Abstract
The pathogenesis of chronic thromboembolic pulmonary hypertension (CTEPH) is unknown. Histopathologic studies revealed that pulmonary vasculature lesions similar to idiopathic pulmonary arterial hypertension (PAH) existed in CTEPH patients as well. It’s well-known that genetic predisposition plays an important role in the mechanism of PAH. So we hypothesized that PAH-causing gene mutation might exist in some CTEPH patients and act as a background to facilitate the development of CTEPH. In this study, we analyzed 7 PAH-causing genes including BMPR2, ACVRL1, ENG, SMAD9, CAV1, KCNK3, and CBLN2 in 49 CTEPH patients and 17 patients recovered from pulmonary embolism (PE) but without pulmonary hypertension(PH). The results showed that the nonsynonymous mutation rate in CTEPH patients is significantly higher than that in PE without PH patients (25 out of 49 (51%) CTEPH patients vs. 3 out of 17 PE without PH patients (18%); p = 0.022). Four CTEPH patients had the same point mutation in ACVRL1 exon 10 (c.1450C>G), a mutation approved to be associated with PH in a previous study. In addition, we identified two CTEPH associated SNPs (rs3739817 and rs55805125). Our results suggest that PAH-causing gene mutation might play an important role in the development of CTEPH.
Introduction
Chronic thromboembolic pulmonary hypertension (CTEPH) constitutes Group 4 of the Dana Point classification of pulmonary hypertension (PH) and is considered an uncommon sequel of acute pulmonary thromboembolism (PE). The cumulative incidences of CTEPH range between 1.5% and 5.1% of survived PE [1,2,3]. Why these patients who have survived an acute PE continue to develop CTEPH is largely unknown.
The characteristics of CTEPH is the presence of unresolved thromboemboli undergoing fibrotic organisation. Although CTEPH is understood as a thromboembolic disorder, neither classical plasmatic risk factors for venous thromboembolism nor defects in fibrinolysis are found to be associated with CTEPH [4]. And secondary remodeling processes are believed to occur in distal pulmonary vascular bed of CTEPH patients, which is histologically indistinguishable from idiopathic pulmonary arterial hypertension (PAH) [5]. The pathophysiological mechanism of PAH is, increasingly clear, that contributing factors (such as hypoxia, drugs, toxins, inflammation, etc.) involving the action of vasoconstrictive and remodeling processes on a background of genetic predisposition [6]. Genetic predisposition caused by heterogeneous germline mutation of the bone morphogenetic protein type Ⅱ receptor (BMPR2) gene (BMPR2) has been found to account for approximately 75% of patients with heritable PAH, and up to 25% of idiopathic PAH [7]. Other genes which have been reported to be involved in the PAH pathogenesis so far are: partners in the transforming growth factor (TGF)-β signaling pathway including activin receptor-like kinase type 1 gene (ACVRL1) [8], endoglin gene (ENG) [9], SMAD9 gene (SMAD9) [10], and genes coded for caveolin-1 (CAV1) [11], potassium channel subfamily K, member 3 (KCNK3) [12], cerebellin 2 (CBLN2) [13]. The inheritance pattern of PAH, which is highly variable and with incomplete expression, implies the existence of environmental modifiers with the capability of modulating disease susceptibility. Since CTEPH share similar histopathological vascular lesions to idiopathic PAH and hereditary PAH, we hypothesized that mutations in heritable or idiopathic PAH genes might be found in patients with CTEPH. And the initial PE event might play as a trigger for facilitating the development of CTEPH.
In the present study, we analyzed the genetic variations in CTEPH patients and patients recovered from PE but without PH to explore whether mutations in known PAH-causing genes played a role in the development of CTEPH.
Materials and Methods
Patients
Forty-nine CTEPH patients and 17 PE without PH patients were tested of gene mutation for BMPR2, ACVRL1, ENG, SMAD9, CAV1, KCNK3, and CBLN2. All study subjects belonged to the Chinese Han population, and were among patients admitted to the Center for Pulmonary Vascular Diseases of Fuwai Hospital from Oct. 2012 to Mar. 2015. CTEPH was diagnosed as: (1) a mean pulmonary arterial pressure ≥ 25 mmHg with a pulmonary capillary wedge pressure ≤ 15 mmHg based on right heart catheterization; (2) ventilation/perfusion lung scan and/ or computed tomographic pulmonary angiogram, and pulmonary angiography confirmed chronic thromboembolic obstruction. Patients recovered from PE and without PH were defined as (1) normal presentation of echocardiography and computed tomographic pulmonary angiogram during at least 1-year follow-up; (2) undergoing at least 6 months of therapeutic anticoagulation. Detected point mutations were screened in 120 unrelated healthy Chinese individuals to determine polymorphisms. Before participation, all study subjects signed written information consent. The study was approved by the Human Ethics Committee of Fuwai Hospital and conformed to the 1975 Declaration of Helsinki.
Molecular studies
Genomic DNA was extracted from whole blood with a standard phenol-chloroform protocol. Direct DNA sequencing were performed to detect point mutations and small insertions/ deletions within coding regions and the flanking intron sequences. PCR primers were designed to amplify the coding regions and the intron/exon boundaries of all candidate genes, e.g. BMPR2, ACVRL1, ENG, SMAD9, CAV1, KCNK3, and CBLN2. The sequences of each pair of primers and PCR conditions are displayed in Table 1. The coding DNA sequence 1 of CBLN2 could not amplified by PCR because of technical problem. PCR products were analyzed by using an ABI 3730 XL (Applied Biosystems, Carlsbad, CA, USA). All results were compared with the reference sequences of the known PAH-associated genes including BMPR2 (GenBank accession no. NM_001204.6), ACVRL1 (GenBank accession no. NM_000020.2), ENG (GenBank accession no. NM_000118.2), SMAD9 (GenBank accession no. NM_001127217.2), CAV1 (GenBank accession no. NM_001753.4), KCNK3 (GenBank accession no. NM_002246.2), and CBLN2 (GenBank accession no. NM_182511.3). The standard nomenclature recommended by the Human Genome Variation Society (www.hgvs.org/mutnomen) was employed to number mutations. Frequencies of single nucleotide polymorphisms (SNPs) of all candidate genes were compared to those previously reported in the HapMap database (http://hapmap.ncbi.nih.gov/index.html.en).
