Abstract
Beckwith-Wiedemann syndrome (BWS) is a human stem cell disorder, and individuals with this disease have a substantially increased risk (~800-fold) of developing tumors. Epigenetic silencing of β2-spectrin (β2SP, encoded by SPTBN1), a SMAD adaptor for TGF-β signaling, is causally associated with BWS; however, a role of TGF-β deficiency in BWS-associated neoplastic transformation is unexplored. Here, we have reported that double-heterozygous Sptbn1+/– Smad3+/– mice, which have defective TGF-β signaling, develop multiple tumors that are phenotypically similar to those of BWS patients. Moreover, tumorigenesis-associated genes IGF2 and telomerase reverse transcriptase (TERT) were overexpressed in fibroblasts from BWS patients and TGF-β–defective mice. We further determined that chromatin insulator CCCTC-binding factor (CTCF) is TGF-β inducible and facilitates TGF-β–mediated repression of TERT transcription via interactions with β2SP and SMAD3. This regulation was abrogated in TGF-β–defective mice and BWS, resulting in TERT overexpression. Imprinting of the IGF2/H19 locus and the CDKN1C/KCNQ1 locus on chromosome 11p15.5 is mediated by CTCF, and this regulation is lost in BWS, leading to aberrant overexpression of growth-promoting genes. Therefore, we propose that loss of CTCF-dependent imprinting of tumor-promoting genes, such as IGF2 and TERT, results from a defective TGF-β pathway and is responsible at least in part for BWS-associated tumorigenesis as well as sporadic human cancers that are frequently associated with SPTBN1 and SMAD3 mutations.
Introduction
Beckwith-Wiedemann syndrome (BWS) is a stem cell overgrowth disorder with an estimated prevalence of 1 in 14,000 (1). Its phenotypic expression includes omphalocele, organomegaly, adrenal cytomegaly, hemihyperplasia, and macrosomia, with considerable clinical heterogeneity (2). It is also associated with an 800-fold increased risk of childhood neoplasms, including Wilms tumor, hepatoblastoma, pancreatic tumor, lymphoma, adrenocortical carcinoma, and optic nerve glioma (3). Patients with BWS can develop multiple tumor types within the same organ simultaneously, an example including the cooccurrence of a mesenchymal hamartoma, capillary hemangioma hepatoblastoma, and cholangiocarcinoma within the liver of one patient (4, 5). These events are suggestive of the multipotentiality of neoplastic transformation and imply dysfunctional processes as stem cells differentiate into mature adult cell types (6). Yet mechanistic insight into downstream effector pathways that lead to transformation and an integrated analysis from mouse models to human disease for BWS remain ill defined.
The molecular etiology of this stem cell disorder is complex and involves alterations in the expression of multiple imprinted growth-regulatory genes on chromosome 11p15, especially IGF2 and H19, whose expressions are regulated by DNA methylation and the chromatin insulator CCCTC-binding factor (CTCF) (7–10). Loss of imprinting (LOI) of IGF2 leads to an expanded progenitor-cell compartment and increases expression of progenitor-cell markers in colon cancer models (11, 12). Similarly, LOI of IGF2 in BWS is specifically associated with cancer risk and leads to the expansion of nephrogenic progenitor cells in Wilms tumors (13). CTCF is an 11 zinc-finger protein that binds more than 20,000 sites in the human genome. Genome-wide assays have shown that CTCF links chromatin domains through long-range interactions between distal genomic regions and support a crucial role of CTCF in chromatin conformation and organization (14). CTCF-mediated enhancer blocking is a constitutive action that can be modulated by DNA methylation and by additional cofactors that bind in the vicinity of CTCF-binding regions. On chromosome 11p15, methylation of CTCF-binding sites at the imprinting control region (ICR) of the IGF2/H19 locus on the parental allele results in loss of enhancer blocking and leads to inappropriate expression of imprinted genes in BWS (7–10). CTCF is directly involved in the transcriptional regulation of various key factors of cellular growth, apoptosis, quiescence, senescence, and differentiation, such as c-MYC, telomerase reverse transcriptase (TERT), the retinoblastoma (RB) family, cyclin-dependent kinase inhibitor 2A (CDKN2A), and TP53, suggesting its role as a tumor suppressor (15). However, the specific role that CTCF plays in BWS remains unclear.
Chromosome 11p15 genes KCNQ1OT1 and CDKN1C are also implicated in BWS. Loss of maternal methylation of KCNQ1OT1 is observed in patients with BWS (16, 17). CDKN1C (p57kip2) is a maternally expressed imprinted gene encoding a cyclin-dependent kinase inhibitor that regulates prenatal development and postnatal growth (18, 19). CDKN1C mutations reported in BWS are either nonsense or missense mutations localized to the cyclin-dependent kinase-binding domain; both types of mutations result in the loss of protein function, increased proliferation, and an increased risk of developing cancer (20). Although 40% of patients with inherited BWS bear mutations in CDKN1C, these are rare (~5%) in the general BWS patient population (21, 22), suggesting other underlying mechanisms. Similarly, despite the suggested roles for IGF2 and other genes on chromosome 11p15 in BWS, they cannot serve as a single effector molecule; therefore, other mechanisms that coordinate epigenetic derepression of these imprinted genes must exist (10).
TGF-β serves as an essential regulator of cell polarity, growth, differentiation, and lineage specificity as well as a tumor suppressor pathway in multiple cell types (23). Defective TGF-β signaling is implicated in multiple cancers owing to the frequent somatic mutations in, or deregulation of, its components, such as SMAD3, SMAD4, and TGF-β receptors 1 and 2 (TGFBR1 and TGFBR2). SMADs are the intracellular mediators of TGF-β signaling (24, 25), and their function is modulated by adaptor proteins, such as the SMAD anchor for receptor activation, filamin, microtubules, and β2-spectrin (β2SP, encoded by SPTBN1) (26–28). TGF-β–activated SMADs also orchestrate specific histone modifications and chromatin remodeling to activate their transcriptional targets. Such examples include SMAD2-mediated p300 recruitment and acetylation of H3K9 and H3K18 at promoters for endogenous SMAD-target genes in a ligand-induced manner (29). In addition, TGF-β1 has been demonstrated as inhibiting telomerase activity, and conversely, TGF-β1–induced arrest of cell growth can be overcome by the activation of human TERT, the protein catalytic subunit of telomerase (30). Telomerase is important for the long-term proliferation potential of human stem cells and cancer cells and for normal tissue renewal (31). While malignant transformation from normal cells often is associated with activation of telomerase and subsequent telomere maintenance, the function of TERT in activating stem cells may be independent of telomerase activity and remains unexplored in BWS (32).
As previously demonstrated, epigenetic silencing of β2SP, a SMAD3/4 scaffolding protein, could be a causal factor in BWS (33). To determine whether this stem cell disorder is caused by a process dependent upon TGF-β or secondary to TGF-β–independent cytoskeletal effects of β2SP, we crossbred Sptbn1+/– and Smad3+/– mice to generate Sptbn1+/– Smad3+/– mutant mice, resulting in mice deficient in TGF-β signaling. Indeed, tumor incidence is synergistically increased in the double-heterozygous mutant Sptbn1+/– Smad3+/– mice compared with the single-heterozygous mutant Sptbn1+/– mice, indicating that a partial loss of 2 of the essential components of the TGF-β pathway exacerbates the BWS-like phenotype. Further, the RNA analyses identified an induction of Tert in Sptbn1+/– Smad3+/– mouse embryonic fibroblasts (MEFs), due likely to the disruption of the epigenetic regulatory system, particularly the chromosomal networking protein CTCF. This finding suggests that dysregulated telomerase expression may be part of the molecular basis of tumor development in BWS patients as well as in sporadic cancers. Thus, we show that the TGF-β–mediated β2SP/SMAD3/CTCF complex regulates telomerase activity and is part of a pathway suppressing the switch to tumorigenesis in BWS-associated cancers and, potentially, sporadic tumors.
Results
Sptbn1+/– Smad3+/– mice develop multiple tumors and phenocopy the features of BWS.
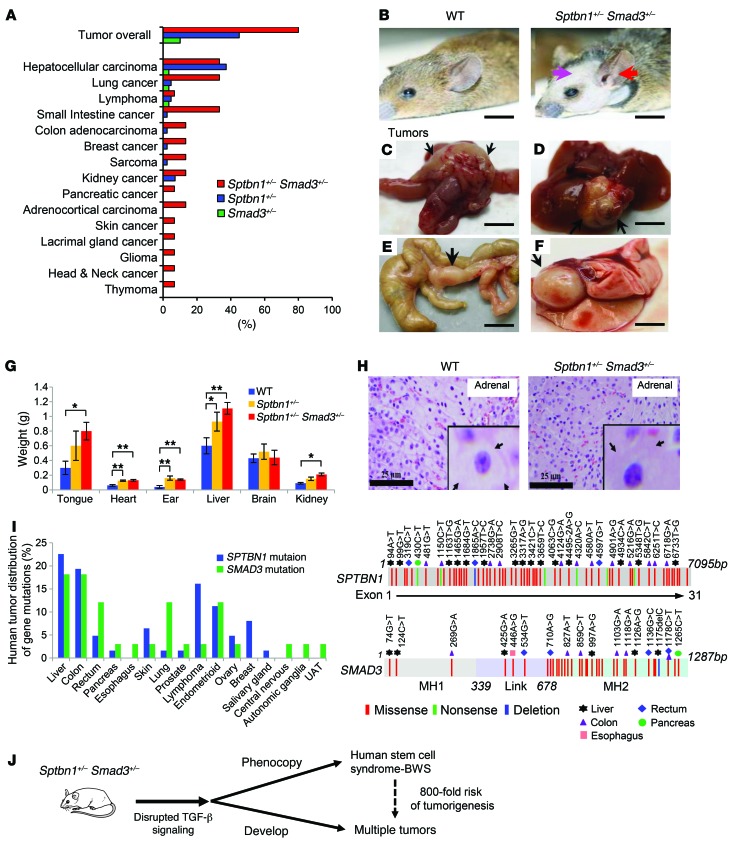
As previously reported, β2SP heterozygous mutant mice (Sptbn1+/–) phenocopy BWS, and epigenetic silencing of this gene may be a causal factor in BWS (33). To determine whether defective TGF-β signaling is involved in this human stem cell syndrome, the tumor incidence in 3 different strains of TGF-β–deficient mice was measured. The double-heterozygous Sptbn1+/– Smad3+/– mice had a significantly higher tumor incidence of 80% compared with the single-heterozygous Sptbn1+/– (40%) or Smad3+/– (10%) mice (Figure 1, A–F, and Supplemental Tables 1 and 2; supplemental material available online with this article; doi:10.1172/JCI80937DS1). These observations indicate that a partial loss of both of these crucial components of the TGF-β pathway synergistically enhances tumorigenesis.
Figure 1. Sptbn1+/– Smad3+/– mice develop multiple tumors and phenocopy features of BWS.
(A) Tumor incidence in double Sptbn1+/– Smad3+/– (n = 15) and in single Sptbn1+/– (n = 43) and Smad3+/– (n = 30) mice. (B) Anterior linear ear lobe crease (red arrow) and frontal balding (pink arrow) are nontumorigenic hallmarks of BWS. Sptbn1+/– Smad3+/–mice spontaneously develop multiple primary cancers (black arrows) by 12 months of age, including (C) colon adenocarcinoma, (D) hepatocellular carcinoma, (E) small bowel adenocarcinoma, and (F) lung adenocarcinoma. Scale bars: 10 mm. (G) Enlargement of organs in Sptbn1+/– Smad3+/– mice at 4 months of age. Error bars are shown as SD. n = 3. *P < 0.05; **P < 0.01, 1-way ANOVA with post-hoc Bonferroni’s test. (H) H&E-stained sections reveal adrenal cytomegaly in the fetal adrenal cortex of mutant Sptbn1+/– Smad3+/– mice not seen in wild-type mice. The arrows point to an adrenal cortical cell with enlarged granular eosinophilic cytoplasm and large hyperchromatic nuclei in Sptbn1+/– Smad3+/– mice compared with wild-type mice. Scale bars: 25 μm; original magnification, ×60 (insets). (I) Somatic mutations in SPTBN1 and SMAD3 occur frequently in multiple human cancers. The contribution of tumor types (left panel) and the location of somatic mutations (right panel) were summarized on the basis of the data set from COSMIC. (J) The diagram shows that Sptbn1+/– Smad3+/– mice phenocopy features of BWS and develop multiple tumors.
