Abstract
In patients with CKD, muscle wasting is common and is associated with morbidity and mortality. Mechanisms leading to loss of muscle proteins include insulin resistance, which suppresses Akt activity and thus stimulates protein degradation via the ubiquitin-proteasome system. However, the specific factors controlling CKD-induced suppression of Akt activity in muscle remain undefined. In mice with CKD, the reduction in Akt activity in muscle exceeded the decrease in upstream insulin receptor substrate-1–associated phosphatidylinositol 3–kinase activity, suggesting that CKD activates other pathways that suppress Akt. Furthermore, a CKD-induced increase uncovered caspase-3 activity in muscle in these mice. In C2C12 muscle cells, activated caspase-3 cleaves and activates Rho-associated protein kinase 1 (ROCK1), which enhances the activity of phosphatase and tensin homolog (PTEN) and reduces Akt activity. Notably, constitutive activation of ROCK1 also led to increased caspase-3 activity in vitro. In mice with either global ROCK1 knockout or muscle-specific PTEN knockout, CKD-associated muscle proteolysis was blunted. These results suggest ROCK1 activation in CKD and perhaps in other catabolic conditions can promote loss of muscle protein via a negative feedback loop.
Keywords: chronic kidney disease, metabolism, cell signaling
It has been known for decades that CKD and its complications (e.g., metabolic acidosis, excess glucocorticoid production plus inflammation and/or impaired insulin/IGF-1 signaling) stimulate the loss of muscle proteins, thereby contributing to CKD-induced morbidity and mortality.1–3 Mechanisms for the losses of muscle protein have focused on a common group of biochemical and transcriptional adaptations that result in muscle atrophy. For example, expression of the muscle-specific, E3 ubiquitin ligases, Atrogin-1/MAFbx and MuRF-1, has been associated with activation of muscle proteolysis in the ubiquitin-proteasome system (UPS).4–7 Moreover, defects in insulin/IGF-1 signaling in muscle are recognized as a major mechanism causing protein losses.8,9
Another protease that participates in muscle protein catabolism is caspase-3. Activation of caspase-3 increases the degradation of muscle proteins in two ways: first, caspase-3 cleaves actomyosin and myofibrillar proteins producing substrates that are degraded in the UPS.10 Second, activated caspase-3 cleaves specific subunits of the 19S proteasome particle, and this in turn stimulates proteolysis in the 26S proteasome.11 When caspase-3 is inhibited by overexpression of XIAP in muscle, muscle wasting is suppressed in mouse models of CKD.12 Caspase-3 also affects signal transduction because caspase-3 activation in Jurkat T cells stimulates apoptosis through an interaction between caspase-3 and Rho-associated kinase 1 (ROCK1). The interaction involves cleavage of an inhibitory Cys domain at the C-terminus of ROCK1, producing an increase in ROCK1 activity.13,14 Activated ROCK1 reportedly stimulates cell migration, autophagy, differentiation, and apoptosis in different types of cells or tissues.15–19 In cultured embryonic kidney cells, activated ROCK1 phosphorylates phosphatase and tensin homolog (PTEN) creating phosphatase activity.20,21 The action of PTEN reduces the level of phosphatidylinositol (3,4,5)-trisphosphate and impairs Akt activity. This is relevant to CKD-induced muscle protein losses because we have found that activated PTEN stimulates muscle proteolysis in the UPS.22,23
We examined the kinase activity of Akt in a mouse model of CKD and found it was unexpectedly lower than the kinase activity of phosphatidylinositol 3–kinase (PI3K). We also uncovered a new, CKD-induced pathway that stimulates muscle protein wasting via activation of caspase-3, which stimulates ROCK1 kinase activity. ROCK1 in turn stimulates PTEN activity, which suppresses the kinase activity of Akt and stimulates muscle protein degradation. Our results provide a new mechanism contributing to the CKD-induced muscle protein degradation and suggest therapeutic targets for blocking the muscle wasting.
Results
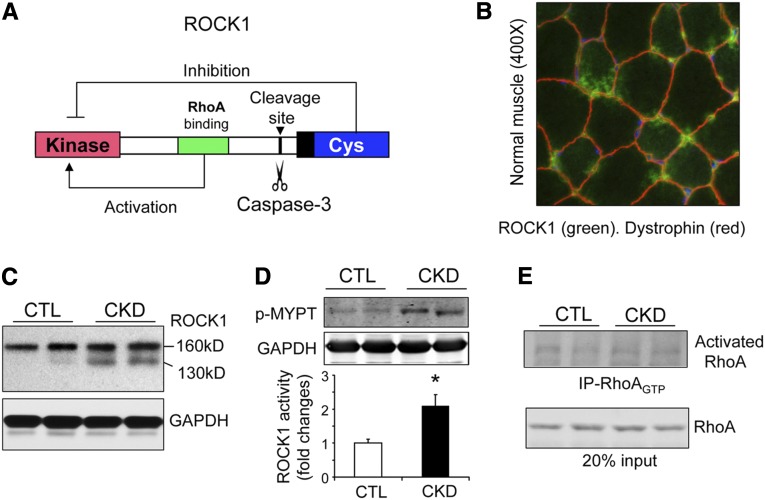
ROCK1 Is Activated in Skeletal Muscles of Mice with CKD
Mammalian ROCK1 can be activated by either a small GTPase, which binds the Rho domain, or by removal of the inhibitory Cys domain.14 In the latter case, caspase-3 recognizes and cleaves the DETD-conserved consensus sequence at amino acid position 1113 of ROCK1 (Figure 1A). When ROCK1 is activated in epithelial or endothelial cells by proteolytic cleavage, the cells undergo apoptosis and death.19 In skeletal muscle of normal mice, we found abundant expression of ROCK1 (Figure 1B). In skeletal muscles of mice with CKD, there was an expression of the intact ROCK1 protein (160 kD) plus an increase in the cleaved 130-kD fragment of activated ROCK1 (Figure 1C). To assess the physiologic relevance of ROCK1 cleavage, we measured ROCK1 activity in muscles using the MYPT1 substrate.24 Notably, there was a 2.6-fold increase in ROCK1 activity (P<0.05) in muscles of CKD mice versus results from muscles of control mice (Figure 1D). Because RhoA GTPase is an upstream activator of ROCK1,14 we also measured RhoA activity; there was no evidence of increased RhoA activity (Figure 1E). We conclude that ROCK1 is activated in muscles of CKD mice principally by cleavage of ROCK1.
Figure 1.
ROCK1 is activated in muscles of mice with CKD. (A) A structural diagram of ROCK1. When RhoA is bound to the Rho domain (green), it activates ROCK1. Alternatively, caspase-3 can remove the inhibitory Cys domain (blue) to activate ROCK1 constitutively. (B) Immunofluorescent staining of ROCK1 in myofibers of a normal mouse. Original magnification ×400. (C) ROCK1 protein levels were assessed by western blots in skeletal muscles of control (CTL) and CKD mice. Note the 130-kD fragment of cleaved ROCK1 is present in muscle of CKD mice. (D) ROCK1 activity assay revealed that the phosphorylation of MYPT (p-MYPT) increased in muscles of CKD mice (*P<0.01; n=5). (E) RhoA activity assay demonstrated that CKD does not activate RhoA in muscle.