Table 1. The primer sequences used for PCR amplification of coding sequences of 7 pulmonary arterial hypertension-associated genes.
| CDS | Forward (5’-3’)Reverse(5’-3’) | Annealing Tm (°C) | Product length (bp) |
|---|---|---|---|
| BMPR2 | |||
| CDS 1 | CACCGAAGCGAAACTTAAGG | 55 | 786 |
| AAGGCGATTTCCCTGGAAG | |||
| CDS 2 | GTCATTCGGATAAGACAAA | 55 | 335 |
| TTTAACATACTCCCATGTCC | |||
| CDS 3 | CCCCCCATGAAATGTCTTTG | 55 | 459 |
| GCCTGGCTTCAACCTTGAAT | |||
| CDS 4 | AGGAGCACATCTACTTGGTGTTTT | 58 | 542 |
| AGTGGCATGGAAAGGGGTAG | |||
| CDS 5 | CTCCCAGAATTTGGCTTTCA | 55 | 454 |
| GTGCCTAGAATAGGCCTTGAC | |||
| CDS 6 | CTGGGTCTGGTAGGAGCTTCA | 55 | 532 |
| CGAGGCTGGTCCTGAACTCT | |||
| CDS 7 | CCTTTCCATCCCTTCCTCTC | 58 | 494 |
| CGTGGGAAAGCTCTTTCTGT | |||
| CDS 8 | GAGTTGAAATTCCGATTTCTCTT | 58 | 463 |
| CCAAGCTGGTCTCGAACTCT | |||
| CDS 9 | TCAGGAAGGGCATTTTATAGGT | 55 | 486 |
| TGCATCCTGCTGCTAATAATGT | |||
| CDS 10 | ATGTGCCTGAAGGGGATGAA | 58 | 396 |
| TTGTGGCATTAGGCAACTCC | |||
| CDS 11 | TCCGTAATCCTTGAAGCCTAA | 55 | 525 |
| GCAGATTTCATCTTGCACTTGT | |||
| CDS 12A | CATTTTTCAGTAGGCTTAATTCAC | 55 | 792 |
| CTGCTGTCCAGTTGCTTCTAC | |||
| CDS 12B | CAGGACTCACGCCAAGTACTG | 55 | 718 |
| CGGGTGTCCTCACCAATAAAC | |||
| CDS 12C | GGCAGCAAGCACAAATCAAA | 55 | 684 |
| CGCCTCAAATGGATCATTTAC | |||
| CDS 13 | TGTCTGGCATTATGAATTTCAAG | 55 | 747 |
| CCTTAAGAAACTGGTCCAAACTG | |||
| ENG | |||
| CDS 1 | CGGTCATACCACAGCCTTCAT | 60 | 684 |
| CGTCGCTGCACAGCATTCT | |||
| CDS 2 | GCCGTTAGCTCATGTCAAGTC | 55 | 305 |
| TGCCTTGGAGCTTCCTCTG | |||
| CDS 3 | ACAGAGCAGGCAGGGAGAGT | 58 | 336 |
| AGACCCTGACCCACAGAGATG | |||
| CDS 4 | ATTCTCAGCTCCGGCCTCT | 60 | 414 |
| GAACTGTGGCACAGCGTGTC | |||
| CDS 5 | ATCTTTGGCTGTGGGTGAGG | 55 | 401 |
| GGTGGGGCTTTATAAGGGAC | |||
| CDS 6 | ACCTGGCCAGGTAAGAGTGC | 58 | 387 |
| CACTCCTGCTGCGTCTTCTG | |||
| CDS 7 | CGAGCTGAGCTGAAGGACAA | 55 | 428 |
| ACAGAGGTGCTTCACCAACAGT | |||
| CDS 8 | CAGCATTGTGGCATCCTTCG | 60 | 861 |
| GCCAGCTCAGGGAGCATTTA | |||
| CDS 9 | GGGGAATGGCTGTGACTT | 55 | 405 |
| GGAGACTAAGCCAACCAATG | |||
| CDS 10 | CCATTGGTTGGCTTAGTCTC | 55 | 301 |
| CTGCTCCGGTCATACAGAAG | |||
| CDS 11 | AGAGTCAGGCAACTCCACAG | 58 | 384 |
| CCTGAGCAATGCCTTCTCT | |||
| CDS 12 | GACTCAGGGGTGGGAACTC | 55 | 496 |
| GGCCACATGCCTGATTAAG | |||
| CDS 13 | AGCTACGAAGCGGTGGAGAT | 55 | 289 |
| CCCTTGCCATGTGCTATGTG | |||
| CDS 14A | AGCCCAGTGAAGCCTCTGA | 60 | 466 |
| TGGCAAGTGGTCTGTCTCCT | |||
| CDS 14B | CAGCCACTGGCTTGGAACA | 60 | 646 |
| CCTGGATGCTGCTACTGTTCA | |||
| ACVRL1 | |||
| CDS 1 | CAGCGGCTGTCACACTTCAT | 58 | 249 |
| CACTCTCCAGTCAAGCTCCTACA | |||
| CDS 2 | TTCTGAGGGAAGGATGACTGA | 58 | 560 |
| CAGACCACTCTGCCAGTTAGAT | |||
| CDS 3 | GGATCTAACTGGCAGAGTGGT | 58 | 386 |
| CCACCGGCTCTAATCTCTG | |||
| CDS 4 | GAGTGAGGAGCTTGCAGTGAC | 55 | 295 |
| ACCGCCTGTGATTCCAGTAG | |||
| CDS 5 | AGCGCAGCATCAAGATGG | 55 | 309 |
| AATGTCTGGAGGTCTGCAAACT | |||
| CDS 6 | CCACCCCCAGACCTAGCTTA | 55 | 545 |
| TCTTGCGGAGGAGAACTGAA | |||
| CDS 7 | CCAGGTCTGCTCTGTGAAGTG | 55 | 520 |
| CTGGCTCCACAGGCTGATT | |||
| CDS 8 | CTCAGGGGTAGCGTGTCCA | 58 | 344 |
| AGGCCTCAGACACAAGTTCC | |||
| CDS 9 | GGCCATCCTCCTCATCTTCT | 58 | 582 |
| CTGGCTTGGCACTCTGACTC | |||
| KCNK3 | |||
| CDS 1 | GACGATGAAGCGGCAGAAC | 55 | 433 |
| GCTGAACTCGGGTTTCTGC | |||
| CDS 2A | TGGACCCAGACCCACAAG | 55 | 575 |
| TCATGAAGCGCAGCACC | |||
| CDS 2B | CAGTACGTGGCCTTCAGCTT | 55 | 684 |
| AGCAGGCACAGTCGGAGAT | |||
| CBLN2 | |||
| CDS 2 | GGCAGTGTTGCTGGTTATCC | 55 | 293 |
| AGCCTCAGAGACCAGGTGAA | |||
| CDS 3 | CAAGCTGCCGGTTCTCTATT | 55 | 622 |
| GAAGGAGGCAGTGCTGAAGTTA | |||
| SMAD 9 | |||
| CDS 1 | TGTGGCCTCTTATGCACTCC | 55 | 614 |
| TGATTGGACAGCTGCCTCAT | |||
| CDS 2 | GTTCCCAAGGGGAAAAACAG | 55 | 559 |
| TCCAGGGTAACTGCTTCAAAAT | |||
| CDS 3 | AGGCTCTCAGAACAACCAGTTT | 55 | 423 |
| CAAAACAGCAGGCCAGTACA | |||
| CDS 4 | GAGCGGCATGCTTGGTCTT | 55 | 515 |
| TGCTTTCCCCAGTGGTAATG | |||
