Abstract
The maintenance of skeletal muscle mass is critical for sustaining health; however, the mechanisms responsible for muscle loss with aging and chronic diseases, such as diabetes and obesity, are poorly understood. We found that expression of a member of the AMPK-related kinase family, the SNF1-AMPK-related kinase (SNARK, also known as NUAK2), increased with muscle cell differentiation. SNARK expression increased in skeletal muscles from young mice exposed to metabolic stress and in muscles from healthy older human subjects. The regulation of SNARK expression in muscle with differentiation and physiological stress suggests that SNARK may function in the maintenance of muscle mass. Consistent with this hypothesis, decreased endogenous SNARK expression (using siRNA) in cultured muscle cells resulted in increased apoptosis and decreased cell survival under conditions of metabolic stress. Likewise, muscle-specific transgenic animals expressing a SNARK dominant-negative inactive mutant (SDN) had increased myonuclear apoptosis and activation of apoptotic mediators in muscle. Moreover, animals expressing SDN had severe, age-accelerated muscle atrophy and increased adiposity, consistent with sarcopenic obesity. Reduced SNARK activity, in vivo and in vitro, caused downregulation of the Rho kinase signaling pathway, a key mediator of cell survival. These findings reveal a critical role for SNARK in myocyte survival and the maintenance of muscle mass with age.
Keywords: Aging, Muscle Biology
Introduction
Skeletal muscle is essential for maintaining health due to its significant mass, large contribution to whole-body metabolism, and critical role in mobility. Age-induced loss of muscle mass and function, or sarcopenia, results in loss of mobility, reduced quality of life, and is one of the strongest predictors of morbidity and mortality (1, 2). As such, sarcopenia represents a rising clinical, social, and economic burden on our increasingly aging population, with the number of people suffering from sarcopenia expected to increase 4-fold to 200 million by 2050 (3). The health toll of sarcopenia is compounded by sharply climbing rates of obesity, as these two conditions exacerbate one another (4), giving rise to the term “sarcopenic obesity” (5). In addition, many age- and obesity-related chronic diseases, such as cancer, heart disease, and diabetes, can negatively impact muscle mass; the muscle loss that accompanies these conditions leads to impaired prognosis and strongly predicts mortality (6–8). Therefore, determining the molecular mechanisms that lead to the maintenance or loss of muscle mass under conditions of aging and in response to metabolic stressors such as obesity is of high priority and has implications for the treatment of several chronic diseases.
The AMPK-related kinases are substrates of the tumor suppressor protein, LKB1. Of these, AMPK itself has been investigated extensively and is established as an important regulator of skeletal muscle metabolism and adaptation in response to stress (9, 10), whereas very little is known about the function of other AMPK-related kinases in muscle. The SNF1-AMPK-related kinase, SNARK (also known as NUAK2), has been implicated in the regulation of critical cellular processes, including metabolism (11) and cell motility (12), and its expression is regulated by disease states in humans, such as cancer (13) and obesity (14). Homozygous deletion of SNARK in mice is lethal at an embryonic stage, suggesting an essential role for SNARK in development (15). In contrast to AMPK, the specific functions of SNARK in skeletal muscle remain largely unknown. Here, using both in vitro and in vivo models, we establish that SNARK plays a protective, antiapoptotic role in myocyte survival in response to physiological stressors by suppressing cell death pathways. These studies reveal what we believe to be a novel role for SNARK in the maintenance of muscle mass with age and obesity.
Results
SNARK increases with metabolic stress and regulates myocyte survival in vitro.
