Abstract
Colorectal cancer metastasis to the liver is a major cause of cancer-related death; however, the genes and pathways that govern this metastatic colonization event remain poorly characterized. Here, using a large-scale in vivo RNAi screen, we identified liver and red blood cell pyruvate kinase (PKLR) as a driver of metastatic liver colonization. PKLR expression was increased in liver metastases as well as in primary colorectal tumors of patients with metastatic disease. Evaluation of a murine liver colonization model revealed that PKLR promotes cell survival in the tumor core during conditions of high cell density and oxygen deprivation by increasing glutathione, the primary endogenous antioxidant. PKLR negatively regulated the glycolytic activity of PKM2, the major pyruvate kinase isoenzyme known to regulate cellular glutathione levels. Glutathione is critical for metastasis, and we determined that the rate-limiting enzyme of glutathione synthesis, GCLC, becomes overexpressed in patient liver metastases, promotes cell survival under hypoxic and cell-dense conditions, and mediates metastatic liver colonization. RNAi-mediated inhibition of glutathione synthesis impaired survival of multiple colon cancer cell lines, and pharmacological targeting of this metabolic pathway reduced colonization in a primary patient-derived xenograft model. Our findings highlight the impact of metabolic reprogramming within the niche as metastases progress and suggest clinical potential for targeting this pathway in colorectal cancer.
Introduction
Colorectal cancer is the third leading cause of cancer death in the United States (1). While surgical resection of localized disease can be curative, mortality is primarily caused by metastatic progression, with the liver being the primary site of relapse. Only 10 percent of patients diagnosed with liver metastatic colorectal cancer live beyond 5 years (2, 3). This poor long-term survival outlook highlights the limitations of standard cytotoxic therapies used, such as 5-fluorouracil and oxaliplatin. While recent nuanced changes to therapeutic regimens and the addition of targeted therapies have modestly improved the management of metastatic disease (3), there is a great need for therapies with significant efficacy against liver metastatic colonization and progression.
To clinically impact colorectal cancer, the process of liver colonization must be better characterized (4–6). Metastatic colonization, the formation of micrometastases and subsequent macrometastases, has been observed to be a significant rate-limiting step during metastatic liver growth (7). This selective pressure may be in part attributable to the liver microenvironment, which is marked by hypoxic regions, a unique circulatory architecture, and significant metabolic activity (8). The genes and pathways most suitable for therapeutic inhibition would be those whose expression is necessary for colonization events. Furthermore, it remains unclear whether cancer cells use different mechanisms for early or late colonization events (9). Late colonization events, which contribute to the continued progression of macrometastases, represent a stage with significant clinical need, given that patients who develop liver metastases exhibit a poor prognosis and that approximately 20 percent of patients present with distant metastatic disease at the time of diagnosis (1).
The identification of key regulators necessitates a systematic, functional approach to characterize the colonization process. Large-scale in vivo RNAi screens have enabled the unbiased characterization of pathological and developmental processes (10). Furthermore, the application of these screens under pathological conditions, as well as under conditions of normal growth, has allowed for the identification of genes with specificity to the disease process being studied. Here, we used a large-scale RNAi drop-out screen (54,591 total hairpins targeting 14,095 human genes) to identify candidate genes required for colonization of the liver by multiple colon cancer cell lines. Our interest in identifying key regulators and pathways led us to focus on kinases as important cell signaling molecules. We found that liver and red blood cell pyruvate kinase (PKLR) promotes colon cancer cell metastatic colonization of the liver, while not promoting basal growth in culture. Analysis of PKLR expression in patient samples revealed a significant association with metastatic disease in multiple data sets. PKLR promotes cell survival in the tumor core, a phenotype that can be recapitulated in vitro under conditions of high cell density and hypoxia. Under these conditions, PKLR is required to maintain levels of the major endogenous antioxidant, glutathione, and support cancer cell survival. PKLR negatively regulates the glycolytic activity of the predominant pyruvate kinase expressed in cancer, PKM2, which is known to regulate glycolytic flux in order to generate reducing power required for antioxidant responses (11). Suppressing glutathione synthesis via RNAi or small-molecule inhibition suppressed metastatic colonization, suggesting potential for clinical utility. Our findings reveal the use of multiple pyruvate kinase isozymes by colon cancer for enhanced regulatory control of glycolysis and antioxidant generation. Furthermore, the microenvironment conditions that render colon cancer cells vulnerable to cell death highlight the metabolically demanding conditions that exist during liver colonization and reveal potential for therapeutic targeting of these pathways.
Results
Large-scale in vivo shRNA screen identifies promoters of liver colonization.
To study colorectal cancer colonization in the liver molecularly, an in vivo shRNA screen (12) was performed to identify regulatory genes (Figure 1A). Three human colorectal cancer cell lines, representing common mutational subtypes (Kras wild-type/mutant, Braf wild-type/mutant, MSS/MSI) seen in patients, were transduced with subpools of shRNA-encoding lentiviral particles totaling 54,591 total hairpins targeting 14,095 human genes. These cell populations were then inoculated into a total of 204 mice by direct liver injection for the selection of cells capable of colonizing the liver. In parallel, cells split from these same cell populations were grown in culture to select for cells capable of basal growth and survival. Once tumors developed, processing of genomic shRNA inserts from liver tumors, cultured cells, and the original parental population was performed to allow for quantification of shRNA genomic sequence depletion. Loss of shRNA representation from tumors would suggest that depletion of the genes targeted suppressed the ability of colon cancer cells to colonize the liver.
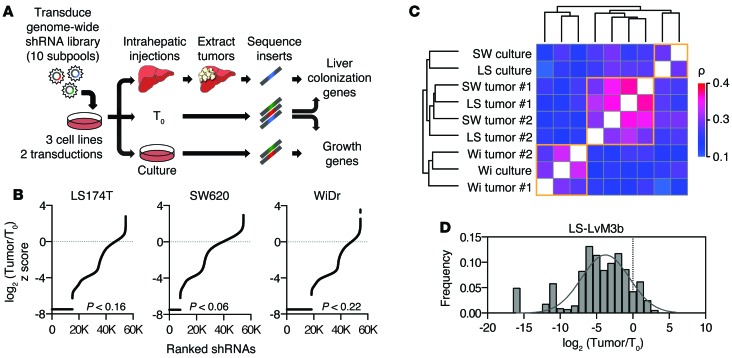
Figure 1. Large-scale RNAi screen for promoters of liver metastasis.
(A) Schematic of pooled shRNA library drop-out screen. (B) Overview of shRNA depletion in liver tumors. shRNAs were ranked based on peak median absolute deviation normalized z score of log2 (Tumor/T0). shRNAs absent in tumors are depicted by the black lines. P values for absent shRNAs were derived by bootstrapping with 1,000 random samplings. (C) Correlation matrix of shRNA-depletion profiles of samples using Spearman’s correlation coefficient. Clustering was performed in R using Euclidean distance and complete agglomeration method. SW, SW620; LS, LS174T; Wi, WiDr. (D) Distribution of shRNA depletion in tumors from a secondary library using top-scoring shRNAs that targeted each of the 556 top genes.
Analysis of shRNA profiling in tumors and cultured cells revealed significant depletion of shRNAs across all cell lines (Figure 1B and Supplemental Figure 1A; supplemental material available online with this article; doi:10.1172/JCI83587DS1). For the majority of samples, liver tumor profiles displayed high similarity, despite differing parental cell line origins (Figure 1C). To identify broadly relevant metastasis regulators, genes were considered as putative promoters of liver colonization if at least two shRNAs targeting a given gene were depleted in tumors derived from all 3 cell lines and in both independent transduction replicates. This analysis yielded 556 candidate promoters of liver colonization (Supplemental Figure 1B). Using similar criteria, 719 genes appeared to be required for survival in cultured cells, while 187 genes appeared to be required in both in vivo and in vitro conditions (Supplemental Figure 1, C and D). To confirm that the analyzed shRNAs were effective in suppressing colonization, a secondary library was generated using top-scoring shRNAs that targeted the 556 candidate genes. This library was transduced into a highly metastatic in vivo–selected colon cell line, LS-LVM3b (13). These populations were then subjected to the experimental procedures performed for the primary screen. Analysis revealed that, of these shRNAs, 89% were markedly depleted from tumors (Figure 1D).
Given the off-target limitations of RNAi (14), we applied additional scoring metrics to select candidate genes most suitable for thorough validation. To better rank the candidate hits, we scored all genes from the primary genome-scale liver colonization screen by applying the established RIGER algorithm, which accounts for variability in number of shRNAs for each gene (15). These scores were used to generate gene percentile scores, providing a continuous scale to assess the relative importance of genes. As expected, the top 556 candidate genes were markedly enriched among the top percentile scores, providing confidence in the candidate liver colonization gene list (Supplemental Figure 1E). Interestingly, creatine kinase, brain-type (CKB), which we previously identified as a promoter of the initiation phase of liver colonization (13), scored as a promoter by the RIGER algorithm (78th percentile) in this independent screen, despite being targeted by only 2 hairpins in the primary screen.
PKLR promotes liver colonization but not tumor growth.
Our interest in uncovering critical regulatory mechanisms led us to focus on kinases. We assessed the gene percentile scores of kinases from the 556 candidate promoters. The top-scoring kinases included many known regulators of colorectal cancer progression, including IGF1R (16) and DDR2 (17), which displayed high gene percentile scores in the screen performed in cultured cells, consistent with their known roles in in vitro proliferation (Figure 2A). Surprisingly, one of the top-scoring genes identified was PKLR, which had been commonly believed to be expressed only in liver, kidney, and red blood cells (18). Interestingly, PKLR displayed a low gene percentile score under culture conditions, suggesting a selective requirement for this gene during liver colonization.