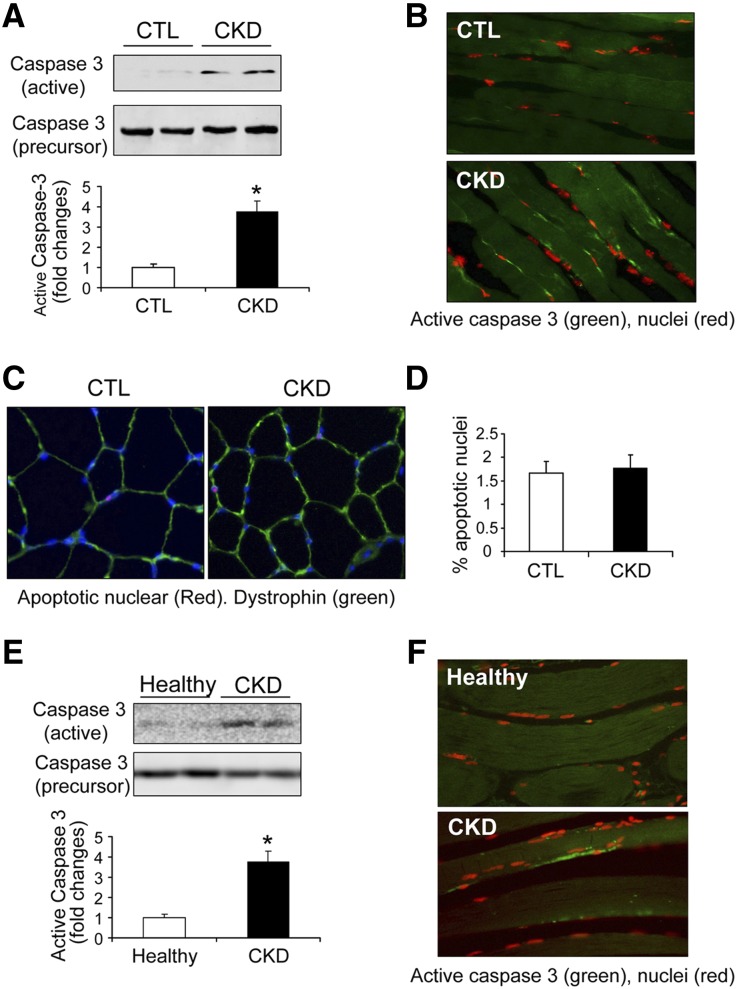
CKD Stimulates Caspase-3 in Skeletal Muscle To Activate ROCK1
Previously, we uncovered that CKD activates caspase-3 in muscles of rats leading to a significant increase in the 19-kD, active fragment of caspase-3.10,25 In these experiments, the 19-kD active fragment of caspase-3 was present in muscles of mice with CKD (Figure 2A). The presence of caspase-3 activity was confirmed by immunofluorescent staining of active caspase-3 in myofibers of muscles in mice with CKD (Figure 2B). These results demonstrate that active caspase-3 is located only in the cytoplasm and not in the nuclei of myofibers indicating that activated caspase-3 does not stimulate apoptosis. The absence of apoptosis was verified by negative results of a terminal deoxynucleotidyl transferase–mediated digoxigenin-deoxyuridine nick-end labeling (TUNEL) assay in myofibers (Figure 2, C and D).
Figure 2.
CKD activates caspase-3 in skeletal muscles. (A) Representative western blots of active caspase-3 (19 kD) and caspase-3 precursor (32 kD) in muscles from control (CTL) and CKD mice. CKD increases active caspase-3 by 3.7-fold (*P<0.01; n=5). (B) Immunofluorescent staining of active caspase-3 (green) in muscles of CTL and CKD mice. Nuclei were visualized with propidium iodide (red). (C) and (D) TUNEL assay of myofibers of CTL and CKD mice. Apoptotic nuclei (arrow) were rarely seen in muscles of CTL and CKD mice. Blue represents DAPI, and green represents dystrophin staining. Apoptotic nuclei were not significantly increased in muscles of mice with CKD versus CTL results (n=5). (E) Representative western blots of active caspase-3 and caspase-3 precursor in human muscle samples. The active caspase-3 was significantly increased in muscles of patients with CKD (*P<0.05; n=3–5). (F) Immunofluorescent staining of active caspase-3 (green) in muscles of healthy patients and patients with CKD. Red staining (propidium iodide) indicates nuclei.
To determine if caspase-3 is activated in muscles of patients with CKD, we examined western blots of samples of abdominal muscles obtained from CKD and healthy individuals of similar ages and sexes. Activated caspase-3 was detected in muscles from patients with CKD versus control subjects (Figure 2E). Immunofluorescence microscopy confirmed that active caspase-3 was mainly located in myofibers (Figure 2F). Therefore, both in rodent models of CKD and in patients with CKD, caspase-3 is activated in skeletal muscles.
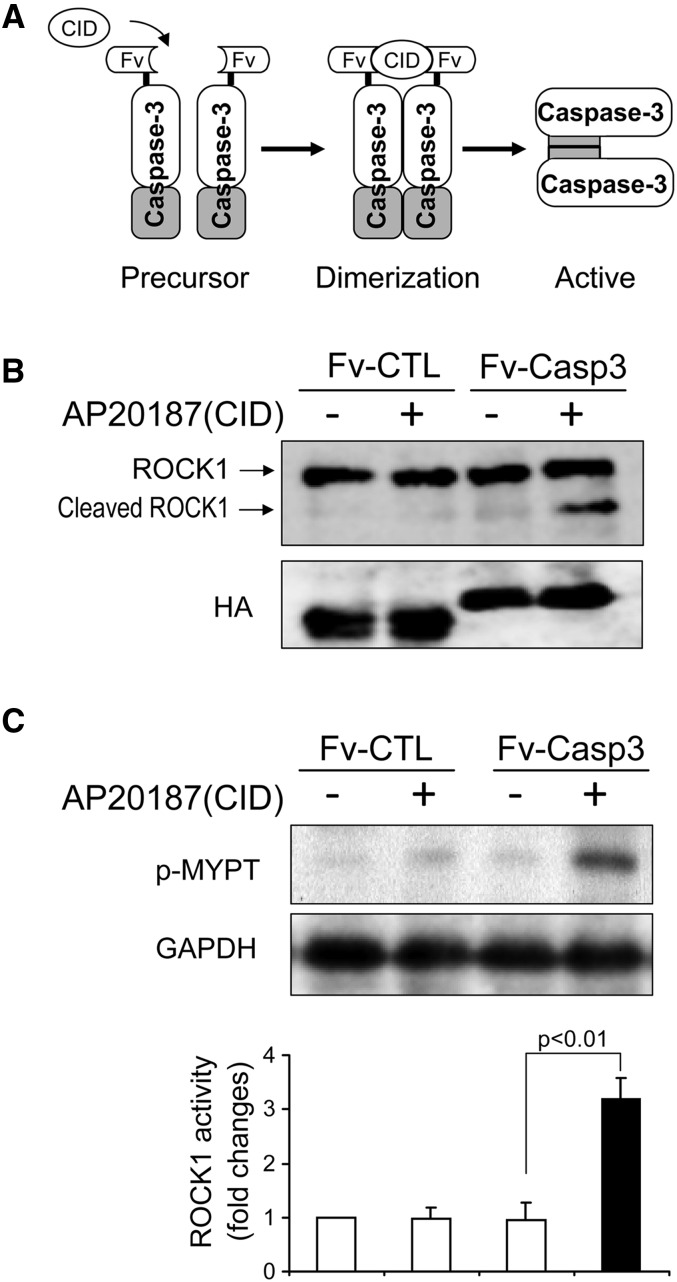
To ascertain if there is linkage between activated caspase-3 and ROCK1 activity, we examined C2C12 myoblasts. First, we activated caspase-3 in C2C12 myoblasts by transfecting them with a plasmid that expresses inducible activity of caspase-3 (M-Fv-Casp 3).26 In this strategy, a caspase-3 precursor protein is fused with a protein that serves as a chemical binding domain. When a chemical inducer is present, it stimulates formation of dimers of the precursor protein to form active caspase-3 (Figure 3A). This strategy can avoid the potential multiple effects of apoptosis inducers. In C2C12 myoblasts, being transfected with M-Fv-Casp 3 did not affect ROCK1 activity. However, the chemical inducer, AP20187,27 rapidly activated caspase-3, which cleaved ROCK1, producing the activated, 130-kD form of ROCK1 (Figure 3B). As expected, increased cleavage of ROCK1 raised ROCK1 kinase activity (Figure 3C), suggesting that ROCK1 activation in muscles of mice with CKD is mediated at least in part by caspase-3.
Figure 3.
Caspase-3 activation leads to increased activity of ROCK1. (A) Diagram of a chemical inducible activation caspase-3 (M-Fv-Casp 3). A plasmid expressing a caspase-3 precursor protein fused to a chemically inducible dimerization (CID) domain. The chemical, AP20187, can induce caspase-3 precursors to form a dimmer becoming active caspase-3. Plasmids with deletion caspase-3 precursor deletion (M-Fv-CTL) serve as control (CTL). Both plasmids contain a hemagglutinin tag (HA tag). (B) Addition of AP20187 (CID) triggers an activation of caspase-3, which cleaves ROCK1 to yield an active, 130-kD ROCK1 in C2C12 myoblasts. Western blot of HA tag indicates equal transfection in each group. (C) ROCK1 activity was evaluated by the level of phosphorylated MYPT (p-MYPT) (n=3).
