Abstract
The ADAMTS (a disintegrin-like and metalloproteinase domain with thrombospondin-type 1 motifs) protein superfamily includes 19 secreted metalloproteases and 7 secreted ADAMTS-like (ADAMTSL) glycoproteins. The possibility of functional linkage between ADAMTS proteins and fibrillin microfibrils was first revealed by a human genetic consilience, in which mutations in ADAMTS10, ADAMTS17, ADAMTSL2 and ADAMTSL4 were found to phenocopy rare genetic disorders caused by mutations affecting fibrillin-1 (FBN1), the major microfibril component in adults. The manifestations of these ADAMTS gene disorders in humans and animals suggested that they participated in the structural and regulatory roles of microfibrils. Whereas two such disorders, Weill–Marchesani syndrome 1 and Weill–Marchesani-like syndrome involve proteases (ADAMTS10 and ADAMTS17, respectively), geleophysic dysplasia and isolated ectopia lentis in humans involve ADAMTSL2 and ADAMTSL4, respectively, which are not proteases. In addition to broadly similar dysmorphology, individuals affected by Weill–Marchesani syndrome 1, Weill–Marchesani-like syndrome or geleophysic dysplasia each show characteristic anomalies suggesting molecule-, tissue-, or context-specific functions for the respective ADAMTS proteins. Ectopia lentis occurs in each of these conditions except geleophysic dysplasia, and is due to a defect in the ciliary zonule, which is predominantly composed of FBN1 microfibrils. Together, this strongly suggests that ADAMTS proteins are involved either in microfibril assembly, stability, and anchorage, or the formation of function-specific supramolecular networks having microfibrils as their foundation. Here, the genetics and molecular biology of this subset of ADAMTS proteins is discussed from the perspective of how they might contribute to fully functional or function-specific microfibrils.
Keywords: Fibrillin microfibrils, ADAMTS protease, Connective tissue disorders, Ectopia lentis, Marfan syndrome, Geleophysic dysplasia
Introduction
Extracellular matrix (ECM) macromolecules commonly form supramolecular structures and networks. Examples of these include elastic fibers, proteoglycan aggregates, collagen fibrils and fibrillin microfibrils. Each structure serves specific structural and regulatory functions in the ECM, yet does not work in isolation, leading to the concept of ECM as a dynamic network of interconnected supramolecular aggregates [1]. Fibrillin microfibrils are widely distributed in tissues. They are typically associated with elastic fibers, whose assembly they guide, but they are also found in elastin-free areas, where they function independent of elastin [2,3]. Among their proposed independent roles are conferring structural integrity to tissues, cell–anchorage through Arg–Gly–Asp (RGD) and heparin-binding sites present in each fibrillin, and growth factor regulation via binding to the large latent complexes of transforming growth factor-β (TGFβ) or bone morphogenetic proteins (BMPs) [4–9]. An unusual structure, the ocular zonule, is a cell free, macroscopic, microfibril-based rigging that spans two basement membranes, the lens capsule and the internal limiting membrane of the ciliary body, providing a matrix–matrix anchorage [10–12]. The zonule centers the lens in the optic path and mediates accommodation by transmitting ciliary sphincter contraction and relaxation to the lens. Thus, it could be viewed functionally as a de facto, but not anatomic, “ciliary muscle tendon” comprising microfibrils.
Although microfibrils are built from fibrillins, and indeed, provide the defining context for the functions of fibrillins, they have long been known to contain or be associated with other ECM molecules [13]. Emerging evidence, some of which is presented here and in other reviews in this cluster, could be distilled to suggest that microfibrils provide a “bare bones” scaffold or foundation for orchestrating the subsequent assembly of supramolecular multiprotein structures of even greater complexity. Thus, microfibril functions in various contexts may result from mechanisms beyond fibrillins, i.e., attributable to added components of function-specific microfibrils, such as the a disintegrin-like and metalloprotease domain with thrombospondin type 1 motif (ADAMTS) proteins, latent TGFβ–binding proteins (LTBPs), fibulins, microfibril associated glycoproteins (MAGPs), the versican–hyaluronan proteoglycan complex and fibronectin [14–27]. Some of these components may express their function by way of assisting microfibril formation, composition, anchorage or stability and microfibrils may provide them essential spatial coordinates. Central to this emerging view is the hypothesis that fibrillin microfibrils formed ab initio are not fully functional, and that other molecules, such as those listed above and others, may extend (“specialize”) their functions. Here, we will discuss this hypothesis from the perspective of the ADAMTS proteins.
Fibrillin assembly and microfibril functions: insights from human genetic disorders and animal mutations
Fibrillins are ECM glycoproteins comprising three isoforms in humans, FBN1, FBN2, and FBN3, and two in mice, FBN1 and FBN2 (Fig. 1A) [28,29]. Fibrillin isoforms can form microfibrils by homotypic or heterotypic head-to-tail and lateral self-assembly [30–33]. During this process, microfibrils are thought to incorporate fibrillin isoforms indiscriminately depending on their availability at the time of assembly [30,31,34]. FBN2 and FBN3 mRNA expression is prevalent during embryogenesis and FBN1 mRNA is prevalent after birth; therefore, it is generally accepted that the preponderance of FBN1 in adult microfibrils reflects the relatively abundant postnatal expression of this isoform [35–37]. Other than differential expression of fibrillin genes, determinants of the fibrillin isoform composition of microfibrils are not known.
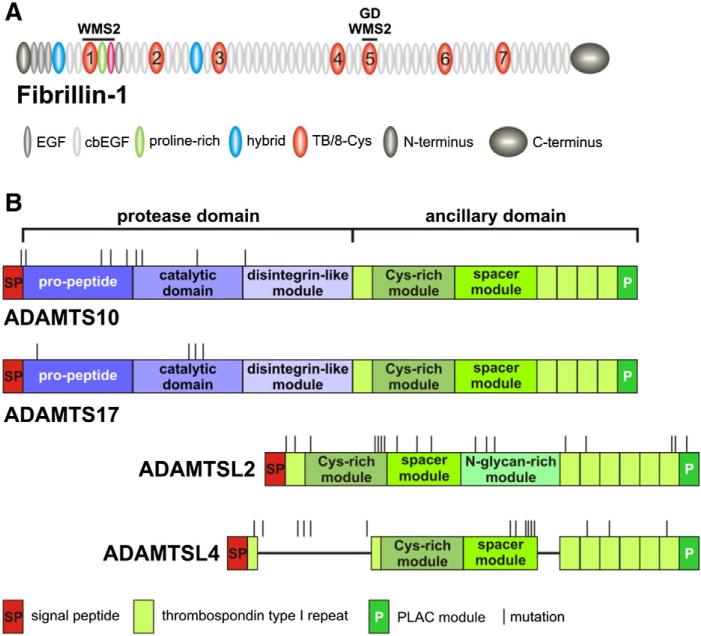
Fig. 1.
Domain organization of FBN1, relevant ADAMTS proteins and localization of disease-causing mutations. A) Domain organization of fibrillin-1. The regions of known mutations causing WMS2 and geleophysic dysplasia (GD) are indicated. In contrast to MFS mutations, which are distributed over the entire protein (not shown), these mutations are localized in distinct domains. TB/8-Cys domains are numbered. B) Domain organization of ADAMTS10, ADAMTS17, ADAMTSL2 and ADAMTSL4. Disease-causing mutations are randomly distributed suggesting they are de facto loss of function mutations. ADAMTSL2 contains a unique N-glycan-rich domain and ADAMTSL4 an unusual split first thrombospondin type 1 repeat present also in ADAMTSL6. cbEGF, calcium-binding EGF-like module; EGF, epidermal growth factor like module, PLAC, protease and lacunin module; TB/8-Cys, transforming growth factor β-binding-like/eight-cysteine domain.