| CDS 5 | TAAGCGGCATTTAGGTCACAC | 58 | 512 |
| TGTCTATCAGAATTGACCGCACA | |||
| CDS 6 | CAGGAAGGTAGAGGCCATGTT | 58 | 731 |
| CTGTAGGGGTTTCACTGCTTTG | |||
| CAV1 | |||
| CDS 1 | TCCACCCCTGCTGAGATGAT | 55 | 498 |
| GGCAGCAGTCGGGATATTTG | |||
| CDS 2 | GTCGGAGCGGTTAGTTCGAT | 55 | 494 |
| GTAACGTTTCTGCCGACTGC | |||
| CDS 3 | TGTGTTCCCAAGTTCCAAGTG | 55 | 780 |
| AGCCAATAAAGCGATGGTTGAT |
CDS: coding DNA sequence.
Thirty CTEPH patients and 14 PE without PH patients who no mutation has been detected by direct DNA sequencing undergone multiple ligation probe amplification (MLPA) analysis for BMPR2, ACVRL1, and ENG by using SALSA MLPA1P093 HHT probe mix kit (MRC-Holland, Amsterdam, the Netherlands). Samples were analyzed using an ABI 3730 Genetic Analyzer, and the results were visualized by the Coffalyser software (MRC-Holland).
Statistical analysis
The demographic and clinical characteristics of different groups were compared using the student’s t tests or the Mann-Whitney U test for continuous variables, and chi-square tests for categorical variables. Differences in the frequencies of targeted gene mutations and the distribution of SNPs between the CTEPH patients and PE without PH patients were evaluated using the Fisher’s exact test. By using the chi-square test, we tested whether the genotype distributions for the studied SNPs were in the Hardy-Weinberg equilibrium. A p value <0.05 were considered as statistical significant. All statistical analyses were performed using the Statistical Package for Social Science (SPSS) 16.0 for Windows (SPSS Inc., Chicago, IL, USA).
Results
Clinical characteristics of the study population
The clinical characteristics of the CTEPH patients and PE without PH patients were displayed on Table 2. The mean±SD age was 51±16 years for CTEPH patients, and 53±18 years for PE without PH patients. Forty-nine percent of CTEPH patients and 41% of PE without PH patients were female. The levels of N-terminal pro-brain natriuretic peptide were significantly higher in CTEPH patients than that in PE without PH patients. While levels of oxygen saturation were significantly lower in CTEPH patients compared with the PE without PH patients.
Table 2. Clinical characteristics of study subjects.
| CTEPH(n = 49) | PE without PH(n = 17) | p value | |
|---|---|---|---|
| Age (years) | 51±16 | 53±18 | 0.646 |
| Female (%) | 24 (49) | 7 (41) | 0.779 |
| BMI (kg/m2) | 22.74±3.26 | 26.13±2.74 | <0.001 |
| NT-proBNP (pg/ml) | 2408.6±1909.7 | 315.6±223.5 | <0.001 |
| SaO2 (%) | 91.14±4.43 | 95.11±3.25 | 0.001 |
| mPAP (mmHg) | 60±14 | ||
| PVR (Wood units) | 14.16±3.48 | ||
| CI (L/min.m2) | 2.53±0.48 | ||
| Previous history of PE | 14 (29) | ||
| Distal (%) | 14 (29) |
CTEPH: chronic thromboembolic pulmonary hypertension; PE: pulmonary embolism; PH: pulmonary hypertension; BMI: body mass index; NT-proBNP: N-terminal pro-brain natriuretic peptide; SaO2: oxygen saturation; mPAP: mean pulmonary artery pressure; PVR: pulmonary vascular resistance; CI: cardiac index.
Mutation rate and distribution
Sixty-six (n = 66) sequences were included for analysis. These sequences were obtained from patients with CTEPH (n = 49) and patients recovered from PE and without PH (n = 17). A total of 40 mutations were identified, including 31 point mutations and 9 large size rearrangements (Table 3). Among them, 5 mutations were synonymous (3 in CTEPH patients, 2 in PE without PH patients).
Table 3. categories of gene mutations in CTEPH patients and PE without PH patients.
| Patient | Point mutation | large size rearrangement | Total | ||
|---|---|---|---|---|---|
| Synonymous | missense | VUS | |||
| CTEPH | 3 | 16 | 9 | 5 | 33 |
| PE without PH | 2 | 1 | 0 | 4 | 7 |
| Total | 5 | 17 | 9 | 9 | 40 |
CTEPH: chronic thromboembolic pulmonary hypertension; PE: pulmonary embolism; PAH: pulmonary arterial hypertension; VUS: variant of unknown significance.