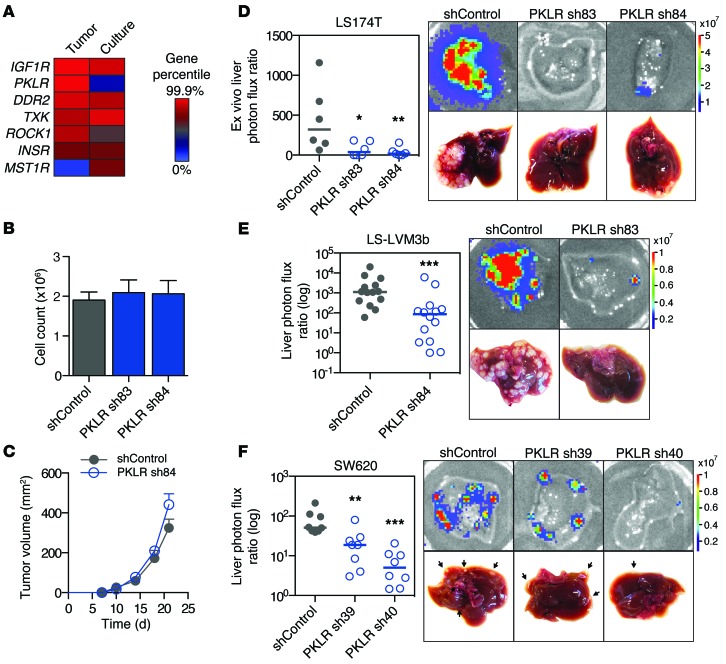
Figure 2. PKLR promotes liver metastatic colonization.
(A) Kinases from the top 556 candidates were ranked by RIGER gene percentile score in liver tumor and cell culture genome-wide screens. (B) 5 × 104 LS174T cells were seeded in triplicate, and viable cells were counted by trypan blue exclusion after 5 days under recommended cell culture conditions. Data shown are from 3 independent experiments (n = 3). (C) 106 LS174T cells were injected into mice subcutaneously, and tumor volume was measured over time (n = 6). (D) 5 × 105 LS174T cells were inoculated by direct liver injection, and liver colonization was measured by ex vivo bioluminescence after 20 days. Representative liver bioluminescence and gross pathology are shown (n = 6). (E) 5 × 105 LS-LVM3b cells were inoculated by portal circulation injection, and metastatic colonization was measured by liver bioluminescence after 20 days (n = 14). Representative liver bioluminescence and gross pathology are shown. (F) 5 × 105 SW620 cells were inoculated by portal circulation injection, and metastatic colonization was measured by liver bioluminescence after 35 days (n = 8). Representative liver bioluminescence and gross pathology are shown. Arrows indicate tumor nodules. Horizontal bars represent the median. *P < 0.05, **P < 0.01, ***P < 0.001, 1-sided Mann Whitney test between indicated sample and shControl.
To confirm these findings, cell lines were generated with additional independent shRNAs not previously used in the aforementioned screens that provided adequate knockdown (13% and 17% relative to control, Supplemental Figure 2A). In vitro proliferation assays confirmed that PKLR depletion did not reduce cell viability in culture (Figure 2B). PKLR depletion also did not reduce subcutaneous tumor growth (Figure 2C). Importantly, in concordance with the in vivo screen results, PKLR-depleted cells were significantly less effective than control cells in colonizing the liver following direct liver inoculation, with many livers remarkably showing an absence of tumor growth (Figure 2D). To validate this colonization defect in the context of metastasis, cells were inoculated via the portal circulation to assess metastatic activity upon hematogenous arrival in the liver. PKLR-depleted cells (expression was knocked down to 9% relative to that in control LS-LVM3b, and expression was knocked down to 58% and 42% relative to that in control in SW620; Supplemental Figure 2, B and C) displayed significantly reduced liver metastatic capacity in metastasis assays in multiple cell lines (Figure 2, E and F). These results reveal that PKLR promotes metastatic liver colonization of colon cancer cells.
PKLR expression is associated with metastatic disease in patients with colorectal cancer.
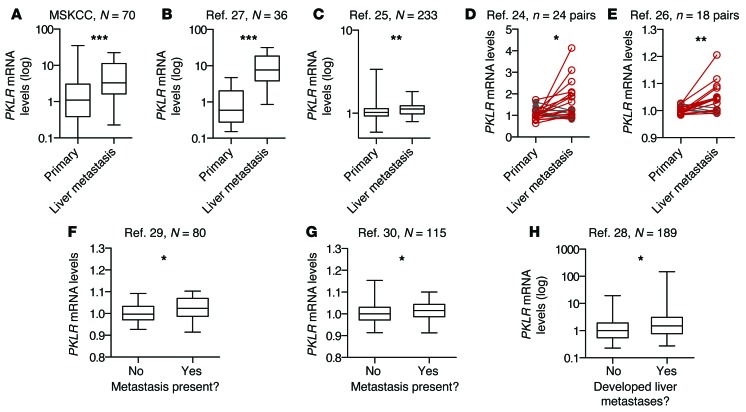
In mammalian cells, pyruvate kinase is encoded by 4 isozymes: M1, M2, liver (PKL), and red blood cell (PKR). While the M1, PKL, and PKR isozymes are described to exhibit tissue-specific expression, the pyruvate kinase M2 isoform is highly expressed across cancer types (19). PKL and PKR have only been described to promote glycolysis in their respective tissues, catalyzing the final rate-limiting step, which involves the transfer of a phosphate group from phosphoenolpyruvate (PEP) to ADP, generating pyruvate and ATP. Although PKLR had originally been described to be solely expressed in liver, kidney, and red blood cells (20), PKL expression has also been observed in epithelial cells in the intestinal epithelium of rats (21, 22). Analysis of the Human Protein Atlas data (23) revealed PKL mRNA expression in normal colon tissue and PKLR protein expression in normal colonic glandular tissue and colorectal cancer tumors (http://www.proteinatlas.org; Supplemental Figure 3). To determine whether PKLR expression is associated with liver metastasis, we obtained patient samples at MSKCC and compared PKLR transcript levels in primary tumors to levels in liver metastases. In 70 samples, PKLR mRNA was significantly upregulated in liver metastases relative to primary tumors (Figure 3A). This upregulation was also observed in multiple independent data sets of unmatched (Figure 3, B and C) and matched (Figure 3, D and E) samples from patients with colorectal cancer (24–27). These samples were from a total of 311 patients. Additionally, in primary tumors, PKLR expression was associated with the presence of metastatic disease (Figure 3, F and G) as well as the development of liver metastases (Figure 3H; refs. 28–30). These tumors totaled 284 in sum. Taken together, PKLR expression in human samples supports a positive role for PKLR in liver metastatic progression in agreement with our experimental data.
Figure 3. PKLR expression is upregulated in human colorectal cancer liver metastases and is associated with metastatic outcomes in primary tumors.
(A) PKLR expression, as measured by quantitative RT-PCR in primary and liver metastasis tumor samples obtained at MSKCC. (B) PKLR expression, as measured by RNA sequencing in primary and liver metastasis tumor samples (27). (C) PKLR expression, as measured by microarray in primary and liver metastasis tumor samples (25). (D and E) PKLR expression, as measured by microarray in matched primary and liver metastasis tumor samples (24, 26). Increased expression is indicated in red, while decreased expression is indicated in gray. (F and G) PKLR expression, as measured by microarray in primary tumor samples of patients with or without metastases present at time of sampling (29, 30). (H) PKLR expression, as measured by microarray in primary tumor samples of patients who were monitored over time for the development of liver metastases (28). Numbers of samples (N) or paired sets (n) are indicated in figure. For box-and-whisker plots, the bounds of the box are the first and third quartiles, the band inside is the median, and the ends of the whiskers are the minimum and maximum values. *P < 0.05, **P < 0.01, ***P < 0.001, 1-sided Mann Whitney test (A–C and F–H), or, where appropriate, paired 1-sided t test for matched samples (D and E).
PKLR promotes cell survival in the metastatic tumor core.
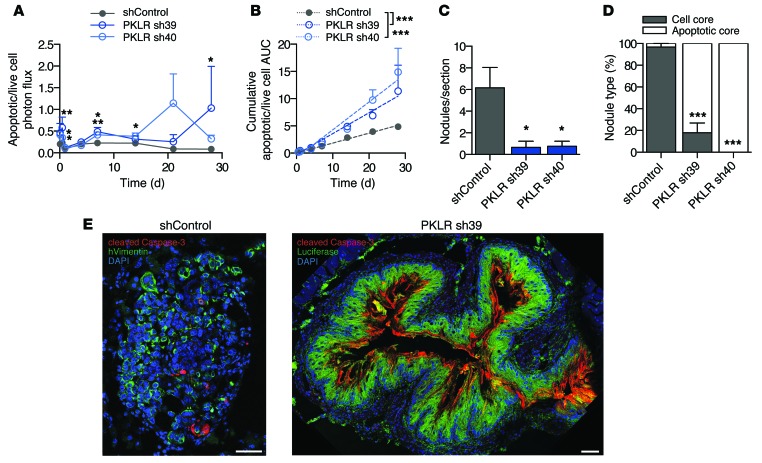
To characterize the metastatic defect, PKLR-depleted cells were assessed for cell death in vivo. To assess cell death, an in vivo caspase-3/7–dependent reporter was used to measure apoptotic cell burden during metastatic liver growth. At early as well as during late time points, the apoptosis signal was significantly increased in PKLR-depleted cells in vivo (Figure 4A). Calculation of cumulative apoptotic burden over time revealed a significantly elevated rate of apoptosis (Figure 4B), indicating that PKLR promotes metastatic cell survival continuously. To confirm these findings, livers were resected and processed to allow for visualization of apoptotic markers. Livers bearing PKLR-depleted cells displayed significantly fewer tumor nodules (Figure 4C), confirming our assessments by gross pathology. Interestingly, while tumor nodules generated from control cells appeared to be highly cellular (Figure 4E), the majority of PKLR-depleted tumor nodule cores appeared devoid of cells and were composed of extracellular cleaved caspase-3 (Figure 4, D and E). While the decreased number of total nodules with PKLR depletion suggests a role in early colonization, enhanced apoptosis in nodule cores suggests a pathophysiological requirement for PKLR during later stages of colonization as well.
Figure 4. PKLR promotes metastatic cell survival in the tumor core.