Assembly of microfibrils is initiated near the cell surface and requires active participation of cells that provide integrins, heparan sulfate proteoglycans or other molecules [7,38]. The precise molecular mechanisms provided by cells to assist microfibril assembly may vary according to the cell type, with at least two distinct, but not mutually exclusive, pathways to microfibril assembly previously identified. In mesenchymal cells such as dermal fibroblasts or vascular smooth muscle cells, microfibril assembly requires the presence of a pre-assembled fibronectin fibril network [26,27]. In retinal pigment epithelial cells, microfibril assembly was independent of fibronectin, but required syndecan-4, consistent with an observed agonistic effect of heparin–sulfate proteoglycans on microfibril assembly [7,38,39]. However, microfibril assembly in human non-pigmented ciliary epithelial cells was fibronectin-dependent, suggesting that even epithelial cells may have different requirements for fibrillin assembly, presumably owing to diversity of their secretomes [34].
Because aortic tissue from patients with Marfan syndrome (MFS), caused by mutations in FBN1 (see below) and Fbn1 mutant mice showed aberrant elastic fibers, microfibrils were first implicated in the formation and maintenance of elastic fibers [40,41]. It was shown that tropoelastin, the monomeric precursor of elastin, directly interacted with fibrillin and that microfibrils coordinated elastic fiber assembly and maturation by providing a scaffold for fibulin-5 and lysyl oxidase [22,42–44]. Unlike elastin, microfibrils have limited elasticity, and thus mediate force transfer in the ciliary zonule and the dermal–epidermal junction, where they anchor microfibrils in the basement membrane via perlecan [45–47]. Microfibrils regulate extracellular growth factor signaling, specifically by conferring latency, or regulating activation of TGFβ and sequestering BMPs in the ECM [8,9,21,48].
Mutations in human FBN1 cause MFS and several rarer fibrillinopathies, such as Weill–Marchesani syndrome (WMS) 2, isolated ectopia lentis, and geleophysic dysplasia [19,49,50] (Fig. 1, Table 1). WMS2 and geleophysic dysplasia are classified as acromelic dysplasias because of their characteristic skeletal manifestations and habitus [51] (Fig. 2). Two in-frame FBN1 deletions, one affecting three domains near the N-terminus, and the other, removing 8 amino acids from TGFβ-binding protein-like/8-cysteine domain (TB) 5, were identified to cause WMS2, whereas mutations causing geleophysic dysplasia were restricted to the TB5 domain (Fig. 1A) [51]. The mechanisms by which FBN1 mutations result in phenotypes that contrast strikingly with MFS (Fig. 2), i.e., WMS2 and geleophysic dysplasia, are not yet fully resolved, but the underlying mechanisms of MFS were advanced considerably in the past decade using mouse genetics. In mice recapitulating a MFS-causing FBN1 mutation, dysregulation (excess) of TGFβ signaling contributed to the pathology of aortic aneurysm and pulmonary emphysema [52,53]. Both phenotypes were ameliorated by neutralizing the active form of TGFβ or by blocking TGFβ signaling pathways with small molecule inhibitors, such as the angiotensin II receptor antagonist Losartan [54]. FBN2 mutations in humans lead to congenital contractural arachnodactyly (Beals syndrome), which has skeletal manifestations resembling MFS, but has no cardiovascular or ocular manifestations [55]. This phenotype, taken together with limb patterning defects and the genetic interaction with Bmp7 in Fbn2 deficient mice supports a role for fibrillin-2 and microfibrils in BMP regulation [56].
Table 1.
Human Mendelian disorders resulting from mutations in ADAMTS proteins. Essentially similar disorders resulting from FBN1 mutations are underlined.
| Mendelian disorder | MIM # | Gene name (chromosomal locus) | Mode of inheritance |
|---|---|---|---|
| Weill–Marchesani syndrome 1 | 277600 | ADAMTS10 (19p13.2) | Autosomal recessive |
| Weill–Marchesani-like syndrome | 613195 | ADAMTS17 (15q26.3) | Autosomal recessive |
| Geleophysic dysplasia 1 | 231050 | ADAMTSL2 (9q34.2) | Autosomal recessive |
| Isolated ectopia lentis 2 | 225100 | ADAMTSL4 (1q21.3) | Autosomal recessive |
| Ectopia lentis et pupillae | 225200 | ||
| Isolated ectopia lentis 1 | 129600 | FBN1 (15q21) | Autosomal dominant |
| Weill–Marchesani syndrome 2 | 608328 | ||
| Geleophysic dysplasia 2 | 614185 | ||
| Marfan syndrome | 154700 | ||
| MASS syndrome (mitral valve, aorta, skin, skeletal) | 604308 | ||
| Acromicric dysplasia | 102370 | ||
| Stiff skin syndrome | 184900 |
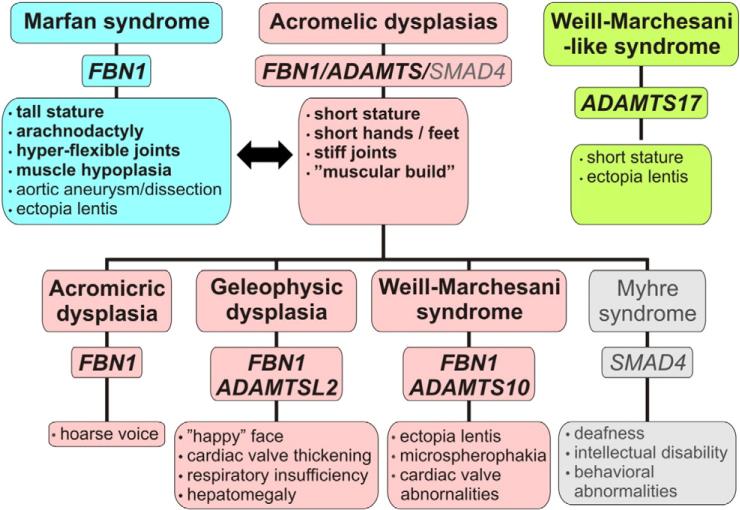
Fig. 2.
Overlapping and differential features in disorders caused by mutations in ADAMTS proteins or FBN1. Musculoskeletal presentations of Marfan syndrome (blue boxes) and the acromelic dysplasias (pink boxes) contrast with each other. Myhre syndrome (gray boxes), an acromelic dysplasia caused by SMAD4 mutations, is also shown for completeness [89]. Weill–Marchesani-like syndrome (green boxes) has clinical overlap with acromelic dysplasias (short stature), WMS and MFS (ectopia lentis).
