknowledge the patients enrolled in this study for their participation, Cruces Hospital Research Unit and the Basque Biobank for Research-OEHUN for their collaboration. Technical and human support provided by General Research Services SGIker [University of the Basque Country (UPV/EHU), Ministry of Economy and Competitiveness (MINECO), Basque Government (GV/EJ), European Regional Development Fund (ERDF), and European Social Fund (ESF)] is gratefully acknowledged. We also thank Dr. Martin for his helpful discussions and for his critical review of the manuscript.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
Support was provided by the Basque Government (www.euskadi.eus/) Etortek IE09-256, Saiotek S-PE10UN82, Plan +Euskadi 09UE09+/57, Saiotek-PE08UN73 and Saiotek-PE09UN64; and by the University of the Basque Country (www.ehu.eus/) UFI 11/20. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Meroni PL, Borghi MO, Raschi E, Tedesco F (2011) Pathogenesis of antiphospholipid syndrome: understanding the antibodies. Nat Rev Rheumatol 7: 330–339. 10.1038/nrrheum.2011.52 [DOI] [PubMed] [Google Scholar]
- 2.Miyakis S, Lockshin MD, Atsumi T, Branch DW, Brey RL, et al. (2006) International consensus statement on an update of the classification criteria for definite antiphospholipid syndrome (APS). J Thromb Haemost 4: 295–306. [DOI] [PubMed] [Google Scholar]
- 3.Ruiz-Irastorza G, Crowther M, Branch W, Khamashta MA (2010) Antiphospholipid syndrome. Lancet 376: 1498–1509. 10.1016/S0140-6736(10)60709-X [DOI] [PubMed] [Google Scholar]
- 4.Levy Y, Almog O, Gorshtein A, Shoenfeld Y (2006) The environment and antiphospholipid syndrome. Lupus 15: 784–790. [DOI] [PubMed] [Google Scholar]
- 5.Sene D, Piette JC, Cacoub P (2009) [Antiphospholipid antibodies, antiphospholipid syndrome and viral infections]. Rev Med Interne 30: 135–141. 10.1016/j.revmed.2008.05.020 [DOI] [PubMed] [Google Scholar]
- 6.Cardenas-Roldan J, Rojas-Villarraga A, Anaya JM (2013) How do autoimmune diseases cluster in families? A systematic review and meta-analysis. BMC Med 11: 73 10.1186/1741-7015-11-73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gregersen PK, Behrens TW (2006) Genetics of autoimmune diseases—disorders of immune homeostasis. Nat Rev Genet 7: 917–928. [DOI] [PubMed] [Google Scholar]
- 8.Heit JA (2008) The epidemiology of venous thromboembolism in the community. Arterioscler Thromb Vasc Biol 28: 370–372. 10.1161/ATVBAHA.108.162545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Biggioggero M, Meroni PL (2010) The geoepidemiology of the antiphospholipid antibody syndrome. Autoimmun Rev 9: A299–304. 10.1016/j.autrev.2009.11.013 [DOI] [PubMed] [Google Scholar]
- 10.Bernales I, Fullaondo A, Marin-Vidalled MJ, Ucar E, Martinez-Taboada V, et al. (2008) Innate immune response gene expression profiles characterize primary antiphospholipid syndrome. Genes Immun 9: 38–46. [DOI] [PubMed] [Google Scholar]
- 11.Bertolaccini ML, Atsumi T, Lanchbury JS, Caliz AR, Katsumata K, et al. (2001) Plasma tumor necrosis factor alpha levels and the -238*A promoter polymorphism in patients with antiphospholipid syndrome. Thromb Haemost 85: 198–203. [PubMed] [Google Scholar]
- 12.Chamorro AJ, Marcos M, Miron-Canelo JA, Cervera R, Espinosa G (2012) Val247Leu beta2-glycoprotein-I allelic variant is associated with antiphospholipid syndrome: systematic review and meta-analysis. Autoimmun Rev 11: 705–712. 10.1016/j.autrev.2011.12.006 [DOI] [PubMed] [Google Scholar]
- 13.Galli M, Finazzi G, Duca F, Norbis F, Moia M (2000) The G1691 —> A mutation of factor V, but not the G20210 —> A mutation of factor II or the C677 —> T mutation of methylenetetrahydrofolate reductase genes, is associated with venous thrombosis in patients with lupus anticoagulants. Br J Haematol 108: 865–870. [DOI] [PubMed] [Google Scholar]
- 14.Hamid C, Norgate K, D'Cruz DP, Khamashta MA, Arno M, et al. (2007) Anti-beta2GPI-antibody-induced endothelial cell gene expression profiling reveals induction of novel pro-inflammatory genes potentially involved in primary antiphospholipid syndrome. Ann Rheum Dis 66: 1000–1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kenet G, Sadetzki S, Murad H, Martinowitz U, Rosenberg N, et al. (2000) Factor V Leiden and antiphospholipid antibodies are significant risk factors for ischemic stroke in children. Stroke 31: 1283–1288. [DOI] [PubMed] [Google Scholar]