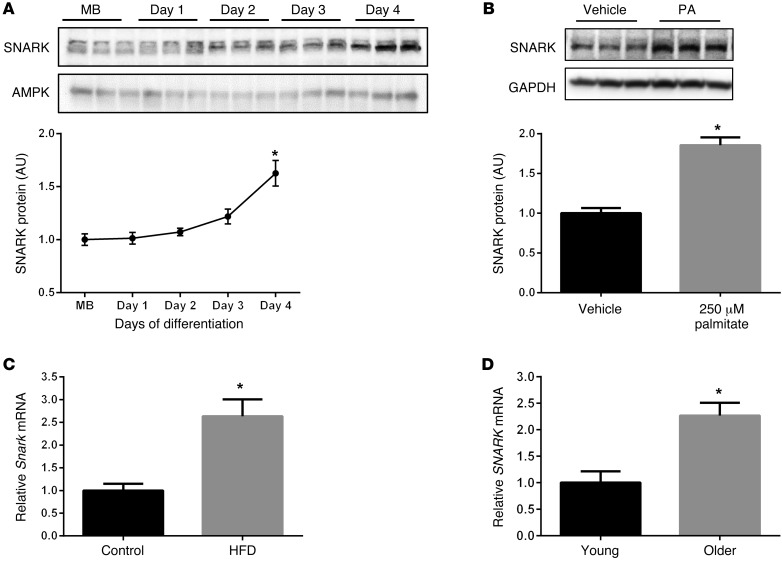
Little is known about SNARK regulation during muscle development or under conditions of metabolic and physiological stress. In C2C12 muscle culture, SNARK protein levels increased by approximately 60% in fully differentiated myotubes (day 4 of differentiation) compared with myoblasts (Figure 1A). Treatment of cultured myotubes with the mild metabolic stressor palmitate (250 μM) for 48 hours induced a 2-fold increase in SNARK protein, demonstrating that SNARK is regulated by metabolic stress in vitro (Figure 1B). Consistent with in vitro data, Snark expression increased in mouse skeletal muscles following consumption of a high-fat diet (58% fat/kcal) for 12 weeks (Figure 1C). SNARK expression was also increased in human skeletal muscle from healthy older (78 ± 5 yr) male subjects compared with that in younger (22 ± 1 yr) male subjects (Figure 1D). These data demonstrate that SNARK expression is increased with muscle differentiation and in response to metabolic stress and aging, raising the possibility that SNARK functions in the preservation of skeletal muscle mass.
Figure 1. SNARK expression in skeletal muscle.
(A) SNARK protein was measured in C2C12 myoblasts (MB) and following 1 to 4 days of exposure to differentiation media (2% horse serum). AMPK is shown as a loading control. (B) Four-day differentiated C2C12 myotubes were exposed to 250 μM palmitate (PA) or vehicle (ethanol in 2% BSA) for 48 hours, and SNARK protein was measured. GAPDH is shown as a loading control. Average results (n = 3) of 3 experimental replicates are shown for A and B. (C) Skeletal muscle Snark expression was measured in C57BL/6 mice that were fed chow (control) or treated with a high-fat diet (HFD) for 12 weeks (n = 8 per group). (D) Skeletal muscle SNARK expression (relative to GAPDH housekeeping gene) was measured in young (age 22 ± 1 yr; n = 8) and older (age 78 ± 5 yr; n = 7) men. *P < 0.05 determined by 1-way ANOVA with Bonferroni post-hoc test (A) and t tests (B–D). Error bars indicate mean ± SEM.
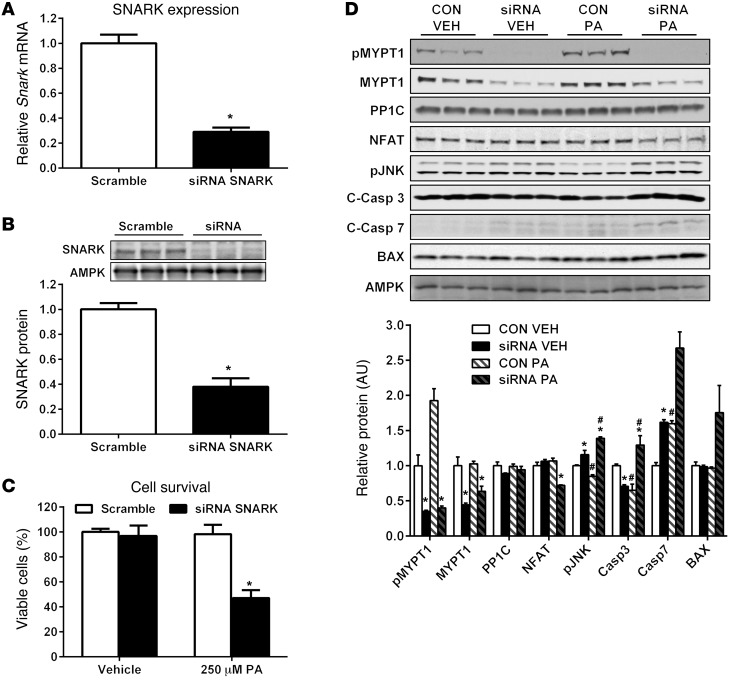
To test the hypothesis that SNARK plays a role in the response of myocytes to stress, SNARK expression was decreased in C2C12 myoblasts using siRNA and cell survival was measured in response to palmitate treatment. There was a 70% reduction of Snark mRNA and a 62% reduction of SNARK protein in siRNA-treated cells compared with that in cells treated with scrambled siRNA (Figure 2, A and B). Cell survival was not compromised in SNARK-knockdown cells following 24 hours of vehicle treatment. In contrast, 24 hours of palmitate exposure decreased cell number by 53% in SNARK-knockdown cells, while cells treated with scrambled siRNA did not undergo palmitate-induced apoptosis (Figure 2C). Thus, our data clearly demonstrate what we believe to be a novel role for SNARK in the survival of myocytes in response to metabolic stress in vitro.