ADAMTS and ADAMTS-like proteins: novel, functionally crucial fibrillin-binding proteins
Nineteen ADAMTS proteases and seven ADAMTS-like proteins constitute a superfamily of glycoproteins which are present in the ECM or remain cell-surface associated after secretion [57]. ADAMTS proteases are characterized by a highly homologous N-terminal protease domain, comprising a pre-propeptide, which is excised by furin, a catalytic module, which binds zinc in the active site, and a disintegrin-like module (Fig. 1B). Their C-terminal ancillary domains have a core region comprising a thrombospondin type 1 repeat (TSR), a cysteine-rich module, a spacer module, and a variable ensemble of additional TSRs and other modules. The ancillary domain is thought to endow substrate recognition, binding and specificity as well as cell-surface or ECM tethering [58,59]. ADAMTS proteases are distinct from the membrane-anchored ADAM proteases, and have little to do with protein ectodomain shedding, which is the principal function of ADAMs [60]. ADAMTS proteases participate in procollagen maturation (ADAMTS2, ADAMTS3), versican turnover during embryogenesis (ADAMTS1, ADAMTS5, ADAMTS9, ADAMTS15, ADAMTS20) and ovulation (ADAMTS1), aggrecan turnover in articular cartilage (ADAMTS4, ADAMTS5), von Willebrand factor proteolysis and hemostasis (ADAMTS13), and in VEGF-C activation during lymphangiogenesis (ADAMTS3) [57,61]. However, specific substrates and functions are unknown for several ADAMTS proteases. ADAMTS-like proteins lack the N-terminal protease domain, but are homologous to the ancillary domain of ADAMTS proteases, and the products of distinct genes (Fig. 1B). Therefore, ADAMTS-like proteins do not possess proteolytic activity.
Genetic consilience between ADAMTS proteins and FBN1 in human genetic disorders
Mutations in a subgroup of ADAMTS proteins cause inherited human genetic disorders, which phenocopy some FBN1 mutations (Fig. 2 and Table 1) [51,62]. ADAMTS10 mutations lead to WMS1, with short stature, brachydactyly and ectopia lentis (dislocation of the lens) being major clinical features [63]. In WMS, the dislocated lens may move anteriorly and block the pupil, resulting in secondary glaucoma due to interference with aqueous humor circulation. Since WMS2, an essentially indistinguishable syndrome, is caused by FBN1 mutations [17,64], a functional relationship between ADAMTS10 and FBN1 emerged, and was validated by studies showing that ADAMTS10 binds FBN1 [17,18]. ADAMTS10 cleaves FBN1 poorly, but enhances microfibril biogenesis. Ectopia lentis in WMS1 also suggested that ADAMTS10 was required for seeding, assembly, or maintaining integrity of the zonule [18]. A WMS-like syndrome in humans results from ADAMTS17 mutations, suggesting it may have mechanistic overlap with ADAMTS10 (Table 1) [65,66]. Both WMS and WMS-like syndrome share short stature and ocular manifestations, including ectopia lentis, but WMS-like syndrome lacks joint stiffness, brachydactyly, and cardiac valve abnormalities. However, there is some crossover in these genotype–phenotype correlations since mutations in ADAMTS17 were recently identified in a case with WMS and mutations in LTBP2, which binds FBN1, have been shown to cause WMS and WMS-like syndrome [67,68]. ADAMTSL2 mutations cause geleophysic dysplasia [69], which presents with short stature, stiff skin, joint contractures, hepatomegaly and tracheo-pulmonary and cardiac anomalies, leading frequently to juvenile death. ADAMTSL4 mutations cause isolated ectopia lentis, ectopia lentis et pupillae, and ectopia lentis combined with craniosynostosis, correlating with the distribution of ADAMTSL4 in many parts of the eye as well as extraocular tissue [16,70,71].
Interestingly, specific dog breeds are affected by mutations in ADAMTS proteins, providing potential animal models for the above human genetic conditions. Canine ADAMTS10 and ADAMTS17 mutations cause open angle glaucoma and ectopia lentis in terriers [65,72,73]. One of the ADAMTSL2 point mutations found in human geleophysic dysplasia causes Musladin–Lueke syndrome (MLS) in beagles via a founder effect [74]. In affected beagles, joint contractures, small stature and stiff skin are prominent. Although MLS resembles geleophysic dysplasia in these respects, pulmonary or cardiac abnormalities are absent and a normal lifespan is reported, in contrast to a significant proportion of affected humans. Adamtsl2 null mice die shortly after birth with severe lung anomalies [75]. Furthermore, Adamts10 inactivation in mice did not precisely phenocopy WMS1; although Adamts10 null mice are slightly smaller in size, they lacked specific skeletal and cardiac anomalies and did not develop ectopia lentis (Wang, Kutz, Apte, manuscript in preparation). These inter-species differences are potentially attributable to the absence of FBN3 in mice, to different expression patterns of the ADAMTS proteins in the three species, and the recent observation that the mouse zonule, in contrast to the human zonule, contains both FBN1 and FBN2 [34,76]. It is a distinct possibility that ADAMTS proteins are deployed slightly differently in each species.
In biochemical and cell culture experiments, purified ADAMTSL4 and ADAMTSL2 directly bound to fibrillin peptides and accelerated microfibril formation when added to cultured fibroblasts [16,18,19,69]. Although not yet implicated in any human or animal genetic disorder, or by engineered genetic inactivation, ADAMTSL6 was shown to be a component of fibrillin microfibrils by light and immunoelectron microscopy [77]. ADAMTSL6 overexpression enhanced the formation of microfibrils, both in cultured cells and in mice [77]. ADAMTS10 colocalization to fibrillin microfibrils in tissues was demonstrated using immunogold labeling and electron microscopy [18]. Neither an interaction nor a functional relationship of ADAMTS17 to fibrillin microfibrils has been reported to date.
A WMS2 mouse model recapitulating an N-terminal, three-domain, in-frame FBN1 deletion found in humans showed reduced bone length and other phenotypes reminiscent of human WMS2, although the eyes of these mice were not examined [17]. Cells from these mice assembled thicker microfibril bundles than wild-type cells, and there was reduced ADAMTSL6 staining of mutant tissues. The authors concluded that this mutation had a distinct effect on microfibrils from that seen in MFS. Cain et al provided biochemical evidence that FBN1 mutations causing WMS2 or geleophysic dysplasia could disrupt fibrillin binding to heparan sulfate proteoglycans [78]. What is not yet clear is how these deletions and mutations affecting FBN1 relate to the ADAMTS protein mutations that result in similar clinical manifestations.
Evidence for growth factor and cell dysregulation in the absence of ADAMTS proteins
Joint contractures and thickened skin, which are found in WMS, geleophysic dysplasia and MLS, are common in fibrosis. Therefore, these disorders could be viewed as belonging to a fibrotic spectrum. Hallmarks of tissue fibrosis include the presence of contractile myofibroblasts, elevated TGFβ signaling, its downstream effects such as enhanced transcription of genes coding for ECM proteins, and ultimately, the deposition of a disorganized collagen-rich ECM [79]. The consequences of these changes are the stiffening of tissues, which impairs organ function.
Potential evidence for ADAMTS involvement in TGFβ signaling came from analysis of fibroblasts isolated from patients with geleophysic dysplasia caused by ADAMTSL2 mutations, which secreted more latent and active TGFβ and showed evidence of enhanced TGFβ signaling [69]. In addition, recombinant ADAMTSL2 protein interacts directly with FBN1, FBN2, and LTBP1 [19,69,75]. Although it is not known if these proteins form a tri-molecular complex or compete with each other in the binding, ADAMTSL2 seems to sit squarely within a context relevant to TGFβ regulation. Interestingly, fibroblasts from patients with geleophysic dysplasia caused by FBN1 mutations were reported to have a reduced amount of microfibrils and a disorganized microfibril network [19]. In the ADAMTSL2 knock-out mouse lung, we observed considerably enhanced FBN2 staining in bronchial microfibrils [75], suggesting that one mechanism by which ADAMTSL2 may work is by regulating the ratio of FBN1 and FBN2 incorporated into microfibrils.