Activated ROCK1 Stimulates PTEN to Diminishing Akt Activity
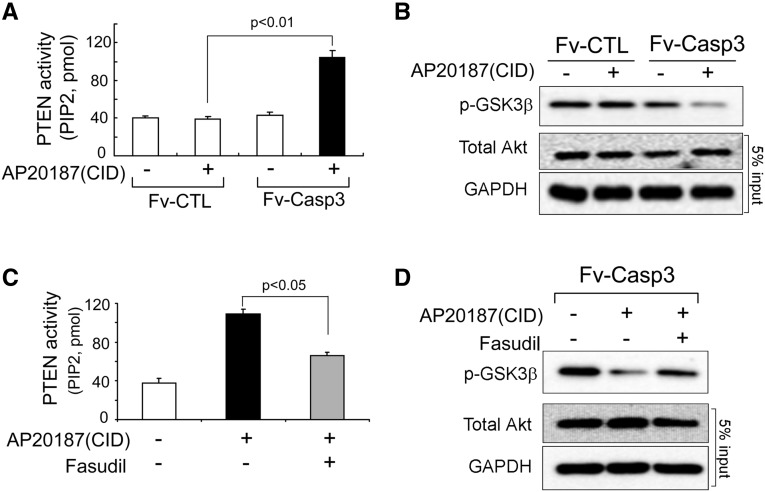
To ascertain if ROCK1 stimulates PTEN activity, we studied C2C12 myoblasts transfected with M-Fv-Casp 3 to activate ROCK1. When the inducer, AP20187, was added, PTEN activity increased causing Akt activity to decrease (Figure 4, A and B). To examine whether ROCK1 mediated the activation of PTEN, we pretreated C2C12 cells with a ROCK1 inhibitor (100 µg fasudil/ml), followed by activation of caspase-3 by adding AP20187. As shown in Figure 4C, fasudil diminished the activation of PTEN resulting in an increase in Akt activity (Figure 4D). Therefore, in skeletal muscle cells, activated caspase-3 may stimulate a pathway from ROCK1 to an increase in PTEN activity that suppresses Akt activity.
Figure 4.
In C2C12 cells, ROCK1 activates PTEN, which impairs Akt activity. (A) PTEN activity assay indicates that caspase-3 activation stimulates PTEN activity (mean±SEM, n=3). (B) Akt activity was measured by the level of phosphorylated GSK3β from cells with or without caspase-3 activation. (C) Increased PTEN activity stimulated by caspase-3 is blunted by the ROCK1 inhibitor (fasudil) (mean±SEM, n=3). (D) Western blot reveals Akt activity is higher after ROCK1 inhibitor was added despite activation of caspase-3.
CKD Suppresses PI3K Activity but Stimulates PTEN Activity in Muscle
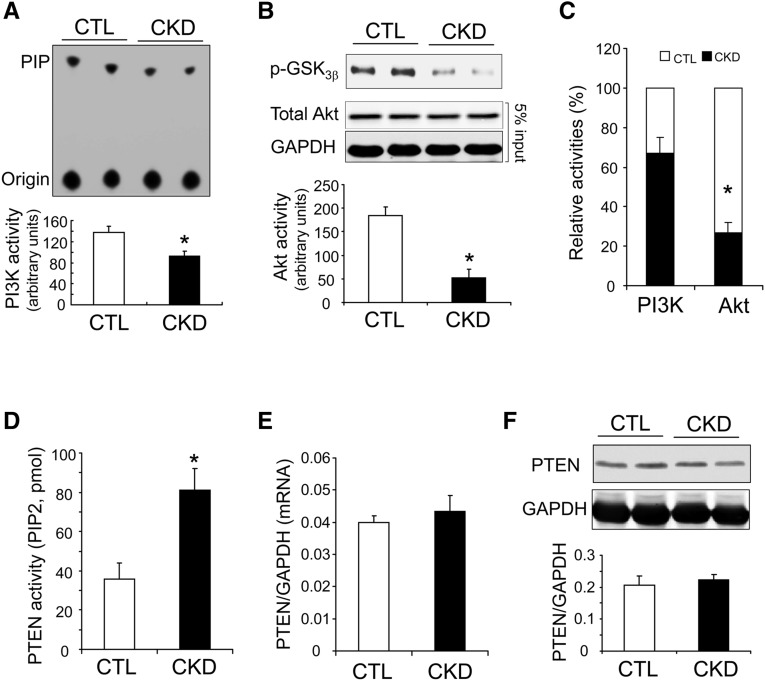
It is well known that CKD causes insulin and IGF-1 resistance resulting in a decrease of Akt activity and a loss of muscle proteins.1,5,28 The latter occurs because low Akt activity stimulates expression of muscle-specific, ubiquitin E3 ligases (MuRF-1 and Atrogin-1/MAFbx), which activate muscle proteolysis in the UPS. What factors reduce the activity of Akt in muscle? One possibility is that suppressed IRS-1–associated PI3K activity lowers the activity of Akt. To evaluate this possibility, we compared the level of IRS-1–associated PI3K activity with that of Akt activity. In response to CKD, we found suppression of both IRS-1–associated PI3K and Akt activities (Figure 5, A and B). However, Akt activity was reduced by >70% versus controls and hence to a greater degree than the loss of IRS-1–associated PI3K activity (39% versus control) (Figure 5C). This result suggests that other mediators are synergistically suppressing Akt activity in muscles of mice with CKD. To evaluate this possibility, we measured the activity of the PTEN because it functions as a negative regulator of PI3K/Akt signaling.22,29 In mice with CKD, there was a 2.3-fold increase in PTEN activity in muscles (Figure 5D). Notably, the decrease was not caused by upregulation of PTEN expression because quantitative RT-PCR and western blotting revealed that both mRNA and protein levels of PTEN in muscles of mice with CKD were not significantly different from the values present in muscles of control mice (Figure 5, E and F). We conclude that diminished Akt activity in muscles of mice with CKD occurred because PTEN phosphatase activity increased.
Figure 5.
CKD stimulates PTEN activity, which contributes to the suppression of Akt in muscles of mice with CKD. (A) In muscles of CKD mice, IRS-1–associated PI3K activity is significantly decreased (*P<0.05; versus controls, n=5). (B) Akt activity assay revealed that CKD suppresses Akt activity in mouse muscles (*P<0.01; versus controls, n=5). (C) CKD suppresses Akt activity more than it lowers IRS-1–associated PI3K activity (*P<0.01 versus controls; n=5). (D) PTEN activity assay indicates that CKD stimulates PTEN activity in muscles (*P<0.05, versus controls, n=5). (E) PTEN mRNA levels (quantitative RT-PCR). (F) PTEN protein level. (F) PTEN protein levels (western blot) were comparable in muscles of CTL mice and mice with CKD (n=5).
Activation of Caspase-3 in Muscle Cells Forms a Negative Feedback Loop that Causes Muscle Protein Wasting
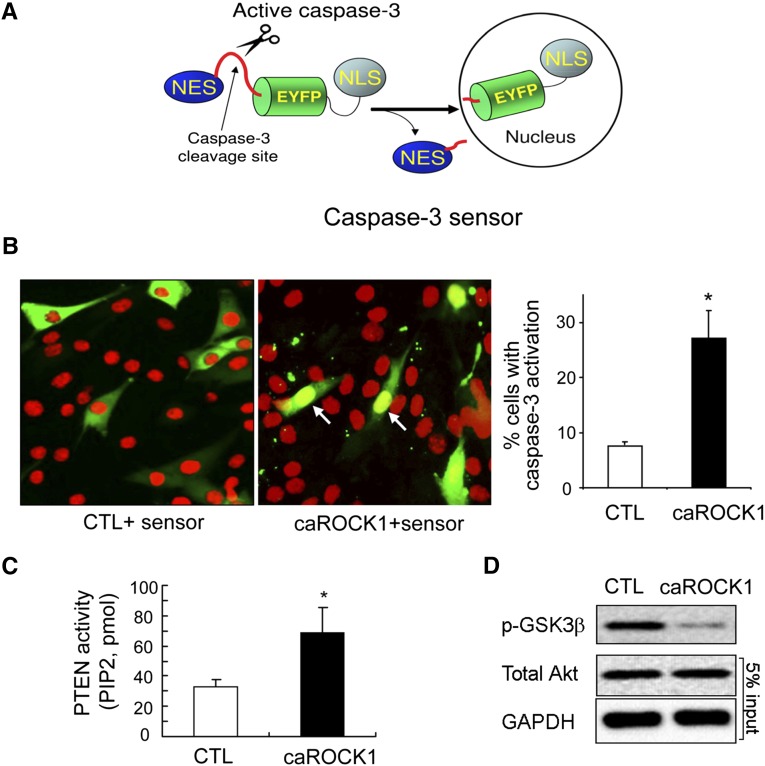
Because low Akt activity can induce apoptosis by activating caspase-3,30 we examined whether activation of ROCK1 can in turn stimulate caspase-3 activity. C2C12 myoblasts were cotransfected with two constructs. The first was an expression vector encoding constitutively activated ROCK1 (caROCK1).31 A second plasmid, the caspase-3 sensor, encodes an enhanced yellow fluorescent protein (EYFP) fusion protein, which translocates to the nucleus when caspase-3 is activated (Figure 6A). The expression of caROCK1 resulted in a 3.2-fold increase in caspase-3 activity versus the results obtained by the expression of the control vector (Figure 6B), indicating constitutively active ROCK1 stimulates caspase-3 activity in C2C12 myoblasts. Caspase-3 activation was accompanied by an increase in PTEN activity but a decrease in Akt activity versus the control results (Figure 6, C and D). Taken together, the results demonstrate that caspase-3 activates ROCK1, which raises PTEN activity, resulting in suppression of Akt. Therefore, these responses form a negative feedback loop that may contribute to loss of muscle proteins.