Sptbn1 homozygous mutant mice are embryonically lethal, and at E11.5, they display a number of abnormalities such as retarded growth, liver and gut hypoplasia, cardiac muscle hypertrophy, and aberrant neural defects (28). Sptbn1+/– Smad3+/– heterozygous mutant mice develop multiple abnormalities characteristic of BWS, including macroglossia, hemihyperplasia, anterior linear ear lobe crease, frontal balding, cardiomegaly, and renal hypertrophy (Figure 1G and Supplemental Figure 1). Additional hallmark features of BWS patients are cytomegaly of the fetal adrenal cortex and hyperplasia of the adrenal gland, and indeed, Sptbn1+/– Smad3+/– mice displayed conspicuous cytomegaly of the adrenal cortex (Figure 1H).
To examine the roles of SMAD3 and SPTBN1 in spontaneous tumorigenesis and to begin to elucidate their roles in BWS-associated tumorigenesis, a broad, unbiased analyses of cases reported in the Cancer Genome Atlas (TCGA; http://cancergenome.nih.gov/) and the Catalogue of Somatic Mutations in Cancer (COSMIC; http://cancer.sanger.ac.uk/cosmic) database was performed. These analyses confirmed that somatic mutations in SPTBN1 and SMAD3 occur in multiple human cancers, including many gastrointestinal cancers (Figure 1I and Supplemental Table 3), which are similar to the malignancies developed in Sptbn1+/– Smad3+/– mice (Figure 1I). Importantly, in the TCGA database, utilized through the cBioPortal (http://www.cbioportal.org), mutations in Sptbn1 (β2SP) and Smad3 have a significant cooccurrence within the same tumors in cervical squamous cell carcinoma, kidney chromophobe, and nasopharyngeal carcinoma (Supplemental Table 4 and refs. 34, 35).
Thus, Sptbn1+/– Smad3+/– mice represent what we believe to be a new genetic and mechanistic model for BWS, and disruption of the TGF-β pathway may be a novel mechanism linking this stem cell disorder to tumorigenesis (Figure 1J).
Disruption of the TGF-β pathway in Sptbn1+/– Smad3+/– mice and BWS cells.
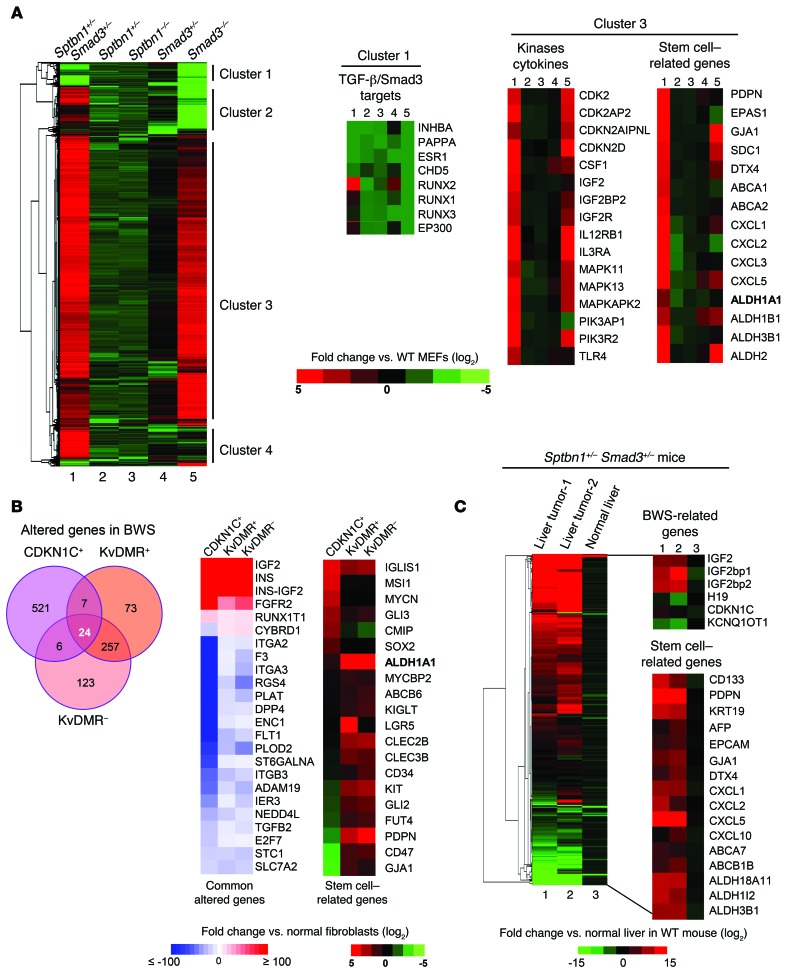
With this model for BWS, the mechanisms though which disruption of TGF-β signaling leads to BWS could be elucidated further. Whole-transcriptome sequencing analyses of MEFs isolated from these 3 TGF-β–deficient strains displayed altered expression patterns in molecular pathways when compared with MEFs from wild-type mice (Figure 2A and Supplemental Table 5). The heat map demonstrates that Sptbn1+/– Smad3+/– MEFs display 2 distinct clusters of differentially regulated genes (clusters 1 and 3) as compared with Sptbn1+/– or Smad3+/– MEFs. As predicted, cluster 1 of repressed genes includes TGF-β/SMAD3-regulated genes Inhba, Pappa, Esr1, Chd5, Runx1, Runx3, and Ep300 (Figure 2A). Cluster 3 of upregulated genes encompasses kinases, cytokines, and growth factors implicated in tumorigenesis including Cdk2, Cdkn2d, Mapk11/13, Pik3ap1, Il12rb1, Csf1, Igf2r, and Tlr4 (Figure 2A). Sptbn1+/– Smad3+/– MEFs also express increased levels of stem cell–associated genes, including Pdpn, Epas1, Cxcl1, and aldehyde dehydrogenase 2 (Aldh2) (Figure 2A). These data suggest that Sptbn1+/– Smad3+/– mice have a unique genetic profile that may facilitate their propensity to develop tumors.
Figure 2. Disruption of TGF-β pathway in Sptbn1+/– Smad3+/– mice and BWS cells.
(A) Heat map comparisons of gene-expression profiles from MEFs from Sptbn1+/– Smad3+/–, Smad3+/–, Sptbn1+/–, Smad3–/–, and Sptbn1–/– versus wild-type MEFs. The mRNA alterations were classified by hierarchical clustering. Representative gene expressions in cluster 1 and cluster 3 are shown (cutoff, P < 0.05). (B) Common altered genes and stemness profiles in the 3 BWS cell lines CDKN1C+, KvDMR+, and KvDMR–. Whole-transcriptome RNA sequencing data of BWS cell lines were analyzed by IPA (cutoff, q value < 0.3). The Venn diagram shows the altered genes in BWS cells (left panel). Twenty-four commonly altered genes are presented on a heat map (right panel). (C) The expression of BWS- and stem cell–associated genes in mouse liver tumors. Two spontaneous liver tumors from 2 individual Sptbn1+/– Smad3+/– mice, normal liver tissue from an Sptbn1+/– Smad3+/– mouse, and normal liver tissue from a wild-type mouse were used for whole-transcriptome RNA sequencing analyses, and the data are displayed as a heat map comparing the gene-expression profiles: liver tumor-1, liver tumor-2, and normal liver from Sptbn1+/– Smad3+/– mice versus normal liver from wild-type mice. The mRNA alterations were classified by hierarchical clustering.
To validate disruption of the TGF-β pathway in Sptbn1+/– Smad3+/– mice, MEFs from these mice as well as wild-type mice were treated with TGF-β1 to examine the gene expression of TGF-β targets. The mRNA expression levels of Inhba, Esr1, Runx2, and Pappa were induced by treatment with TGF-β1 in wild-type MEFs, but not in Sptbn1+/– Smad3+/– MEFs, demonstrating that double-heterozygote MEFs do not respond to TGF-β treatment (Supplemental Figure 2A). Furthermore, the transcriptome data of 2 independent Smad3–/– keratinocytes were downloaded from the Affymetrix mRNA microarray data via the NCBI’s Gene Expression Omnibus (GEO GSE3228) (Supplemental Figure 2B and ref. 36). When normalized with wild-type mouse keratinocytes, most of the TGF-β/SMAD3–regulated genes that were decreased, as presented in Figure 2A, were validated in these Smad3–/– keratinocytes by Ingenuity Pathway Analysis (IPA) (Ingenuity Systems) (Supplemental Figure 2B).
Next, we further characterized the molecular mechanisms relevant to the etiology of BWS and tumor development via whole-transcriptome sequencing of 3 different BWS fibroblastic cell lines established from patients (Figure 2B). The CDKN1C+ cell line was established from a BWS patient with a CDKN1C mutation; the KvDMR+ and KvDMR– cell lines were established from monozygotic twins of BWS patients who had loss of methylation in the KCNQ1OT1 region. The KvDMR+ cells were derived from a BWS patient with the KvDMR molecular defect, and the KvDMR– cells were derived from a BWS patient with an absence of KvDMR molecular defect, but with some clinical signs of BWS.
The RNA sequence analyses revealed that gene-expression profiles in these 3 BWS cell lines were different from that of a fibroblast cell line established from a person without BWS. The CDKN1C mutation resulted in the most striking alterations (Supplemental Figure 3A). Twenty-four genes were identified as being commonly altered in the 3 BWS cell lines, and 2 of these genes, IGF2 and insulin, were markedly upregulated, supporting the important role of IGF2 as an effector in BWS (Figure 2B and refs. 7–10). The commonly repressed genes included some that are related to the TGF-β pathway, such as integrin α2 and α3 (ITGA2 and 3), plasminogen activator (PLAT), neural precursor cell expressed developmentally downregulated 4-like-E3 ubiquitin protein ligase (NEDD4L), and TGF-β2 (TGFB2). A similar gene-expression profile of BWS-related genes was observed in Sptbn1+/– Smad3+/– MEFs, and these altered gene expressions included increased Igf2 and decreased H19. All 3 BWS cell lines expressed increased levels of stem cell genes, including SOX2, ALDH1A1, PDPN, and CD34 (Figure 2B).
IPA analysis of RNA sequencing data from the BWS cell lines generated a TGF-β1 downstream network (Supplemental Figure 3B). We validated TGF-β target gene expression in CDKN1C+ cells by quantitative PCR (Q-PCR) (Supplemental Figure 3C). The majority of TGF-β1–related genes were downregulated in CDKN1C+ BWS cells, and these cells displayed significant alterations in tumorigenesis-related signaling, whereas the KvDMR- BWS cells displayed genetic abnormalities associated with body size and embryonic cell proliferation (Supplemental Table 6).
Next, we performed whole-transcriptome sequencing of 2 spontaneously developing liver tumors from 2 individual Sptbn1+/– Smad3+/– mice, normal liver tissue extracted from a Sptbn1+/– Smad3+/– mouse, and normal liver tissue extracted from a wild-type mouse. The gene-expression profiles of these 2 liver tumors share certain similarities with BWS gene profiles, including a 15-fold increase of Igf2 and a 7-fold decrease of H19 expression levels compared with the normal liver tissues, significantly decreased levels of Cdkn1c and Kcnq1ot1, and increased levels of several stem cell–related genes (Figure 2C). These findings demonstrate that Sptbn1+/– Smad3+/– mice and BWS patients share several key commonalities in the disruption of genes regulated by TGF-β, suggesting potential mechanistic bases underlying their similar phenotypes.