In skin biopsies from beagles with MLS, an extensive collagen network extending from the dermis to the subcutaneous tissue was demonstrated [74]. A skin biopsy from a WMS2 patient showed accumulation of abnormal fibrillin aggregates [17], and skin fibroblasts isolated from WMS1 patients showed thicker bundles of actin filaments than cells from unaffected individuals [63]. This may be indicative of their transformation to myofibroblasts, especially in light of data showing that cells isolated from stiff (fibrotic) lung tissue apparently retained “memory” of their altered microenvironment and continued to behave like myofibroblasts for subsequent passages in vitro [80].
Insights on microfibril assembly and functionalization by ADAMTS proteins from the ciliary zonule and ectopia lentis
Mutations in human ADAMTS10, ADAMTS17, ADAMTSL4, FBN1, and LTBP2 (whose protein does not bind latent TGFβ) cause ectopia lentis, in which the ciliary zonule is not properly formed or maintained, as well as other ocular defects. The fibrillins forming the ciliary zonule are synthesized by non-pigmented ciliary epithelial cells located in the ciliary body and the assembled microfibrils insert into the lens capsule [12,34,81]. Several publications have described the composition and arrangement of the zonule, as well as the process of ciliary zonule growth and attachment [12,34,37,76,82–85]. Shi et al showed that Fbn2 expressed by the non-pigmented ciliary epithelium was the dominant embryonic fibrillin isoform in the mouse and that Fbn1 mRNA replaced Fbn2 mRNA around birth [76]. This mimics the biphasic expression of Fbn1 and Fbn2 mRNAs observed in other mouse tissues [35]. Complementary to this study, it was shown that the genetic ablation of Fbn1 in mice did not result in ectopia lentis, because FBN2 was a normal component of the zonule in adult mice, rats, and hamsters, although not humans [34]. A previous proteomic analysis of microfibrils extracted from human ciliary zonules identified only FBN1, MAGP1 and β-crystallin as “core components” [86]. However, the stringent extraction conditions used in this study may have stripped other proteins, including ADAMTS and ADAMTS-like proteins from the zonule microfibrils.
What is known about the distribution and function of ADAMTS, ADAMTS-like and related proteins in eye development? ADAMTS10 was localized to the human ciliary zonule and present in the vicinity of the ciliary body and the lens epithelium, indicating that ADAMTS10 is likely to be present over the entire length of the zonule [18]. LTBP2 and FBN2 were also shown to be present in the mouse zonule, the latter being the sole zonule fibrillin isoform in the absence of FBN1 [34,83]. In the post-natal human zonule, FBN1 appears to be the sole fibrillin isoform, despite all three isoforms localizing to the area of the prospective zonule in human embryonic eyes and FBN3 being present in the infant zonule [37]. RNA in-situ hybridization suggests that most zonule components (FBN1, FBN2, ADAMTS10, LTBP2) are produced by the non-pigmented ciliary epithelium [18,76,83]. ADAMTSL4 was localized by immunohistochemistry and by Western blotting to the human ciliary body, the lens equator, choroid, and retina [16,87]. However, Adamtsl4 mRNA in-situ hybridization showed the strongest signal in the equatorial lens epithelium, i.e., at the insertion site of the ciliary zonule into the lens capsule (Hubmacher and Apte, unpublished). This suggests that ADAMTSL4 may anchor microfibrils into the lens capsule. It could also be involved in seeding microfibril formation, since the lens and ciliary body are closely approximated during early eye development. The role of ADAMTS17 in the formation and maintenance of microfibrils remains to be established.
The first mouse model for ectopia lentis, an Ltbp2 knock-out, was described recently [83]. The ciliary zonule appeared to form normally in the absence of LTBP2, but was disrupted in eyes of adult mice. LTBP2 co-localized with FBN1 over the entire length of the zonule, similar to ADAMTS10. In the absence of LTBP2, microfibril continuity between the lens and ciliary body was lost, but microfibrils retained their attachment to the non-pigmented ciliary epithelium and the lens capsule. The authors concluded that LTBP2 bundled microfibrils into larger fibers and that its absence compromised zonule integrity. Thus, different zonule components may be required for determining the mechanical properties of the central part of the zonule, and for its anchorage at each end, respectively.
Open questions and future directions
Presently, genetic evidence from a number of models and emerging biochemical data have clarified some aspects of the relationship between ADAMTS proteins and fibrillin microfibrils. Clearly, several ADAMTS proteins can bind to FBN1 and co-localize with microfibrils. There is also evidence supporting their role in accelerating microfibril assembly, and ensuring microfibril longevity. However, several pressing mechanistic questions remain. Does the absence of specific ADAMTS proteins simply result in fewer microfibrils, with rupture of those that remain in the eye and elsewhere, or are they qualitatively different? Does the ensuing tissue microdamage lead to fibrosis in the skin and elsewhere as an epiphenomenon, or is there specific microenvironment dysregulation arising early in each ADAMTS defect? Do ADAMTS proteins only accelerate the assembly of FBN1 microfibrils, or are they also involved in conferring tissue and developmental stage-specific functions to a microfibril scaffold (“specialization”) (Fig. 3), such as in growth factor signaling? Do ADAMTS mutations recapitulate the phenotype of a FBN1 (tissue-specific) mutation by acting only through FBN1, or do acromelic dysplasias reflect the secondary repair responses of connective tissue? A similar concept was first proposed for the aberrant TGFβ activation in the vascular wall upon mutation of different ECM components associated with deposition of latent TGFβ [88]. The fact that the majority of FBN1 mutations lead to MFS, whereas only rare or domain-specific mutations lead to acromelic dysplasias suggests that the explanation is not simply fewer microfibrils, but rather a lack of specific intermolecular interaction between ADAMTS proteins and fibrillins that has profound regulatory consequences (Fig. 3). Do ADAMTS/ADAMTS-like proteins modulate the isoform composition of microfibrils? Preponderance of FBN2 in bronchial microfibrils of Adamtsl2-deficient mice suggests this is a distinct possibility. What is the role of the specific FBN1 domains mutated in geleophysic dysplasia and Weill–Marchesani syndrome and why do these FBN1 mutations not cause MFS? One possibility is that fibrillin microfibrils would serve as the foundation for the proper tethering of ADAMTS proteins at specific points in the ECM (specialization). In that case, the domain-specific mutations in FBN1 would likely impair that localization process. Are microfibrils or associated proteins substrates for ADAMTS10 and ADAMTS17? Studies of ADAMTS10 showed that, if activated by furin, ADAMTS10 has the capacity to cleave FBN1 in vitro [18]. However, since the required furin processing site within ADAMTS10 is sub-optimal, the question arises, how is ADAMTS10 activated in vivo, if at all? Furthermore, the apparently similar roles of ADAMTS-like proteins and ADAMTS10 suggest that ADAMTS10 may function more as an ADAMTS-like protein than a protease. How then, to reconcile the finding that ADAMTS17, which carries a consensus furin recognition sequence, is genetically implicated in a WMS-like disorder?