Point mutation analysis is summarized in Table 4. Portions of these transitions were subsequently analyzed in 120 unrelated healthy Chinese individuals (Table A in S1 File). PolyPhen-2 was employed to predict the effect on protein function. Thirteen mutations were predicted to be probably damaging, 2 mutation was predicted to be possibly damaging, and 2 mutations were predicted to be benign. Three patients had multiple point mutations.
Table 4. Gene mutations in Chinese CTEPH patients and PE without PH patients.
| Patientsnumber | Group | Gene | Location | Nucleotide change | Amino acid change | Mutation type |
|---|---|---|---|---|---|---|
| 54 | CTEPH | BMPR2 | 5’-UTR | c.-93A>G | p.? | VUS |
| 183 | PE without PH | Exon3 | c.292G>A | Glu98Lys | Missense | |
| 101 | CTEPH | Exon11 | c.1569T>C | Thr523Thr | Synonymous | |
| 281 | CTEPH | Exon12 | c.1739G>T* | Gly580Val | Missense | |
| 198 | CTEPH | Exon12 | c.2030T>A* | Let677His | Missense | |
| 275 | CTEPH | Exon12 | c.2006A>G* | Asp669Gly | Missense | |
| 285 | CTEPH | Exon12 | c.2325C>A | Ser775Arg | Missense | |
| 248 | CTEPH | Exon12 | c.2357C>G* | Thr786Ser | Missense | |
| 121 | CTEPH | Exon12 | c.2663T>G | Val888Gly | Missense | |
| 198 | CTEPH | ENG | Intron13 | c.111+62A>G | p.? | VUS |
| 77 | CTEPH | 3’-UTR | c.124+277A>G | p.? | VUS | |
| 248 | CTEPH | ACVRL1 | Exon5 | c.583C>T* | Gln195Trp | Missense |
| 198 | CTEPH | Exon8 | c.1196G>T* | Trp399Leu | Missense | |
| 2 | CTEPH | Exon10 | c.1450C>G* | Arg484Gly | Missense | |
| 191 | CTEPH | Exon10 | c.1450C>G* | Arg484Gly | Missense | |
| 263 | CTEPH | Exon10 | c.1450C>G* | Arg484Gly | Missense | |
| 285 | CTEPH | Exon10 | c.1450C>G* | Arg484Gly | Missense | |
| 274 | CTEPH | KCNK3 | Exon1 | c.92C>A* | Ser31Trp | Missense |
| 117 | PE without PH | Exon2 | c.414C>T | Tyr138Tyr | Synonymous | |
| 262 | CTEPH | CBLN2 | 3’-UTR | c.200+108G>C | ? | VUS |
| 285 | CTEPH | 3’-UTR | c.200+630T>C | ? | VUS | |
| 211 | PE without PH | Exon5 | c.648T>C | Phe216Phe | Synonymous | |
| 285 | CTEPH | SMAD9 | 5’-UTR | c.-98C>T | ? | VUS |
| 278 | CTEPH | Exon2 | c.260T>A* | Leu87Gln | Missense | |
| 285 | CTEPH | Exon3 | c.521A>G* | Asn174Ser | Missense | |
| 235 | CTEPH | CAV1 | 5’-UTR | c.-106A>G | p.? | VUS |
| 235 | CTEPH | 5’-UTR | c.-17T>C | p.? | VUS | |
| 287 | CTEPH | Exon2 | c.76A>G | Lys26Glu | Missense | |
| 263 | CTEPH | Exon3 | c.387A>G | Ala129ALa | Synonymous | |
| 191 | CTEPH | Exon3 | c.462C>T | Thr154Thr | Synonymous | |
| 124 | CTEPH | 3’-UTR | c.341+274A>T | p.? | VUS |
CTEPH: chronic thromboembolic pulmonary hypertension; PE: pulmonary embolism; PH: pulmonary hypertension; UTR: untranslated region; VUS: variant of unknown significance. Numbering is based on +1 as A of the ATG initiation codon.
*: missense mutation predicted to be probably damaging by Polyphen-2 software.
The large size rearrangement analysis is summarized in Table 5. Three patients had more than one large size rearrangements.
Table 5. Large size rearrangement identified in CTEPH patients and PE without PH patients.
| Identification number | Diagnosis | Gene | Rearrangement |
|---|---|---|---|
| 270 | CTEPH | BMPR2 exon 1 | c.1-?_c.76+?del |
| ENG exon 6 | c.690-?_c.816+?del | ||
| ENG exon 11–14 | c.1312-?_c.1977+?del | ||
| 282 | CTEPH | BMPR2 exon 1 | c.1-?_c.76+?del |
| ENG exon 12 | c.1429-?_c.1686+?del | ||
| 187 | PE without PH | ENG exon 14 | c.1742-?_c.1977+?dup |
| 603 | PE without PH | ENG exon 5 | C.524-?_c.689+?dup |
| ENG exon 8 | c.992-?_1134+?dup | ||
| ENG exon 14 | c.1742-?_c.1977+?dup |
Abbreviations are in accord with nomenclature guidelines as recommended by the Human Genome Variation Society. CTEPH: chronic thromboembolic pulmonary hypertension; PE: pulmonary embolism; PH: pulmonary hypertension; del: deletion; dup: duplication; c.: coding DNA where nucleotide 1 is the A of the ATG translation initiation codon.