Figure 6.
Constitutively active ROCK1 (caROCK1) activates caspase-3 and impairs Akt phosphorylation in C2C12 cells. (A) Structural diagram of the caspase-3 sensor, which was used to detect cytosol caspase-3 activation. EYFP was fused with a nuclear export signal (NES) at its N-terminus and a nuclear localization signal (NLS) at the C-terminus. A caspase-3–specific cleavage site was inserted between NES and EYFP. When caspase-3 is activated, it removes NES from EYFP, and the NSL drives EYFP translocation into the nucleus. (B) C2C12 cells were cotransfected with plasmids expressing the caspase-3 sensor or a caROCK1. Cotransfection of galactosidase and the caspase-3 sensor severed as control (CTL). With ROCK1 activation, EYFP was accumulated in the nuclei indicating caspase-3 activation occurred in the cytoplasm. The caspase-3 activity was also evaluated by the percentage of transfected cells exhibiting nuclear EYFP; at least 100 transfected cells were counted in each experiment (*P<0.01, mean±SEM, n=3). Green represents EYFP-fusion protein; red represents propidium iodide stain of nuclei. (C) PTEN activity assay showed that caROCK1 induces PTEN activation (*P<0.05, mean±SEM, n=3). (D) Representative western blot showing diminished Akt activity in cells transfected with caROCK1.
ROCK1 Knockout or Muscle-Specific PTEN Knockout Prevent CKD-Induced Muscle Wasting
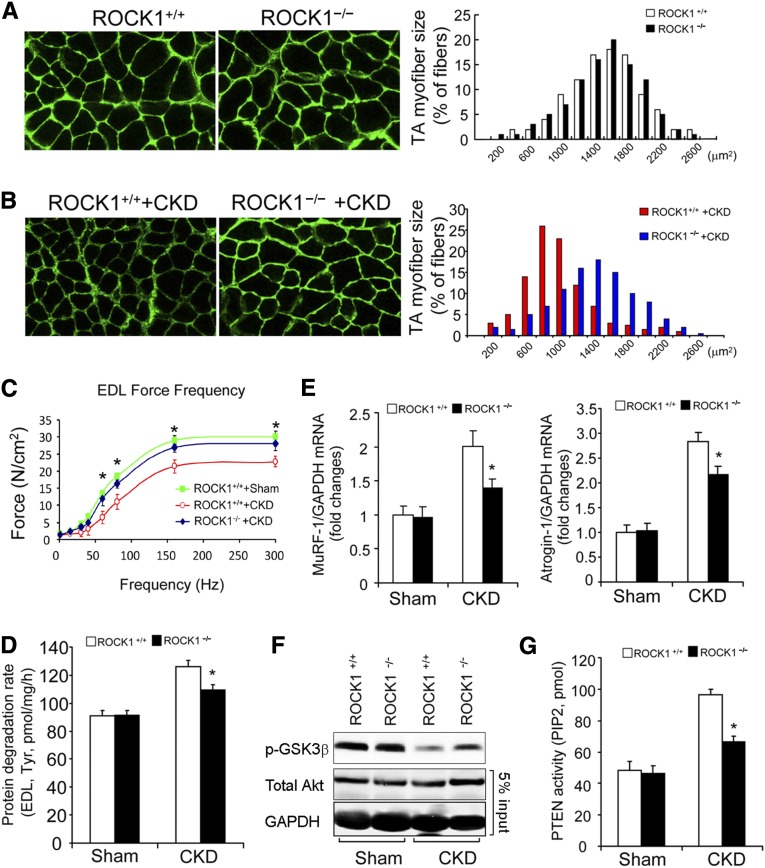
To determine if the negative feedback loop is active in vivo, we studied mice with whole-body knockout (KO) of ROCK1 or with muscle-specific KO of PTEN. ROCK1 KO (ROCK1−/−) mice have been characterized and grow normally.32 Values of BUN and serum creatinine levels in sham-operated, pair-fed ROCK1+/+ or ROCK1−/− mice with CKD confirmed the presence of renal insufficiency (Table 1). In sham-operated, pair-fed, ROCK1−/− mice, the frequency distribution of the cross-sectional areas of myofibers (CSA) in the tibialis anterior (TA) muscles did not differ from results in the TA muscles of ROCK1+/+ mice (Figure 7A). However, the myofiber sizes (CSA) distribution in the TA muscles of ROCK1−/− mice with CKD was shifted to the right compared with results from pair-fed ROCK1+/+ mice with CKD (Figure 7B). The increase in muscle mass of ROCK1−/− mice with CKD was associated with greater contractile strength of the extensor digitorum longus (EDL) muscles from ROCK1−/− mice with CKD (Figure 7C). To uncover why muscle mass increased in ROCK1−/− mice we measured protein degradation: there was no difference in the rates of muscle proteolysis in ROCK1+/+ and ROCK1−/− mice. However, protein degradation in muscles of ROCK1−/− mice with CKD was significantly lower than that in muscles of ROCK1+/+ mice with CKD (Figure 7D). We also found evidence that the UPS was activated because the mRNA of MuRF-1 and Atrogin-1/MAFbx was significantly reduced in muscles of ROCK1−/− mice with CKD versus ROCK1+/+ mice with CKD (Figure 7E). Next, we examined Akt activity in muscles of mice with CKD. As shown in Figure 7F, Akt activity was higher in muscles of ROCK1−/− mice despite the presence of CKD. The mechanism for the improvement in Akt activity is that PTEN activity in muscle of ROCK1−/− mice with CKD was reduced (Figure 7G). In CKD, therefore, activated ROCK1 raises PTEN activity to suppress Akt in muscles; however, but in ROCK1−/− mice muscle protein losses caused by CKD were suppressed.
Table 1.
Serum BUN and creatinine in experimental animals
| Genotype | Serum BUN (mg/dl) | Serum Creatinine (mg/dl) | ||
|---|---|---|---|---|
| Sham | CKD | Sham | CKD | |
| RCOK1+/+ | 30.5±1.7 | 85.4±7.6a | 0.39±0.05 | 1.39±0.12a |
| ROCK1−/− | 32.6±2.7 | 87.9±5.7a | 0.35±0.04 | 1.47±0.16a |
| lox/lox | 27.5±3.1 | 94.8+8.2a | 0.42+0.03 | 1.58+0.21a |
| MPKO | 28.4±3.7 | 89.4+5.5a | 0.37+0.04 | 1.51+0.11a |
Serum BUN and creatinine are shown for the experimental mice; at least 7 animals were examined in each group (n=7–12). Values are presented as mean±SEM.
P<0.01 versus sham.
Figure 7.
ROCK1 null suppresses CKD-induced muscle proteolysis. (A) In the TA muscles of sham-operated, ROCK+/+ and ROCK1 KO mice, the myofibers sizes were assessed as a distribution of the myofibers’ CSA (n=3, approximately 300 myofibers were measured in each mouse). (B) Leftward-shift of distribution of the myofibers’ CSA in TA muscles of ROCK+/+ mice with CKD was prevented in ROCK1−/− mice with CKD (n=5, approximately 300 myofibers in muscle of each mouse were examined). (C) CKD impairs the contractile function of EDL muscles of ROCK+/+ mice. The decreased contractile force caused by CKD was improved in ROCK1−/− mice despite CKD (mean±SEM, *P<0.05, ROCK1−/− +CKD versus ROCK1+/+ +CKD; n=3–5 in each group). (D) Rates of protein degradation, assessed by tyrosine release isolated EDL muscle, were significantly suppressed in ROCK1 KO mice with CKD (mean±SEM; n=5–7 in each group; *P<0.05 versus ROCK+/++CKD). (E) The expressions of Atrogin-1/MAFbx and MuRF-1 (quantitative RT-PCR) were significantly suppressed in muscles of ROCK1−/− mice with CKD (mean±SEM, *P<0.01 versus ROCK+/+with CKD, n=5). (F) Phosphorylation of Akt was examined by western blotting. CKD suppressed Akt activity in muscles of ROCK+/+ mice. This response was blunted in muscles of ROCK1−/− mice with CKD. (G) PTEN activities in the absences or presence of CKD were examined in gastrocnemius muscles of ROCK+/+ and ROCK1−/− mice. The absence of ROCK1 reduced PTEN activity stimulated by CKD (mean±SEM; *P<0.01 versus ROCK+/+ mice with CKD; n=5).