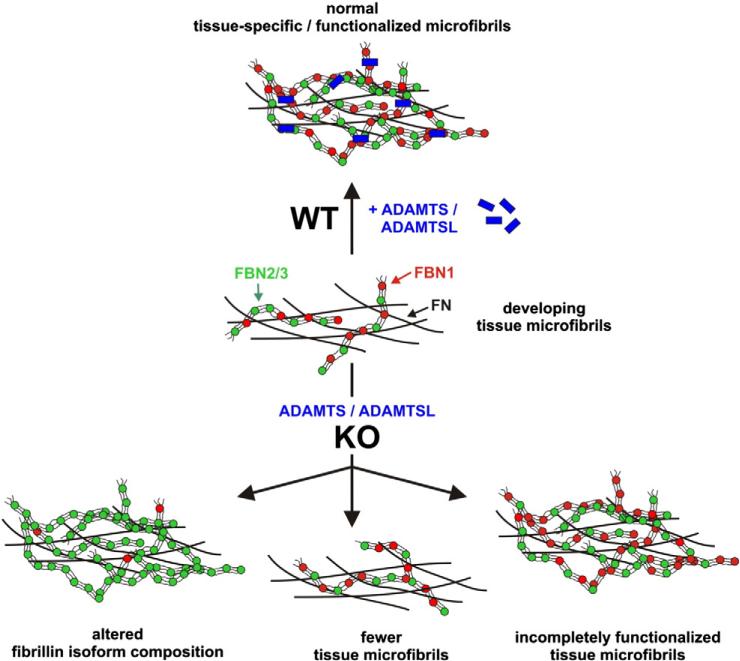
Fig. 3.
Proposed roles of ADAMTS proteins in the formation and specialization of tissue microfibrils. Microfibrils can contain three fibrillin isoforms (FBN1 is shown in red, FBN2 and FBN3 are shown in green as characteristic “beads-on-a-string” structures) and assemble on a fibronectin network. Normally, in the presence of ADAMTS/ADAMTS-like proteins (wild type, WT), microfibrils have a developmental and tissue-specific isoform composition. Although microfibril FBN isoform composition is predominantly determined by differential gene expression, current evidence suggests that absence of one or more ADAMTS/ADAMTS-like proteins (KO) can result in (i) altered fibrillin isoform composition of microfibrils (bottom left), (ii) reduction of the number of microfibrils (bottom center), or (iii) absence of tissue specific functionality due to the missing ADAMTS/ADAMTS-like proteins (bottom right).
These intriguing questions remain unanswered for now, but the required tools, such as animal models, recombinant proteins, antibodies etc. are now at hand or under development. In addition, the explant culture of the anterior chamber of the eye [83], and eye-derived cells in combination with mouse genetic tools make ciliary zonule development tractable to experimental manipulation in vitro and in vivo. New methods for genome editing will expedite ongoing genetic analysis, but for now, the protein chemistry and intermolecular interactions of fibrillins and ADAMTS-like proteins remains a compelling challenge, because of their complex post-translational modification and large size.
Acknowledgment
This work was supported by an award from the National Institutes of Health (EY 021151 to S.A.) and the National Marfan Foundation (Early Investigator Grant to D.H.).
References
- 1.Hynes RO. The extracellular matrix: not just pretty fibrils. Science. 2009;326:1216–9. doi: 10.1126/science.1176009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hubmacher D, Tiedemann K, Reinhardt DP. Fibrillins: from biogenesis of microfibrils to signaling functions. Curr Top Dev Biol. 2006;75:93–123. doi: 10.1016/S0070-2153(06)75004-9. [DOI] [PubMed] [Google Scholar]
- 3.Ramirez F, Sakai LY. Biogenesis and function of fibrillin assemblies. Cell Tissue Res. 2010;339:71–82. doi: 10.1007/s00441-009-0822-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McGowan SE, Holmes AJ, Mecham RP, Ritty TM. Arg–Gly– Asp-containing domains of fibrillins-1 and -2 distinctly regulate lung fibroblast migration. Am J Respir Cell Mol Biol. 2008;38:435–45. doi: 10.1165/rcmb.2007-0281OC. [DOI] [PubMed] [Google Scholar]
- 5.Sakamoto H, Broekelmann T, Cheresh DA, Ramirez F, Rosenbloom J, Mecham RP. Cell-type specific recognition of RGD- and non-RGD-containing cell binding domains in fibrillin-1. J Biol Chem. 1996;271:4916–22. [PubMed] [Google Scholar]
- 6.Cain SA, Baldwin AK, Mahalingam Y, Raynal B, Jowitt TA, Shuttleworth CA, et al. Heparan sulfate regulates fibrillin-1 N-and C-terminal interactions. J Biol Chem. 2008;283:27017–27. doi: 10.1074/jbc.M803373200. [DOI] [PubMed] [Google Scholar]
- 7.Tiedemann K, Batge B, Muller PK, Reinhardt DP. Interactions of fibrillin-1 with heparin/heparan sulfate, implications for microfibrillar assembly. J Biol Chem. 2001;276:36035–42. doi: 10.1074/jbc.M104985200. [DOI] [PubMed] [Google Scholar]
- 8.Sengle G, Charbonneau NL, Ono RN, Sasaki T, Alvarez J, Keene DR, et al. Targeting of bone morphogenetic protein growth factor complexes to fibrillin. J Biol Chem. 2008;283:13874–88. doi: 10.1074/jbc.M707820200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zilberberg L, Todorovic V, Dabovic B, Horiguchi M, Courousse T, Sakai LY, et al. Specificity of latent TGF-beta binding protein (LTBP) incorporation into matrix: role of fibrillins and fibronectin. J Cell Physiol. 2012;227:3828–36. doi: 10.1002/jcp.24094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Davis EC, Roth RA, Heuser JE, Mecham RP. Ultrastructural properties of ciliary zonule microfibrils. J Struct Biol. 2002;139:65–75. doi: 10.1016/s1047-8477(02)00559-2. [DOI] [PubMed] [Google Scholar]
- 11.Bornfeld N, Spitznas M, Breipohl W, Bijvank GJ. Scanning electron microscopy of the zonule of Zinn. I. Human eyes. Albrecht Von Graefes Arch Klin Exp Ophthalmol. 1974;192:117–29. doi: 10.1007/BF00410698. [DOI] [PubMed] [Google Scholar]
- 12.Wheatley HM, Traboulsi EI, Flowers BE, Maumenee IH, Azar D, Pyeritz RE, et al. Immunohistochemical localization of fibrillin in human ocular tissues. Relevance to the Marfan syndrome. Arch Ophthalmol. 1995;113:103–9. doi: 10.1001/archopht.1995.01100010105028. [DOI] [PubMed] [Google Scholar]
- 13.Kielty CM, Sherratt MJ, Marson A, Baldock C. Fibrillin microfibrils. Adv Protein Chem. 2005;70:405–36. doi: 10.1016/S0065-3233(05)70012-7. [DOI] [PubMed] [Google Scholar]
- 14.Ohno-Jinno A, Isogai Z, Yoneda M, Kasai K, Miyaishi O, Inoue Y, et al. Versican and fibrillin-1 form a major hyaluronan-binding complex in the ciliary body. Invest Ophthalmol Vis Sci. 2008;49:2870–7. doi: 10.1167/iovs.07-1488. [DOI] [PubMed] [Google Scholar]
- 15.Isogai Z, Aspberg A, Keene DR, Ono RN, Reinhardt DP, Sakai LY. Versican interacts with fibrillin-1 and links extracellular microfibrils to other connective tissue networks. J Biol Chem. 2002;277:4565–72. doi: 10.1074/jbc.M110583200. [DOI] [PubMed] [Google Scholar]