(A) 5 × 105 SW620 cells were inoculated by portal circulation injection, and apoptotic cell burden in the liver was monitored using DEVD-luciferin bioluminescence relative to live cell bioluminescence over time (n = 5). *P < 0.05, **P < 0.01, 1-sided t test between indicated sample and shControl. (B) Cumulative apoptotic/live cell burden over time was determined by calculating the area under the curve for each mouse. Linear regression lines are shown. ***P < 0.001, ANCOVA testing for difference in slope between indicated sample and shControl. (C) Livers were extracted in the previous experiment after 28 days, and nodules per liver section were quantified (n = 4). The average value from 3 liver sections for each mouse was used. *P < 0.05, 1-sided t test between indicated sample and shControl. (D) For each liver, proportion of the indicated nodule type is shown. ***P < 0.001, 2-sided Fisher’s exact test on total nodule count for each PKLR shRNA compared with shControl. (E) Representative images of nodule types. DAPI (blue) was used to label nuclei, cleaved caspase-3 (red) was used to label apoptosis, and luciferase and human vimentin (green) were used to label cancer cells. Scale bar: 50 μm.
PKLR promotes cell survival under concurrent conditions of high cell density and hypoxia.
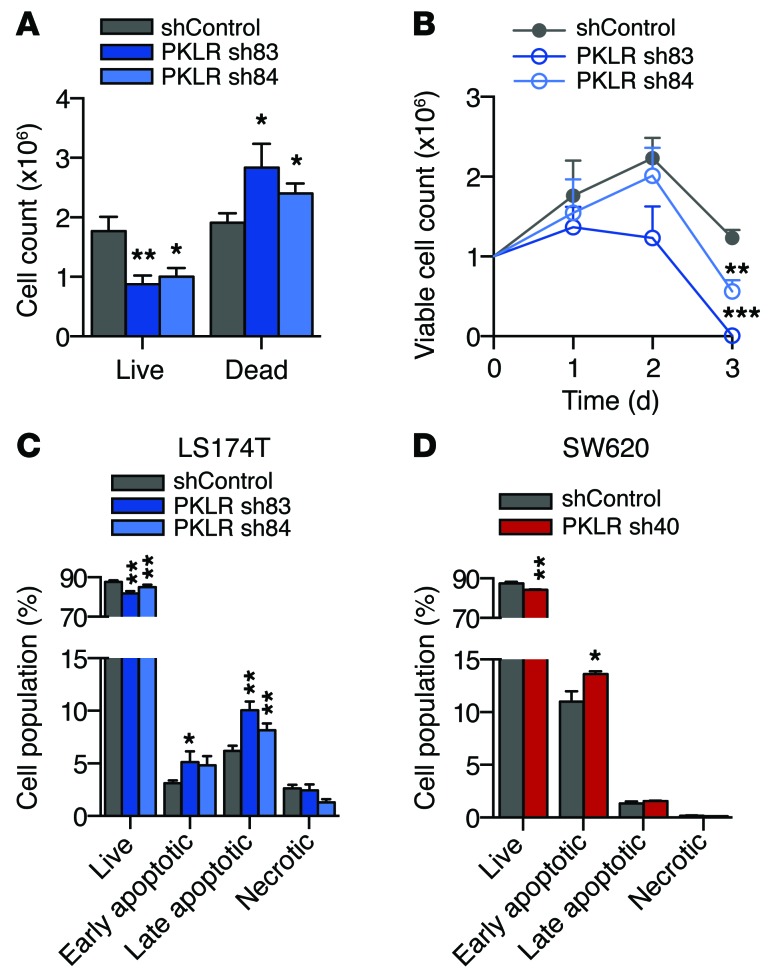
The tumor core represents a unique microenvironment with limited nutrient supply, distinct stromal composition, and altered cell-to-cell architecture (31). Liver metastases are susceptible to stress-induced apoptosis (32) and demonstrate variable angiogenic capacity (33), suggesting that cancer cells farther from the vasculature may be particularly sensitive to nutrient and oxygen deprivation. To identify a phenotypic defect in vitro, PKLR-depleted cells were grown under various conditions individually and were found to exhibit no survival defects under physiologic levels of glucose, hypoxia (1% O2), anoxia (<0.01% O2), high cell density, or low pH (Supplemental Figure 4, A–E). Moreover, PKLR-depleted cells did not exhibit defects in well-established prometastatic phenotypes, including matrigel invasion capacity, anchorage-independent growth, and migration (Supplemental Figure 4, F–H). While individual stressors elicited no phenotypic defect alone, we reasoned that the tumor core features multiple stressors that simultaneously impact cancer cells. To better recapitulate the microenvironment in the tumor core, cells were seeded concurrently at a high cell density and under hypoxia. Under these conditions, PKLR-depleted cells displayed reduced survival — decreased numbers of live cells and concomitant increased numbers of dead cells (Figure 5A). To characterize whether the survival defect occurs continuously or whether nutrient depletion led to a population collapse, cells were assayed daily for viability under high cell density and hypoxia. Cells with reduced PKLR expression began to display a survival defect as early as 1 day under these conditions, indicating that the effect is continuous (Figure 5B). Given the continuous survival defect observed in PKLR-depleted tumors, cells were assessed for apoptosis and necrosis by flow cytometry after 1 day under concurrent high cell density and hypoxia. PKLR-depleted cells exhibited fewer live cells, a greater proportion of early apoptotic cells, and a greater proportion of late apoptotic cells under these conditions (Figure 5, C and D). These results indicate that PKLR functions to enhance cell survival under cell-dense, hypoxic conditions, simulating pathophysiological features pertaining to metastatic tumor growth in the liver.
Figure 5. PKLR promotes survival under conditions of hypoxia and high cell density.
(A) 106 LS174T cells were seeded in triplicate at a density of 1,000 cells per mm2 and were allowed to grow for 5 days in 1% O2. Live and dead cells were quantified using trypan blue exclusion. Data shown are from 4 independent experiments (n = 4). (B) 106 LS174T cells were seeded in triplicate at a density of 1,000 cells per mm2 and allowed to grow in 1% O2. Live cells were quantified using trypan blue exclusion. Data shown are from 3 independent experiments (n = 3). (C) 106 LS174T and 106 (D) SW620 cells were seeded at a density of 1,000 cells per mm2 and were assessed for apoptosis after 24 hours in 1% O2. Experiments were conducted at least twice. *P < 0.05, **P < 0.01, ***P < 0.001, 1-sided t test between indicated sample and shControl.
PKLR negatively regulates PKM2 pyruvate kinase activity.
Interestingly, the in vitro survival defect of PKLR-depleted cells was observed using cell culture media supplemented with excess pyruvate, which can be readily imported into cells (34), arguing against the canonical proglycolytic function of PKLR in these colon cancer cells. Additionally, the M2 isoenzyme of pyruvate kinase has been described to function as the predominant isoform observed in colorectal cancer tissue and is the only isoform encoded by the PKM gene observed to be expressed in the cell lines used (35–37). To determine whether PKLR could be functioning noncanonically, independent of its proglycolytic role, we globally assessed PKL- and PKR-binding partners in colorectal cancer cells using tagged proteins, as commercial and custom antibodies were inadequate for the recognition of endogenous PKLR proteins in cell lysates. Immunoprecipitation of PKL and PKR followed by mass spectrometry revealed similar protein binding profiles and binding to many glycolytic enzymes (Figure 6A and Supplemental Figure 5, A and B), consistent with the role of pyruvate kinase in a glycolytic enzyme complex that allows for efficient lactate production (38). Interestingly, one of the most abundant binding partners of PKLR identified was PKM, with multiple peptides matching to the M2 isoform specifically (Supplemental Figure 5C). Immunoprecipitation of tagged PKL or PKR revealed an association with endogenous PKM2 (Figure 6B). Moreover, immunoprecipitation of endogenous PKM2 revealed PKM2 to associate with endogenous PKL (Figure 6C and Supplemental Figure 5D).
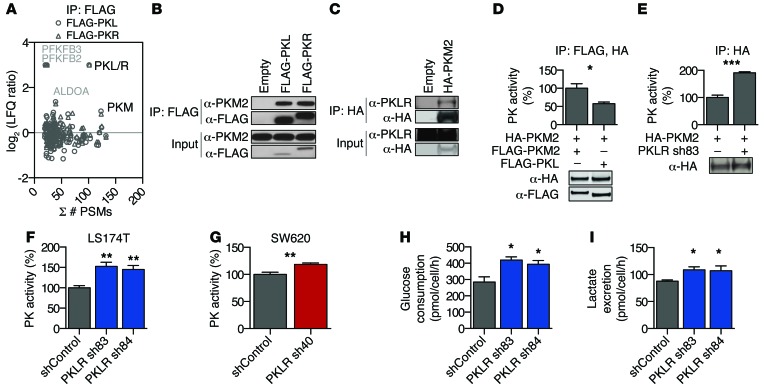
Figure 6. PKLR negatively regulates PKM2 pyruvate kinase activity.
(A) Cell lysates from LS-LVM3b cells with indicated overexpression constructs were subjected to immunoprecipitation using anti-FLAG beads followed by LC-MS/MS. Protein peptide-spectrum matches (PSMs), as compared with label-free quantification (LFQ) of protein enrichment (relative to empty vector immunoprecipitation), are shown. (B) Cell lysates from LS-LVM3b cells were subjected to immunoprecipitation using anti-FLAG beads followed by Western blot analysis. The experiment was conducted at least 3 times. (C) Cell lysates from LS174T cells were subjected to immunoprecipitation using anti-HA beads followed by Western blot analysis. The experiment was conducted at least 3 times. (D) Sequentially immunoprecipitated PKM2 complexes from transfected HEK293T cells (n = 3) were assayed for pyruvate kinase activity. Fusion protein expression was confirmed by Western blotting. The experiment was conducted twice. *P < 0.05, 2-sided t test. (E) Immunoprecipitated PKM2 complexes from LS174T cells (n = 3), subjected to 16 hours under high cell density and hypoxia, were assayed for pyruvate kinase activity. Activity was normalized to immunoprecipitated HA-PKM2, as measured by quantitative Western blotting. The experiment was conducted 3 times. ***P < 0.001, 2-sided t test. (F and G) Pyruvate kinase activity was measured from (F) LS174T and (G) SW620 cell lysates after 24 hours under hypoxic, cell-dense conditions. Activity was normalized to live cells by cell count. Data shown are from 3 biological replicates and 2 independent experiments. **P < 0.01, 1-sided t test between indicated sample and shControl. (H) Glucose uptake rates and (I) lactate excretion rates were determined from conditioned media of LS174T cells grown in high cell density and hypoxia for 24 hours. Data shown are from 3 biological replicates. *P < 0.05, 1-sided t test between indicated sample and shControl.