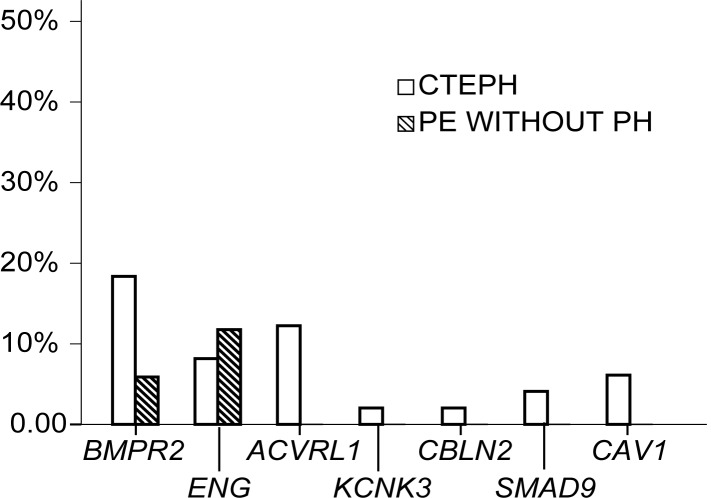
The nonsynonymous mutation rate in CTEPH patients is significantly higher than that in PE without PH patients (25 out of 49 (51%) CTEPH patients vs. 3 out of 17 PE without PH patients (18%); p = 0.022). The mutation rate for individual gene was displayed on Fig 1.
Fig 1. Distribution of nonsynonymous mutations of 7 pulmonary arterial hypertension-causing genes in chronic thromboembolic pulmonary hypertension patients and patients recovered from pulmonary embolism and without pulmonary hypertension.
Polymorphisms
Twelve SNPs which already recorded in the public dbSNP database were detected among all the 66 patients (Table 6). Four single nucleotide changes were considered to be polymorphisms because they were also found in control individuals (Table 6). Genotype distributions of the studied SNPs were all in Hardy-Weinberg equilibrium. We compared the genotype and allele frequencies of these SNPs between the CTEPH patients and the PE without PH patients, and found out that the allele of SNP rs3739817 and SNP rs55805125 were significantly correlated with CTEPH.
Table 6. Outline of SNPs.
| SNPs | Genotype | P value | Allele | P value | OR | |||
|---|---|---|---|---|---|---|---|---|
| rs140683387 | A/A | A/T | T/T | A | T | 1.000 | - | |
| CTEPH | 48(0.98) | 1(0.02) | 0(0) | 1.000 | 97(0.99) | 1(0.01) | ||
| PE without PH | 17(1.0) | 0(0) | 0(0) | 34(1.00) | 0(0) | |||
| rs369291114 | C/C | C/T | T/T | C | T | 1.000 | - | |
| CTEPH | 48(0.98) | 1(0.02) | 0(0) | 1.000 | 97(0.99) | 1(0.01) | ||
| PE without PH | 17(1.0) | 0(0) | 0(0) | 34(1.00) | 0(0) | |||
| rs1061157 | G/G | G/A | A/A | G | A | 0.446 | 2.776(0.36–21.39) | |
| CTEPH | 41(0.84) | 8(0.16) | 0(0) | 0.427 | 90(0.92) | 8(0.08) | ||
| PE without PH | 16(0.94) | 1(0.06) | 0(0) | 33(0.97) | 1(0.03) | |||
| rs148475405 | C/C | C/T | T/T | C | T | 0.450 | 0.347(0.02–5.40) | |
| CTEPH | 48(0.98) | 1(0.02) | 0(0) | 0.452 | 97(0.99) | 1(0.01) | ||
| PE without PH | 16(0.94) | 1(0.06) | 0(0) | 33(0.97) | 1(0.03) | |||
| rs11545664 | G/G | G/A | A/A | G | A | 1.000 | 1.041(0.11–9.67) | |
| CTEPHPE without PH | 46(0.94)16(0.94) | 3(0.06)1(0.06) | 0(0)0(0) | 1.000 | 95(0.97)33(0.97) | 3(0.03)1(0.03) | ||
| rs1800956 | G/G | G/C | C/C | G | C | 1.000 | 0.925(0.26–3.29) | |
| CTEPH | 43(0.88) | 4(0.08) | 2(0.04) | 0.404 | 90(0.92) | 8(0.08) | ||
| PE without PH | 14(0.82) | 3(0.18) | 0(0) | 31(0.91) | 3(0.09) | |||
| rs3739817 | C/C | C/T | T/T | C | T | 0.038 | 0.173(0.03–0.91) | |
| CTEPH | 48(0.98) | 0(0) | 1(0.02) | 0.003 | 96(0.98) | 2(0.02) | ||
| PE without PH | 13(0.76) | 4(0.24) | 0(0) | 30(0.88) | 4(0.12) | |||
| rs55805125 | C/C | C/T | T/T | C | T | 0.016 | 0.087(0.01–0.75) | |
| CTEPH | 48(0.98) | 1(0.02) | 0(0) | 0.014 | 97(0.99) | 1(0.01) | ||
| PE without PH | 13(0.76) | 4(0.24) | 0(0) | 30(0.88) | 4(0.12) | |||
| rs199874575 | C/C | C/T | T/T | C | T | 0.300 | - | |
| CTEPH | 49(1.00) | 0(0) | 0(0) | 0.258 | 98(1.00) | 0(0) | ||
| PE without PH | 16(0.94) | 1(0.06) | 0(0) | 33(0.98) | 1(0.02) | |||
| rs368057934 | C/C | C/T | T/T | C | T | 1.000 | - | |
| CTEPH | 48(0.98) | 1(0.02) | 0(0) | 1.000 | 97(0.99) | 1(0.01) | ||
| PE without PH | 17(1.00) | 0(0) | 0(0) | 34(1.00) | 0(0) | |||
| rs549923058CTEPH | G/G48(0.98) | G/A1(0.02) | A/A0(0) | 1.000 | G97(0.99) | A1(0.01) | ||
| PE without PH | 17(1.00) | 0(0) | 0(0) | 34(1.00) | 0(0) | 1.000 | - | |
| rs79733377 | C/C | C/A | A/A | C | A | 0.450 | 0.347(0.02–5.40) | |
| CTEPH | 48(0.98) | 1(0.02) | 0(0) | 0.452 | 97(0.99) | 1(0.01) | ||
| PE without PH | 16(0.94) | 1(0.06) | 0(0) | 33(0.97) | 1(0.03) | |||
| ENG c.-59C>A | C/C | C/A | A/A | C | A | 1.000 | - | |
| CTEPH | 48(0.98) | 1(0.02) | 0(0) | 1.000 | 97(0.99) | 1(0.01) | ||
| PE without PH | 17(1.00) | 0(0) | 0(0) | 34(1.00) | 0(0) | |||
| CBLN2 exon5 | ||||||||
| c.527A>C | C/C | C/A | A/A | C | A | 0.230 | 1.388(0.84–2.28) | |
| CTEPH | 12(0.24) | 26(0.53) | 11(0.23) | 0.365 | 50(0.51) | 48(0.49) | ||
| PE without PH | 7(0.41) | 8(0.47) | 2(0.12) | 22(0.65) | 12(0.35) | |||
| CBLN2 exon5 | ||||||||
| c.628T>C | T/T | T/C | C/C | T | C | 1.000 | 1.388(0.31–6.22) | |
| CTEPH | 2(0.04) | 4(0.08) | 43(0.88) | 0.704 | 8(0.08) | 90(0.92) | ||
| PE without PH | 0(0) | 2(0.12) | 15(0.88) | 2(0.06) | 32(0.94) | |||
| SMAD9 exon3 | ||||||||
| c.487G>C | G/G | G/C | C/C | G | C | 1.000 | - | |
| CTEPH | 48(0.98) | 1(0.02) | 0(0) | 1.000 | 97(0.08) | 1(0.92) | ||
| PE without PH | 17(0) | 0(0) | 0(0) | 34(1.00) | 0(0) | |||
SNP: single nucleotide polymorphism; CTEPH: chronic thromboembolic pulmonary hypertension; PE: pulmonary embolism; PAH: pulmonary arterial hypertension.