- 16.Gabriel LA, Wang LW, Bader H, Ho JC, Majors AK, Hollyfield JG, et al. ADAMTSL4, a secreted glycoprotein widely distributed in the eye, binds fibrillin-1 microfibrils and accelerates microfibril biogenesis. Invest Ophthalmol Vis Sci. 2012;53:461–9. doi: 10.1167/iovs.10-5955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sengle G, Tsutsui K, Keene DR, Tufa SF, Carlson EJ, Charbonneau NL, et al. Microenvironmental regulation by fibrillin-1. PLoS Genet. 2012;8:e1002425. doi: 10.1371/journal.pgen.1002425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kutz WE, Wang LW, Bader HL, Majors AK, Iwata K, Traboulsi EI, et al. ADAMTS10 protein interacts with fibrillin-1 and promotes its deposition in extracellular matrix of cultured fibroblasts. J Biol Chem. 2011;286:17156–67. doi: 10.1074/jbc.M111.231571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Le Goff C, Mahaut C, Wang LW, Allali S, Abhyankar A, Jensen S, et al. Mutations in the TGFbeta binding-protein-like domain 5 of FBN1 are responsible for acromicric and geleophysic dysplasias. Am J Hum Genet. 2011;89:7–14. doi: 10.1016/j.ajhg.2011.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Massam-Wu T, Chiu M, Choudhury R, Chaudhry SS, Baldwin AK, McGovern A, et al. Assembly of fibrillin microfibrils governs extracellular deposition of latent TGF beta. J Cell Sci. 2010;123:3006–18. doi: 10.1242/jcs.073437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ono RN, Sengle G, Charbonneau NL, Carlberg V, Bachinger HP, Sasaki T, et al. LTBPS and fibulins compete for fibrillin-1 and exhibit exquisite specificities in binding sites. J Biol Chem. 2009;284:16872–81. doi: 10.1074/jbc.M809348200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.El-Hallous E, Sasaki T, Hubmacher D, Getie M, Tiedemann K, Brinckmann J, et al. Fibrillin-1 interactions with fibulins depend on the first hybrid domain and provide an adaptor function to tropoelastin. J Biol Chem. 2007;282:8935–46. doi: 10.1074/jbc.M608204200. [DOI] [PubMed] [Google Scholar]
- 23.Penner AS, Rock MJ, Kielty CM, Shipley JM. Microfibril-associated glycoprotein-2 interacts with fibrillin-1 and fibrillin-2 suggesting a role for MAGP-2 in elastic fiber assembly. J Biol Chem. 2002;277:35044–9. doi: 10.1074/jbc.M206363200. [DOI] [PubMed] [Google Scholar]
- 24.Jensen SA, Reinhardt DP, Gibson MA, Weiss AS. Protein interaction studies of MAGP-1 with tropoelastin and fibrillin-1. J Biol Chem. 2001;276:39661–6. doi: 10.1074/jbc.M104533200. [DOI] [PubMed] [Google Scholar]
- 25.Sabatier L, Djokic J, Fagotto-Kaufmann C, Chen M, Annis DS, Mosher DF, et al. Complex contributions of fibronectin to initiation and maturation of microfibrils. Biochem J. 2013;456:283–95. doi: 10.1042/BJ20130699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sabatier L, Chen D, Fagotto-Kaufmann C, Hubmacher D, McKee MD, Annis DS, et al. Fibrillin assembly requires fibronectin. Mol Biol Cell. 2009;20:846–58. doi: 10.1091/mbc.E08-08-0830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kinsey R, Williamson MR, Chaudhry S, Mellody KT, McGovern A, Takahashi S, et al. Fibrillin-1 microfibril deposition is dependent on fibronectin assembly. J Cell Sci. 2008;121:2696–704. doi: 10.1242/jcs.029819. [DOI] [PubMed] [Google Scholar]
- 28.Corson GM, Charbonneau NL, Keene DR, Sakai LY. Differential expression of fibrillin-3 adds to microfibril variety in human and avian, but not rodent, connective tissues. Genomics. 2004;83:461–72. doi: 10.1016/j.ygeno.2003.08.023. [DOI] [PubMed] [Google Scholar]
- 29.Hubmacher D, Reinhardt D. Microfibrils and fibrillin. In: Mecham RP, editor. The Extracellular Matrix; an Overview. First ed. Springer-Verlag; Berlin Heidelberg: 2011. pp. 233–65. [Google Scholar]
- 30.Charbonneau NL, Dzamba BJ, Ono RN, Keene DR, Corson GM, Reinhardt DP, et al. Fibrillins can co-assemble in fibrils, but fibrillin fibril composition displays cell-specific differences. J Biol Chem. 2003;278:2740–9. doi: 10.1074/jbc.M209201200. [DOI] [PubMed] [Google Scholar]
- 31.Lin G, Tiedemann K, Vollbrandt T, Peters H, Batge B, Brinckmann J, et al. Homo- and heterotypic fibrillin-1 and -2 interactions constitute the basis for the assembly of microfibrils. J Biol Chem. 2002;277:50795–804. doi: 10.1074/jbc.M210611200. [DOI] [PubMed] [Google Scholar]
- 32.Marson A, Rock MJ, Cain SA, Freeman LJ, Morgan A, Mellody K, et al. Homotypic fibrillin-1 interactions in microfibril assembly. J Biol Chem. 2005;280:5013–21. doi: 10.1074/jbc.M409029200. [DOI] [PubMed] [Google Scholar]
- 33.Ashworth JL, Kelly V, Wilson R, Shuttleworth CA, Kielty CM. Fibrillin assembly: dimer formation mediated by amino-terminal sequences. J Cell Sci. 1999;112:3549–58. doi: 10.1242/jcs.112.20.3549. [DOI] [PubMed] [Google Scholar]
- 34.Beene LC, Wang LW, Hubmacher D, Keene DR, Reinhardt DP, Annis DS, et al. Nonselective assembly of fibrillin 1 and fibrillin 2 in the rodent ocular zonule and in cultured cells: implications for marfan syndrome. Invest Ophthalmol Vis Sci. 2013;54:8337–44. doi: 10.1167/iovs.13-13121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang H, Hu W, Ramirez F. Developmental expression of fibrillin genes suggests heterogeneity of extracellular micro-fibrils. J Cell Biol. 1995;129:1165–76. doi: 10.1083/jcb.129.4.1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mariencheck MC, Davis EC, Zhang H, Ramirez F, Rosenbloom J, Gibson MA, et al. Fibrillin-1 and fibrillin-2 show temporal and tissue-specific regulation of expression in developing elastic tissues. Connect Tissue Res. 1995;31:87–97. doi: 10.3109/03008209509028396. [DOI] [PubMed] [Google Scholar]
- 37.Hubmacher D, Reinhardt DP, Plesec T, Schenke-Layland K, Apte SS. Human eye development is characterized by coordinated expression of fibrillin isoforms. Invest Ophthalmol Vis Sci. 2014;55:7934–44. doi: 10.1167/iovs.14-15453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Baldwin AK, Cain SA, Lennon R, Godwin A, Merry CL, Kielty CM. Epithelial–mesenchymal status influences how cells deposit fibrillin microfibrils. J Cell Sci. 2014;127:158–71. doi: 10.1242/jcs.134270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dzamba BJ, Keene DR, Isogai Z, Charbonneau NL, Karaman-Jurukovska N, Simon M, et al. Assembly of epithelial cell fibrillins. J Invest Dermatol. 2001;117:1612–20. doi: 10.1046/j.0022-202x.2001.01588.x. [DOI] [PubMed] [Google Scholar]
- 40.Baldwin AK, Simpson A, Steer R, Cain SA, Kielty CM. Elastic fibres in health and disease. Expert Rev Mol Med. 2013;15:e8. doi: 10.1017/erm.2013.9. [DOI] [PubMed] [Google Scholar]