PKM2 is essential for aerobic glycolysis observed in cancer cells, which leads to preferential lactate production and decreased mitochondrial metabolism (36). Previous studies have demonstrated an advantage conferred by decreasing PKM2 glycolytic activity in cancer cells, resulting in a shift toward alternative pathways that promote biosynthetic processes (39, 40) and intracellular reducing power under oxidizing conditions such as hypoxic stress (11). Given the differing biochemical properties of pyruvate kinase isozymes (38), PKL could function to alter PKM2 pyruvate kinase activity. Indeed, immunoprecipitated PKM2 complexes associated with PKL exhibited reduced pyruvate kinase activity relative to PKM2-only complexes (Figure 6D), which corresponded to decreased glycolytic enzyme complex formation (Supplemental Figure 6A). Conversely, immunoprecipitated PKM2 complexes from PKLR-depleted cells grown in cell-dense, hypoxic conditions exhibited enhanced pyruvate kinase activity relative to complexes from control cells (Figure 6E), consistent with an observed increase in glycolytic enzyme complex formation (Supplemental Figure 6, B and C). Additionally, cell lysates from PKLR-depleted cells displayed an increase in total pyruvate kinase activity (Figure 6, F and G) as well as a decreased PEP-to-pyruvate ratio following incubation under hypoxic, cell-dense conditions (Supplemental Figure 6D). To determine whether altered pyruvate kinase activity contributes to glycolytic deregulation, glycolytic flux was assessed under conditions of hypoxia and high cell density. PKLR depletion resulted in increased glucose consumption as well as lactate excretion (Figure 6, H and I). Our findings reveal that, in colon cancer cells, PKLR expression can be used to negatively regulate PKM2 pyruvate kinase activity and deregulate glycolytic metabolism to allow for cancer progression.
PKLR increases glutathione levels.
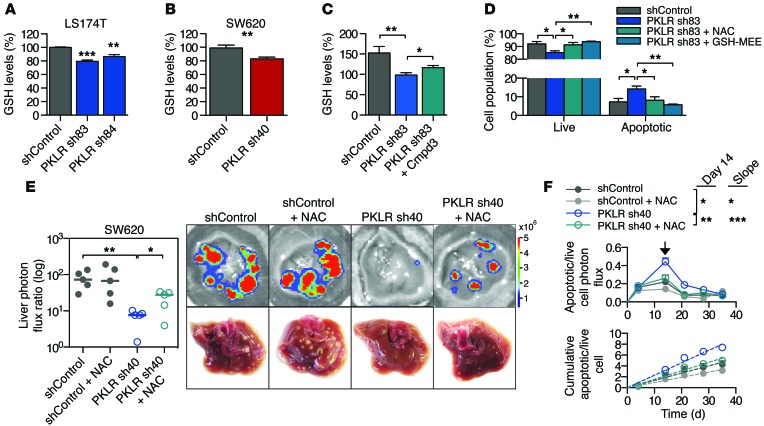
Decreased PKM2 activity can be advantageous for cancer cells, as glycolysis is critical for regulating a metabolic shift toward increased biosynthetic processes and intracellular reducing power (11, 39–41). Additionally, conditions of hypoxia and high cell density each are known to affect cellular redox status (11, 42, 43). To determine whether the survival defect could be due to an impaired antioxidant response, PKLR-depleted cells were assessed for depletion of intracellular glutathione, the main endogenous antioxidant produced in cells, under hypoxic, cell-dense conditions. Indeed, PKLR-depleted cells demonstrated decreased glutathione levels (Figure 7, A and B). To demonstrate that PKLR functions through PKM2 to elicit a change in glutathione levels, PKLR-depleted cells were treated with a PKM2 small-molecule inhibitor (ref. 44 and Supplemental Figure 7A). Treatment with the PKM2 small-molecule inhibitor partially restored glutathione levels under hypoxic, cell-dense conditions (Figure 7C). To confirm that glutathione depletion is responsible for the cell survival defect, PKLR-depleted cells incubated in hypoxic and cell-dense conditions were incubated with the antioxidant N-acetyl cysteine (NAC), a prodrug to a glutathione precursor, and glutathione monoethylester, a membrane-permeable GSH analog. Treatment with either of these compounds rescued cell viability and decreased apoptosis in vitro (Figure 7D and Supplemental Figure 7B). Consistent with these in vitro findings, treatment of mice bearing PKLR-depleted cells with NAC partially rescued metastatic liver colonization capacity (Figure 7E), while nearly completely suppressing apoptosis and apoptotic rate in vivo (Figure 7F).
Figure 7. PKLR increases glutathione levels.
(A and B) Glutathione levels were measured from (A) LS174T and (B) SW620 cell lysates in quadruplicate using ThiolTracker after 24 hours under hypoxic, cell-dense conditions. Data shown are from 2 independent experiments. **P < 0.01, ***P < 0.001, 1-sided t test between indicated sample and shControl. (C) 106 LS174T cells were seeded in quadruplicate at a density of 1,000 cells per mm2, incubated with DMSO or 50 μM compound 3 for 3 hours in 1% O2, and then assessed for glutathione levels. Data shown are from 3 independent experiments. *P < 0.05, **P < 0.01, 1-sided t test. (D) 106 LS174T cells were seeded in triplicate at a density of 1,000 cells per mm2, incubated with indicated compounds, and assessed for apoptosis after 24 hours in 1% O2. The experiment was conducted twice. *P < 0.05, **P < 0.01, 1-sided t test. GSH-MEE, glutathione monoethylester. (E) 5 × 105 SW620 cells were inoculated by portal circulation injection, mice were treated with NAC or control drinking water, and metastatic colonization was measured by liver bioluminescence after 35 days (n = 5). Representative liver bioluminescence and gross pathology are shown. *P < 0.05, **P < 0.01, 1-sided Mann Whitney test between indicated samples. Horizontal bars represent the median. (F) Apoptotic cell burden from the experiment in E was monitored by DEVD-luciferin liver bioluminescence relative to total live cell bioluminescence over time (top). Day 14 is indicated with arrow. *P < 0.05, **P < 0.01, 1-sided t test. Cumulative apoptotic/live cell burden over time was determined by calculating the area under the curve for each mouse (bottom). Linear regression lines are shown (dashed lines). *P < 0.05, ***P < 0.001, ANCOVA for difference in slope.
GCLC as therapeutic target for metastatic liver colonization.
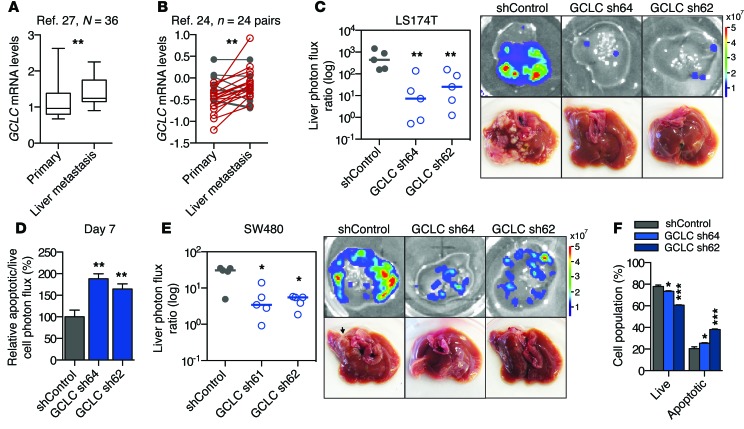
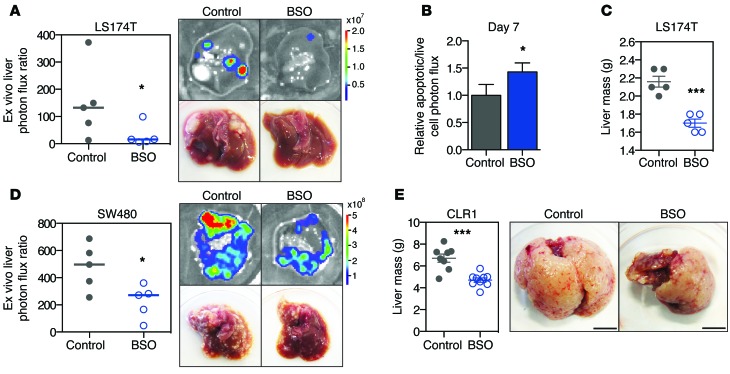
Given the susceptibility of liver metastases to glutathione depletion, we reasoned that glutathione synthesis might serve as a critical pathway used for metastatic progression. GCLC, the catalytic subunit of glutamate cysteine ligase, is responsible for the rate-limiting reaction of glutathione synthesis (45). Consistent with our experimental observations suggesting a role for glutathione in colorectal cancer progression, GCLC expression levels were significantly higher in human colorectal cancer liver metastases relative to primary tumors (Figure 8, A and B, and Supplemental Figure 8A). Using GCLC-depleted cells (expression was knocked down to 27% and 13% relative to control levels in LS174T cells and to 41% and 26% relative to control levels in SW480 cells; Supplemental Figure 8, B and C), functional studies revealed GCLC to promote metastatic liver colonization (Figure 8, C and E), suppress apoptosis in vivo (Figure 8D), and promote cell survival under hypoxic, cell-dense conditions (Figure 8F and Supplemental Figure 8D), consistent with the observed phenotypes seen upon PKLR depletion. Additionally, GCLC depletion did not significantly alter proliferation under optimal cell culture conditions (Supplemental Figure 8E). These findings suggested that small molecules that block glutathione synthesis may exhibit metastasis-suppressive effects (46). To test this hypothesis, colon cancer cells were injected into the portal circulation of mice that were provided l-buthionine-(S,R)-sulfoximine (BSO), a micromolar inhibitor of glutathione synthesis, in their drinking water. Therapeutic delivery of BSO suppressed metastatic colonization of colon cancer cell lines and increased cancer cell apoptosis in vivo (Figure 9, A–D). Importantly, BSO also suppressed metastatic colonization by a patient-derived primary colon cancer graft (Figure 9E). These results highlight the importance of cancer cell glutathione levels for metastatic survival in the liver and identify this metabolic pathway for potential therapeutic targeting.