Figure 2. SNARK protects against palmitate-induced apoptosis in myocytes.
C2C12 myoblasts were treated with scrambled or SNARK siRNA to generate SNARK-knockdown cells. (A) Snark mRNA (relative to B2m housekeeping gene) (control: n = 5 wells, siRNA: n = 6 wells) and (B) protein levels were measured, with AMPK shown as a loading control. Average protein levels were quantified from n = 6 wells per condition. Following 24 hours of transfection with siRNA, scrambled (CON) or SNARK siRNA (siRNA) cells were treated with vehicle (VEH; ethanol in 2% BSA) or 250 μM palmitate for 24 hours. (C) Viable cells in each treatment group were quantified using the MTT proliferation assay. The average results from 3 independent experiments (n = 6 wells per condition per experiment) are shown. (D) Western blotting was performed on cell lysates to analyze MYPT1/Rho kinase and apoptotic signaling. AMPK is shown as a loading control. The average protein levels were quantified from 3 wells per group and are representative of 3 independent experiments. C-Casp 3, cleaved caspase-3. *P < 0.05 vs. CON; #P < 0.05 vs. VEH of same treatment group determined by t tests (A and B) and 2-way ANOVA with Bonferroni post-hoc testing (C and D). Error bars indicate mean ± SEM.
To determine the mechanism by which SNARK regulates survival in muscle cells, we investigated RhoA signaling, which has been shown to be critical for survival in multiple cell types (16). SNARK’s only identified substrate to date, MYPT1, is a key mediator of RhoA signaling in response to myosin remodeling, and MYPT1 is also a substrate of the Rho kinase (ref. 17). In addition, SNARK has been shown to bind directly to the RhoA regulator, myosin phosphatase Rho-interacting protein (18), indicating that SNARK could interact with RhoA signaling at multiple levels. The specific phosphorylation site for SNARK on MYPT1 has not yet been identified (19), therefore we examined phosphorylation of MYPT1 on the Rho kinase site T696 in C2C12 myocytes. SNARK knockdown was associated with a severe blunting of MYPT1 phosphorylation at T696 (Figure 2D). Interestingly, MYPT1 phosphorylation was increased by 2-fold in control cells in response to palmitate exposure, but this increase was abolished in SNARK-knockdown cells, suggesting that Rho kinase signaling through MYPT1 is involved in the protection of myocytes from metabolic stress–induced apoptosis. Protein levels of the MYPT1-binding partner, protein phosphatase 1 (PP1C), were not altered by SNARK knockdown or palmitate exposure. Increased stress-induced apoptosis and blunted Rho kinase signaling in SNARK-knockdown cells were associated with reduced protein expression of the RhoA target NFAT, which is an important regulator of hypertrophy and survival in cardiac and skeletal muscle (refs. 20, 21, and Figure 2D). In addition, increased apoptosis in SNARK-knockdown cells was accompanied by hyperactivation of the apoptotic regulator, JNK, and cleavage (activation) of the cell death proteases caspase-3 and caspase-7 (Figure 2D).
Generation of muscle-specific SNARK transgenic mice.
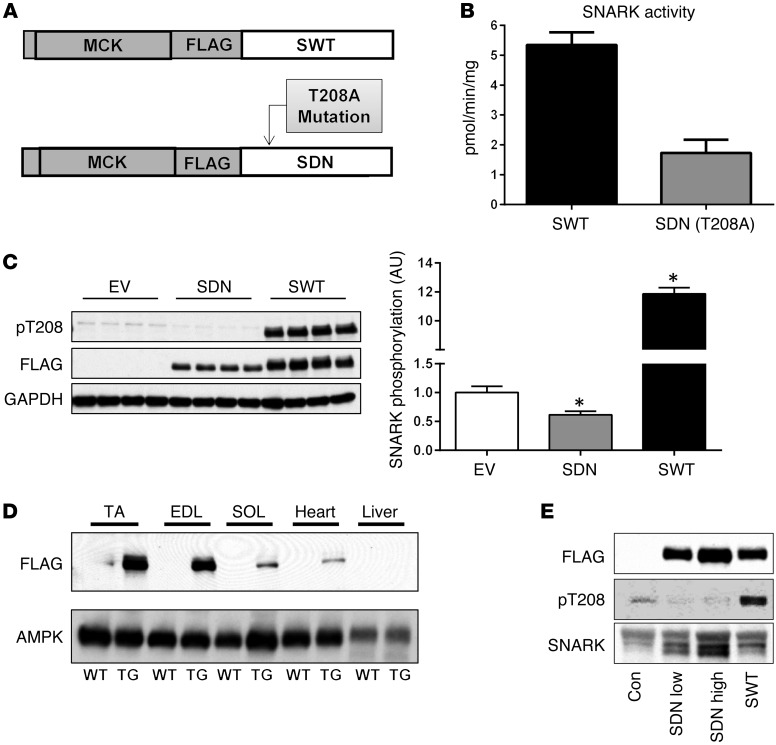