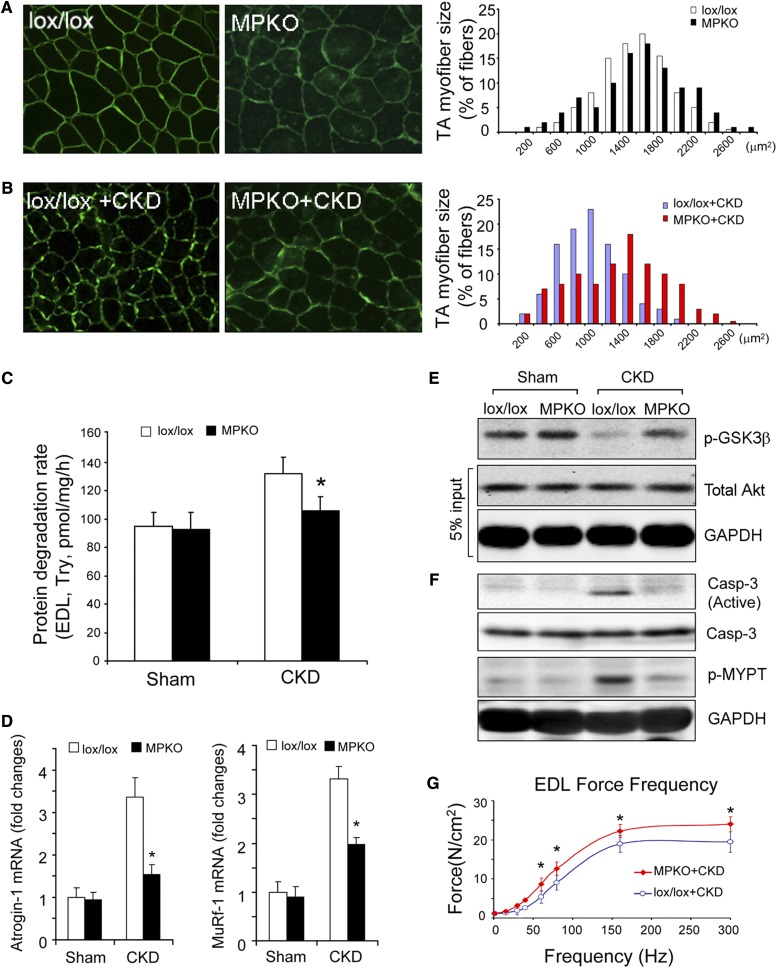
To examine how PTEN activity in muscles of mice with CKD affects protein losses, we studied mice with muscle-specific phosphatase and tensin homolog knockout (MPKO). The generation and characteristics of the MPKO mice have been reported previously.29 In control mice (lox/lox, Cre–), the frequency distribution of myofiber sizes (CSA) in TA muscles was not statistically different from muscles of MPKO (lox/lox, Cre+) mice (Figure 8A). However, when CKD was present, the frequency distribution of CSA myofibers in muscles of MPKO mice was shifted to the right versus results in the control (lox/lox, Cre–) mice (Figure 8B). These results indicate that PTEN deletion in muscle blocks the protein losses even in the presence of CKD.
Figure 8.
MPKO prevents CKD-induced muscle wasting. (A) The distribution of the myofibers' CSA in TA muscles of lox/lox (control) or MPKO mice were not different identical (n=3, approximately 300 myofibers in each mouse were measured). (B) There was a rightward shift of the CSA distribution in TA muscles of MPKO mice with CKD versus results in lox/lox mice with CKD (n=5, approximately 300 myofibers in each mouse were examined). (C) Rates of protein degradation in EDL muscles of lox/lox and MPKO mice indicated that deletion of PTEN blocked the increase in muscle proteolysis that is stimulated by CKD (n=5 in each group). (D) The mRNA levels of Atrogin-1/MAFbx and MuRF-1 in TA muscles were examined by quantitative RT-PCR. The increased expressions of Atrogin-1/MAFbx and MuRF-1 in muscles lox/lox mice with CKD were blocked in muscles of MPKO mice with CKD (mean±SEM; *P<0.01 versus lox/lox +CKD, n=5). (E) Akt activities were examined in muscles of lox/lox and MPKO mice with or without CKD. In muscles of MPKO mice with CKD, the Akt activity was higher than that of lox/lox mice with CKD. (F) Levels of active caspase-3 and ROCK1 activities were measured in muscles of lox/lox and MPKO mice with or without CKD. MPKO suppresses caspase-3 and ROCK1 activation in muscles of mice with CKD. (G) Reduced contractile force of the EDL muscle caused by CKD was improved in MPKO mice with CKD (mean±SEM; *P<0.05 versus lox/lox +CKD; n=5 in each group).
An explanation for this finding is that protein degradation in muscles of MPKO mice with CKD was depressed when compared with muscles of the control (lox/lox, Cre–) mice with CKD (Figure 8C). Reduced muscle proteolysis in MPKO mice with CKD involves suppression of the mRNA of both Atrogin-1/MAFbx and MuRF-1 (Figure 8D). The decrease in the mRNA can be attributed to an increase in Akt activity, even in mice with CKD (Figure 8E). In fact, higher levels of Akt activity were associated with significant decreases in the activities of both caspase-3 and ROCK1 (Figure 8F). With the improvements in muscle protein metabolism, contractile strength also increases in the EDL muscles from MPKO mice with CKD (Figure 8G). Therefore, an increase in Akt activity in muscles of MPKO mice will suppress the activation of caspase-3 resulting in reduced ROCK1 activation and interruption of the negative feedback loop. As outlined in Figure 9, the decrease in ROCK1 activity prevents muscle wasting in mice with CKD.
Figure 9.
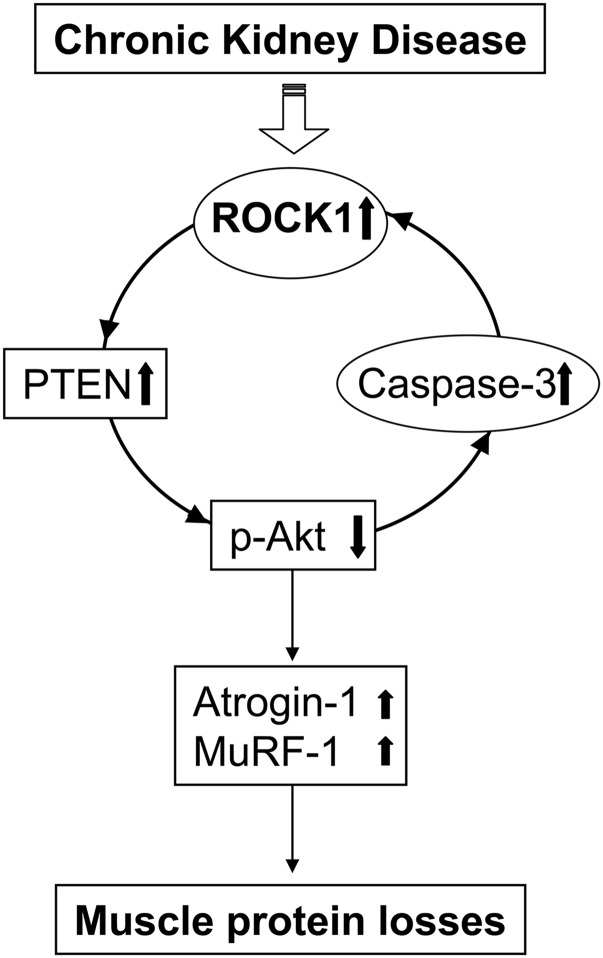
In muscle of mice with CKD, caspase-3/ROCK1/PTEN forms a negative feedback loop that increases muscle proteolysis. The negative feedback loop consists of activation of caspase-3, which activates ROCK1, leading to stimulation of PTEN activity. The increase in PTEN activity suppresses Akt, which stimulates caspase-3 to form a negative feedback loop.