β2SP interacts with SMAD3 in a TGF-β–dependent manner.
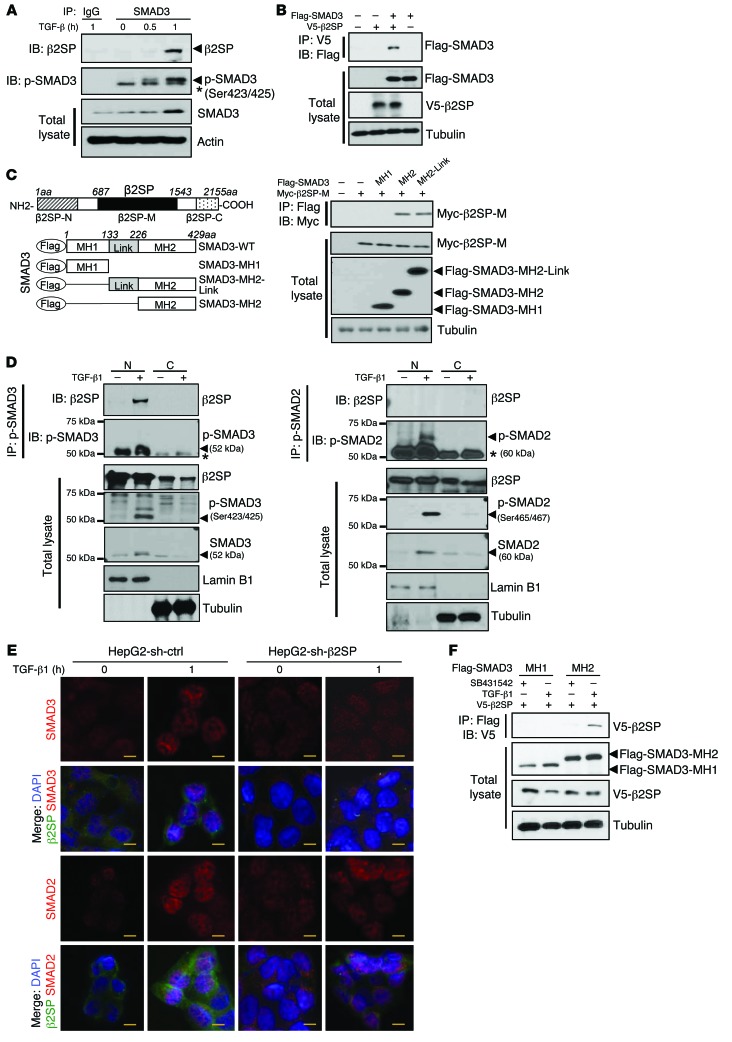
Previous reports demonstrated that β2SP is an adaptor protein for SMAD3/SMAD4 activation in response to TGF-β stimulation (28), but precisely how β2SP interacts with SMADs in the TGF-β pathway has not been elucidated. To further investigate a detailed mechanism for TGF-β regulation of β2SP and SMAD3, we looked for physical interactions between β2SP and SMAD3. In HepG2 human liver cancer cells (with an intact TGF-β pathway), endogenous β2SP associated with SMAD3 after TGF-β1 treatment (Figure 3A). Moreover, in SNU398 cells, which express low levels of endogenous β2SP, ectopically expressed β2SP and SMAD3 interacted (Figure 3B). We also found, with the use of expression plasmids for the β2SP central domain and SMAD3 deletion proteins, that the β2SP central domain interacts with the SMAD3 MAD homology 2 (MH2) domain (Figure 3C). In addition, only TGF-β–induced, phosphorylated SMAD3, but not phosphorylated SMAD2, was observed to associate with β2SP in the nucleus (Figure 3D and Supplemental Figure 4A), suggesting that the β2SP/p-SMAD3 complex predominantly functions in the nucleus in response to TGF-β. β2SP knockdown via shRNA significantly inhibited SMAD3, but not SMAD2 nuclear translocation, supporting the role of the β2SP/p-SMAD3 complex in TGF-β–induced nuclear translocation of SMAD3 (Figure 3E and refs. 28, 37). Treatment with SB431542, a selective TGF-β inhibitor, completely blocked the interaction between β2SP and SMAD3 (Figure 3F), furthering the hypothesis that β2SP interacts with SMAD3 in a TGF-β–dependent manner. KvDMR+ BWS cells had reduced β2SP and limited TGF-β–stimulated SMAD3 nuclear translocation, but transient transfection of full-length β2SP plasmid rescued this defect (Supplemental Figure 4B). These findings strongly suggest that β2SP is required for proper SMAD3 function in BWS cells and that β2SP cooperates with SMAD3 to transcriptionally regulate SMAD3 targets in the nucleus (Supplemental Figure 4C).
Figure 3. β2SP is required for SMAD3 nuclear translocation in BWS cells.
(A) Interaction of β2SP and SMAD3 is TGF-β dependent. HepG2 cells were treated with 200 pM TGF-β1 for the indicated times. Cell lysates were immunoprecipitated with a SMAD3 antibody and immunoblotted with the indicated antibodies. Asterisk designates nonspecific bands. (B) β2SP interacts with SMAD3. SNU398 cells were cotransfected with the indicated plasmids. Cell lysates were immunoprecipitated with a V5 antibody and immunoblotted with indicated antibodies. (C) β2SP interacts with SMAD3 MH2 domain. SNU398 cells were cotransfected with indicated plasmids. Cell lysates were immunoprecipitated with a Myc antibody and immunoblotted with indicated antibodies. (D) Interaction of β2SP and p-SMAD3 in cell nucleus. Cell lysates were isolated as nuclear (N) and cytoplasmic (C) compartments. HepG2 cells were treated with 200 pM TGF-β for 2 hours. Cell lysates were immunoprecipitated with a p-SMAD3 antibody or a p-SMAD2 antibody and immunoblotted with indicated antibodies. Asterisk designates IgG heavy chain bands. (E) Knockdown of β2SP compromises TGF-β–induced SMAD3, but not SMAD2, nuclear translocation. HepG2 shRNA control cells and shRNA-β2SP stable cells were treated with 200 PM TGF-β for 1 hour. Immunofluorescent staining was performed to detect β2SP, SMAD3, or SMAD2. Scale bars: 40 μm. (F) TGFBR1 inhibitor SB431542 blocks the interaction of β2SP and SMAD3. SNU398 cells were cotransfected with indicated plasmids and were treated with 5 μM TGFBR1 inhibitor SB431542 overnight. Cell lysates were immunoprecipitated with a Flag antibody and immunoblotted with indicated antibodies. Data are representative of 3 (A, B, and D), 2 (C), and 1 (F) independent experiments.
TGF-β/SMAD3/β2SP upregulates CTCF posttranscriptionally.
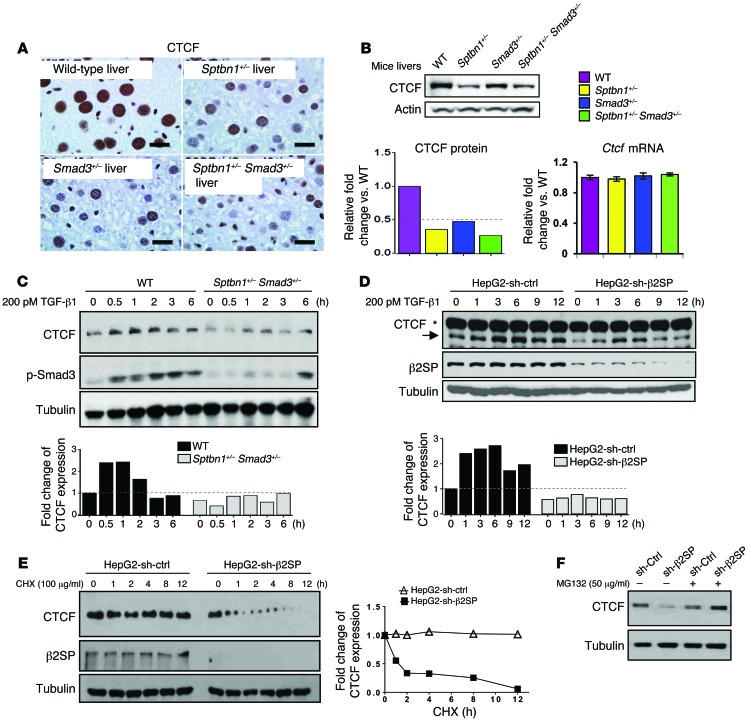
Multiple proteins have been implicated in BWS, including IGF2, CDKN1C, CTCF, and KCNQ1OT1. We performed a comprehensive expression analysis of these proteins in tissues from Sptbn1+/–, Smad3+/–, and Sptbn1+/– Smad3+/–mice compared with wild-type tissues. The results revealed markedly lower CTCF levels in all mutant mouse livers compared with the wild-type liver tissue (Figure 4A). Western blot analysis confirmed the lower CTCF protein levels in mutant mouse livers, while Q-PCR revealed no change in mRNA levels (Figure 4B). Also, Sptbn1+/– Smad3+/– MEFs had lower levels of CTCF than wild-type MEFs (Figure 4C). Similarly, CTCF levels in the cell lines from BWS patients were significantly lower than in normal hepatocytes (Supplemental Figure 5A). Importantly, TGF-β1 stimulation increased the levels of CTCF in HepG2 cells, but both basal and TGF-β1–induced expressions of CTCF were attenuated in HepG2 cells after knockdown with sh-β2SP (Figure 4D). Knockdown of β2SP or SMAD3 decreased CTCF protein stability (Figure 4E and Supplemental Figure 5B), and this downregulation of CTCF was inhibited by MG132, a 26S proteasome inhibitor (Figure 4F and Supplemental Figure 5C). Notably, CTCF mRNA levels were not affected by TGF-β1 treatment in MEFs or HepG2 cells, suggesting that TGF-β/SMAD3/β2SP regulates CTCF levels posttranscriptionally (Supplemental Figure 5, D and E). Moreover, overexpression of ectopic β2SP and SMAD3 significantly increased CTCF levels in HepG2 cells treated with TGF-β1, but not in cells treated with the TGF-β receptor 1 (TGFBR1) inhibitor SB431542 (Supplemental Figure 6A). This difference suggests that β2SP/SMAD3 also increases CTCF protein levels in a TGF-β–dependent manner.
Figure 4. TGF-β/SMAD3/β2SP upregulates CTCF.
(A) Decreased CTCF levels in Sptbn1+/–, Smad3+/–, and Sptbn1+/– Smad3+/– mouse livers were observed. CTCF levels were detected by immunohistochemical analysis in wild-type, Sptbn1+/–, Smad3+/–, and Sptbn1+/– Smad3+/– mouse livers. Scale bars: 20 μm. (B) CTCF protein expression levels but not mRNA levels were decreased in Sptbn1+/–, Smad3+/–, and Sptbn1+/– Smad3+/– mouse livers. CTCF protein levels were detected by immunoblotting analysis. Ctcf mRNA levels were detected by Q-PCR. Error bars are shown as SD. (C and D) TGF-β increased CTCF protein expression levels in Sptbn1+/– Smad3+/– MEFs (C) and in β2SP-knockdown HepG2 cells (D). MEFs or HepG2 cells were treated with 200 pM TGF-β1 for the indicated times. Cell lysates were immunoblotted with the indicated antibodies. The density of CTCF and the integrated optical density were measured. Asterisk designates nonspecific bands. (E) Knockdown β2SP decreased CTCF protein stability. HepG2–sh-Ctrl or HepG2–sh-β2SP cells were treated with 100 μg/ml cycloheximide (CHX) for the indicated times. The density of CTCF and the integrated optical density were measured. The turnover of CTCF is indicated graphically. (F) β2SP-mediated CTCF downregulation was proteasome dependent. HepG2–sh-Ctrl or HepG2–sh-β2SP cells were treated with or without 50 μg/ml MG132 for 6 hours. Cell lysates were immunoblotted with CTCF antibodies. Data are representative of 3 (B–E) and 2 (F) independent experiments.