Based on our finding that SNARK expression is modulated during muscle development and in response to metabolic stress and aging (Figure 1) and our data demonstrating a role for SNARK in myocyte survival (Figure 2), we aimed to determine the effects of SNARK activity on muscle structure and function in vivo. Therefore, we generated lines of muscle-specific SNARK transgenic mice that overexpress either SNARK (SWT mice) or a SNARK dominant-negative (kinase-inactive) mutant (SDN mice) under the control of the muscle creatine kinase (MCK) promoter. The SDN construct was generated by mutating the LKB1 activation site on human SNARK from Thr to Ala (Figure 3A), generating a full-length SNARK protein with greatly reduced phosphotransferase activity against the AMARA substrate peptide (Figure 3B). Overexpression of the SDN construct decreased endogenous SNARK phosphorylation at its activating site, demonstrating its ability to reduce cellular SNARK activity (Figure 3C). Western blotting of FLAG expression in multiple tissues revealed transgene expression in various skeletal muscles and a small amount of expression in the heart, as expected using the MCK promoter (Figure 3D). FLAG immunohistochemistry revealed a perinuclear or nuclear localization of transgenes in the skeletal muscles of SWT and SDN mice (Supplemental Figure 1; supplemental material available online with this article; doi:10.1172/JCI79197DS1). Heart morphology and function were examined and found to be normal in SNARK transgenic mice (Supplemental Figure 1). There was no FLAG expression in nonmuscle tissues, including the liver (Figure 3D), demonstrating muscle-specific expression of SNARK transgenes. Two lines of SDN mice generated from independent founders that expressed lower levels (SDN-Low mice) and higher levels (SDN-High mice) of dominant-negative SNARK in muscle were studied to determine the “dose-response” effect of reduced SNARK activity in muscle, while data from two independent lines of mice with similar levels of wild-type SNARK overexpression were pooled (SWT mice) to determine the effect of increased SNARK activity in muscle (Figure 3E).
Figure 3. Generation of muscle-specific SNARK transgenic mice.
(A) Transgenic mice with muscle-specific overexpression of FLAG-tagged wild-type SNARK (SWT) or a dominant-negative inactive SNARK mutant (T208A; SDN) were generated using the MCK promoter. (B) Phosphotransferase activity of SWT and SDN constructs against the AMARA peptide was assessed by immunoprecipitation of FLAG from 293 HEK overexpression lysates using 32P-γ-ATP. n = 5 per group. (C) The dominant-negative nature of the SNARK dominant-negative construct was verified by measuring endogenous SNARK phosphorylation at its activating site T208 in 293 HEK cells transfected with PCDNA3.1 empty vector (EV), SNARK dominant-negative mutant (SDN), or wild-type human SNARK (SWT). n = 4 per group. (D) A FLAG immunoblot demonstrating the expression of SNARK transgene in the TA, extensor digitorum longus (EDL), soleus (SOL), and heart muscles compared with wild-type littermates, and the absence of expression in livers of SNARK transgenic mice. AMPK is shown as a loading control. (E) Relative expression levels of transgene and endogenous protein in the muscles of SNARK transgenic mice and littermate controls using FLAG, phospho-SNARK T208, and total SNARK antibodies. Two lines of transgenic mice overexpressing different levels of dominant-negative SNARK (SDN-Low and SDN-High) were studied, and data from two independent lines of SWT mice with similar expression levels were pooled for analysis. *P < 0.05 vs. control determined by 1-way ANOVA and Bonferroni post-hoc testing. Error bars indicate mean ± SEM.
SNARK-inactive transgenic mice have age- and dose-dependent muscle atrophy.