Figure 8. GCLC, the rate-limiting enzyme in glutathione synthesis, promotes liver metastatic colonization.
(A and B) GCLC expression as measured in (A) unmatched (27) and (B) matched primary and liver metastasis tumor samples (24). Increased GCLC expression is indicated in red; decreased expression is indicated in gray. Numbers of samples (N) or paired sets (n) are indicated in figure. For the box-and-whisker plot, the bounds of the box are the first and third quartiles, the band inside is the median, and the ends of the whiskers are the minimum and maximum values. **P < 0.01, 1-sided Mann Whitney test (A), paired 1-sided t test for matched samples (B). (C) 5 × 105 LS174T cells were inoculated by portal circulation injection, and metastatic colonization was measured by liver bioluminescence after 21 days. Representative liver bioluminescence and gross pathology are shown (n = 5). **P < 0.01, 1-sided Mann Whitney test between indicated sample and shControl. Horizontal bars represent the median. (D) DEVD-luciferin bioluminescence relative to live cell bioluminescence was measured in the liver at day 7 (n = 5). **P < 0.01, 1-sided t test between indicated sample and shControl. (E) 5 × 105 SW480 cells were inoculated by portal circulation injection, and metastatic colonization was measured by liver bioluminescence after 30 days. Representative liver bioluminescence and gross pathology are shown (n = 5). *P < 0.05, 1-sided Mann Whitney test between indicated sample and shControl. Horizontal bars represent the median. (F) 106 LS174T cells were seeded at a density of 1,000 cells per mm2 and were assessed for apoptosis after 24 hours in 1% O2. *P < 0.05, ***P < 0.001, 1-sided t test between indicated sample and shControl unless otherwise indicated.
Figure 9. Targeting of glutathione synthesis as a therapeutic strategy.
(A) 5 × 105 LS174T cells were inoculated by portal circulation injection, and mice were provided drinking water containing 20 mM BSO. Metastatic colonization was measured by liver bioluminescence after 21 days (n = 5). Representative liver bioluminescence and gross pathology are shown. *P < 0.05, 1-sided Mann Whitney test between indicated sample and shControl. (B) DEVD-luciferin bioluminescence relative to live cell bioluminescence was measured in livers at day 7 from mice in A (n = 5). *P < 0.05, 1-sided t test. (C) Liver tumor mass was measured in mice in A after 21 days. ***P < 0.001, 1-sided t test. (D) 5 × 105 SW480 cells were inoculated by portal circulation injection, and mice were provided drinking water containing BSO. Metastatic colonization was measured by liver bioluminescence after 35 days (n = 5). Representative liver bioluminescence and gross pathology are shown. *P < 0.05, 1-sided Mann Whitney test between indicated sample and shControl. (E) 7 × 105 CLR1 primary cells were inoculated by portal circulation injection, and mice were provided drinking water containing 20 mM BSO. Liver tumor mass was measured after 28 days (n = 8). Representative gross pathology is shown. Scale bar: 1 cm. ***P < 0.001, 1-sided t test between indicated sample and shControl.
Discussion
Glycolytic deregulation has proven to be critical for cancer progression. The activity of PKM2, a central regulator of glycolysis, is tightly regulated to balance the need for high-energy compounds and the need for anabolic molecules required for cancer cell growth, proliferation, and antioxidant defense (11, 36, 39, 47, 48). Cancer cells take advantage of this regulation — PKM2 activity can be inhibited through destabilized subunit interactions and resultant loss of homotetramer structure, which is required for maximum enzymatic activity (48). Our findings reveal that colon cancer cells use an additional means of PKM2 regulation, namely expression of an additional isoenzyme, PKL. While PKL has been shown to promote glycolysis in hepatocytes, in which it is the only pyruvate kinase isoenzyme expressed (49), we found that PKL negatively regulates pyruvate kinase activity in colon cancer cells by antagonizing PKM2 and allows for maintenance of the primary endogenous antioxidant glutathione. This enables metastatic cell survival, particularly in the tumor core. Tumors are inherently cell dense, and liver parenchyma is a highly cellular organ; thus, as oxygen concentration decreases near the tumor core, the core is especially vulnerable to the combination of cell density and hypoxia. Additionally, death in the tumor core could be detrimental to tumors as a whole, since dying cells could recruit immune effectors that would impact progression of remaining cells in the tumor periphery. While these conditions in the core may be relevant to other sites of metastasis or contexts during cancer progression, we observe these conditions as an important selective pressure during colorectal cancer liver colonization.
The advantageous expression of liver-specific pyruvate kinase in cancer cells, which allows for metastatic growth in the liver, suggests that Paget’s “seed and soil” hypothesis can be explained in part by cancer cell utilization of pathways endogenous to host cell types within the metastatic niche (50). The PKL isozyme, as the sole enzyme for glycolytic pyruvate production and regulation of biosynthetic pathways, is essential for hepatocytes. This importance is highlighted by regulation of PKL activity and expression in response to glucagon, insulin, and dietary carbohydrates (49). Since hepatocytes, such as those lining the portal circulation, experience significant oxidative stress as a result of hypoxemic blood supply and xenobiotic metabolism (8), antioxidant supply and regulated glutathione regeneration is critical for hepatocyte survival and function. Previously, we have shown that the initiation of colon cancer metastatic colonies requires colon cancer cells to metabolically convert liver-derived creatine to phosphocreatine for use as an energetic source during low-energy states (13). While early metastatic colonization can be achieved through utilization of organ-specific nutrients, such as creatine and phosphocreatine, cancer cell activation of a metastatic pathway endogenous to the liver niche appears to contribute to continued progression of metastatic colonies.
Colon cancer cells modulate glutathione levels to allow for survival in the liver through expression of PKLR. The importance of glutathione levels is highlighted by further independent enhancement of glutathione levels in liver metastases through an increase in transcript-level expression of GCLC, the rate-limiting reaction in glutathione synthesis. While GCLC function and PKLR function appear to be critical regulatory steps that are broadly used by colon cancer metastases, as suggested by clinical mRNA expression association, other mechanisms may be additionally used to ensure sustained glutathione levels, such as activation of hepatocyte glutathione efflux to allow for the import of glutathione precursors from plasma (51). Additionally, the utility of glutathione is not limited to colon cancer or liver colonization, as glutathione appears critical for other cancer types and stages of progression (11, 52). Nevertheless, sufficient glutathione levels are important for metastatic cell survival and represent a potential target for clinical therapies.
We found that therapeutic inhibition of glutathione synthesis can suppress liver colonization by increasing cancer cell apoptosis, suggesting that targeting this pathway has the potential to induce cell death within metastatic nodules. While our studies showed efficacy at doses that mice tolerated well, the development of more potent inhibitors of GCLC may allow for enhanced efficacy (53) and suitability for clinical use. Given that small-molecule activation of PKM2 is being investigated as a strategy to suppress tumor growth (54), our findings suggest that glycolytic activation might display additional apoptotic effects in liver metastasis. The poor survival of patients with metastatic disease and the need for therapies specific to metastasis warrants further development of metabolic modulation as a therapeutic approach.
Methods
Additional details are provided in the Supplemental Methods.
Cell culture.
LS174T, LS-LVM3b, SW620, WiDr, and SW480 cell lines were propagated as previously described (13) and were obtained from ATCC, except for the in vivo–selected LS-LVM3b line, which was derived in the laboratory. These lines were selected to represent the mutational spectrum most commonly seen in patients (Kras wild-type/mutant, Braf wild-type/mutant, MSS/MSI). Cells in culture were routinely tested for mycoplasma contamination. Standard cell culture conditions for these cell lines are considered to be cell seeding at a density of approximately 50 to 100 cells per mm2, growth at 37°C and 5% CO2, and dissociation and dilution of cells before reaching confluency.
Stable and transfected cell lines.
Virus was generated using the ViraSafe lentiviral packaging system (Cell Biolabs). shRNA plasmids used were obtained from the Sigma-Aldrich TRC library. Indicated shRNAs (Sigma-Aldrich) are as follows: shControl (SHC002), PKLR sh83 (TRCN0000006383), PKLR sh84 (TRCN0000006384), PKLR sh39 (TRCN0000199139), PKLR sh40 (TRCN0000194740), GCLC sh64 (TRCN0000333564), and GCLC sh62 (TRCN0000344862). Because shRNAs displayed varying efficacy depending on the cell line used, multiple shRNAs were tested for target mRNA depletion, and the best shRNAs were used for experiments. To generate overexpression, N-terminal FLAG tag or HA tag was added to cDNA of PKL/PKR or PKM2, respectively, and cloned into pBabe-Puro or pBabe-Hygro expression vector. Transduction was performed with 8 μg/ml polybrene. Transfection of HEK293T cells was performed using Lipofectamine 2000 (Invitrogen).
shRNA screening.