Clinical characteristics of CTEPH patients in mutation carriers and noncarriers
There is no significance of gender, age, previous PE history, and hemodynamic parameters between the mutation carriers and noncarriers CTEPH patients. However, BMI in mutation carriers are lower than the noncarriers (Table 7). Detailed information for each patient was displayed on Table B in S1 File.
Table 7. Comparison of clinical characteristics of CTEPH patients in nonsynonymous mutation carriers and noncarriers.
| Variable | Mutation carrier(n = 19) | Noncarrier(n = 30) | p value |
|---|---|---|---|
| Age (years) | 51±15 | 51±17 | 0.918 |
| Female (%) | 9 (53) | 14 (47) | 0.684 |
| BMI (kg/m2) | 21.55±2.53 | 23.78±3.56 | 0.022 |
| NT-proBNP (pg/ml) | 2311.0±2245.8 | 2474.9±1685.2 | 0.776 |
| SaO2 (%) | 92.23±3.44 | 90.53±4.80 | 0.188 |
| mPAP (mmHg) | 59±16 | 61±12 | 0.640 |
| PVR (Wood units) | 13.29±2.64 | 14.09±3.04 | 0.345 |
| CI (L/min.m2) | 2.68±0.43 | 2.52±0.44 | 0.239 |
| Previous history of PE (%) | 5 (26) | 9 (30) | 0.781 |
| Distal (%) | 5 (26) | 8 (27) | 0.978 |
CTEPH: chronic thromboembolic pulmonary hypertension; BMI: body mass index; NT-proBNP: N-terminal pro-brain natriuretic peptide; mPAP: mean pulmonary artery pressure; PVR: pulmonary vascular resistance; CI: cardiac index; PE: pulmonary thromboembolism.
Discussion
The present study is the first to investigate the association between the genotype of PAH-causing genes and the development of CTEPH. We screened 7 known PAH-causing genes in 49 Chinese CTEPH patients and 17 PE without PH patients. We found that CTEPH patients had a higher frequency of mutations in PAH-causing genes compared with PE without PH patients. And we also found two previously identified SNPs (rs3739817 and rs55805125) were significantly correlated with CTEPH.
The underlying mechanism of CTEPH is largely unknown. Since the characterization of CTEPH is the presence of unresolved thromboemboli undergoing fibrotic organization and secondary remodeling processes of the pulmonary vascular bed [4], previous researches have focused on the imbalance between thrombosis and fibrinolysis. However, no correlation has been found between CTEPH and the classical thromboembolic risk factors such as the deficiency of antithrombin, protein C and protein S, the gene mutation of prothrombin, factor V Leiden, and hyperhomocysteinaemia [14]. In one research, the investigators found that fibrin derived from CTEPH patients was resistant to lysis compared with the healthy controls [15]. But a recent study showed that fibrin resistance to lysis occurred in PAH other than CTEPH and, to a smaller extent, in prior PE without PH patients [16]. These findings imply that CTEPH might develop in a more insidious way other than abnormal coagulation and fibrinolysis, especially in those patients who develop CTEPH after the first episode of PE. This assumption seems plausible if we take it into consideration that only a small part of PE patients developed CTEPH, though the percentage of patients with residual pulmonary thrombi was as high as 52% after 11 months of PE diagnosis [17]. In a series of histopathologic studies in CTEPH patients, researchers reported that there were not only the thrombi obstructions in large vessels, but lesions similar to PH including plexogenic lesions were observed under microscopy [18,19,20]. And these typical plexogenic lesions distributed in both of the no-flow lung tissues and normal-flow lung tissues [20]. These findings suggest that CTEPH might develop in a mechanism similar to PAH.
It’s acknowledged that gene mutation play a pivotal role in PAH. The inheritance pattern is complex due to the incomplete penetrance. The penetrance is influenced by environmental factors such as hormone, appetite-suppressant drugs, concurrent inflammation, reactive oxygen species formation [7]. So we hypothesized that genetic predisposition for PAH might exist in some CTEPH patients, and the PE event might act as a second genetic hit within the pulmonary vasculature to promote the mutation gene expression, which lead to endothelial cell proliferation and finally development of CTEPH. BMPR2 was identified as the first PAH-causing gene in 2000 by two teams of investigators. After that, AVCRL1 (or ALK1) and ENG were reported as the PAH-causing genes in 2001 and 2004 respectively [8,9]. These three genes are all belong to the transforming growth factor-beta (TGF-β) receptor superfamily. The TGF-β superfamily comprises a large series of cytokine growth factors that controls a host of cellular functions, including proliferation, migration, differentiation, apoptosis, and extracellular matrix secretion and deposition. In 2009, several teams identified mutation in SMAD9, another member of TGF-β superfamily, in patients with PAH [7]. In 2012, a mutation in CAV1 (codes for caveolin-1, a membrane protein of caveolae abundant in the endothelium and other cells of the lung) was identified in hereditary PAH patients [11]. In 2013, two novel genes, KCNK3 (the gene encoding potassium channel subfamily K, member 3) and CBLN2 (codes for cerebellin 2) were detected in hereditary and idiopathic PAH patients [12,13].