CTCF interacts with β2SP and SMAD3 in a TGF-β–dependent manner.
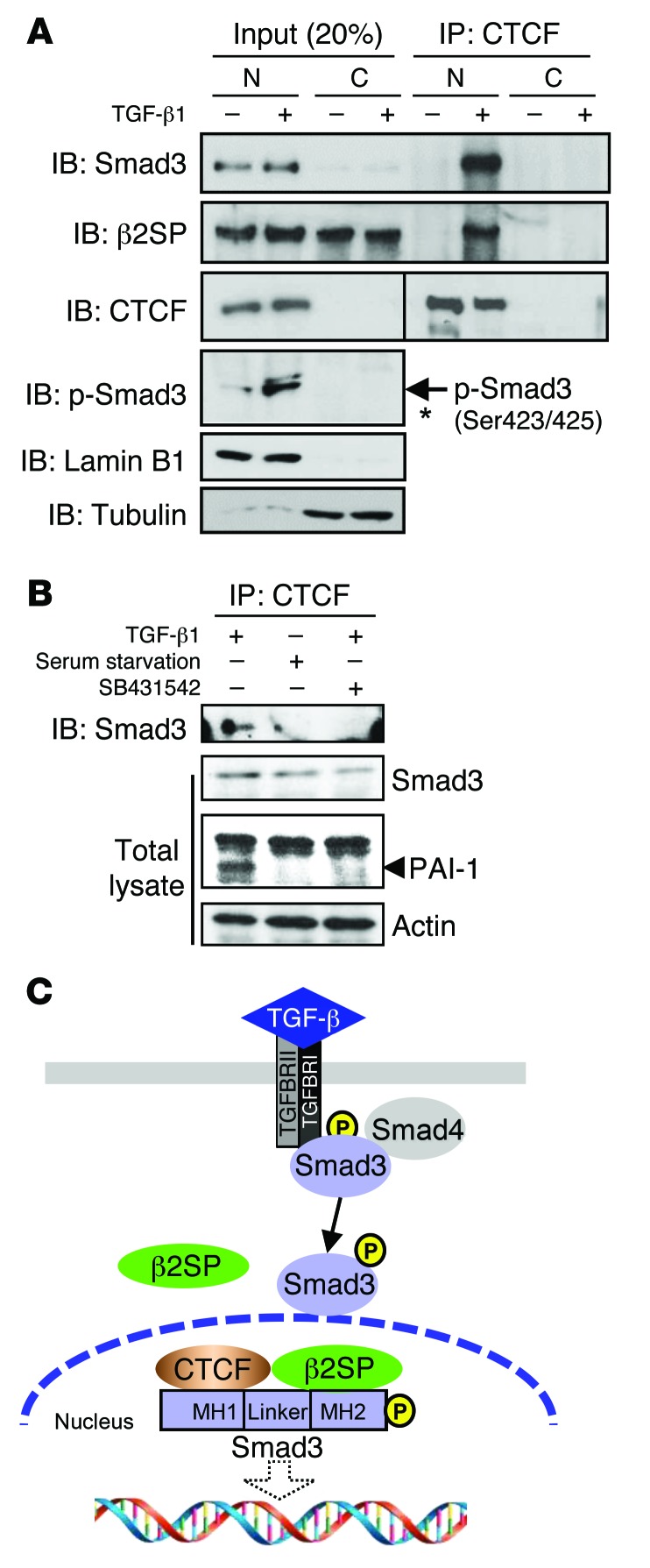
Next, we examined the interaction of CTCF, β2SP, and SMAD3. We found that nuclear CTCF interacted with β2SP and SMAD3 in TGF-β1–treated cells (Figure 5A). We further found that binding of endogenous CTCF with SMAD3 was completely disrupted by treatment with SB431542 or by serum starvation (Figure 5B), indicating that the interaction between CTCF and SMAD3 is TGF-β dependent. CTCF interacted with the MH1 domain of SMAD3 (Supplemental Figure 6B), and the interactions between ectopic β2SP and SMAD3 with CTCF were dramatically reduced with SB431542 treatment (Supplemental Figure 6C). Immunofluorescent microscopy detected TGF-β–induced nuclear translocation of CTCF and β2SP in the HCC cell line SNU475 (Supplemental Figure 6D). Taken together, these results support the concept that the formation of a β2SP/SMAD3/CTCF protein complex in the nucleus is TGF-β dependent (Figure 5C).
Figure 5. β2SP/SMAD3 interact with CTCF in cell nucleus.
(A) CTCF interacts with β2SP/SMAD3 in cell nucleus. HepG2 cells were treated with 200 pM TGF-β for 2 hours. Cell lysates were isolated as nuclear (N) and cytoplasmic (C) compartments and were immunoprecipitated (IP) with a CTCF antibody and immunoblotted (IB) with indicated antibodies. Asterisk designates nonspecific bands. (B) The interaction of CTCF and SMAD3 is TGF-β dependent. SNU398 cells were cotransfected with the indicated plasmids and were cultured with serum-free medium or treated with 5 μm SB431542 overnight. Then cells were treated with 200 pM TGF-β1 for 2 hours and cell lysates were immunoprecipitated with a CTCF antibody. Data are representative of 2 (A), and 3 (B) independent experiments. (C) Proposed model of the complex of β2SP/SMAD3/CTCF in the cell nucleus responding to TGF-β.
Dysfunction of the β2SP/SMAD3/CTCF complex increases TERT levels in BWS and in Sptbn1+/– Smad3+/– mice.
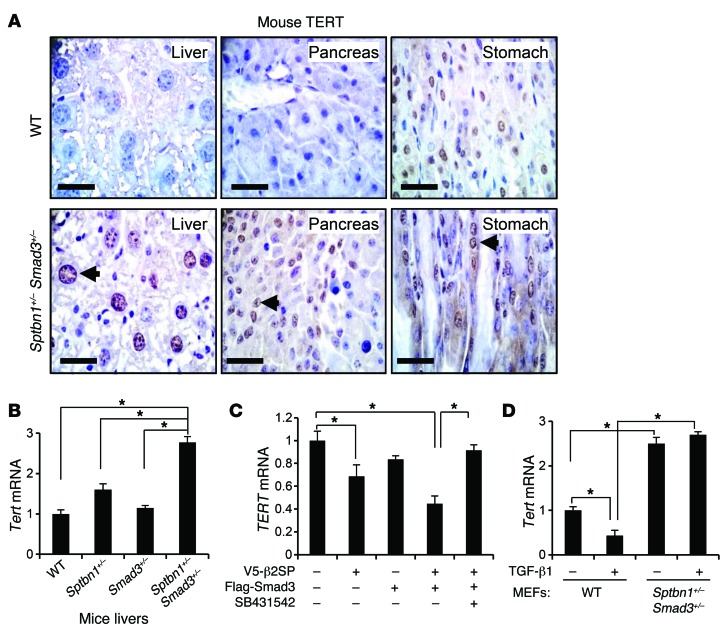
CTCF is directly involved in the transcriptional regulation of various key factors that control cellular growth, apoptosis, quiescence, senescence, and differentiation, such as TERT, c-MYC, RB family proteins, CDKN2A, and p53 (15). We observed a marked increase in TERT and c-MYC expression in kidney tumors from BWS patients (Supplemental Figure 7A). Increased TERT expression was also frequently found in several different types of tissues from Sptbn1+/– Smad3+/– mice, suggesting that the same genetic alteration occurs in both the BWS tumors and the murine TGF-β/SMAD3 disruption tumorigenesis model (Figure 6A). Increased Tert and Myc mRNA expression levels were confirmed in Sptbn1+/– Smad3+/– mouse liver tissues by Q-PCR analysis (Figure 6B and Supplemental Figure 7B). Overexpression of ectopic β2SP and SMAD3 significantly decreased TERT mRNA levels in SNU398 cells, which express low levels of endogenous β2SP. The TGFBR1 inhibitor SB431542 blocked this effect, suggesting that β2SP/SMAD3 decreases TERT mRNA levels in a TGF-β–dependent manner (Figure 6C). Moreover, unlike in wild-type MEFs and normal human fibroblasts, increased TERT mRNA expression was not reduced by TGF-β treatment in either Sptbn1+/– Smad3+/– MEFs or CDKN1C+ BWS cells (Figure 6D and Supplemental Figure 7C).
Figure 6. Increased TERT levels in Sptbn1+/– Smad3+/– mice.
(A) Increased levels of TERT in liver, pancreas, and stomach of Sptbn1+/– Smad3+/– mice were observed. Representative immunohistochemical staining of mouse TERT in wild-type or Sptbn1+/– Smad3+/– mouse organs. The arrows point to increased expression levels of TERT in Sptbn1+/– Smad3+/– mice. Scale bars: 20 μm. (B) Increased mRNA expression levels of Tert in Sptbn1+/–, Smad3+/–, and Sptbn1+/– Smad3+/– mouse livers were observed. *P < 0.001, 1-way ANOVA with post-hoc Bonferroni’s test. (n = 3). (C) β2SP and SMAD3 decrease TERT mRNA expression levels in a TGF-β–dependent manner. SNU398 cells were cotransfected with ectopic V5-β2SP and Flag-SMAD3 for 24 hours. Cells were then treated with 50 μM of TGFBR1 inhibitor SB431542 overnight. *P < 0.01, 1-way ANOVA with post-hoc Bonferroni’s test. (n = 3). (D) Reduction of TGF-β–induced Tert expression levels was observed in wild-type MEFs, but not in Sptbn1+/– Smad3+/– MEFs. Cells were treated with 200 pM TGF-β for 2 hours. Tert mRNA levels were detected by Q-PCR. *P < 0.001, 1-way ANOVA with post-hoc Bonferroni’s test. (n = 3). Error bars are shown as SD.
β2SP/SMAD3/CTCF complex transcriptionally regulates TERT.
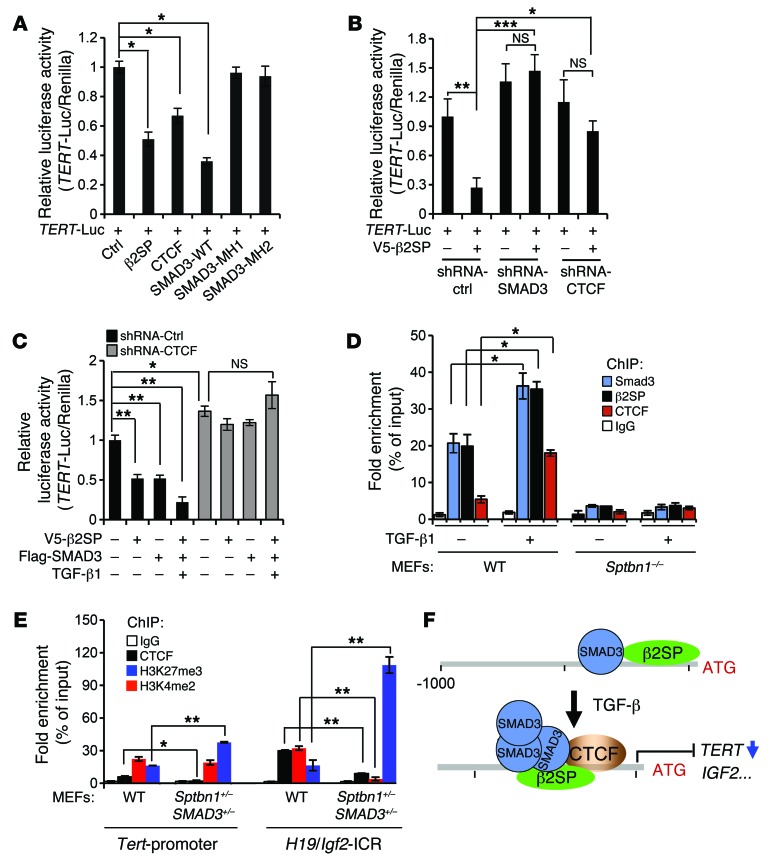
Overexpression of β2SP, CTCF, or wild-type SMAD3, but not SMAD3 MH1 or MH2 fragments, decreased TERT transcriptional activity (Figure 7A). Knockdown of β2SP, SMAD3, or CTCF increased TERT mRNA expression levels and compromised TGF-β–induced reduction of TERT expression (Supplemental Figure 7D). Importantly, we found that TGF-β/Smad3/β2SP–induced repression of TERT transcriptional activity was blocked by knockdown of SMAD3 or CTCF (Figure 7B and Supplemental Figure 8, A and B), suggesting that β2SP regulates TERT transcriptional activity through the SMAD3 pathway and that CTCF participates in and is required for TGF-β–mediated TERT transcriptional regulation (Figure 7, B and C). Collectively, these results suggest that interactions and cooperation among SMAD3, β2SP, and CTCF are required for TERT transcriptional regulation.