10 subpools of lentiviral particles from the TRC1 human shRNA pooled library, consisting of 71,444 shRNA clones representing 14,523 genes, were transduced into luciferase-expressing LS174T, WiDr, and SW620 colon cancer cells. Two transductions for each cell line were performed as biological replicates at a low titer (MOI <1) to reduce the likelihood of multiple shRNAs in a single cell. 48 hours after transduction, transduced cells were selected with puromycin for 48 hours to remove untransduced cells. After antibiotic selection, the remaining cells were allowed to recover for a week prior to subsequent experiments. A portion of the selected cells was set aside, and genomic DNA was extracted. This was the reference pool of genomic DNA prior to the selective pressure of liver colonization. A second population of cells was used for in vivo experiments, while a third was kept in culture for the duration of the in vivo experiment. For direct liver injections, an average of 7 mice were inoculated with the 800,000 cells from transduced populations, allowing for a 783× depth of coverage. After 3 to 4 weeks, when tumors had developed, as measured by bioluminescence, mice were sacrificed, and tumors were resected. Cells from the various conditions were processed by isolation of genomic DNA using the DNeasy Kit (Qiagen). A total of 5 μg of genomic DNA isolated from tumors from different mice was pooled, and PCR amplification of shRNA inserts was performed using 10 reaction tubes and pooled. A first amplification was performed using touchdown PCR (Fwd_rd1: TGGACTATCATATGCTTACCGTAACT; Rev_rd1: AAAGAGGATCTCTGTCCCTGT). The PCR product from all 10 reactions (~350 bp) was gel purified and pooled. Subsequently, a second round of amplification was performed with Illumina sequencing-specific primers (Fwd_rd2: AATGATACGGCGACCACCGAGATCTACACTCTTTCCCTACACGACGCTCTTCCGATCTGTATTCTTGGCTTTATATATCTTGTGGAAAGGAC, Rev_rd2: CAAGCAGAAGACGGCATACGAGCTCTTCCGATCTGGATGAATACTGCCATTTGTCTCGAGGTCGA). The subsequent PCR product was gel purified (~300 bp) in preparation for high-throughput sequencing on an Illumina HiSeq2000. Once the sequencing results were obtained, the data were filtered to normalize samples by total reads and to remove shRNAs that were not included in all reference samples. 54,591 shRNA clones representing 14,095 genes were used in the final data analysis. For each sample, shRNA ratios to the reference cell line were calculated, and z scores were calculated using peak median absolute deviation (15) to normalize for the global loss of shRNAs under these experimental conditions. Given the effects of off-target RNAi silencing, genes were scored as hits only if at least two shRNAs in each cell line were absent from the final tumor samples in both independent transduction replicates. The secondary library was generated by cloning a top-scoring shRNA from the genome-wide screen into plko.1 and producing a viral library that was subjected to the experimental procedures listed above. Gene scoring by the RIGER algorithm was performed using the weighted-sum method on shRNAs in all cell lines. Gene ranking was averaged across cell lines, which was then used to calculate gene percentile. To identify kinases, genes from the top 556 hits were selected if annotated as kinases by GO annotation KINASE_ACTIVITY (GO: 0016301) and mean depletion in the secondary screen was greater than two scores. These kinases were then assessed for their gene percentile score by RIGER.
Animal studies.
NOD/SCID male mice (The Jackson Laboratory), aged 6 weeks, were used in experiments involving direct liver injections and LS-LVM3b cells. NOD/SCID gamma male mice (The Jackson Laboratory), aged 6 to 8 weeks, were used in all other experiments. Cells inoculated by direct liver and subcutaneous injection were mixed with Matrigel (Corning) at a 1:1 ratio prior to injection. For portal circulation injections, cells were injected into the spleen followed by removal of the spleen. Animals were excluded from studies if inoculated cells did not arrive in the liver. In vivo bioluminescence was generated using d-luciferin substrate (Perkin Elmer) and measured by normalization to bioluminescence signal immediately following cell inoculation. VivoGlo Caspase-3/7 Substrate (Promega) was used as previously described to measure the apoptosis in vivo (13). The bioluminescence signal obtained using this substrate was normalized to the bioluminescence signal obtained with d-luciferin to calculate relative apoptotic cell burden. NAC treatment was performed by providing 40 mM drinking water that was pH matched to control water and replacing it twice weekly. BSO treatment studies were performed by supplying BSO in drinking water at 20 mM, a dose previously demonstrated to display no significant toxicities in mice (55), and replacing it twice per week. Mice were randomized following cancer cell inoculation prior to treatment. Blinding was not performed.
Patient-derived primary colon cancer graft.
CLR1 was derived from the liver metastasis of a subject with recurrent rectal adenocarcinoma with a KRAS G12 mutation. Within 2 hours of surgical resection, tumor tissue not needed for diagnostic purposes was implanted subcutaneously into 2 NSG mice at the Antitumor Assessment Core. When the tumor reached 1,000 mm3, mice were euthanized, tumors were excised, and 100-mm3 pieces of tumor tissue were reimplanted into another set of mice at The Rockefeller University. To maintain the xenograft line, the subcutaneous tumor was passaged each time the tumor reached 1,000 mm3. The cells used for the splenic injection of CLR1 had been passaged subcutaneously 6 times previously. To obtain a single-cell suspension for injection, mice were euthanized and subcutaneous tumors were excised and subjected to a series of mechanical and enzymatic separations to obtain a single-cell suspension. Cells of mouse origin were depleted from the single-cell suspension via magnetic-activated cell sorting using the Mouse Depletion Kit (Miltenyi Biotec), leaving only cells derived from human colorectal cancer.
Quantitative RT-PCR.
For cell lines, RNA was extracted using the Total RNA Isolation Kit (Norgen Biotek). For clinical samples, RNA was obtained from MSKCC and blinded until after quantitation was performed. cDNA was generated using SuperScript III (Invitrogen). Fast SYBR Green Master Mix (Life Technologies) was used to analyze samples on Applied Biosystems 7900HT. Expression was normalized to HPRT expression. Primers sequences are as follows: HPRT-F, GACCAGTCAACAGGGGACAT; HPRT-R, CCTGACCAAGGAAAGCAAAG; PKLR-F, TGGGAAAACTGGGTGGGATGGATG; PKLR-R, GAAGGAAGCAGCCGGGGATTTGAC; GCLC-F, GACCCATGGAGGTGCAATTA; GCLC-R, AACCTTTGACAGTGGAATGAGA.
Clinical analysis.
The following GEO data sets were used for analysis: GSE41258, GSE50760, GSE6988, GSE14297, GSE18105, GSE27854, and GSE14095. RNA-sequencing data were normalized to total reads per sample. Affymetrix microarrays were analyzed for PKLR expression using probe 210451_at.
Immunofluorescence imaging.
Livers were fixed in 4% paraformaldehyde, submerged in 30% sucrose, and frozen in OCT. Livers were sliced on a tissue microtome into 10-μm sections and permeabilized with methanol/acetone on slides. Slides were stained with the following antibodies: anti-luciferase (1:100, Pierce, clone Luc17), anti–cleaved caspase-3 (1:1,000, Cell Signaling, 9661), and anti-hVimentin (1:40, Vector Laboratories, clone V9). DAPI was added prior to mounting. Slides were imaged on a Leica TCS SP5 system at ×40.
Hypoxia and high cell density assay.
Hypoxia was performed using 1% O2/5% CO2/94% N2 mixed gas (Praxair) in a Modular Incubator Chamber (Billups-Rothernberg). High cell density was defined as a seeding density of 1,000 cells per mm2. Experiments were performed in 6-well or 12-well plates using 2 ml and 1 ml of media, respectively.
Flow cytometry.
All experiments were done under conditions of high cell density and hypoxia for 24 hours. Apoptosis was assessed using the GFP-certified Apoptosis/Necrosis Detection Kit (Enzo Life Sciences). Annexin V+ cells were considered apoptotic, and Annexin V–, 7-AAD– cells were considered live. Compound 3 (44), 5-(2,5-Dimethyl-pyrrol-1-yl)-2-hydroxy-benzoic acid, was purchased from Matrix Scientific. For rescue experiments, NAC was pH adjusted to 7.4 and used fresh at 2.5 mM. Glutathione-Reduced Ethyl Ester (Sigma-Aldrich) was used at 1 mM. For glutathione measurements, ThiolTracker Violet (Life Technologies) was used according to the manufacturer’s instructions at 10 μM and analyzed in combination with 7-AAD to exclude dead cells. Cells were analyzed on LSRII (BD). Analysis was performed on FlowJo (Tree Star). Data shown are from at least 3 biological replicates and at least two independent experiments.
Coimmunoprecipitation and Western blotting.
Cells were lysed using 50 mM Tris, pH 7.5, 150 mM NaCl, 1 mM EDTA, 1% NP40, protease inhibitor (Roche), and phosphoSTOP (Roche). Immunoprecipitation was performed using anti-FLAG M2 magnetic beads (Sigma-Aldrich) or anti-HA magnetic beads (Pierce) for 1 hour at 4°C. For FLAG-based immunoprecipitation, beads were washed twice with wash buffer containing 1 M NaCl, followed by a wash with wash buffer containing 50 mM NaCl. For HA-based immunoprecipitation, beads were washed twice with lysis buffer. Elution was performed using either 3× FLAG peptide (Sigma-Aldrich) or HA peptide (Pierce). For immunoprecipitation from transfected HEK293T cells, cells were transfected with plasmids encoding HA-PKM2, FLAG-PKM2, or FLAG-PKL using Lipofectamine 2000 (Invitrogen). 48 hours later, cells were lysed with lysis buffer containing 100 μM fructose 1,6-bisphosphate and incubated with anti-FLAG M2 magnetic beads (Sigma-Aldrich) for 1 hour at 4°C. Beads were washed twice with lysis buffer, and elution was performed using 3× FLAG peptide (Sigma-Aldrich). Eluate was then incubated with anti-HA magnetic beads (Pierce) for 1 hour at 4°C. Beads were washed twice with lysis buffer, and elution was performed using HA peptide (Pierce). For Western blotting, samples were denatured, separated by SDS-PAGE, transferred to PVDF membrane (Pierce or Millipore), blocked, and probed with primary antibody. The following antibodies were used: anti-FLAG (1:1,000; Sigma-Aldrich, clone M2), anti-PKM2 (1:1,000; Cell Signaling, D78A4), anti-PKLR (1:75; Santa Cruz, clone E2), and anti-HA (1:1,000; Cell Signaling, 6E2). For PKM2 and HA detection, fluorescent secondary antibodies (1:10,000; Li-Cor IRDye800CW and IRDye680RD) were used and detected on an Odyssey SA Imaging System (Li-Cor). For PKLR and FLAG detection, antibodies were chemiluminescently detected using horseradish peroxidase–conjugated secondary antibodies (1:10,000), ECL2 Western Blotting Substrate (Pierce), and the SRX-101A (Konica Minolta) developer, according to the manufacturer’s instructions.