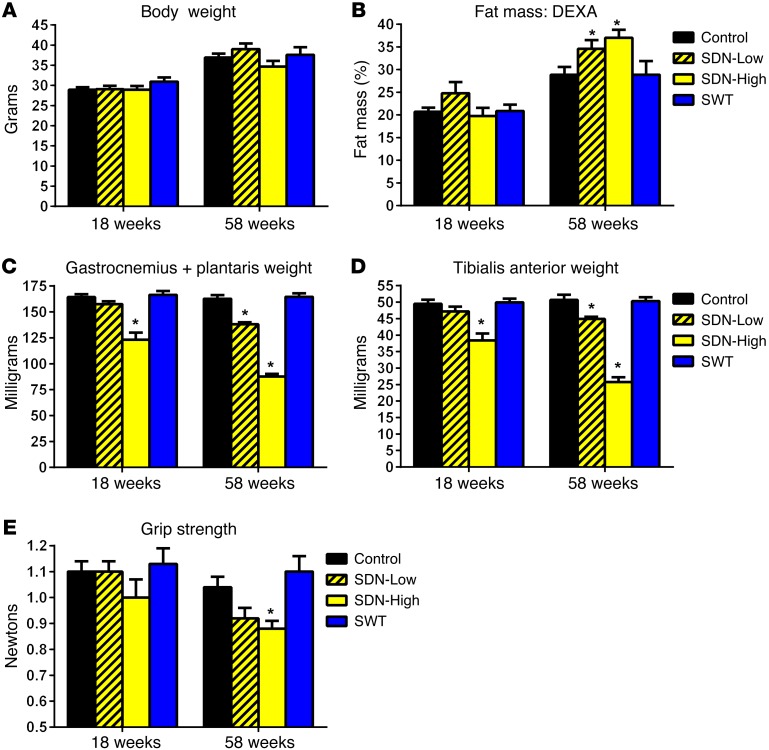
SNARK transgenic mice were studied at approximately 18 and 58 weeks of age to determine the interactive effects of SNARK activity and age on muscle phenotype. Body weights were similar among SNARK transgenic mice and littermate controls in both age groups (Figure 4A), and the percentage of fat mass increased in all groups with age (Figure 4B). However, the percentage of fat mass was 6% and 8% higher, respectively, in SDN-Low and SDN-High mice at 58 weeks of age (Figure 4B), demonstrating an exacerbation of age-dependent increases in adiposity in SDN mice. In addition, we observed a dramatic loss of muscle mass in SDN mice that was dependent on the expression level of dominant-negative SNARK (low vs. high) and age (18 week vs. 58 week). SDN-Low mice had an approximately 15% and SDN-High mice had an approximately 50% reduction in the masses of the gastrocnemius/plantaris and tibialis anterior (TA) muscle complexes by 58 weeks of age (Figure 4, C and D). No age-induced muscle atrophy was observed in SWT mice or control littermates by 58 weeks of age, indicating that mutation of the LKB1-activating site was responsible for muscle atrophy in SDN mice, rather than SNARK overexpression per se. Reduced muscle mass in SDN mice was accompanied by loss of forelimb grip strength at 58 weeks of age, whereas grip strength was maintained in SWT mice and littermate controls (Figure 4E). Our data demonstrate that SDN mice undergo severe, early-onset muscle atrophy, combined with age-induced increases in adiposity, or sarcopenic obesity. Thus, we have identified what we believe to be a novel role for SNARK in the maintenance of muscle mass with age.
Figure 4. Age- and dose-dependent loss of muscle mass in SDN mice.
(A) Body weight and (B) body composition were assessed in young adult (18-week-old) and older (58-week-old) SNARK transgenic mice and wild-type littermates (Control). DEXA, dual-energy X-ray absorptiometry. (C) The gastrocnemius/plantaris and (D) TA muscle complexes were weighed, and (E) forelimb grip strength was measured. For all panels, n = 20 for control, n = 13 for SDN-Low, n = 14 for SDN-High, n = 15 for SWT. *P < 0.05 vs. control of the same age group determined by 1-way ANOVA and Bonferroni post-hoc testing. Error bars indicate mean ± SEM.