Discussion
In cultured C2C12 muscle cells and mouse muscles we have uncovered a novel pathway that begins with activation of caspase-3, which stimulates ROCK1 activity, and is followed by an increase in PTEN phosphatase activity. The latter suppresses Akt activity and thereby accelerates muscle protein catabolism. In cultured C2C12 muscle cells, we used a novel strategy to activate caspase-3 to cleave and activate ROCK1 (Figure 3). An increase in ROCK1 activity stimulated PTEN activity and suppressed Akt activity (Figure 9). A decrease in Akt activity is a documented stimulus of caspase-3 activity producing a negative feedback loop. We also demonstrated that disrupting this pathway in mouse muscles restored Akt activity and prevented loss of muscle protein. Specifically, when we inhibited ROCK1 or PTEN genetically in mice, the activity of Akt in muscles increased to suppress the expressions of Atrogin-1/MAFbx and MuRF-1. These events resulted in reduced muscle proteolysis. The increase in muscle mass also improved muscle contractile strength (Figures 7 and 8). Therefore, the negative feedback loop amplifies CKD-induced muscle proteolysis. It is tempting to speculate that similar events may be active in other catabolic conditions associated with insulin resistance and caspase-3 activation. If the negative feedback loop persists, a persistent activation of caspase-3 as could occur with the development of inflammation or worsening of CKD would lead to ongoing activation of muscle protein wasting. However, if inflammation is corrected or dialysis is instituted to remove toxins or exercise is undertaken to raise IGF-1 responses in muscle, caspase-3 activity would decrease and PTEN would be reduced causing a rise in Akt activity. The latter reduces activity of the UPS and promotes muscle growth.
In disorders associated with inflammation, caspase-3 frequently causes apoptosis in epithelial or endothelial cells. In skeletal muscles, however, we found that caspase-3 activation does not cause apoptosis. Instead, caspase-3 remained in the cytoplasm and did not enter the nuclei of myofibers from patients or mice with CKD (Figure 2). We confirmed the absence of apoptosis when we found a negative TUNEL assay. In summary, in CKD-induced muscle wasting, caspase-3 activation in muscle fibers stimulates an increase in proteolysis rather than apoptosis/cell death.
How does activated caspase-3 affect muscle wasting? It interacts with the UPS in two ways: first, in response to CKD or diabetes, activated caspase-3 cleaves myofibrillar proteins to produce substrates for the UPS.10,25 Second, activated caspase-3 cleaves specific subunits of the 19S particle in the 26S proteasome leading to increased proteolytic activity of the 26S proteasome.11 The present results uncover another influence of caspase-3 on muscle wasting: caspase-3 stimulates ROCK1 activity, which activates PTEN, resulting in suppressed Akt activity. Low Akt actvity in turn stimulates Atrogin-1 and MuRF-1 expressions leading to muscle wasting. Therefore, caspase-3 activation initiates a coordinated program of muscle proteolysis and atrophy.
ROCK1 activity is regulated by small G proteins, such as RhoA,33 to exert detrimental effects in pathophysiologic conditions, including obesity, cardiovascular diseases, renal fibrosis, and diabetes.34–36 Recently, ROCK1 in mouse models of diabetic nephropathy has been identified as a mechanism causing mitochondrial fission and apoptosis in endothelial cells and podocytes.32 There are other reports that ROCK1 triggered death-inducing signals in many cells but not in skeletal muscles.14 The demonstration that ROCK1 is activated by caspase-3 is relevant because it elucidates an essential role for ROCK1 in facilitating CKD-induced muscle proteolysis. It is tempting to speculate that inhibition of ROCK1 could improve muscle protein metabolism in patients with CKD. Other mechanisms could adversely affect muscle atrophy, including mitochondrial dysfunction. This is suggested because ROCK1 stimulates mitochondrial fragmentation, a process that can significantly contribute to muscle wasting.37 Alternatively, CKD-associated muscle protein loss can be initiated by inflammation that stimulates caspase-3, which is associated with the development of CKD-induced insulin resistance.38,39 Inflammatory cytokines affect insulin signaling by interfering with the metabolism of IRS-1-PI3K and Akt,40,41 or they can stimulate NF-kB or Stat3 transcription eventually, increase the expression of genes involved in programmed atrophy”. Stimulating and eventually, increase the expression of genes involved in programmed atrophy.42,43 By uncovering that CKD-associated ROCK1 activation initiates a negative feedback loop to suppress Akt, we present evidence for a new pathway causing muscle wasting.
Our goal in creating models of uremia has been to study the genesis and consequences of abnormal metabolic responses in CKD. Consequently, the CKD model studied here should have some similarities to characteristic abnormalities present in patients with advanced CKD. Because mice and rats rapidly excrete acid and are resistant to the accumulation of waste products, we increased the precursors of acid/waste products, namely high dietary protein. This strategy has been in use since the 1930s,44 and the model of CKD we study in mice develops metabolic acidosis (serum HCO3=16 mM) with a reasonably stable BUN and serum creatinine.42,45–47 We recognize that feeding a high protein diet to the rodent model of CKD could influence changes in muscle protein metabolism. Therefore, an interaction resulting from the high protein diet could influence the results of our studies.
In summary, we have identified that caspase-3 activates ROCK1 to initiate a negative feedback loop contributing to CKD-induced muscle protein losses. Inhibition or deletion of ROCK1 or of PTEN aborts this negative feedback loop. These observations could suggest therapeutic strategies inhibiting ROCK1. Alternatively, inhibitors of caspase-3 or PTEN could be developed, but they would require more extensive evaluations.
Concise Methods
Animal Models and Muscle Protein Degradation Measurement
Mice were housed with 12-hour light-dark cycles, and all animal procedures were approved by the Baylor College of Medicine Institutional Animal Care and Use Committee. Characteristics of the ROCK1−/− mice have been described previously.32 MPKO mice were obtained by crossing loxp-flanked PTEN mice (The Jackson Laboratory, Bar Harbor, ME) with MCK-Cre mice (The Jackson Laboratory). Lox/lox plus Cre+ mice were designated as MPKO mice, and lox/lox plus Cre– mice served as controls. Two-stage nephrectomy of MPKO or lox/lox, control, male mice under anesthesia was used to create CKD.42 Briefly, approximately 75% of the right kidney was removed, and bleeding was stopped by Vetbond tissue adhesive (3M, St. Paul, MN). One week later the left kidney was removed, and the mice were fed a 24% protein diet for 1 week to minimize death from uremia. Subsequently, mice were fed a 40% protein diet (Harlan Teklab, Indianapolis, IN) for 3 weeks. Sham-operated control mice were pair-fed with CKD mice using the same diets. To confirm CKD, aortic blood obtained from anesthetized mice was used to measure BUN and serum creatinine (Cayman Chemical, Ann Arbor, MI). Under anesthesia, TA muscles were collected for histologic analysis, and EDL and soleus muscles were used for protein degradation measurement as previous described.23 Gastrocnemius muscles were removed and frozen in liquid nitrogen and stored at –80°C until mRNA and protein were measured. Muscle biopsies were obtained from healthy subjects and patients with CKD as previously described.42
Muscle Contractile Function Analysis
EDL muscles were dissected and placed in a chamber containing a Krebs-Ringer buffer (pH 7.4) and equilibrated for 10 min at 35–37°C. At a physiologic optimal length, EDL muscles were stimulated by a Grass S48 stimulator (Grass Technologies, Warwick, RI) with a series of functional frequency (10, 30, 40, 60, 80, 160, 300Hz). The peak isometric force was measured with 3 min between stimulations. The results were normalized with the weights of EDL muscle.48
Muscle Histologic Examination Sand Immunostaining
TA muscles were fixed at resting length and imbedded in HistoPrep Frozen Embedding Media (Fisher, Pittsburgh, PA), and 5-μm sections were stained for ROCK1 or the active, cleaved caspase-3 (Cell Signaling Technology, Beverly, MA). Active caspase-3 in muscle was detected by immunofluorescent staining with an active caspase-3 antibody followed by Alex 488-secondary antibody (Life Technologies, Carlsbad, CA). Myofibers were outlined by staining of dystrophin (Santa Cruz Technology, Santa Cruz, CA), and nuclei were visualized by DAPI or PI. To determine the CSA, we used Image Pro software (Silver Spring, MD) and assessed sizes of 300 myofibers in muscles of each animal. The negative control was normal rabbit lgG instead of the primary antibody. TUNEL assays were performed following the manufacturer’s protocol (EMD Millipore, Billerica, MA).