- 41.Pereira L, Lee SY, Gayraud B, Andrikopoulos K, Shapiro SD, Bunton T, et al. Pathogenetic sequence for aneurysm revealed in mice underexpressing fibrillin-1. Proc Natl Acad Sci U S A. 1999;96:3819–23. doi: 10.1073/pnas.96.7.3819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Trask TM, Trask BC, Ritty TM, Abrams WR, Rosenbloom J, Mecham RP. Interaction of tropoelastin with the amino-terminal domains of fibrillin-1 and fibrillin-2 suggests a role for the fibrillins in elastic fiber assembly. J Biol Chem. 2000;275:24400–6. doi: 10.1074/jbc.M003665200. [DOI] [PubMed] [Google Scholar]
- 43.Choudhury R, McGovern A, Ridley C, Cain SA, Baldwin A, Wang MC, et al. Differential regulation of elastic fiber formation by fibulin-4 and -5. J Biol Chem. 2009;284:24553–67. doi: 10.1074/jbc.M109.019364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hirai M, Ohbayashi T, Horiguchi M, Okawa K, Hagiwara A, Chien KR, et al. Fibulin-5/DANCE has an elastogenic organizer activity that is abrogated by proteolytic cleavage in vivo. J Cell Biol. 2007;176:1061–71. doi: 10.1083/jcb.200611026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tiedemann K, Sasaki T, Gustafsson E, Gohring W, Batge B, Notbohm H, et al. Microfibrils at basement membrane zones interact with perlecan via fibrillin-1. J Biol Chem. 2005;280:11404–12. doi: 10.1074/jbc.M409882200. [DOI] [PubMed] [Google Scholar]
- 46.Wang MC, Lu Y, Baldock C. Fibrillin microfibrils: a key role for the interbead region in elasticity. J Mol Biol. 2009;388:168–79. doi: 10.1016/j.jmb.2009.02.062. [DOI] [PubMed] [Google Scholar]
- 47.Thurmond F, Trotter J. Morphology and biomechanics of the microfibrillar network of sea cucumber dermis. J Exp Biol. 1996;199:1817–28. doi: 10.1242/jeb.199.8.1817. [DOI] [PubMed] [Google Scholar]
- 48.Ramirez F, Rifkin DB. Extracellular microfibrils: contextual platforms for TGFbeta and BMP signaling. Curr Opin Cell Biol. 2009;21:616–22. doi: 10.1016/j.ceb.2009.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Robinson PN, Arteaga-Solis E, Baldock C, Collod-Beroud G, Booms P, De Paepe A, et al. The molecular genetics of Marfan syndrome and related disorders. J Med Genet. 2006;43:769–87. doi: 10.1136/jmg.2005.039669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sadiq MA, Vanderveen D. Genetics of ectopia lentis. Semin Ophthalmol. 2013;28:313–20. doi: 10.3109/08820538.2013.825276. [DOI] [PubMed] [Google Scholar]
- 51.Le Goff C, Cormier-Daire V. The ADAMTS(L) family and human genetic disorders. Hum Mol Genet. 2011;20:R163–7. doi: 10.1093/hmg/ddr361. [DOI] [PubMed] [Google Scholar]
- 52.Neptune ER, Frischmeyer PA, Arking DE, Myers L, Bunton TE, Gayraud B, et al. Dysregulation of TGF-beta activation contributes to pathogenesis in Marfan syndrome. Nat Genet. 2003;33:407–11. doi: 10.1038/ng1116. [DOI] [PubMed] [Google Scholar]
- 53.Habashi JP, Judge DP, Holm TM, Cohn RD, Loeys BL, Cooper TK, et al. Losartan, an AT1 antagonist, prevents aortic aneurysm in a mouse model of Marfan syndrome. Science. 2006;312:117–21. doi: 10.1126/science.1124287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Habashi JP, Doyle JJ, Holm TM, Aziz H, Schoenhoff F, Bedja D, et al. Angiotensin II type 2 receptor signaling attenuates aortic aneurysm in mice through ERK antagonism. Science. 2011;332:361–5. doi: 10.1126/science.1192152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Callewaert BL, Loeys BL, Ficcadenti A, Vermeer S, Landgren M, Kroes HY, et al. Comprehensive clinical and molecular assessment of 32 probands with congenital contractural arachnodactyly: report of 14 novel mutations and review of the literature. Hum Mutat. 2009;30:334–41. doi: 10.1002/humu.20854. [DOI] [PubMed] [Google Scholar]
- 56.Arteaga-Solis E, Gayraud B, Lee SY, Shum L, Sakai LY, Ramirez F. Regulation of limb patterning by extracellular microfibrils. J Cell Biol. 2001;154:275–81. doi: 10.1083/jcb.200105046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Apte SS. A disintegrin-like and metalloprotease (reprolysin-type) with thrombospondin type 1 motif (ADAMTS) super-family: functions and mechanisms. J Biol Chem. 2009;284:31493–7. doi: 10.1074/jbc.R109.052340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tortorella M, Pratta M, Liu RQ, Abbaszade I, Ross H, Burn T, et al. The thrombospondin motif of aggrecanase-1 (ADAMTS-4) is critical for aggrecan substrate recognition and cleavage. J Biol Chem. 2000;275:25791–7. doi: 10.1074/jbc.M001065200. [DOI] [PubMed] [Google Scholar]
- 59.Foulcer SJ, Nelson CM, Quintero MV, Kuberan B, Larkin J, Dours-Zimmermann MT, et al. Determinants of versican-V1 proteoglycan processing by the metalloproteinase ADAMTS5. J Biol Chem. 2014;289:27859–73. doi: 10.1074/jbc.M114.573287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Blobel CP. ADAMs: key components in EGFR signalling and development. Nat Rev Mol Cell Biol. 2005;6:32–43. doi: 10.1038/nrm1548. [DOI] [PubMed] [Google Scholar]
- 61.Porter S, Clark IM, Kevorkian L, Edwards DR. The ADAMTS metalloproteinases. Biochem J. 2005;386:15–27. doi: 10.1042/BJ20040424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hubmacher D, Apte SS. Genetic and functional linkage between ADAMTS superfamily proteins and fibrillin-1: a novel mechanism influencing microfibril assembly and function. Cell Mol Life Sci. 2011;68:3137–48. doi: 10.1007/s00018-011-0780-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Dagoneau N, Benoist-Lasselin C, Huber C, Faivre L, Megarbane A, Alswaid A, et al. ADAMTS10 mutations in autosomal recessive Weill–Marchesani syndrome. Am J Hum Genet. 2004;75:801–6. doi: 10.1086/425231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Faivre L, Gorlin RJ, Wirtz MK, Godfrey M, Dagoneau N, Samples JR, et al. In frame fibrillin-1 gene deletion in autosomal dominant Weill–Marchesani syndrome. J Med Genet. 2003;40:34–6. doi: 10.1136/jmg.40.1.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Farias FH, Johnson GS, Taylor JF, Giuliano E, Katz ML, Sanders DN, et al. An ADAMTS17 splice donor site mutation in dogs with primary lens luxation. Invest Ophthalmol Vis Sci. 2010;51:4716–21. doi: 10.1167/iovs.09-5142. [DOI] [PubMed] [Google Scholar]