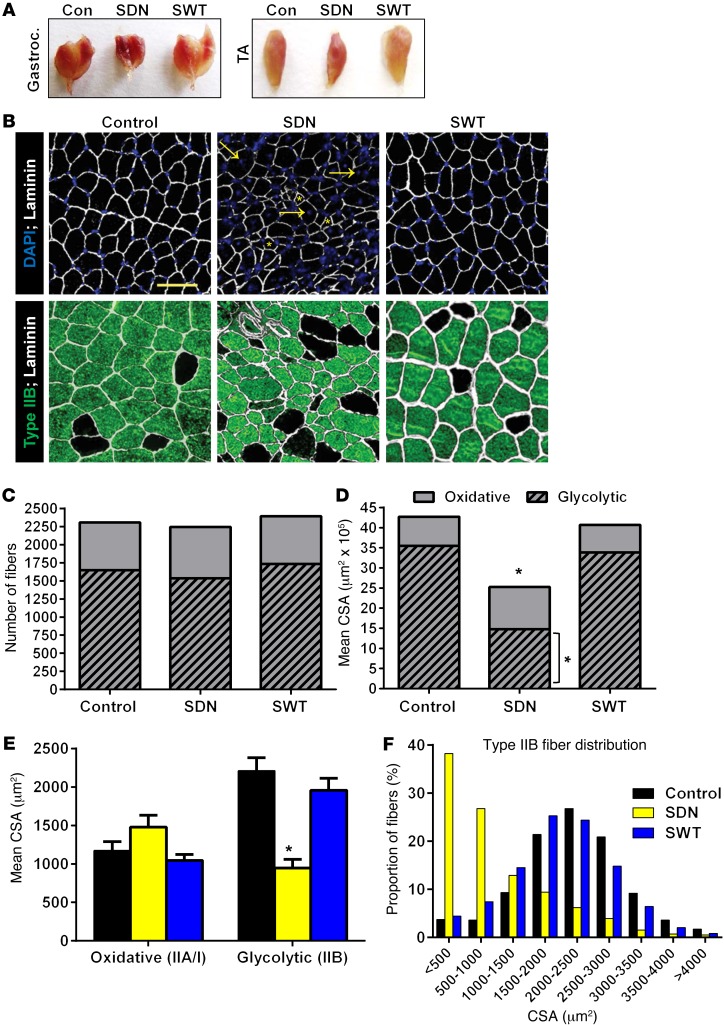
SNARK-inactive transgenic mice have atrophy of type IIB fibers.
To investigate the loss of muscle mass in SDN mice and the effect of SNARK activity on muscle phenotype, we performed a detailed analysis of muscle morphology in SDN and SWT mice and littermate controls at 58 weeks of age. Comprehensive histological analysis of SDN mice was limited to the SDN-High line, which had a similar, but more pronounced, muscle-wasting phenotype compared with that of the SDN-Low line. Gross morphology of the gastrocnemius and TA muscles demonstrated a marked reduction in muscle size and a reddish appearance of muscles in SDN mice compared with those in SWT mice and littermate controls (Figure 5A). Immunofluorescent analysis of myofiber morphology of TA muscles revealed small, atrophic fibers combined with regenerating fibers containing centralized nuclei in SDN mice, with no detectable centralized nuclei in sections from SWT or control mice (Figure 5B). Quantification of the number of glycolytic (type IIB) and oxidative (type IIA/I) muscle fibers in TA cross sections revealed that the total number of fibers and the relative number of oxidative and glycolytic fibers were similar among all genotypes (Figure 5C). However, consistent with reduced muscle weight (Figure 4D), total muscle cross-sectional area (CSA) was reduced by 41% in SDN mice, compared with that in littermate controls (Figure 5D). Decreased muscle CSA in SDN mice was due to a decrease in the relative area of glycolytic, but not oxidative, fibers (Figure 5D), with glycolytic fibers in SDN mice having a 57% lower mean CSA compared with that in controls (Figure 5E). A striking change in glycolytic fiber size distribution was also observed in SDN mice, with 38% of fibers having a CSA of less than 500 μm2 in SDN mice compared with less than 5% of fibers in SWT mice and controls (Figure 5F). These data demonstrate that muscle loss in SDN mice is a result of type IIB/glycolytic muscle fiber atrophy. The reason for type IIB–specific fiber atrophy in SDN mice is unknown; however, the specific atrophy of glycolytic fibers is also the primary cause of sarcopenia in aging humans (22, 23).
Figure 5. Muscle morphology in SNARK transgenic mice.
(A) Images of gastrocnemius and TA muscles taken from 58-week-old SDN mice, SWT mice, and wild-type littermates. (B) Cross sections (6 μm) were taken from TA muscles, muscle sections were stained with anti-laminin (white), and nuclei were visualized with DAPI (blue). Glycolytic fibers were stained with anti–myosin heavy chain IIB (green). Compared with those of wild-type and SWT mice, SDN muscles displayed small, atrophic fibers (yellow, asterisks) and large fibers with centralized nuclei (yellow, arrows) indicating muscle regeneration. Original magnification, ×200; scale bar: 50 μm. (C) The number of glycolytic (type IIB) and oxidative (unstained) fibers were counted for each section, and (D) the CSA of each fiber was measured. (E) Mean CSA was calculated for oxidative and glycolytic fibers from each genotype, and (F) the size distribution of glycolytic fibers was plotted. *P < 0.05 vs. control determined by 1-way ANOVA and Bonferroni post-hoc testing. Data for all panels were quantified from sections taken from n = 5 animals per group. Error bars indicate mean ± SEM.