- 16.Lopez-Pedrera C, Cuadrado MJ, Herandez V, Buendia P, Aguirre MA, et al. (2008) Proteomic analysis in monocytes of antiphospholipid syndrome patients: deregulation of proteins related to the development of thrombosis. Arthritis Rheum 58: 2835–2844. 10.1002/art.23756 [DOI] [PubMed] [Google Scholar]
- 17.Potti A, Bild A, Dressman HK, Lewis DA, Nevins JR, et al. (2006) Gene-expression patterns predict phenotypes of immune-mediated thrombosis. Blood 107: 1391–1396. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 18.Sebastiani GD, Galeazzi M, Tincani A, Scorza R, Mathieu A, et al. (2003) HLA-DPB1 alleles association of anticardiolipin and anti-beta2GPI antibodies in a large series of European patients with systemic lupus erythematosus. Lupus 12: 560–563. [DOI] [PubMed] [Google Scholar]
- 19.Vazquez-Del Mercado M, Garcia-Cobian TA, Munoz Valle JF, Torres-Carrillo N, Martin-Marquez BT, et al. (2007) Genotype Ser413/Ser of PAI-2 polymorphism Ser413/Cys is associated with anti-phospholipid syndrome and systemic lupus erythematosus in a familial case: comparison with healthy controls. Scand J Rheumatol 36: 206–210. [DOI] [PubMed] [Google Scholar]
- 20.Vega-Ostertag ME, Pierangeli SS (2007) Mechanisms of aPL-mediated thrombosis: effects of aPL on endothelium and platelets. Curr Rheumatol Rep 9: 190–197. [DOI] [PubMed] [Google Scholar]
- 21.Lopez-Pedrera C, Buendia P, Cuadrado MJ, Siendones E, Aguirre MA, et al. (2006) Antiphospholipid antibodies from patients with the antiphospholipid syndrome induce monocyte tissue factor expression through the simultaneous activation of NF-kappaB/Rel proteins via the p38 mitogen-activated protein kinase pathway, and of the MEK-1/ERK pathway. Arthritis Rheum 54: 301–311. [DOI] [PubMed] [Google Scholar]
- 22.Sherer Y, Shoenfeld Y (2006) Mechanisms of disease: atherosclerosis in autoimmune diseases. Nat Clin Pract Rheumatol 2: 99–106. [DOI] [PubMed] [Google Scholar]
- 23.Del Papa N, Guidali L, Sala A, Buccellati C, Khamashta MA, et al. (1997) Endothelial cells as target for antiphospholipid antibodies. Human polyclonal and monoclonal anti-beta 2-glycoprotein I antibodies react in vitro with endothelial cells through adherent beta 2-glycoprotein I and induce endothelial activation. Arthritis Rheum 40: 551–561. [DOI] [PubMed] [Google Scholar]
- 24.Fischetti F, Durigutto P, Pellis V, Debeus A, Macor P, et al. (2005) Thrombus formation induced by antibodies to beta2-glycoprotein I is complement dependent and requires a priming factor. Blood 106: 2340–2346. [DOI] [PubMed] [Google Scholar]
- 25.George J, Harats D, Gilburd B, Afek A, Levy Y, et al. (1999) Immunolocalization of beta2-glycoprotein I (apolipoprotein H) to human atherosclerotic plaques: potential implications for lesion progression. Circulation 99: 2227–2230. [DOI] [PubMed] [Google Scholar]
- 26.Matsuura E, Kobayashi K, Lopez LR (2009) Atherosclerosis in autoimmune diseases. Curr Rheumatol Rep 11: 61–69. [DOI] [PubMed] [Google Scholar]
- 27.Zhang DW, Lagace TA, Garuti R, Zhao Z, McDonald M, et al. (2007) Binding of proprotein convertase subtilisin/kexin type 9 to epidermal growth factor-like repeat A of low density lipoprotein receptor decreases receptor recycling and increases degradation. J Biol Chem 282: 18602–18612. [DOI] [PubMed] [Google Scholar]
- 28.Folsom AR, Peacock JM, Boerwinkle E (2009) Variation in PCSK9, low LDL cholesterol, and risk of peripheral arterial disease. Atherosclerosis 202: 211–215. 10.1016/j.atherosclerosis.2008.03.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Abboud S, Karhunen PJ, Lutjohann D, Goebeler S, Luoto T, et al. (2007) Proprotein convertase subtilisin/kexin type 9 (PCSK9) gene is a risk factor of large-vessel atherosclerosis stroke. PLoS One 2: e1043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Levy Z, Rachmani R, Trestman S, Dvir A, Shaish A, et al. (2003) Low-dose interferon-alpha accelerates atherosclerosis in an LDL receptor-deficient mouse model. Eur J Intern Med 14: 479–483. [DOI] [PubMed] [Google Scholar]
- 31.Linsel-Nitschke P, Gotz A, Erdmann J, Braenne I, Braund P, et al. (2008) Lifelong reduction of LDL-cholesterol related to a common variant in the LDL-receptor gene decreases the risk of coronary artery disease—a Mendelian Randomisation study. PLoS One 3: e2986 10.1371/journal.pone.0002986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Martinelli N, Girelli D, Lunghi B, Pinotti M, Marchetti G, et al. (2010) Polymorphisms at LDLR locus may be associated with coronary artery disease through modulation of coagulation factor VIII activity and independently from lipid profile. Blood 116: 5688–5697. 10.1182/blood-2010-03-277079 [DOI] [PubMed] [Google Scholar]