mRNA Preparation and Expression Analysis
Total RNA was extracted using TRIzol (Sigma-Aldrich, St. Louis, MO) and precipitated in isopropanol. cDNA was synthesized using iScript cDNA Synthesis Kit (Bio-Rad, Hercules, CA). SYBR Green Real-time quantitative PCR was performed with CFX96 System (Bio-Rad). The primer sequences for mouse Atrogin-1, MuRF-1, and GAPDH have been reported.23
Cell Culture and Western Blot Analyses
C2C12 myoblasts were maintained in DMEM with 10% FBS (HyClone, Logan, UT), penicillin (200 units/ml), and streptomycin (50 μg/ml) (Life Technologies). Plasmid DNA was introduced into cells using Invitrogen transfection system (Life Technologies). The inducible, activated caspase-3 (M-Fv-Casp 3) and control plasmids (without the precursor caspase-3 insert, M-Fv-CTL) were obtained from Addgene (Cambridge, MA). caROCK1 and caspase-3 sensor plasmids were kindly provided by Dr. Jiang Chang.31 For western blotting, lysates of C1C12 cells were prepared in RIPA buffer (20 mM Tris, pH 7.5, 5 mM EDTA, 150 mM NaCl, 1% NP40, 0.5% Na-deoxycholate, 0.025% SDS, 1 mM Na-orthovanadate, 10 mM NaF, 25 µM β-glycerophosphate) containing protease inhibitors (Roche Diagnostics, Indianapolis, IN). Skeletal muscle lysates were prepared from approximately 50 mg of muscle by homogenizing in 0.5 ml RIPA buffer. After centrifugation at 15,000 g for 15 minutes at 4°C, the supernatants were subjected to western blotting as previously described.23
ROCK1, PTEN, PI3K, and AKT Activity Assays
To measure total Rho-kinase activity, cultured cells or muscle samples were lysed in RIPA buffer (containing 0,1 mM EGTA, 0,1 M DTT, 10 mM MgCl2) and added to 500 ng of MYPT1 (654–880; EMD Millipore) plus ATP (1mM). The mixtures are incubated for 30 minutes at 30°C before the reaction is stopped by adding 2× Laemmli Sample Buffer (Sigma-Aldrich). The samples are subjected to western blot using an anti–phospho-MYPT1 (Thr-850; EMD Millipore) at 1:500 dilution. GAPDH is used as the loading control. PTEN activity was measured by immunoprecipitating 500 µg of muscle lysate with anti-PTEN antibody (Cell Signaling Technology, Cambridge, MA) overnight, followed by incubation with protein A/G-agarose for 1 hour at 4°C. Precipitates were washed five times, and enzyme activity was determined using PTEN activity ELISA kit (Echelon Biosciences, Salt Lake City, UT). For PI3K kinase activity, the gastrocnemius muscles (approximately 100 mg) were homogenized, and tissue lysate were immunoprecipitated with 4 μg anti–IRS-1 with protein A/G (25 μl) agarose beads (Santa Cruz Biotechnology) for overnight at 4°C. After washing, the beads were resuspended in 50 μl of PI3K reaction buffer containing 15 μg of phosphatidylinositol (Sigma-Aldrich). Reactions were performed as described previously.22 The assay of Akt activity assay was performed as per the manufacturer’s instruction (Cell Signal Technology).
Statistical Analyses
Results are presented as mean±SEM. For experiments comparing two groups, we analyzed results by the Student–Newman–Keuls tests. When more than two groups were compared, we analyzed the result by ANOVA followed by Tukey's test; P<0.05 was considered statistically significant.
Disclosures
None.
Acknowledgments
This work was supported by National Institutes of Health grants (5RO1-AR063686 to Z.H. and R37-DK 37175 to W.E.M.) and the National Natural Science Foundation of China (NSFC81470970 to J.X.). Hui Peng was supported by NSFC81470955 and by China Scholarship Council. These experiments were partially supported by Dr. and Mrs. Harold Selzman.
H.P., J.C., R.Y., Z.H., and J.X. carried out experiments, study design, and data analysis, F.D., W.E.M., Y.W., and Z.H. interpreted the data, and W.M., J.X., and Z.H. were involved in writing the article. All authors had final approval of the submitted versions.
Footnotes
Published online ahead of print. Publication date available at www.jasn.org.
References
- 1.Kalantar-Zadeh K, Cano NJ, Budde K, Chazot C, Kovesdy CP, Mak RH, Mehrotra R, Raj DS, Sehgal AR, Stenvinkel P, Ikizler TA: Diets and enteral supplements for improving outcomes in chronic kidney disease. Nat Rev Nephrol 7: 369–384, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kopple JD, Wang H, Casaburi R, Fournier M, Lewis MI, Taylor W, Storer TW: Exercise in maintenance hemodialysis patients induces transcriptional changes in genes favoring anabolic muscle. J Am Soc Nephrol 18: 2975–2986, 2007 [DOI] [PubMed] [Google Scholar]
- 3.Cheung WW, Paik KH, Mak RH: Inflammation and cachexia in chronic kidney disease. Pediatr Nephrol 25: 711–724, 2010 [DOI] [PubMed] [Google Scholar]
- 4.Zheng B, Ohkawa S, Li H, Roberts-Wilson TK, Price SR: FOXO3a mediates signaling crosstalk that coordinates ubiquitin and atrogin-1/MAFbx expression during glucocorticoid-induced skeletal muscle atrophy. FASEB J 24: 2660–2669, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sandri M, Sandri C, Gilbert A, Skurk C, Calabria E, Picard A, Walsh K, Schiaffino S, Lecker SH, Goldberg AL: Foxo transcription factors induce the atrophy-related ubiquitin ligase atrogin-1 and cause skeletal muscle atrophy. Cell 117: 399–412, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mitch WE, Goldberg AL: Mechanisms of muscle wasting. The role of the ubiquitin-proteasome pathway. N Engl J Med 335: 1897–1905, 1996 [DOI] [PubMed] [Google Scholar]
- 7.Bodine SC, Latres E, Baumhueter S, Lai VK, Nunez L, Clarke BA, Poueymirou WT, Panaro FJ, Na E, Dharmarajan K, Pan ZQ, Valenzuela DM, DeChiara TM, Stitt TN, Yancopoulos GD, Glass DJ: Identification of ubiquitin ligases required for skeletal muscle atrophy. Science 294: 1704–1708, 2001 [DOI] [PubMed] [Google Scholar]
- 8.Ding H, Gao XL, Hirschberg R, Vadgama JV, Kopple JD: Impaired actions of insulin-like growth factor 1 on protein synthesis and degradation in skeletal muscle of rats with chronic renal failure. Evidence for a postreceptor defect. J Clin Invest 97: 1064–1075, 1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gao Y, Ordas R, Klein JD, Price SR: Regulation of caspase-3 activity by insulin in skeletal muscle cells involves both PI3-kinase and MEK-1/2. J Appl Physiol (1985) 105: 1772–1778, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Du J, Wang X, Miereles C, Bailey JL, Debigare R, Zheng B, Price SR, Mitch WE: Activation of caspase-3 is an initial step triggering accelerated muscle proteolysis in catabolic conditions. J Clin Invest 113: 115–123, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang XH, Zhang L, Mitch WE, LeDoux JM, Hu J, Du J: Caspase-3 cleaves specific 19 S proteasome subunits in skeletal muscle stimulating proteasome activity. J Biol Chem 285: 21249–21257, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hu J, Du J, Zhang L, Price SR, Klein JD, Wang XH: XIAP reduces muscle proteolysis induced by CKD. J Am Soc Nephrol 21: 1174–1183, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sebbagh M, Renvoizé C, Hamelin J, Riché N, Bertoglio J, Bréard J: Caspase-3-mediated cleavage of ROCK I induces MLC phosphorylation and apoptotic membrane blebbing. Nat Cell Biol 3: 346–352, 2001 [DOI] [PubMed] [Google Scholar]
- 14.Julian L, Olson MF: Rho-associated coiled-coil containing kinases (ROCK): Structure, regulation, and functions. Small GTPases 5: e29846, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gilkes DM, Xiang L, Lee SJ, Chaturvedi P, Hubbi ME, Wirtz D, Semenza GL: Hypoxia-inducible factors mediate coordinated RhoA-ROCK1 expression and signaling in breast cancer cells. Proc Natl Acad Sci U S A 111: E384–E393, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 16.Mleczak A, Millar S, Tooze SA, Olson MF, Chan EY: Regulation of autophagosome formation by Rho kinase. Cell Signal 25: 1–11, 2013 [DOI] [PubMed] [Google Scholar]