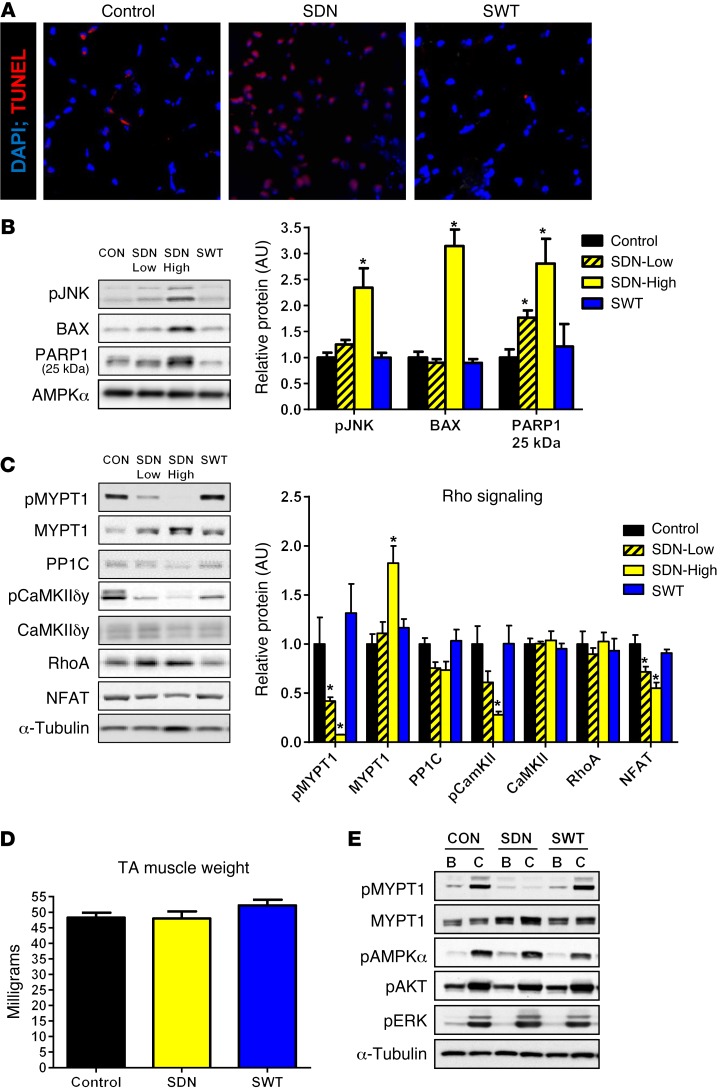
SNARK-inactive transgenic mice have increased apoptotic signaling and impaired Rho kinase signaling.
To determine the mechanisms that may lead to muscle atrophy in SDN mice, we measured several known mediators of protein synthesis and degradation at the protein (Supplemental Figure 2) and mRNA levels (Supplemental Figure 3). Signaling through the BMP pathway, which is a positive regulator of muscle mass (24), tended to be increased in SDN mice, demonstrating that impaired BMP signaling was not responsible for muscle atrophy in SDN mice (Supplemental Figure 2). In addition, expression levels of the key ubiquitin ligases, FBXO32 and TRIM63, were lower in SDN mice at 52 weeks of age (Supplemental Figure 3). However, the expression of the ubiquitin ligases TRAF6 and MUSA1, which have been implicated in muscle atrophy (25–27), were elevated in 8-week-old SDN-High animals (Supplemental Figure 4). Elevated ubiquitin ligase expression in young (8-week-old) SDN-High animals was accompanied by increased global protein ubiquitination (Supplemental Figure 4) and occurred prior to the onset of severe muscle atrophy in SDN mice, indicating that increased protein degradation and activation of the ubiquitin proteasome system may be early defects that initiate muscle loss in SDN mice. An imbalance of catabolic/anabolic signaling (28), leading to increased myonuclear apoptosis, is postulated to be responsible for pathological muscle atrophy in several disease states and with aging (29). Therefore, we measured several markers of catabolism and cellular apoptosis to determine whether dysregulation of survival signaling may be a mechanism for muscle atrophy in SDN mice. Diffuse TUNEL staining was visible in muscle cross sections from SDN mice, while very few TUNEL-positive nuclei were apparent in the muscles of control and SWT animals, indicating increased apoptosis in SDN mice (Figure 6A). Basal activation of the apoptosis-inducing kinase JNK was elevated 2.3-fold and protein levels of the proapoptotic BCL-2 family member BAX were elevated 3-fold in SDN-High mice compared with those in controls (Figure 6B). Consistent with enhanced apoptotic signaling in SDN mice, muscle levels of cleaved (25-kDa) PARP1 — a hallmark of cellular apoptosis (30) — were 1.8- and 2.8-fold higher in SDN-Low and SDN-High mice compared with those in littermate controls (Figure 6B). Proteins that mediate autophagy during muscle atrophy, beclin-1 and autophagocytosis associated protein (ATG3 and ATG12), were increased in the muscles of SDN-High mice (Supplemental Figure 2). Thus, muscle atrophy in 52-week-old SDN mice was accompanied by upregulation of catabolic signaling and enhanced myonuclear apoptosis, identifying SNARK as a regulator of catabolic/apoptotic signaling in skeletal muscle.