Label-free quantitation liquid chromatography/tandem mass spectrometry and analysis.
Coimmunoprecipitated and 3xFLAG eluted proteins were trypsinized in solution overnight. Peptides were desalted using home-made Empore C18 columns prior to being analyzed by liquid chromatography/tandem mass spectrometry (LC-MS/MS) (Dionex 3000 HPLC coupled to Orbitrap XL, Thermo Scientific). The mass spectrometer was operated in a high/low mode. Peptides were separated at 300 nl per minute using a gradient increasing from 10% B to 45% B in 120 minutes (A: 0.1% formic acid; B: acetonitrile/0.1% formic acid). Generated LC-MS/MS data were queried against Uniprots complete Human Proteome (August 2013) and quantitated using MaxQuant 1.5.0.30. In short, the peptide-spectrum match false discovery rate was set to 1%, while the protein false discovery rate was set to 1%. A total of 1,140 proteins was matched. Match between runs was used for the label-free quantitation. Generated label-free quantitation values were analyzed using Perseus 1.5.0.9. All label-free quantitation values were log2 transformed and filtered, with the requirement that a given protein displayed at least 5 matching peptides and 3 times as many peptide-spectrum matches. This resulted in 109 proteins. Absent label-free quantitation values were replaced by values one order of magnitude less.
Pyruvate kinase assay.
106 LS174T cells were seeded in triplicate in 6-well plates in 1% O2 for 16 hours. Immunoprecipitation was performed as described above for tagged proteins. Following elution of immunoprecipitated pyruvate kinase complex, pyruvate kinase activity was measured using a lactate dehydrogenase–coupled assay (39). Each reaction was performed under the following conditions: 50 mM Tris, pH 7.5, 100 mM KCl, 10 mM MgCl2, 1 mM PEP, 1 mM ADP, 200 μM NADH, 8 units lactate dehydrogenase in 50% glycerol, and 100 μM FBP. Activity was normalized to immunoprecipitated HA-tagged PKM2. The assay was monitored for pyruvate-dependent conversion of NADH to NAD+ by fluorescence on Biotek Synergy Neo. For whole-cell lysates, a Pyruvate Kinase Activity Assay (Biovision) was used according to the manufacturer’s instructions by fluorescent measurements. Activity was normalized to the relative live cell population, as measured by live cell count.
Metabolite measurement assay.
For pyruvate, PEP, and ATP assays, 3 × 106 cells were seeded in 60-mm plates in triplicate and grown under 1% O2 for 24 hours in recommended media, without supplemental sodium pyruvate. Cells were washed with PBS containing 100 μM phloretin and lysed and deproteinized using perchloric acid precipitation (Biovision). Pyruvate, PEP, and ATP Assay Kits (Biovision) were used by fluorometric methods on Synergy Neo (Biotek) to quantify intracellular metabolite levels. For lactate and glucose assays, 106 cells were seeded in 6-well plates in triplicate and grown under 1% O2 for 24 hours when conditioned media was sampled. Levels of glucose were measured using the Amplex Red Glucose Assay (Invitrogen), and levels of lactate were measured using the Lactate Assay Kit (Biovision). Metabolite consumption/excretion per cell per hour was calculated as previously described (40).
Statistics.
Sample size in mouse experiments was chosen based on the biological variability observed with a given genotype. Nonparametric tests were used when normality could not be assumed. Mann Whitney test and t test were used when comparing independent shRNAs to shControl. One-tailed tests were used when a difference was predicted to be in one direction; otherwise, a two-tailed test was used. Bootstrapping was performed by sampling with replacement in R. Analysis of covariance (ANCOVA) testing was performed using aov() command in R. A P value less than 0.05 was considered significant. Error bars represent SEM unless otherwise indicated.
Study approval.
Approval for the study was obtained through the MSKCC Institutional Review Board/Privacy Board (protocol 10-018A), the MSKCC Institutional Animal Care and Use Committee (protocol 04-03-009), The Rockefeller University Institutional Review Board (protocol STA-0681), and The Rockefeller University Institutional Animal Care and Use Committee (protocol 15783-H). Written consent was obtained from all human participants who provided samples for patient-derived xenografts.
Author contributions
AN, JML, and SFT designed the research. AN, JML, RM, EMW, and FYM performed the experiments. ZZ, PBP, LS, YYJ, and ES obtained, curated, and provided access to clinical samples and patient-derived grafts. AN and SFT wrote the manuscript.
Supplementary Material
Acknowledgments
We thank members of our laboratory for helpful comments and discussion. We thank the Proteomics Resource Center (H. Molina, director; J. Fernandez) for mass spectrometry profiling and the Genomics Resource Center (C. Zhou, director) for high-throughput sequencing. We thank the Bio-Imaging Resource Center (A. North, director), the Flow Cytometry Resource Center (S. Mazel, director), and the High-Throughput and Spectroscopy Resource Center (F. Glickman, director) for technical assistance and resources. We thank A. Hosios and J. Mayers for advice on experiments. We thank J. Friedman for tissue microtome use. This work was supported by the Irma T. Hirschl/Monique Weill-Caulier Trust, the Starr Cancer Consortium, the Leona M. and Harry B. Helmsley Charitable Trust, and the NIH Director’s New Innovator Award (award 1DP2OD006506-01). This work was supported in part by grant 8 UL1 TR000043 from the National Center for Research Resources and the National Center for Advancing Translational Sciences/NIH. A. Nguyen was supported by a Medical Scientist Training Program grant from the National Institute of General Medical Sciences of the NIH (award T32GM07739) to the Weill Cornell/Rockefeller/Sloan-Kettering Tri-Institutional MD-PhD Program. J.M. Loo is an A*STAR National Science Scholar.
Footnotes
Jia Min Loo’s present address is: Cancer Therapeutics and Stratified Oncology, Genome Institute of Singapore, Singapore.
Conflict of interest: The authors have declared that no conflict of interest exists.
Reference information:J Clin Invest. 2016;126(2):681–694. doi:10.1172/JCI83587.
References
- 1.Siegel R, Ma J, Zou Z, Jemal A. Cancer statistics, 2014. CA Cancer J Clin. 2014;64(1):9–29. doi: 10.3322/caac.21208. [DOI] [PubMed] [Google Scholar]
- 2.Ferlay J, et al. Cancer incidence mortality worldwide: Sources, methods major patterns in GLOBOCAN 2012. Int J Cancer. 2014;136(5):E359–E386. doi: 10.1002/ijc.29210. [DOI] [PubMed] [Google Scholar]
- 3.Davies JM, Goldberg RM. Treatment of metastatic colorectal cancer. Semin Oncol. 2011;38(4):552–560. doi: 10.1053/j.seminoncol.2011.05.009. [DOI] [PubMed] [Google Scholar]
- 4.Talmadge JE, Fidler IJ. AACR centennial series: the biology of cancer metastasis: historical perspective. Cancer Res. 2010;70(14):5649–5669. doi: 10.1158/0008-5472.CAN-10-1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gupta GP, Massague J. Cancer metastasis: building a framework. Cell. 2006;127(4):679–695. doi: 10.1016/j.cell.2006.11.001. [DOI] [PubMed] [Google Scholar]
- 6.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 7.Luzzi KJ, et al. Multistep nature of metastatic inefficiency: dormancy of solitary cells after successful extravasation and limited survival of early micrometastases. Am J Pathol. 1998;153(3):865–873. doi: 10.1016/S0002-9440(10)65628-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jungermann K, Kietzmann T. Zonation of parenchymal and nonparenchymal metabolism in liver. Annu Rev Nutr. 1996;16:179–203. doi: 10.1146/annurev.nu.16.070196.001143. [DOI] [PubMed] [Google Scholar]
- 9.Sullivan WJ, Christofk HR. The metabolic milieu of metastases. Cell. 2015;160(3):363–364. doi: 10.1016/j.cell.2015.01.023. [DOI] [PubMed] [Google Scholar]
- 10.Beronja S, et al. RNAi screens in mice identify physiological regulators of oncogenic growth. Nature. 2013;501(7466):185–190. doi: 10.1038/nature12464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Anastasiou D, et al. Inhibition of pyruvate kinase M2 by reactive oxygen species contributes to cellular antioxidant responses. Science. 2011;334(6060):1278–1283. doi: 10.1126/science.1211485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Root DE, Hacohen N, Hahn WC, Lander ES, Sabatini DM. Genome-scale loss-of-function screening with a lentiviral RNAi library. Nat Methods. 2006;3(9):715–719. doi: 10.1038/nmeth924. [DOI] [PubMed] [Google Scholar]
- 13.Loo JM, et al. Extracellular metabolic energetics can promote cancer progression. Cell. 2015;160(3):393–406. doi: 10.1016/j.cell.2014.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kaelin WG., Jr Molecular biology. Use and abuse of RNAi to study mammalian gene function. Science. 2012;337(6093):421–422. doi: 10.1126/science.1225787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Luo B, et al. Highly parallel identification of essential genes in cancer cells. Proc Natl Acad Sci U S A. 2008;105(51):20380–20385. doi: 10.1073/pnas.0810485105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Reinmuth N, et al. Impact of insulin-like growth factor receptor-I function on angiogenesis, growth, and metastasis of colon cancer. Lab Invest. 2002;82(10):1377–1389. doi: 10.1097/01.LAB.0000032411.41603.C2. [DOI] [PubMed] [Google Scholar]