Figure 7. β2SP/SMAD3/CTCF complex transcriptionally regulates TERT.
(A) β2SP decreases TERT transcriptional activity. A luciferase reporter containing TERT promoter region (–1,000 base pairs) was cotransfected with indicated plasmids into SNU398 cells. *P < 0.001, 1-way ANOVA with post-hoc Bonferroni’s test. (n = 3). (B) β2SP regulates TERT transcriptional activity through SMAD3 and/or CTCF. HepG2–sh-Ctrl, HepG2–sh-SMAD3, or HepG2–sh-CTCF cells were cotransfected with indicated plasmids. *P < 0.05; **P < 0.01; ***P < 0.001, 1-way ANOVA with post-hoc Bonferroni’s test. (n = 3). (C) CTCF is required for TGF-β/β2SP/SMAD3-mediated TERT transcriptional activity. HepG2–sh-Ctrl or HepG2–sh-CTCF cells were cotransfected with indicated plasmids. Cells were treated with 200 pM TGF-β1 for 2 hours. *P < 0.05; **P < 0.001, 1-way ANOVA with post-hoc Bonferroni’s test. (n = 3). (D) Binding ability of SMAD3 and CTCF on the TERT promoter region is β2SP dependent. ChIP assays were performed. *P < 0.001 versus indicated, 1-way ANOVA with post-hoc Bonferroni’s test. (n = 3). (E) Decreased binding abilities of CTCF, H3K27me3, and increased binding abilities of H3k4me2 on the Tert and Igf2 promoter regions were observed in Sptbn1+/– Smad3+/– MEFs. *P < 0.05; **P < 0.001, 1-way ANOVA with post-hoc Bonferroni’s test. (n = 3). Error bars are shown as SD. (F) Proposed model of the role of the β2SP/SMAD3/CTCF complex in the regulation of TERT transcriptional activity.
SMAD3 transcriptionally regulates its targets via the SMAD-binding element (SBE) motif, 5′-GTCT-3′, or its complement, 5′-AGAC-3′ (38). To further investigate the role of β2SP/SMAD3 in the regulation of TERT, 4 evolutionarily conserved SBE motifs within –1,000 base pairs of the promoter region of TERT or Tert were identified (Supplemental Figure 9A). Through ChIP assays, significant enrichment of SMAD3 was found on 2 SBE motifs in the human TERT promoter, –335AGAC–332 and –264AGAC–261, and on 1 SBE motif in the mouse Tert promoter, –368GTCTAGAC–361 (Figure 7D and Supplemental Figure 9B). Furthermore, TGF-β1 treatment increased both β2SP and SMAD3 binding on the TERT promoter in normal hepatocytes, but not in the β2SP-negative BWS hepatoblastoma cells KvDMR+T (where T indicates hepatoblastoma) (Supplemental Figure 9C and ref. 33). TGF-β1 treatment also increased β2SP- and SMAD3-binding activities on human and mouse PAI-1 promoters, suggesting that the β2SP/SMAD3 complex is also present on the SMAD-binding regions of this TGF-β–induced gene (Supplemental Figure 9D).
We also identified several evolutionarily conserved CTCF-binding motifs (CCCTC) from 100 base pairs of exon 1 to –1,000 base pairs of the promoter region of TERT or Tert (Supplemental Figure 9A and refs. 39, 40). TGF-β1 treatment dramatically increased CTCF binding on the TERT promoter (–150CCCTCs–79) in normal hepatocytes and on the Tert promoter (–298GTGCGCCCCCGTTACT–278) in wild-type MEFs (Figure 7D and Supplemental Figure 9E). Furthermore, this TGF-β1–mediated increase in CTCF binding on the TERT promoter was lower in both SNU398 and BWS hepatoblastoma cells compared with that in normal hepatocytes, suggesting that these CTCF-binding activities depend on the intact TGF-β/β2SP pathway (Supplemental Figure 9F). Importantly, Smad3 and CTCF binding on the Tert promoter are completely lost in Sptbn1–/– MEFs, suggesting that β2SP is necessary for TERT transcriptional regulation by a β2SP/SMAD3/CTCF complex (Figure 7D).
CTCF is involved in multiple aspects of epigenetic regulation, including genomic imprinting in BWS (14). ChIP assays revealed significantly decreased CTCF-binding activities on the Tert and Igf2 promoters in MEFs from Sptbn1+/– Smad3+/– mice as compared with wild-type MEFs (Figure 7E). Moreover, the inactive transcriptional methylation marker H3K27me3 was increased and the active marker H3K4me2 was decreased on the TERT and IGF2 promoters in Sptbn1+/– Smad3+/– MEFs, suggesting that this dual knockdown may increase the tumorigenicity via epigenetic modulation of TERT, IGF2, and possibly other cancer genes. Strikingly, expression of Tert, Igf2, and Myc were increased in Sptbn1+/–, Smad3+/–, and Sptbn1+/– Smad3+/– mouse livers and in 4 tumor cell lines established from individual tumors (3 hepatocarcinomas and 1 lymphoma) in Sptbn1+/– Smad3+/– mice, signifying that β2SP/SMAD3 may also cooperate with CTCF in regulating IGF2 and MYC gene expression (Supplemental Figure 10, A and B).
In summary, these results support the notions that the β2SP/SMAD3/CTCF complex is critical for regulation of TERT transcription and that a defective TGF-β pathway in human BWS derepresses TERT and potentially other tumor-promoting genes (e.g., IGF2) implicated in BWS-associated tumorigenesis (Figure 7F).
Dysfunction of the β2SP/SMAD3/CTCF complex increases stem-like properties and enhances tumorigenesis of tumor-initiating cell.
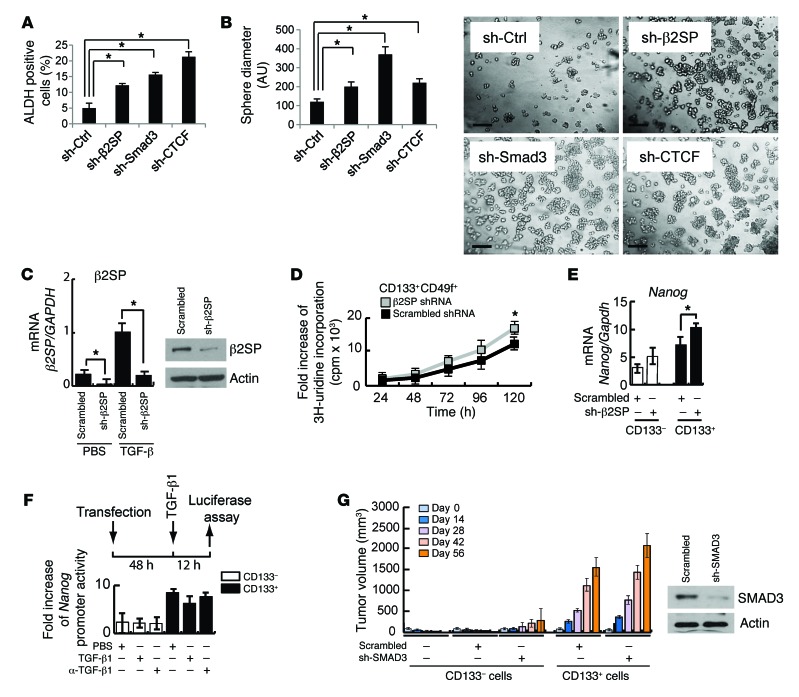
Whole-transcriptome RNA sequencing and Q-PCR analyses revealed that increased levels of stem cell–associated genes were observed in Sptbn1+/– Smad3+/– MEFs and 3 BWS cell lines compared with wild-type MEFs or a fibroblast cell line established from a non-BWS individual, respectively (Figure 2, A and B, and Supplemental Figure 11). These stemness genes included ALB, KRT19, CD133, NANOG, OCT4, SOX2, EPCAM, and AFP (Supplemental Figure 11). To determine the roles of the TGF-β–mediated β2SP/SMAD3/CTCF complex in stem cell biological function, each of the 3 elements was stably knocked down in HepG2 cells via lentiviral vectors, thus creating individual cell lines. Compared with shRNA-control (shRNA-Ctrl) cells, knockdown of any element of the β2SP/SMAD3/CTCF complex resulted in an increase of ALDH+ cell populations, a hallmark of stemness (Figure 8A). Importantly, ALDH1A1 expression also was increased in all 3 cell lines derived from BWS patients (Figure 2B). Further, knockdown of β2SP, SMAD3, or CTCF in HepG2 cells resulted in an increase in sphere formation, further supporting a role of these elements in regulating stemness (Figure 8B).
Figure 8. Dysfunction of the β2SP/SMAD3/CTCF complex increases stem-like properties and promotes TIC tumorigenesis.
(A) Increased ALDH population in β2SP, SMAD3, or CTCF knockdown cells. The positive ALDH cells were isolated from HepG2–sh-Ctrl, HepG2–sh-β2SP, HepG2–sh-SMAD3, or HepG2–sh-CTCF cells and then measured by flow cytometry. Bar graph data represent percentages of ALDH-positive cells. *P < 0.001, 1-way ANOVA with post-hoc Bonferroni’s test. (B) Knockdown of β2SP, SMAD3, or CTCF increases sphere formation. HepG2–sh-Ctrl, HepG2–sh-β2SP, HepG2–sh-SMAD3, or HepG2–sh-CTCF cells were cultured in serum-free DMEM/F12 medium with growth factors (10 ng/ml of EGF and FGF) for 6 days. Quantification of the spheres is shown. *P < 0.001, 1-way ANOVA with post-hoc Bonferroni’s test. Representative images of spheres were taken at day 6. Scale bars: 5 μm. (C) β2SP knockdown efficiency is shown in CD133+CD49f+ TICs isolated from liver tumors of alcohol-fed HCV Ns5a Tg mice. β2SP protein levels are effectively silenced by transduction of lentiviral shRNA in TICs (insets). *P < 0.01, Student’s t test. (D) β2SP knockdown increases CD133+CD49f+ TIC proliferation. Cell proliferation rates were determined by measuring 3H-uridine incorporation. *P < 0.05, Student’s t test (vs. scrambled control). (E) β2SP knockdown increases Nanog expression levels in CD133+ TICs. The mRNA expression levels of Nanog were measured by Q-PCR. *P < 0.05, Student’s t test. (F) Nanog promoter activity is higher in CD133+ TICs, while TGF-β stimulation does not significantly inhibit Nanog promoter activity in these TICs. Nanog promoter luciferase assays were performed. (G) SMAD3 knockdown enhanced subcutaneous tumor growth of TICs in a xenograft NOG mouse model. SMAD3 protein levels are effectively silenced by transduction of lentiviral shRNA in TICs (insets). Error bars are shown as SD. Each result shown is representative of 3 independent experiments (A–G).