- 66.Morales J, Al-Sharif L, Khalil DS, Shinwari JM, Bavi P, Al-Mahrouqi RA, et al. Homozygous mutations in ADAMTS10 and ADAMTS17 cause lenticular myopia, ectopia lentis, glaucoma, spherophakia, and short stature. Am J Hum Genet. 2009;85:558–68. doi: 10.1016/j.ajhg.2009.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Haji-Seyed-Javadi R, Jelodari-Mamaghani S, Paylakhi SH, Yazdani S, Nilforushan N, Fan JB, et al. LTBP2 mutations cause Weill–Marchesani and Weill–Marchesani-like syndrome and affect disruptions in the extracellular matrix. Hum Mutat. 2012;33:1182–7. doi: 10.1002/humu.22105. [DOI] [PubMed] [Google Scholar]
- 68.Hirani R, Hanssen E, Gibson MA. LTBP-2 specifically interacts with the amino-terminal region of fibrillin-1 and competes with LTBP-1 for binding to this microfibrillar protein. Matrix Biol. 2007;26:213–23. doi: 10.1016/j.matbio.2006.12.006. [DOI] [PubMed] [Google Scholar]
- 69.Le Goff C, Morice-Picard F, Dagoneau N, Wang LW, Perrot C, Crow YJ, et al. ADAMTSL2 mutations in geleophysic dysplasia demonstrate a role for ADAMTS-like proteins in TGF-beta bioavailability regulation. Nat Genet. 2008;40:1119–23. doi: 10.1038/ng.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Christensen AE, Fiskerstrand T, Knappskog PM, Boman H, Rodahl E. A novel ADAMTSL4 mutation in autosomal recessive ectopia lentis et pupillae. Invest Ophthalmol Vis Sci. 2010;51:6369–73. doi: 10.1167/iovs.10-5597. [DOI] [PubMed] [Google Scholar]
- 71.Ahram D, Sato TS, Kohilan A, Tayeh M, Chen S, Leal S, et al. A homozygous mutation in ADAMTSL4 causes autosomal-recessive isolated ectopia lentis. Am J Hum Genet. 2009;84:274–8. doi: 10.1016/j.ajhg.2009.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ahonen SJ, Kaukonen M, Nussdorfer FD, Harman CD, Komaromy AM, Lohi H. A novel missense mutation in ADAMTS10 in Norwegian Elkhound primary glaucoma. PLoS One. 2014;9:e111941. doi: 10.1371/journal.pone.0111941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Gould D, Pettitt L, McLaughlin B, Holmes N, Forman O, Thomas A, et al. ADAMTS17 mutation associated with primary lens luxation is widespread among breeds. Vet Ophthalmol. 2011;14:378–84. doi: 10.1111/j.1463-5224.2011.00892.x. [DOI] [PubMed] [Google Scholar]
- 74.Bader HL, Ruhe AL, Wang LW, Wong AK, Walsh KF, Packer RA, et al. An ADAMTSL2 founder mutation causes Musladin–Lueke Syndrome, a heritable disorder of beagle dogs, featuring stiff skin and joint contractures. PLoS One. 2010;5:e12817. doi: 10.1371/journal.pone.0012817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hubmacher D, Wang LW, Mecham RP, Reinhardt DP, Apte SS. Adamtsl2 deletion results in bronchial fibrillin microfibril accumulation and bronchial epithelial dysplasia: a novel mouse model providing insights on geleophysic dysplasia. Dis Model Mech. 2015 Mar 11; doi: 10.1242/dmm.017046. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Shi Y, Tu Y, De Maria A, Mecham RP, Bassnett S. Development, composition, and structural arrangements of the ciliary zonule of the mouse. Invest Ophthalmol Vis Sci. 2013;54:2504–15. doi: 10.1167/iovs.13-11619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Tsutsui K, Manabe RI, Yamada T, Nakano I, Oguri Y, Keene DR, et al. A disintegrin and metalloproteinase with thrombospondin motifs-like-6 (ADAMTSL-6) is a novel extracellular matrix protein that binds to fibrillin-1 and promotes fibrillin-1 fibril formation. J Biol Chem. 2010;285:4870–82. doi: 10.1074/jbc.M109.076919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Cain SA, McGovern A, Baldwin AK, Baldock C, Kielty CM. Fibrillin-1 mutations causing Weill–Marchesani syndrome and acromicric and geleophysic dysplasias disrupt heparan sulfate interactions. PLoS One. 2012;7:e48634. doi: 10.1371/journal.pone.0048634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Klingberg F, Hinz B, White ES. The myofibroblast matrix: implications for tissue repair and fibrosis. J Pathol. 2013;229:298–309. doi: 10.1002/path.4104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Balestrini JL, Chaudhry S, Sarrazy V, Koehler A, Hinz B. The mechanical memory of lung myofibroblasts. Integr Biol. 2012;4:410–21. doi: 10.1039/c2ib00149g. [DOI] [PubMed] [Google Scholar]
- 81.Hanssen E, Franc S, Garrone R. Synthesis and structural organization of zonular fibers during development and aging. Matrix Biol. 2001;20:77–85. doi: 10.1016/s0945-053x(01)00122-6. [DOI] [PubMed] [Google Scholar]
- 82.Shi Y, Tu Y, Mecham RP, Bassnett S. Ocular phenotype of Fbn2-null mice. Invest Ophthalmol Vis Sci. 2013;54:7163–73. doi: 10.1167/iovs.13-12687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Inoue T, Ohbayashi T, Fujikawa Y, Yoshida H, Akama TO, Noda K, et al. Latent TGFbeta binding protein-2 is essential for the development of ciliary zonule microfibrils. Hum Mol Genet. 2014;23:5672–82. doi: 10.1093/hmg/ddu283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Mir S, Wheatley HM, Hussels IE, Whittum-Hudson JA, Traboulsi EI. A comparative histologic study of the fibrillin microfibrillar system in the lens capsule of normal subjects and subjects with Marfan syndrome. Invest Ophthalmol Vis Sci. 1998;39:84–93. [PubMed] [Google Scholar]
- 85.Traboulsi EI, Whittum-Hudson JA, Mir SH, Maumenee IH. Microfibril abnormalities of the lens capsule in patients with Marfan syndrome and ectopia lentis. Ophthalmic Genet. 2000;21:9–15. [PubMed] [Google Scholar]
- 86.Cain SA, Morgan A, Sherratt MJ, Ball SG, Shuttleworth CA, Kielty CM. Proteomic analysis of fibrillin-rich microfibrils. Proteomics. 2006;6:111–22. doi: 10.1002/pmic.200401340. [DOI] [PubMed] [Google Scholar]
- 87.Chandra A, Jones M, Cottrill P, Eastlake K, Limb GA, Charteris DG. Gene expression and protein distribution of ADAMTSL-4 in human iris, choroid and retina. Br J Ophthalmol. 2013;97:1208–12. doi: 10.1136/bjophthalmol-2013-303353. [DOI] [PubMed] [Google Scholar]
- 88.Horiguchi M, Ota M, Rifkin DB. Matrix control of transforming growth factor-beta function. J Biochem. 2012;152:321–9. doi: 10.1093/jb/mvs089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Le Goff C, Mahaut C, Abhyankar A, Le Goff W, Serre V, Afenjar A, et al. Mutations at a single codon in Mad homology 2 domain of SMAD4 cause Myhre syndrome. Nat Genet. 2012;44:85–8. doi: 10.1038/ng.1016. [DOI] [PubMed] [Google Scholar]