- 17.Badiola I, Villace P, Basaldua I, Olaso E. Downregulation of discoidin domain receptor 2 in A375 human melanoma cells reduces its experimental liver metastasis ability. Oncol Rep. 2011;26(4):971–978. doi: 10.3892/or.2011.1356. [DOI] [PubMed] [Google Scholar]
- 18.Luo W, Semenza GL. Emerging roles of PKM2 in cell metabolism and cancer progression. Trends Endocrinol Metab. 2012;23(11):560–566. doi: 10.1016/j.tem.2012.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mazurek S, Boschek CB, Hugo F, Eigenbrodt E. Pyruvate kinase type M2 and its role in tumor growth and spreading. Semin Cancer Biol. 2005;15(4):300–308. doi: 10.1016/j.semcancer.2005.04.009. [DOI] [PubMed] [Google Scholar]
- 20.Imamura K, Tanaka T. Multimolecular forms of pyruvate kinase from rat and other mammalian tissues. I. Electrophoretic studies. J Biochem. 1972;71(6):1043–1051. doi: 10.1093/oxfordjournals.jbchem.a129852. [DOI] [PubMed] [Google Scholar]
- 21.Osterman J, Fritz PJ. Pyruvate kinase isozymes from rat intestinal mucosa. Characterization and the effect of fasting and refeeding. Biochemistry. 1974;13(8):1731–1736. doi: 10.1021/bi00705a027. [DOI] [PubMed] [Google Scholar]
- 22.Domingo M, Einig C, Eigenbrodt E, Reinacher M. Immunohistological demonstration of pyruvate kinase isoenzyme type L in rat with monoclonal antibodies. J Histochem Cytochem. 1992;40(5):665–673. doi: 10.1177/40.5.1374093. [DOI] [PubMed] [Google Scholar]
- 23.Uhlen M, et al. Towards a knowledge-based Human Protein Atlas. Nat Biotechnol. 2010;28(12):1248–1250. doi: 10.1038/nbt1210-1248. [DOI] [PubMed] [Google Scholar]
- 24.Ki DH, et al. Whole genome analysis for liver metastasis gene signatures in colorectal cancer. Int J Cancer. 2007;121(9):2005–2012. doi: 10.1002/ijc.22975. [DOI] [PubMed] [Google Scholar]
- 25.Sheffer M, et al. Association of survival and disease progression with chromosomal instability: a genomic exploration of colorectal cancer. Proc Natl Acad Sci U S A. 2009;106(17):7131–7136. doi: 10.1073/pnas.0902232106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stange DE, et al. Expression of an ASCL2 related stem cell signature and IGF2 in colorectal cancer liver metastases with 11p15.5 gain. Gut. 2010;59(9):1236–1244. doi: 10.1136/gut.2009.195701. [DOI] [PubMed] [Google Scholar]
- 27.Kim SK, et al. A nineteen gene-based risk score classifier predicts prognosis of colorectal cancer patients. Mol Oncol. 2014;8(8):1653–1666. doi: 10.1016/j.molonc.2014.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Watanabe T, et al. Gene expression signature and response to the use of leucovorin, fluorouracil and oxaliplatin in colorectal cancer patients. Clin Transl Oncol. 2011;13(6):419–425. doi: 10.1007/s12094-011-0676-z. [DOI] [PubMed] [Google Scholar]
- 29.Matsuyama T, et al. MUC12 mRNA expression is an independent marker of prognosis in stage II and stage III colorectal cancer. Int J Cancer. 2010;127(10):2292–2299. doi: 10.1002/ijc.25256. [DOI] [PubMed] [Google Scholar]
- 30.Kikuchi A, et al. Identification of NUCKS1 as a colorectal cancer prognostic marker through integrated expression and copy number analysis. Int J Cancer. 2013;132(10):2295–2302. doi: 10.1002/ijc.27911. [DOI] [PubMed] [Google Scholar]
- 31.Quail DF, Joyce JA. Microenvironmental regulation of tumor progression and metastasis. Nat Med. 2013;19(11):1423–1437. doi: 10.1038/nm.3394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bao S, et al. Periostin potently promotes metastatic growth of colon cancer by augmenting cell survival via the Akt/PKB pathway. Cancer Cell. 2004;5(4):329–339. doi: 10.1016/S1535-6108(04)00081-9. [DOI] [PubMed] [Google Scholar]
- 33.Vermeulen PB, et al. Liver metastases from colorectal adenocarcinomas grow in three patterns with different angiogenesis and desmoplasia. J Pathol. 2001;195(3):336–342. doi: 10.1002/path.966. [DOI] [PubMed] [Google Scholar]
- 34.Garcia CK, Goldstein JL, Pathak RK, Anderson RG, Brown MS. Molecular characterization of a membrane transporter for lactate, pyruvate, and other monocarboxylates: implications for the Cori cycle. Cell. 1994;76(5):865–873. doi: 10.1016/0092-8674(94)90361-1. [DOI] [PubMed] [Google Scholar]
- 35.Bluemlein K, Gruning NM, Feichtinger RG, Lehrach H, Kofler B, Ralser M. No evidence for a shift in pyruvate kinase PKM1 to PKM2 expression during tumorigenesis. Oncotarget. 2011;2(5):393–400. doi: 10.18632/oncotarget.278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Christofk HR, et al. The M2 splice isoform of pyruvate kinase is important for cancer metabolism and tumour growth. Nature. 2008;452(7184):230–233. doi: 10.1038/nature06734. [DOI] [PubMed] [Google Scholar]
- 37.Thangaraju M, Carswell KN, Prasad PD, Ganapathy V. Colon cancer cells maintain low levels of pyruvate to avoid cell death caused by inhibition of HDAC1/HDAC3. Biochem J. 2009;417(1):379–389. doi: 10.1042/BJ20081132. [DOI] [PubMed] [Google Scholar]
- 38.Mazurek S. Pyruvate kinase type M2: a key regulator of the metabolic budget system in tumor cells. Int J Biochem Cell Biol. 2011;43(7):969–980. doi: 10.1016/j.biocel.2010.02.005. [DOI] [PubMed] [Google Scholar]
- 39.Christofk HR, Vander Heiden MG, Wu N, Asara JM, Cantley LC. Pyruvate kinase M2 is a phosphotyrosine-binding protein. Nature. 2008;452(7184):181–186. doi: 10.1038/nature06667. [DOI] [PubMed] [Google Scholar]
- 40.Lunt SY, et al. Pyruvate kinase isoform expression alters nucleotide synthesis to impact cell proliferation. Mol Cell. 2015;57(1):95–107. doi: 10.1016/j.molcel.2014.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gerriets VA, et al. Metabolic programming and PDHK1 control CD4+ T cell subsets and inflammation. J Clin Invest. 2015;125(1):194–207. doi: 10.1172/JCI76012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sheta EA, Trout H, Gildea JJ, Harding MA, Theodorescu D. Cell density mediated pericellular hypoxia leads to induction of HIF-1alpha via nitric oxide and Ras/MAP kinase mediated signaling pathways. Oncogene. 2001;20(52):7624–7634. doi: 10.1038/sj.onc.1204972. [DOI] [PubMed] [Google Scholar]
- 43.Bertout JA, Patel SA, Simon MC. The impact of O2 availability on human cancer. Nat Rev Cancer. 2008;8(12):967–975. doi: 10.1038/nrc2540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vander Heiden MG, et al. Identification of small molecule inhibitors of pyruvate kinase M2. Biochem Pharmacol. 2010;79(8):1118–1124. doi: 10.1016/j.bcp.2009.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Griffith OW. Biologic and pharmacologic regulation of mammalian glutathione synthesis. Free Radic Biol Med. 1999;27(9–10):922–935. doi: 10.1016/s0891-5849(99)00176-8. [DOI] [PubMed] [Google Scholar]
- 46.Schulze A, Harris AL. How cancer metabolism is tuned for proliferation and vulnerable to disruption. Nature. 2012;491(7424):364–373. doi: 10.1038/nature11706. [DOI] [PubMed] [Google Scholar]
- 47.Chaneton B, et al. Serine is a natural ligand and allosteric activator of pyruvate kinase M2. Nature. 2012;491(7424):458–462. doi: 10.1038/nature11540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324(5930):1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yamada K, Noguchi T. Nutrient and hormonal regulation of pyruvate kinase gene expression. Biochem J. 1999;337(Pt 1):1–11. [PMC free article] [PubMed] [Google Scholar]
- 50.Fidler IJ. The pathogenesis of cancer metastasis: the ‘seed and soil’ hypothesis revisited. Nat Rev Cancer. 2003;3(6):453–458. doi: 10.1038/nrc1098. [DOI] [PubMed] [Google Scholar]
- 51.Obrador E, Benlloch M, Pellicer JA, Asensi M, Estrela JM. Intertissue flow of glutathione (GSH) as a tumor growth-promoting mechanism: interleukin 6 induces GSH release from hepatocytes in metastatic B16 melanoma-bearing mice. J Biol Chem. 2011;286(18):15716–15727. doi: 10.1074/jbc.M110.196261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Harris IS, et al. Glutathione and thioredoxin antioxidant pathways synergize to drive cancer initiation and progression. Cancer Cell. 2015;27(2):211–222. doi: 10.1016/j.ccell.2014.11.019. [DOI] [PubMed] [Google Scholar]
- 53.Tokutake N, Hiratake J, Katoh M, Irie T, Kato H, Oda J. Design, synthesis and evaluation of transition-state analogue inhibitors of Escherichia coli gamma-glutamylcysteine synthetase. Bioorg Med Chem. 1998;6(10):1935–1953. doi: 10.1016/S0968-0896(98)00142-4. [DOI] [PubMed] [Google Scholar]
- 54.Anastasiou D, et al. Pyruvate kinase M2 activators promote tetramer formation and suppress tumorigenesis. Nat Chem Biol. 2012;8(10):839–847. doi: 10.1038/nchembio.1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Watanabe T, et al. A novel model of continuous depletion of glutathione in mice treated with L-buthionine (S,R)-sulfoximine. J Toxicol Sci. 2003;28(5):455–469. doi: 10.2131/jts.28.455. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.