In previous studies on the effects of TGF-β signaling in tumor-initiating stem-like cells, knockdown of β2SP promoted CD133+CD49+ tumor-initiating cell (TIC) tumorigenesis in alcohol-fed HCV Ns5a Tg (NOG) mice, revealing that this complex is involved in some aspects of liver cancer stem cell biology (41). Here, knockdown of β2SP increased proliferation of TICs (Figure 8, C and D), and NANOG expression levels were significantly increased in CD133+ cells compared with shRNA-Ctrl (Figure 8E). Using luciferase reporter assays, a significantly higher level of Nanog transcriptional activity was detected in CD133+ cells than in CD133– cells. However, Nanog transcriptional activity was not significantly suppressed by TGF-β stimulation, suggesting defective TGF-β signaling in TICs (Figure 8F). Finally, to test whether suppressed TGF-β/SMAD3 signaling further induces oncogenesis, CD133+ and CD133– TICs with or without SMAD3 knockdown were transplanted into NOG mice. Growth of the TIC-derived tumors was accelerated with SMAD3 knockdown, demonstrating that inhibition of TGF-β signaling enhances oncogenicity of TICs (Figure 8G).
Discussion
Several TGF-β signaling components are bona fide tumor suppressors with the ability to constrain cell growth and inhibit cancer development in its early stages. Inactivation of at least one of these components (such as the TGF-β receptors [TGFBR1, TGFBR2], SMAD2, or the common mediator SMAD4) occurs in almost all gastrointestinal tumors (42, 43). For instance, TGFBR2 is mutated in up to 30% of colon cancers, and TGFBR1 is mutated in 15% of biliary cancers (44). SMAD4 is deleted in up to 60% of pancreatic cancers and is mutated in hereditary juvenile polyposis coli (45, 46). Loss of β2SP, a SMAD3/4 adaptor protein, is observed in human hepatocellular cancers (33) and causes the spontaneous development of hepatocellular cancers in mice. However, the specific roles that TGF-β–activated β2SP/SMAD3 plays in suppressing the development of human cancers are unclear. We found that double-heterozygous Sptbn1+/– Smad3+/– mutant mice exhibit phenotypes that are similar to those of humans with BWS, including a markedly heightened tumor incidence. A key clinical feature of human BWS is adrenal cytomegaly. Adrenal cytomegaly is also observed in the Sptbn1+/– Smad3+/– mutants and implies dysfunctional homeostatic mechanisms as stem/progenitor cells divide, differentiate, and maintain expanding zones in the adrenal glands (47–49). Our study points to β2SP as an essential scaffolding protein required for TGF-β signaling, including p-SMAD3 nuclear translocation and cooperation with the chromosome insulator CTCF to regulate oncogenic genes such as TERT. More importantly, we showed that CTCF is a TGF-β–dependent protein. Thus, in double-heterozygous Sptbn1+/– Smad3+/– mutant mice, partial loss of β2SP/SMAD3 disrupts TGF-β signaling, reduces CTCF expression, and leads to derepression of Tert, Myc, and IGF2. These results all lead to clinical characteristics of BWS and tumorigenesis (Figure 9).
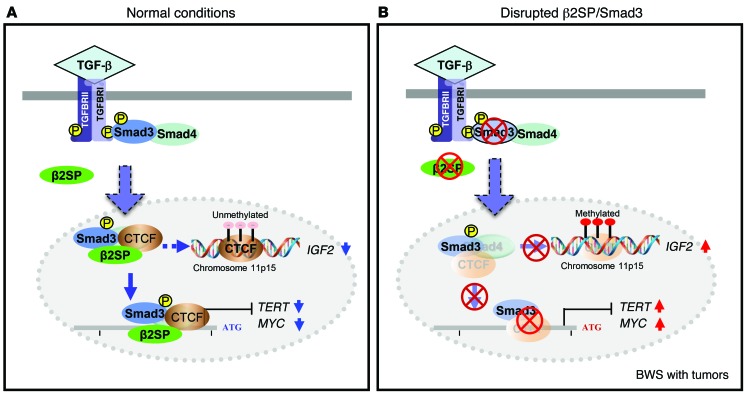
Figure 9. Schematic model of disrupted TGF-β signaling and BWS tumorigenesis.
(A) The model demonstrates that, under normal conditions, β2SP is an essential SMAD3 adaptor protein required for key events in the propagation of TGF-β signaling and docking SMAD3 and CTCF to DNA, thus either affecting chromosome 11p15-imprinted IGF2 genes or directly suppressing TERT and MYC transcription. (B) In human BWS, disruption of β2SP signaling through an aberrant interaction of β2SP/SMAD3/CTCF in the cell nucleus results in increased expression of IGF2 from chromosome 11p15 and directly activates TERT and MYC transcription, causing large organs and multiple cancers in patients with BWS.
The molecular etiology of BWS is complex and involves alterations in the expression of multiple imprinted growth-regulatory genes on chromosome 11p15, including the loss of H19, overexpression of IGF2, and dysfunction of CDKN1C and KCNQ1 (9, 17, 22). The following transgenic mouse models are frequently used for studying mechanisms of BWS: transgenic chimeric mice with IGF2 overexpression (9), mice with H19 deletion (50), mice with deletion of the maternally inherited Igf2r, H19/Igf2r double-heterozygote mice (51, 52), Kcnq1 mutant mice (53), mice with loss-of-function mutations in p57KIP2 (54), and mice with deletions of the murine homolog of the glypican-3 gene (55). However, none of these murine models phenocopy all the common features of BWS, suggesting that they do not recapitulate some of the fundamental genetic and molecular deficiencies that result in this genetic syndrome.
Genome-wide analysis enables predictive modeling of genetic pathways driving many cancers, genetic diseases, and human syndromes. We performed whole-transcriptome sequencing for 3 BWS cell lines and 4 MEFs from TGF-β–deficient mice. We used IPA bioinformatic tools to investigate the roles of the TGF-β signaling pathway in BWS. Importantly, Sptbn1+/– Smad3+/– MEFs exhibited unique genetic alterations, suggesting that the disruption of the physical interaction between β2SP and SMAD3 and the consequent deregulation of the chromatin regulatory network is a molecular mechanism that contributes to phenotypes of BWS, particularly to tumor development.
Interestingly, the tumors in human BWS are mostly embryonic, including nephroblastoma, hepatoblastoma, and pancreatoblastoma, whereas those in our murine model were mainly carcinomas or other adult neoplasms, even though they still appeared to have some common features of BWS embryonic tumors (hepatoblastomas and hepatocellular cancer are often considered indistinguishable in rodent livers). Our results from the whole-transcriptome sequencing analyses demonstrate the diversity of genetic alterations in BWS. The mice that developed tumors could have had disruptions of signaling pathways, chromosome instability, or increasing gene transcriptional regulation, eventually causing oncogene amplification or somatic mutations and the development of adult-type tumors. Our results show that both Igf2 and Tert mRNA expression were increased in Sptbn1+/– Smad3+/– mouse tissues, suggesting that disrupted TGF-β signaling may interrupt the CTCF functions of regulating imprinted H19/IGF2 on chromosome 11p15 and directly regulating its targets, such as TERT and MYC (Figure 9).
Our results are indicative of dysfunctional regulation of the Tert promoter by SMAD3, β2SP, and CTCF, leading to increased enzymatic telomerase activity in BWS tumors and in Sptbn1+/– Smad3+/– mice. They also show that activation of the SMAD3/β2SP/CTCF complex of the TERT promoter is dependent on the TGF-β signaling pathway. It is also possible that activation of the SMAD3/β2SP/CTCF complex on the TERT promoter region may cooperate with MYC activation because MYC also promotes TERT activity through the TERT promoter E-box region (Supplemental Figure 9A). Thus, it is conceivable that biologically significant TERT and MYC repression occurs through β2SP interactions with SMAD3 and CTCF. MYC activation and telomerase dysfunction have been shown to play prominent roles in early hepatocellular carcinoma initiation (56). Our studies indicate that Tert activated together with Myc may be functionally responsible for the spontaneous cancers that developed in the Sptbn1+/– Smad3+/– mice. Importantly, enhancing the effects of β2SP and/or SMAD3 may provide a new strategy of targeting telomerase activity in anticancer therapy for lethal foregut cancers such as hepatocellular carcinoma and pancreatic cancer. In addition, monitoring TERT and TGF-β signaling pathway components could provide insight into BWS as well as insight into temporal events, from dysplasia to carcinoma formation.
Methods
Cell culture, transfection, and shRNA-mediated silencing.
Human liver cancer cell lines HepG2 (ATCC, HB8065), SNU475 (ATCC, CRL-2236), and SNU398 (ATCC, CRL-2233) were cultured in DME/F12 medium (Sigma-Aldrich, D5671) and supplemented with 10% FBS (Sigma-Aldrich, F2442). The human BWS cell lines CDKN1C+, KvDMR–, KvDMR+, KvDMR+T, UPD+NT, UPD+T, UPD+1, UPD+2, and normal fibroblasts were developed by R. Weksberg. The BWS cells and MEFs (Sptbn1+/–, Sptbn1–/–, Smad3+/–, and Sptbn1+/– Smad3+/–) were cultured in DME medium, and mRNAs were extracted for whole-transcriptome RNA sequencing analyses. For transfection, HepG2 and SNU398 cells were transfected with MYC-β2SP-Mid and Flag-SMAD3 plasmids using Lipofectamine 2000 (Invitrogen). TGF-β1 (Sigma-Aldrich, T1654) was added to create a final concentration of 200 pM, and cultures were incubated for 60 minutes. SB431542 (S4317), MG132 (M7449), and cycloheximide (C7698) were purchased from Sigma-Aldrich. Lentiviral particles containing shRNA of β2SP (sc-36551), SMAD3 (sc-38376), CTCF (sc-35125), or control shRNA (sc-108080) were purchased from Santa Cruz Biotechnology Inc. and were used to infect HepG2 cells.
Generation of transgenic mice and tumorigenesis analysis.
Sptbn1–/– and Sptbn1+/– mutant mice were generated as previously described (28, 33). For generation of Sptbn1+/– Smad3+/– mice, Sptbn1+/– mice were mated with Smad3+/– mice. Genotypes were determined by PCR. A total of 15 Sptbn1+/– Smad3+/– mice were generated. Eleven of these Sptbn1+/– Smad3+/– mice spontaneously developed multiple primary cancers by 12 months of age. A total of 43 Sptbn1+/– mice were generated, and 19 of these spontaneously developed primary cancers by 12 months of age. A total of 30 Smad3+/– mice were generated, and 3 of these spontaneously developed primary cancers by 12 months of age. HCV Ns5a Tg mice were provided by Ratna Ray (Saint Louis University, Saint Louis, Missouri, USA) (41). The TICs maintained both the transformed phenotype in vitro after thawing and tumorigenecity in NOG mice after transplantation. These cells were infected with a lentiviral vector expressing a shRNA that targets SMAD3 (Open Biosystems). These cells were then assessed for NANOG expression and promoter activity by Q-PCR and transfection-reporter assay, respectively. We determined the cell proliferation rate as assessed by incorporation of 3H-uridine in 96-well plates. The TICs and control cells stably transduced with SMAD3 shRNA or scrambled shRNA were transplanted subcutaneously into NOG mice with the Matrigel beads (Invitrogen), and the tumor formation and growth were monitored every 2 weeks (41).
Whole-transcriptome sequencing and IPA analyses.