- 17.Chang J, Xie M, Shah VR, Schneider MD, Entman ML, Wei L, Schwartz RJ: Activation of Rho-associated coiled-coil protein kinase 1 (ROCK-1) by caspase-3 cleavage plays an essential role in cardiac myocyte apoptosis. Proc Natl Acad Sci U S A 103: 14495–14500, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Amin E, Dubey BN, Zhang SC, Gremer L, Dvorsky R, Moll JM, Taha MS, Nagel-Steger L, Piekorz RP, Somlyo AV, Ahmadian MR: Rho-kinase: Regulation, (dys)function, and inhibition. Biol Chem 394: 1399–1410, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wickman G, Julian L, Olson MF: How apoptotic cells aid in the removal of their own cold dead bodies. Cell Death Differ 19: 735–742, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li Z, Dong X, Wang Z, Liu W, Deng N, Ding Y, Tang L, Hla T, Zeng R, Li L, Wu D: Regulation of PTEN by Rho small GTPases. Nat Cell Biol 7: 399–404, 2005 [DOI] [PubMed] [Google Scholar]
- 21.Meili R, Sasaki AT, Firtel RA: Rho Rocks PTEN. Nat Cell Biol 7: 334–335, 2005 [DOI] [PubMed] [Google Scholar]
- 22.Hu Z, Lee IH, Wang X, Sheng H, Zhang L, Du J, Mitch WE: PTEN expression contributes to the regulation of muscle protein degradation in diabetes. Diabetes 56: 2449–2456, 2007 [DOI] [PubMed] [Google Scholar]
- 23.Xu J, Li R, Workeneh B, Dong Y, Wang X, Hu Z: Transcription factor FoxO1, the dominant mediator of muscle wasting in chronic kidney disease, is inhibited by microRNA-486. Kidney Int 82: 401–411, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Feng J, Ito M, Ichikawa K, Isaka N, Nishikawa M, Hartshorne DJ, Nakano T: Inhibitory phosphorylation site for Rho-associated kinase on smooth muscle myosin phosphatase. J Biol Chem 274: 37385–37390, 1999 [DOI] [PubMed] [Google Scholar]
- 25.Song YH, Li Y, Du J, Mitch WE, Rosenthal N, Delafontaine P: Muscle-specific expression of IGF-1 blocks angiotensin II-induced skeletal muscle wasting. J Clin Invest 115: 451–458, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fan L, Freeman KW, Khan T, Pham E, Spencer DM: Improved artificial death switches based on caspases and FADD. Hum Gene Ther 10: 2273–2285, 1999 [DOI] [PubMed] [Google Scholar]
- 27.Wang Y, He H, Srivastava N, Vikarunnessa S, Chen YB, Jiang J, Cowan CW, Zhang X: Plexins are GTPase-activating proteins for Rap and are activated by induced dimerization. Sci Signal 5: ra6, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dong J, Ikizler TA: New insights into the role of anabolic interventions in dialysis patients with protein energy wasting. Curr Opin Nephrol Hypertens 18: 469–475, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hu Z, Wang H, Lee IH, Modi S, Wang X, Du J, Mitch WE: PTEN inhibition improves muscle regeneration in mice fed a high-fat diet. Diabetes 59: 1312–1320, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee SW, Dai G, Hu Z, Wang X, Du J, Mitch WE: Regulation of muscle protein degradation: Coordinated control of apoptotic and ubiquitin-proteasome systems by phosphatidylinositol 3 kinase. J Am Soc Nephrol 15: 1537–1545, 2004 [DOI] [PubMed] [Google Scholar]
- 31.Zhang YM, Bo J, Taffet GE, Chang J, Shi J, Reddy AK, Michael LH, Schneider MD, Entman ML, Schwartz RJ, Wei L: Targeted deletion of ROCK1 protects the heart against pressure overload by inhibiting reactive fibrosis. FASEB J 20: 916–925, 2006 [DOI] [PubMed] [Google Scholar]
- 32.Wang W, Wang Y, Long J, Wang J, Haudek SB, Overbeek P, Chang BH, Schumacker PT, Danesh FR: Mitochondrial fission triggered by hyperglycemia is mediated by ROCK1 activation in podocytes and endothelial cells. Cell Metab 15: 186–200, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Furukawa N, Ongusaha P, Jahng WJ, Araki K, Choi CS, Kim HJ, Lee YH, Kaibuchi K, Kahn BB, Masuzaki H, Kim JK, Lee SW, Kim YB: Role of Rho-kinase in regulation of insulin action and glucose homeostasis. Cell Metab 2: 119–129, 2005 [DOI] [PubMed] [Google Scholar]
- 34.Soga J, Noma K, Hata T, Hidaka T, Fujii Y, Idei N, Fujimura N, Mikami S, Maruhashi T, Kihara Y, Chayama K, Kato H, Liao JK, Higashi Y, ROCK Study Group : Rho-associated kinase activity, endothelial function, and cardiovascular risk factors. Arterioscler Thromb Vasc Biol 31: 2353–2359, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhou L, Liu F, Huang XR, Liu F, Chen H, Chung AC, Shi J, Wei L, Lan HY, Fu P: Amelioration of albuminuria in ROCK1 knockout mice with streptozotocin-induced diabetic kidney disease. Am J Nephrol 34: 468–475, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Peng H, Luo P, Li Y, Wang C, Liu X, Ye Z, Li C, Lou T: Simvastatin alleviates hyperpermeability of glomerular endothelial cells in early-stage diabetic nephropathy by inhibition of RhoA/ROCK1. PLoS ONE 8: e80009, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Romanello V, Guadagnin E, Gomes L, Roder I, Sandri C, Petersen Y, Milan G, Masiero E, Del Piccolo P, Foretz M, Scorrano L, Rudolf R, Sandri M: Mitochondrial fission and remodelling contributes to muscle atrophy. EMBO J 29: 1774–1785, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kalantar-Zadeh K, Stenvinkel P, Pillon L, Kopple JD: Inflammation and nutrition in renal insufficiency. Adv Ren Replace Ther 10: 155–169, 2003 [DOI] [PubMed] [Google Scholar]
- 39.Wang XH, Mitch WE: Mechanisms of muscle wasting in chronic kidney disease. Nat Rev Nephrol 10: 504–516, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li YP, Niu A, Wen Y: Regulation of myogenic activation of p38 MAPK by TACE-mediated TNFα release. Front Cell Dev Biol 2: 21, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Doyle A, Zhang G, Abdel Fattah EA, Eissa NT, Li YP: Toll-like receptor 4 mediates lipopolysaccharide-induced muscle catabolism via coordinate activation of ubiquitin-proteasome and autophagy-lysosome pathways. FASEB J 25: 99–110, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang L, Pan J, Dong Y, Tweardy DJ, Dong Y, Garibotto G, Mitch WE: Stat3 activation links a C/EBPδ to myostatin pathway to stimulate loss of muscle mass. Cell Metab 18: 368–379, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang G, Jin B, Li YP: C/EBPβ mediates tumour-induced ubiquitin ligase atrogin1/MAFbx upregulation and muscle wasting. EMBO J 30: 4323–4335, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chanutin A, Ludewig S: Experimental renal insufficiency produced by partial nephrectomy. Arch Intern Med 64: 756–766, 1939 [Google Scholar]
- 45.May RC, Kelly RA, Mitch WE: Mechanisms for defects in muscle protein metabolism in rats with chronic uremia. Influence of metabolic acidosis. J Clin Invest 79: 1099–1103, 1987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bailey JL, Zheng B, Hu Z, Price SR, Mitch WE: Chronic kidney disease causes defects in signaling through the insulin receptor substrate/phosphatidylinositol 3-kinase/Akt pathway: Implications for muscle atrophy. J Am Soc Nephrol 17: 1388–1394, 2006 [DOI] [PubMed] [Google Scholar]
- 47.Thomas SS, Dong Y, Zhang L, Mitch WE: Signal regulatory protein-α interacts with the insulin receptor contributing to muscle wasting in chronic kidney disease. Kidney Int 84: 308–316, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Durham WJ, Aracena-Parks P, Long C, Rossi AE, Goonasekera SA, Boncompagni S, Galvan DL, Gilman CP, Baker MR, Shirokova N, Protasi F, Dirksen R, Hamilton SL: RyR1 S-nitrosylation underlies environmental heat stroke and sudden death in Y522S RyR1 knockin mice. Cell 133: 53–65, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]