- 33.Tincani A, Allegri F, Sanmarco M, Cinquini M, Taglietti M, et al. (2001) Anticardiolipin antibody assay: a methodological analysis for a better consensus in routine determinations—a cooperative project of the European Antiphospholipid Forum. Thromb Haemost 86: 575–583. [PubMed] [Google Scholar]
- 34.Harris EN, Pierangeli SS (2002) Revisiting the anticardiolipin test and its standardization. Lupus 11: 269–275. [DOI] [PubMed] [Google Scholar]
- 35.Wong RC, Australasian aCL Working Party (2004) Consensus guidelines for anticardiolipin antibody testing. Thromb Res 114: 559–571. [DOI] [PubMed] [Google Scholar]
- 36.Brandt JT, Triplett DA, Alving B, Scharrer I (1995) Criteria for the diagnosis of lupus anticoagulants: an update. On behalf of the Subcommittee on Lupus Anticoagulant/Antiphospholipid Antibody of the Scientific and Standardisation Committee of the ISTH. Thromb Haemost 74: 1185–1190. [PubMed] [Google Scholar]
- 37.Wisløff F, Jacobsen EM, Liesthttpl S (2002). Laboratory diagnosis of the antiphospholipid syndrome. Thromb Res 108: 263–271. [DOI] [PubMed] [Google Scholar]
- 38.Barrett JC, Fry B, Maller J, Daly MJ (2005) Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics 21: 263–265. [DOI] [PubMed] [Google Scholar]
- 39.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, et al. (2007) PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 81: 559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hochberg YBY (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society Series B (Methodological) 57: 12. [Google Scholar]
- 41.Pfaffl MW, Horgan GW, Dempfle L (2002) Relative expression software tool (REST) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res 30: e36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Boyle AP, Hong EL, Hariharan M, Cheng Y, Schaub MA, Kasowski M, et al. (2012) Annotation of functional variation in personal genomes using RegulomeDB. Genome Res, 22, 1790–1797. 10.1101/gr.137323.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Westra HJ, Peters MJ, Esko T, Yaghootkar H, Schurmann C, Kettunen J, et al. (2013) Systematic identification of trans eQTLs as putative drivers of known disease associations. Nat Genet, 45, 1238–1243. 10.1038/ng.2756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Brown MS, Goldstein JL (1979) Receptor-mediated endocytosis: insights from the lipoprotein receptor system. Proc Natl Acad Sci U S A 76: 3330–3337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hobbs HH, Brown MS, Goldstein JL (1992) Molecular genetics of the LDL receptor gene in familial hypercholesterolemia. Hum Mutat 1: 445–466. [DOI] [PubMed] [Google Scholar]
- 46.Goldstein JL, Brown MS (1979) The LDL receptor locus and the genetics of familial hypercholesterolemia. Annu Rev Genet 13: 259–289. [DOI] [PubMed] [Google Scholar]
- 47.Stanic AK, Stein CM, Morgan AC, Fazio S, Linton MF, et al. (2006) Immune dysregulation accelerates atherosclerosis and modulates plaque composition in systemic lupus erythematosus. Proc Natl Acad Sci U S A 103: 7018–7023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Swadzba J, Sanak M, Iwaniec T, Dziedzina S, Musial J (2006) Valine/Leucine247 polymorphism of beta2-glycoprotein I in patients with antiphospholipid syndrome: lack of association with anti-beta2-glycoprotein I antibodies. Lupus 15: 218–222. [DOI] [PubMed] [Google Scholar]
- 49.Yasuda S, Atsumi T, Matsuura E, Kaihara K, Yamamoto D, et al. (2005) Significance of valine/leucine247 polymorphism of beta2-glycoprotein I in antiphospholipid syndrome: increased reactivity of anti-beta2-glycoprotein I autoantibodies to the valine247 beta2-glycoprotein I variant. Arthritis Rheum 52: 212–218. [DOI] [PubMed] [Google Scholar]
- 50.Full LE, Ruisanchez C, Monaco C (2009) The inextricable link between atherosclerosis and prototypical inflammatory diseases rheumatoid arthritis and systemic lupus erythematosus. Arthritis Res Ther 11: 217 10.1186/ar2631 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOC)
(DOC)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.