Whole-transcriptome RNA sequencing was performed and analyzed at the MD Anderson Cancer Center DNA core facility (See Supplemental Methods). Gene-expression profiling data including human BWS cell lines (CDKN1C+, KvDMR–, KvDMR+), MEFs (Sptbn1+/–, Sptbn1–/–, Smad3+/–, and Sptbn1+/– Smad3+/–), and 2 mouse liver tumors (spontaneously developing liver tumors from 2 individual Sptbn1+/– Smad3+/– mice), have been deposited in the NCBI’s Gene Expression Omnibus (GEO GSE69737 and GSE69752). We identified differentially expressed mRNAs using DESeq with the standard comparison mode between the 2 experimental conditions with multiple testing corrections (cutoff: adjusted P value < 0.05). The mRNA alterations were classified by hierarchical clustering. Heat map comparisons of 4 gene-expression profiles from Sptbn1+/– Smad3+/–, Smad3+/–, Sptbn1+/–, and Sptbn1–/– MEFs versus wild-type MEFs were generated. The representative gene expressions in cluster 1 and cluster 3 are shown (cutoff, P < 0.05). The candidate signatures identified were uploaded into IPA for pathway analysis. Ingenuity’s knowledge base is a repository of molecular interactions, regulatory events, gene-to-phenotype associations, and chemical knowledge that provides the building blocks for pathway construction.
Luciferase assays.
For transcriptional reporter assays, HepG2–sh-Ctrl or HepG2–sh-β2SP cells were seeded at a density of 1 × 104 cells per well in 24-well dishes. The next day, the cells were cotransfected with a luciferase reporter containing a TERT promoter region (–1,000 base pairs) and V5-β2SP wild-type and/or Flag-SMAD3 plasmids using Lipofectamine 2000. After 24 hours of transfection, the cells were treated with 200 pM TGF-β1 for 4 hours. In all transfections, the expression plasmid Renilla (Promega) served as an internal control to correct for transfection efficiency. The cells were extracted using 100 μl of luciferase cell culture lysis reagent. Ten microliters of cell extract were used for measuring Renilla enzyme activity. Twenty microliters of cell extract were used for the luciferase assay using a luciferase assay kit according to the manufacturer’s instructions (Promega). After subtracting the background (no transfection control), the luciferase activity was normalized to Renilla activity (AU) for each sample, and fold changes were calculated.
Immunoblotting, immunoprecipitation, and immunohistochemical and immunofluorescence analyses.
HepG2, SNU398, or 293T cells were harvested, lysed with lysis buffer (50 mM Tris-HCl, pH 7.5, 0.15 M NaCl, 1% NP-40, 1 mM EDTA), Complete Protease Inhibitor Cocktail (Roche Applied Science), 1 mM PMSF, 1 mM NaF, and 1 mM sodium orthovanadate, and sonicated. Nuclear and cytoplasmic proteins were prepared (Supplemental Methods). The antibodies were used for immunoblotting and immunoprecipitation analyses (Supplemental Methods).
For immunohistochemical analyses, mouse tissues were fixed in 10% formalin and embedded in paraffin in accordance with standard procedures. Sections were stained with CTCF (Cell Signaling Technology, 2899) and TERT (NOVUS, NB100-317) antibodies. Diaminobenzidine was used as a chromogen, and hematoxylin was used for counterstaining. Sections from a human BWS patient kidney tumor sample were prepared and processed for immunohistochemical analysis using TERT antibodies (NOVUS, NB100-317).
For immunofluorescence analyses, HepG2 shRNA control cells and shRNA-β2SP stable cells were prepared and seeded onto chamber slides at 2 × 104 cells per well for 12 hours. Cells were then treated with 200 pM TGF-β for time points. Immunofluorescent staining was performed to detect β2SP and SMAD3. Confocal images were acquired using either of the ×40 objectives on an IX81 Olympus microscope.
Quantitative reverse-transcription–PCR analyses.
Total RNA was extracted with the use of TRIzol reagent (Invitrogen) according to the manufacturer’s instructions. Reverse transcription was performed by using a SuperScript III First-Strand Kit (Invitrogen). Each cDNA (10 ng) was amplified in triplicate with the iQ SYBR Green Supermix PCR Kit (Bio-Rad Laboratories) for 40 cycles on a Bio-Rad system (iCycler Thermal Cycler). The sequences used for the analysis are listed in the Supplemental Methods.
ChIP assays.
ChIP assays were performed using EZ ChIP Assay Kit (catalog P-2002, Millipore) following the manufacturer’s instructions. MEFs (Sptbn1+/+ and Sptbn1–/–), BWS cells, and normal human hepatocytes (Liver Tissue Cell Distribution System, University of Pittsburgh, Pittsburgh, Pennsylvania, USA, and University of Minnesota, Minneapolis, Minnesota, USA) were washed, cross-linked with formaldehyde (1% final concentration), and sonicated on ice to fragment the chromatin into an average length of 500 bp to 1 kb. The lysates were diluted using chromatin dilution buffer. SMAD3 (9523, Cell Signaling), β2SP (customized antibody from bioSythesis), CTCF (2899, Cell Signaling), H3K27me3 (GAM-9205, QIAGEN), and H3K4me2 (GAM-3203, QIAGEN) antibodies were used to immunoprecipitate the respective antigens at 4°C overnight. For each ChIP assay, a fraction of the input chromatin (10%) was also processed for DNA purification; a mock immunoprecipitation with a neutral, unrelated IgG (2729, Cell Signaling) antiserum was carried out in parallel. With the DNA isolated at the end of the ChIP analysis, Q-PCR was performed using the TERT promoter–specific primers. The PCR primers for the ChIP assay are detailed in the Supplemental Methods.
Flow cytometry assays.
CD133+CD49f+ TICs were separated from CD133–CD49f+ or CD133–CD49f– cells by FACS from a CD45– fraction of tumor tissue cell suspension from the animal models (41). HepG2–sh-Ctrl, HepG2–sh-β2SP, HepG2–sh-SMAD3, or HepG2–sh-CTCF cells were assessed for ALDH activity utilizing the Aldefluor Assay Kit (catalog 01700, StemCell Technologies Inc.) following the manufacturer’s instructions. ALDH-positive cells were measured by FACScan flow cytometry.
Spheroid formation assays.
HepG2–sh-Ctrl, HepG2–sh-β2SP, HepG2–sh-SMAD3, or HepG2–sh-CTCF cells were cultured in serum-free DMEM/F12 medium with growth factors (10 ng/ml of EGF and FGF) for 6 days. 1 × 104 cells per well mixed with 2% Matrigel-containing media were plated on a Matrigel precoated chamber slide. The images of spheres (×10) were taken at day 6. Quantification of the spheres was determined by measuring spheres’ diameters.
Statistics.
Differences between 2 groups were evaluated using 2-tailed Student’s t tests. For multiple comparisons, 1-way ANOVA with post-hoc Bonferroni’s test was used. Results are expressed as mean ± SD unless otherwise indicated. For all statistical analyses, P < 0.05 was considered statistically significant.
Study approval.
All animal experiments were performed according to the guidelines for the care and use of laboratory animals and were approved by the institutional biomedical research ethics committee of The University of Texas MD Anderson Cancer Center for Biological Sciences.
Author contributions
JC performed and supervised most of the in vitro experiments and some in vivo experiments. JC, JSC, YJG, NAM, JZ, and XS performed whole-transcriptome RNA sequencing analysis. ZXY, YSJ, KO, and JC performed immunofluorescence analysis for CTCF and β2SP as well as Smad2 and Smad3 nuclear translocation. JC, YJG, and AG performed coimmunoprecipitation assays. ZXY, MJF, and AR performed immunohistochemical analysis. AM and SL performed spheroid formation assays. KM and HT performed CD133+CD49f+ TIC isolation assays in alcohol-fed HCV Ns5a Tg mice and performed TIC tumor growth assays in a xenograft NOG mouse model. SC and RW provided human BWS cells. NMM, SK, HFH, SL, BM, JW, AYW, MJ, MD, PM, WLH, MJF, JWS, and HT provided intellectual input and edited the manuscript. LM and JC designed and coordinated the research, analyzed the data, and wrote the manuscript.
Supplementary Material
Acknowledgments
We thank Wilma S. Jogunoori, Sai-Ching Jim Yeung, Deborah Berry, Bhaskar V.S. Kallakury, and Shareen J. Iqbal for technical support. We also thank Waun Ki Hong, Robert C. Bast, and Eduardo M. Sotomayor for critical review and helpful comments on the manuscript. This research was supported by NIH grants R01CA106614 (to L. Mishra), R01CA042857 (to L. Mishra), R01AA023146 (to L. Mishra), P01 CA130821 (to L. Mishra), R01AA018857 (to K. Machida), RC2 AA019392 (to H. Tsukamoto and L. Mishra), P50AA011999 (to H. Tsukamoto), U01AA018663 (to H. Tsukamoto), RSG-12-177-01-MPC (to K. Machida), and IRG-58-007-48 (to K. Machida). Funding was also received from the American Cancer Society (to K. Machida), the Medical Research Service of the Department of Veterans Affairs (to H. Tsukamoto), the University of Texas MD Anderson Multidisciplinary Research Program (to L. Mishra), Science & Technology Acquisition and Retention Funding (to L. Mishra), P30 DK56338 (to L. Mishra and Mary Estes), and P30 CA016672 (to L. Mishra and R. DePinho).
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Reference information:J Clin Invest. 2016;126(2):527–542. doi:10.1172/JCI80937.