In the present study, we identified 25 nonsynonymous point mutations and 5 large size rearrangements in 19 out of 49 CTEPH patients, including 9 mutations in BMPR2, 5 mutations in ENG, 6 mutations in ACVRL1, 1 mutation in KCNK3, 2 mutations in CBLN2, 3 mutations in SMAD9, and 4 mutations in CAV1. While in 17 PE without PH patients, 1 patient with BMPR2 point mutation and 2 patients with 4 large size rearrangements of ENG were identified. BMPR2 is the most studied PAH-causing gene. It comprises 13 exons and harbors four distinct functional domains. Over 300 BMPR2 unique mutations have been reported so far [21], which distribute in all 13 exons. Six missense mutation of BMPR2 identified in CTEPH patients in the present study were confined to exon 12, which codes for the cytoplasmic tail. Four of these 6 missense mutations were predicted to be probably damaging by Polyphen-2 software. Surprisingly, we found one mutation in exon 3 in PE without PH patients. Exon 3 codes for an extracellular binding ligand. But the mutation was predicted to be possible damaging by Polyphen-2 software with a score of 0.515. A thorough history and clinical examination did not reveal any sign for PH in this patient or his family. All 6 missense mutations in ACVRL1 detected in CTEPH were predicted to be probably damaging with scores of 1.0. The mutations were located in exon 5, 8, 10. ACVRL1 exon 5 codes for the glycine- serine domain, and exons 6–10 code for the kinase domain, all are critical for signaling activity of ACVRL1. Four CTEPH patients had the same point mutation in ACVRL1 exon 10 (c.1450C>G), a variant which had been observed cosegregated with PH in a hereditary hemorrhagic telangiectasia kindred [8]. And histologic assessment had demonstrated that the affected patients had characteristic features of end-stage plexogenic PH similar to those seen in patients with idopathic PH.
The SMAD signal pathway is most strongly associated with driving the differentiation state in development. And the SMAD9 variants are more convincingly associated with PAH among all SMADs according to previous researches [7]. In the present study, we found 2 SMAD9 missense mutations, which were predicted to be probably damaging. KCNK3 encodes an pH-sensitive potassium channel protein that contains two pore-forming P domains. KCNK3 channels are major contributors to the resting potential in human pulmonary-artery smooth muscle cells. Gurney AM, et al [22] suggested that the pH-sensitive channels could be responsible for the modulatory effects of pH on hypoxic pulmonary vasoconstriction. In the present study, we found a missense mutation in KCNK3 exon 1 in one CTEPH patient. The mutation was predicted to be probably damaging with a score of 1.0. It is plausible that the KCNK3 mutation might play an important role in CTEPH after the initial PE-induced hypoxia in pulmonary vasculature. CAV1 encodes caveolin-1, which is the predominant member of three proteins (caveolin-1, caveolin-2, and caveolin-3) that coat the flask-like invaginations of the plasma membrane known as caveolae. Signaling cascades relevant to PAH such as the TGF-β superfamily, nitric oxide pathway, and G-protein coupled receptors rely heavily on proper caveolar function [23]. We identified one missense mutation in CAV1 exon2, which was predicted to be possible damaging.
In addition, we found mutations in untranslated regions and introns of BMPR2, ENG, SMAD9, CAV1, and CBLN2 (encodes cerebellin 2, which highly expressed in lungs of PAH patients). And we found a significant association between the genotypes of 2 SNPs rs3739817 (ENG exon8) and rs55805125 (ACVRL1 exon 7) with CTEPH. Although there was no encoding amino acid changes resulting from these mutations and polymorphisms, they may modify gene expression, or cause aberrant pre-mRNA splicing due to alternations in sequences recognized by various splicing factors.
Lack of functional analyses of these mutations is the main limitation of the present study. But previous study provided a convincing evidence for the association between one specific mutation in ACVRL1 exon 10 with PH. Besides, the relatively small sample of patients hindered a thorough mapping of PAH-causing genes variants in CTEPH patients.
Conclusions
Based on direct sequencing and MLPA method, we identified 25 nonsynonymous point mutations, 5 large size rearrangements, and 2 CTEPH associated SNPs of 7 PAH-causing genes (BMPR2, ACVRL1, ENG, SMAD9, CAV1, KCNK3, and CBLN2) out of 49 CTEPH patients. Among them, one specific mutation in ACVRL1 exon 10 shared by 4 CTEPH patients had been approved to be associated with PH in a previous study. The high frequency of mutations of PAH-causing genes might be associated with CTEPH in Chinese population.
Supporting Information
(RAR)
Acknowledgments
The authors thankfully acknowledge the staff members of the ward of Pulmonary Vascular Disease at Fuwai Hospital for their assistance in data collection.
Abbreviations
- BMPR2
bone morphogenetic protein type Ⅱ receptor
- CTEPH
chronic thromboembolic pulmonary hypertension
- MLPA
multiple ligation probe amplification
- PAH
pulmonary arterial hypertension
- PE
pulmonary thromboembolism
- PH
pulmonary hypertension
- SNP
single nucleotide polymorphism
- TGF
transforming growth factor
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This study was supported by a grant from National Key Technology R&D Program in the 12th Five year Plan of China (2011BA11B17). ZHL recieved the funding.