References
- 1.Pettenati MJ, Haines JL, Higgins RR, Wappner RS, Palmer CG, Weaver DD. Wiedemann-Beckwith syndrome: presentation of clinical and cytogenetic data on 22 new cases and review of the literature. Hum Genet. 1986;74(2):143–154. doi: 10.1007/BF00282078. [DOI] [PubMed] [Google Scholar]
- 2.Weksberg R, Shuman C, Beckwith JB. Beckwith-Wiedemann syndrome. Eur J Hum Genet. 2010;18(1):8–14. doi: 10.1038/ejhg.2009.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.DeBaun MR, Tucker MA. Risk of cancer during the first four years of life in children from The Beckwith-Wiedemann Syndrome Registry. J Pediatr. 1998;132(3 pt 1):398–400. doi: 10.1016/s0022-3476(98)70008-3. [DOI] [PubMed] [Google Scholar]
- 4.Hadzic N, Finegold MJ. Liver neoplasia in children. Clin Liver Dis. 2011;15(2):443–462. doi: 10.1016/j.cld.2011.03.011. [DOI] [PubMed] [Google Scholar]
- 5.Keller RB, Demellawy DE, Quaglia A, Finegold M, Kapur RP. Methylation status of the chromosome arm 19q MicroRNA cluster in sporadic and androgenetic-biparental mosaicism-associated hepatic mesenchymal hamartoma. Pediatr Dev Pathol. 2015;18(3):218–227. doi: 10.2350/15-01-1600-OA.1. [DOI] [PubMed] [Google Scholar]
- 6.Feinberg AP, Ohlsson R, Henikoff S. The epigenetic progenitor origin of human cancer. Nat Rev Genet. 2006;7(1):21–33. doi: 10.1038/nrg1748. [DOI] [PubMed] [Google Scholar]
- 7.Murrell A, et al. An association between variants in the IGF2 gene and Beckwith-Wiedemann syndrome: interaction between genotype and epigenotype. Hum Mol Genet. 2004;13(2):247–255. doi: 10.1093/hmg/ddh013. [DOI] [PubMed] [Google Scholar]
- 8.Ishihara K, Oshimura M, Nakao M. CTCF-dependent chromatin insulator is linked to epigenetic remodeling. Mol Cell. 2006;23(5):733–742. doi: 10.1016/j.molcel.2006.08.008. [DOI] [PubMed] [Google Scholar]
- 9.Sun FL, Dean WL, Kelsey G, Allen ND, Reik W. Transactivation of Igf2 in a mouse model of Beckwith-Wiedemann syndrome. Nature. 1997;389(6653):809–815. doi: 10.1038/39797. [DOI] [PubMed] [Google Scholar]
- 10.Choufani S, Shuman C, Weksberg R. Molecular findings in Beckwith-Wiedemann syndrome. Am J Med Genet C Semin Med Genet. 2013;163C(2):131–140. doi: 10.1002/ajmg.c.31363. [DOI] [PubMed] [Google Scholar]
- 11.Sakatani T, et al. Loss of imprinting of Igf2 alters intestinal maturation and tumorigenesis in mice. Science. 2005;307(5717):1976–1978. doi: 10.1126/science.1108080. [DOI] [PubMed] [Google Scholar]
- 12.Harper J, Burns JL, Foulstone EJ, Pignatelli M, Zaina S, Hassan AB. Soluble IGF2 receptor rescues Apc(Min/+) intestinal adenoma progression induced by Igf2 loss of imprinting. Cancer Res. 2006;66(4):1940–1948. doi: 10.1158/0008-5472.CAN-05-2036. [DOI] [PubMed] [Google Scholar]
- 13.Ravenel JD, et al. Loss of imprinting of insulin-like growth factor-II (IGF2) gene in distinguishing specific biologic subtypes of Wilms tumor. J Natl Cancer Inst. 2001;93(22):1698–1703. doi: 10.1093/jnci/93.22.1698. [DOI] [PubMed] [Google Scholar]
- 14.Handoko L, et al. CTCF-mediated functional chromatin interactome in pluripotent cells. Nat Genet. 2011;43(7):630–638. doi: 10.1038/ng.857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fiorentino FP, Giordano A. The tumor suppressor role of CTCF. J Cell Physiol. 2012;227(2):479–492. doi: 10.1002/jcp.22780. [DOI] [PubMed] [Google Scholar]
- 16.Bliek J, et al. Increased tumour risk for BWS patients correlates with aberrant H19 and not KCNQ1OT1 methylation: occurrence of KCNQ1OT1 hypomethylation in familial cases of BWS. Hum Mol Genet. 2001;10(5):467–476. doi: 10.1093/hmg/10.5.467. [DOI] [PubMed] [Google Scholar]
- 17.Weksberg R, et al. Tumor development in the Beckwith-Wiedemann syndrome is associated with a variety of constitutional molecular 11p15 alterations including imprinting defects of KCNQ1OT1. Hum Mol Genet. 2001;10(26):2989–3000. doi: 10.1093/hmg/10.26.2989. [DOI] [PubMed] [Google Scholar]
- 18.Matsuoka S, et al. p57KIP2, a structurally distinct member of the p21CIP1 Cdk inhibitor family, is a candidate tumor suppressor gene. Genes Dev. 1995;9(6):650–662. doi: 10.1101/gad.9.6.650. [DOI] [PubMed] [Google Scholar]
- 19.Tsugu A, et al. Expression of p57(KIP2) potently blocks the growth of human astrocytomas and induces cell senescence. Am J Pathol. 2000;157(3):919–932. doi: 10.1016/S0002-9440(10)64605-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bourcigaux N, Gaston V, Logie A, Bertagna X, Le Bouc Y, Gicquel C. High expression of cyclin E and G1 CDK and loss of function of p57KIP2 are involved in proliferation of malignant sporadic adrenocortical tumors. J Clin Endocrinol Metab. 2000;85(1):322–330. doi: 10.1210/jcem.85.1.6303. [DOI] [PubMed] [Google Scholar]
- 21.Lam WW, et al. Analysis of germline CDKN1C (p57KIP2) mutations in familial and sporadic Beckwith-Wiedemann syndrome (BWS) provides a novel genotype-phenotype correlation. J Med Genet. 1999;36(7):518–523. [PMC free article] [PubMed] [Google Scholar]
- 22.Li M, et al. Imprinting status of 11p15 genes in Beckwith-Wiedemann syndrome patients with CDKN1C mutations. Genomics. 2001;74(3):370–376. doi: 10.1006/geno.2001.6549. [DOI] [PubMed] [Google Scholar]
- 23.Massague J, Blain SW, Lo RS. TGFβ signaling in growth control, cancer, and heritable disorders. Cell. 2000;103(2):295–309. doi: 10.1016/S0092-8674(00)00121-5. [DOI] [PubMed] [Google Scholar]
- 24.Moses HL, Serra R. Regulation of differentiation by TGF-β. Curr Opin Genet Dev. 1996;6(5):581–586. doi: 10.1016/S0959-437X(96)80087-6. [DOI] [PubMed] [Google Scholar]
- 25.Shi Y, Massague J. Mechanisms of TGF-β signaling from cell membrane to the nucleus. Cell. 2003;113(6):685–700. doi: 10.1016/S0092-8674(03)00432-X. [DOI] [PubMed] [Google Scholar]
- 26.Wu G, et al. Structural basis of Smad2 recognition by the Smad anchor for receptor activation. Science. 2000;287(5450):92–97. doi: 10.1126/science.287.5450.92. [DOI] [PubMed] [Google Scholar]
- 27.Mishra L, Marshall B. Adaptor proteins and ubiquinators in TGF-β signaling. Cytokine Growth Factor Rev. 2006;17(1–2):75–87. doi: 10.1016/j.cytogfr.2005.09.001. [DOI] [PubMed] [Google Scholar]
- 28.Tang Y, Katuri V, Dillner A, Mishra B, Deng CX, Mishra L. Disruption of transforming growth factor-β signaling in ELF β-spectrin-deficient mice. Science. 2003;299(5606):574–577. doi: 10.1126/science.1075994. [DOI] [PubMed] [Google Scholar]
- 29.Ross S, Cheung E, Petrakis TG, Howell M, Kraus WL, Hill CS. Smads orchestrate specific histone modifications and chromatin remodeling to activate transcription. EMBO J. 2006;25(19):4490–4502. doi: 10.1038/sj.emboj.7601332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stampfer MR, Garbe J, Levine G, Lichtsteiner S, Vasserot AP, Yaswen P. Expression of the telomerase catalytic subunit, hTERT, induces resistance to transforming growth factor β growth inhibition in p16INK4A(–) human mammary epithelial cells. Proc Natl Acad Sci U S A. 2001;98(8):4498–4503. doi: 10.1073/pnas.071483998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cong Y, Shay JW. Actions of human telomerase beyond telomeres. Cell Res. 2008;18(7):725–732. doi: 10.1038/cr.2008.74. [DOI] [PubMed] [Google Scholar]
- 32.Choi J, et al. TERT promotes epithelial proliferation through transcriptional control of a Myc- and Wnt-related developmental program. PLoS Genet. 2008;4(1): doi: 10.1371/journal.pgen.0040010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yao ZX, et al. Epigenetic silencing of β-spectrin, a TGF-β signaling/scaffolding protein in a human cancer stem cell disorder: Beckwith-Wiedemann syndrome. J Biol Chem. 2010;285(46):36112–36120. doi: 10.1074/jbc.M110.162347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gao J, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6(269): doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cerami E, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2(5):401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bae DS, Blazanin N, Licata M, Lee J, Glick AB. Tumor suppressor and oncogene actions of TGFβ1 occur early in skin carcinogenesis and are mediated by Smad3. Mol Carcinog. 2009;48(5):441–453. doi: 10.1002/mc.20482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Piek E, et al. Functional characterization of transforming growth factor β signaling in Smad2- and Smad3-deficient fibroblasts. J Biol Chem. 2001;276(23):19945–19953. doi: 10.1074/jbc.M102382200. [DOI] [PubMed] [Google Scholar]
- 38.Massague J, Seoane J, Wotton D. Smad transcription factors. Genes Dev. 2005;19(23):2783–2810. doi: 10.1101/gad.1350705. [DOI] [PubMed] [Google Scholar]
- 39.Plasschaert RN, et al. CTCF binding site sequence differences are associated with unique regulatory and functional trends during embryonic stem cell differentiation. Nucleic Acids Res. 2014;42(2):774–789. doi: 10.1093/nar/gkt910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nakahashi H, et al. A genome-wide map of CTCF multivalency redefines the CTCF code. Cell Rep. 2013;3(5):1678–1689. doi: 10.1016/j.celrep.2013.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen CL, et al. Reciprocal regulation by TLR4 and TGF-β in tumor-initiating stem-like cells. J Clin Invest. 2013;123(7):2832–2849. doi: 10.1172/JCI65859. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 42.Majumdar A, et al. Hepatic stem cells and transforming growth factor β in hepatocellular carcinoma. Nat Rev Gastroenterol Hepatol. 2012;9(9):530–538. doi: 10.1038/nrgastro.2012.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bhowmick NA, et al. TGF-β signaling in fibroblasts modulates the oncogenic potential of adjacent epithelia. Science. 2004;303(5659):848–851. doi: 10.1126/science.1090922. [DOI] [PubMed] [Google Scholar]
- 44.Markowitz S, et al. Inactivation of the type II TGF-β receptor in colon cancer cells with microsatellite instability. Science. 1995;268(5215):1336–1338. doi: 10.1126/science.7761852. [DOI] [PubMed] [Google Scholar]
- 45.van Hattem WA, et al. Large genomic deletions of SMAD4, BMPR1A and PTEN in juvenile polyposis. Gut. 2008;57(5):623–627. doi: 10.1136/gut.2007.142927. [DOI] [PubMed] [Google Scholar]
- 46.Hahn SA, et al. Allelotype of pancreatic adenocarcinoma using xenograft enrichment. Cancer Res. 1995;55(20):4670–4675. [PubMed] [Google Scholar]
- 47.Walczak EM, Hammer GD. Regulation of the adrenocortical stem cell niche: implications for disease. Nat Rev Endocrinol. 2015;11(1):14–28. doi: 10.1038/nrendo.2014.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Simon DP, Hammer GD. Adrenocortical stem and progenitor cells: implications for adrenocortical carcinoma. Mol Cell Endocrinol. 2012;351(1):2–11. doi: 10.1016/j.mce.2011.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Martin YC, Danaher EB, May CS, Weininger D, Van Drie JH. Strategies in drug design based on 3D-structures of ligands. Prog Clin Biol Res. 1989;291:177–181. [PubMed] [Google Scholar]
- 50.Leighton PA, Ingram RS, Eggenschwiler J, Efstratiadis A, Tilghman SM. Disruption of imprinting caused by deletion of the H19 gene region in mice. Nature. 1995;375(6526):34–39. doi: 10.1038/375034a0. [DOI] [PubMed] [Google Scholar]
- 51.Leighton PA, Saam JR, Ingram RS, Stewart CL, Tilghman SM. An enhancer deletion affects both H19 and Igf2 expression. Genes Dev. 1995;9(17):2079–2089. doi: 10.1101/gad.9.17.2079. [DOI] [PubMed] [Google Scholar]
- 52.Eggenschwiler J, Ludwig T, Fisher P, Leighton PA, Tilghman SM, Efstratiadis A. Mouse mutant embryos overexpressing IGF-II exhibit phenotypic features of the Beckwith-Wiedemann and Simpson-Golabi-Behmel syndromes. Genes Dev. 1997;11(23):3128–3142. doi: 10.1101/gad.11.23.3128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fitzpatrick GV, Soloway PD, Higgins MJ. Regional loss of imprinting and growth deficiency in mice with a targeted deletion of KvDMR1. Nat Genet. 2002;32(3):426–431. doi: 10.1038/ng988. [DOI] [PubMed] [Google Scholar]
- 54.Zhang P, et al. Altered cell differentiation and proliferation in mice lacking p57KIP2 indicates a role in Beckwith-Wiedemann syndrome. Nature. 1997;387(6629):151–158. doi: 10.1038/387151a0. [DOI] [PubMed] [Google Scholar]
- 55.Cano-Gauci DF, et al. Glypican-3-deficient mice exhibit developmental overgrowth and some of the abnormalities typical of Simpson-Golabi-Behmel syndrome. J Cell Biol. 1999;146(1):255–264. doi: 10.1083/jcb.146.1.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sharpless NE, DePinho RA. Telomeres, stem cells, senescence, and cancer. J Clin Invest. 2004;113(2):160–168. doi: 10.1172/JCI20761. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.