References
- 1.Becattini C, Agnelli G, Pesavento R, Silingardi M, Poggio R, et al. (2006) Incidence of chronic thromboembolic pulmonary hypertension after a first episode of pulmonary embolism. Chest 130: 172–175. [DOI] [PubMed] [Google Scholar]
- 2.Pengo V, Lensing AW, Prins MH, Marchiori A, Davidson BL, et al. (2004) Incidence of chronic thromboembolic pulmonary hypertension after pulmonary embolism. N Engl J Med 350: 2257–2264. [DOI] [PubMed] [Google Scholar]
- 3.Ribeiro A, Lindmarker P, Johnsson H, Juhlin-Dannfelt A, Jorfeldt L (1999) Pulmonary embolism: one-year follow-up with echocardiography doppler and five-year survival analysis. Circulation 99: 1325–1330. [DOI] [PubMed] [Google Scholar]
- 4.Lang I (2010) Advances in understanding the pathogenesis of chronic thromboembolic pulmonary hypertension. Br J Haematol 149: 478–483. 10.1111/j.1365-2141.2010.08142.x [DOI] [PubMed] [Google Scholar]
- 5.Delcroix M, Vonk Noordegraaf A, Fadel E, Lang I, Simonneau G, et al. (2013) Vascular and right ventricular remodelling in chronic thromboembolic pulmonary hypertension. Eur Respir J 41: 224–232. 10.1183/09031936.00047712 [DOI] [PubMed] [Google Scholar]
- 6.Tuder RM, Archer SL, Dorfmuller P, Erzurum SC, Guignabert C, et al. (2013) Relevant issues in the pathology and pathobiology of pulmonary hypertension. J Am Coll Cardiol 62: D4–12. 10.1016/j.jacc.2013.10.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Soubrier F, Chung WK, Machado R, Grunig E, Aldred M, et al. (2013) Genetics and genomics of pulmonary arterial hypertension. J Am Coll Cardiol 62: D13–21. 10.1016/j.jacc.2013.10.035 [DOI] [PubMed] [Google Scholar]
- 8.Trembath RC, Thomson JR, Machado RD, Morgan NV, Atkinson C, et al. (2001) Clinical and molecular genetic features of pulmonary hypertension in patients with hereditary hemorrhagic telangiectasia. N Engl J Med 345: 325–334. [DOI] [PubMed] [Google Scholar]
- 9.Chaouat A, Coulet F, Favre C, Simonneau G, Weitzenblum E, et al. (2004) Endoglin germline mutation in a patient with hereditary haemorrhagic telangiectasia and dexfenfluramine associated pulmonary arterial hypertension. Thorax 59: 446–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huang Z, Wang D, Ihida-Stansbury K, Jones PL, Martin JF (2009) Defective pulmonary vascular remodeling in Smad8 mutant mice. Hum Mol Genet 18: 2791–2801. 10.1093/hmg/ddp214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Austin ED, Ma L, LeDuc C, Berman Rosenzweig E, Borczuk A, et al. (2012) Whole exome sequencing to identify a novel gene (caveolin-1) associated with human pulmonary arterial hypertension. Circ Cardiovasc Genet 5: 336–343. 10.1161/CIRCGENETICS.111.961888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ma L, Roman-Campos D, Austin ED, Eyries M, Sampson KS, et al. (2013) A novel channelopathy in pulmonary arterial hypertension. N Engl J Med 369: 351–361. 10.1056/NEJMoa1211097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Germain M, Eyries M, Montani D, Poirier O, Girerd B, et al. (2013) Genome-wide association analysis identifies a susceptibility locus for pulmonary arterial hypertension. Nat Genet 45: 518–521. 10.1038/ng.2581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lang IM, Pesavento R, Bonderman D, Yuan JX (2013) Risk factors and basic mechanisms of chronic thromboembolic pulmonary hypertension: a current understanding. Eur Respir J 41: 462–468. 10.1183/09031936.00049312 [DOI] [PubMed] [Google Scholar]
- 15.Morris TA, Marsh JJ, Chiles PG, Auger WR, Fedullo PF, et al. (2006) Fibrin derived from patients with chronic thromboembolic pulmonary hypertension is resistant to lysis. Am J Respir Crit Care Med 173: 1270–1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Miniati M, Fiorillo C, Becatti M, Monti S, Bottai M, et al. (2010) Fibrin resistance to lysis in patients with pulmonary hypertension other than thromboembolic. Am J Respir Crit Care Med 181: 992–996. 10.1164/rccm.200907-1135OC [DOI] [PubMed] [Google Scholar]
- 17.Nijkeuter M, Hovens MM, Davidson BL, Huisman MV (2006) Resolution of thromboemboli in patients with acute pulmonary embolism: a systematic review. Chest 129: 192–197. [DOI] [PubMed] [Google Scholar]
- 18.Haythe J (2012) Chronic thromboembolic pulmonary hypertension: a review of current practice. Prog Cardiovasc Dis 55: 134–143. 10.1016/j.pcad.2012.07.005 [DOI] [PubMed] [Google Scholar]
- 19.Yi ES, Kim H, Ahn H, Strother J, Morris T, et al. (2000) Distribution of obstructive intimal lesions and their cellular phenotypes in chronic pulmonary hypertension. A morphometric and immunohistochemical study. Am J Respir Crit Care Med 162: 1577–1586. [DOI] [PubMed] [Google Scholar]
- 20.Moser KM, Bloor CM (1993) Pulmonary vascular lesions occurring in patients with chronic major vessel thromboembolic pulmonary hypertension. Chest 103: 685–692. [DOI] [PubMed] [Google Scholar]
- 21.Ma L, Chung WK (2014) The genetic basis of pulmonary arterial hypertension. Hum Genet 133: 471–479. 10.1007/s00439-014-1419-3 [DOI] [PubMed] [Google Scholar]
- 22.Gurney AM, Osipenko ON, MacMillan D, McFarlane KM, Tate RJ, et al. (2003) Two-pore domain K channel, TASK-1, in pulmonary artery smooth muscle cells. Circ Res 93: 957–964. [DOI] [PubMed] [Google Scholar]
- 23.Mathew R, Huang J, Shah M, Patel K, Gewitz M, et al. (2004) Disruption of endothelial-cell caveolin-1alpha/raft scaffolding during development of monocrotaline-induced pulmonary hypertension. Circulation 110: 1499–1506. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(RAR)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.