Figure 6. SNARK regulates muscle apoptosis and Rho kinase signaling.
(A) Apoptosis was visualized in TA cross sections from 58-week-old SDN mice, SWT mice, and wild-type littermates using TUNEL staining (red) to detect apoptotic nuclei. DAPI (blue) was used as a control nuclear stain. Original magnification, ×200. Data are representative of n = 4 animals per group. (B) Protein levels of key mediators of cellular apoptosis and (C) Rho kinase signaling were measured in muscles of SDN and SWT mice as well as wild-type littermates at 58 weeks of age. n = 5–6 per group. (D) In situ contraction experiments were performed on SDN, SWT, and control mice at 15 weeks of age, prior to the onset of muscle atrophy in SDN. Muscle weights were recorded. n = 3 per group. (E) Western blotting for contraction-regulated phospho-proteins was performed on lysates generated from basal (B) and contracted (C) legs (shown in adjacent lanes) from each animal. AMPKα and α-tubulin blotting are included as loading controls. pJNK, phosphorylated JNK; BAX, Bcl-2–associated X protein; PARP1 25 kDa, cleaved poly(ADP-ribose) polymerase 25-kDa fragment; AMPK, AMP-activated protein kinase; MYPT, myosin phosphatase-targeting subunit; CaMKII, Ca2+/calmodulin-activated protein kinase; NFAT, nuclear factor of activated T cells; AKT, protein kinase B. *P < 0.05 vs. control determined by 1-way ANOVA and Bonferroni post-hoc testing. Error bars indicate mean ± SEM.
Consistent with our in vitro data implicating altered Rho kinase signaling as a mechanism for increased myocellular apoptosis with loss of SNARK function, basal phosphorylation of MYPT1 at T696 was 60% and 90% lower in SDN-Low and SDN-High mice, respectively, compared with littermate controls (Figure 6C). Blunted MYPT1 phosphorylation occurred, despite an 80% increase in MYPT1 protein in SDN-High mice, suggesting that dominant-negative SNARK can inhibit Rho kinase signaling in response to MYPT1. Protein levels of the MYPT1-binding partner, PP1C, tended (P = 0.1) to be decreased in SDN-Low and SDN-High mice (Figure 6C), demonstrating further dysregulation of the myosin phosphatase complex by dominant-negative SNARK. Blunted signaling through the Rho kinase pathway in SDN mice is further supported by our observation of reduced activation (T286 phosphorylation) of the known RhoA activator, CaMKII, and reduced protein expression of the CaMKII/RhoA target NFAT (Figure 6C).
We hypothesized that severely blunted Rho kinase signaling through the SNARK substrate MYPT1 may contribute to increased apoptosis and muscle atrophy in 58-week-old SDN animals. To determine whether impaired Rho kinase signaling precedes loss of muscle mass in SDN mice, we measured contraction-stimulated MYPT1 phosphorylation in TA muscles of young (15-week-old) SNARK transgenic animals, prior to the onset of muscle atrophy (Figure 6D). MYPT1 phosphorylation was increased by muscle contraction in control and SWT mice, representing what we believe to be a novel contraction-regulated phosphorylation site and suggesting that myosin phosphatase may be involved in the response of skeletal muscle to exercise. However, consistent with data from older (58-week-old) animals, MYPT1 T696 phosphorylation was blunted in SDN mice (Figure 6E). In contrast, other exercise-activated proteins (AMPK, AKT, and ERK) were robustly and normally phosphorylated by contraction in SDN mice, demonstrating that impaired MYPT1 phosphorylation at T696 is a specific and primary defect in SDN mice that occurs prior to the onset of muscle atrophy. Thus, our data demonstrate that loss of SNARK expression (in vitro) or function (in vivo) results in altered Rho kinase signaling and enhanced stress-induced myocellular apoptosis that results in severe muscle atrophy in vivo.
Discussion
The combination of increased fat mass and reduced muscle mass that occurs with aging, known as sarcopenic obesity, is becoming increasingly common and imparts a higher risk for morbidity and mortality than either condition alone. Furthermore, loss of muscle mass is a pathological condition that coincides with obesity-related diseases, including diabetes and cancer, and contributes to impaired prognosis and increased mortality. Given the scope of the population that may be affected by muscle loss that accompanies physiological stress (obesity, aging) or chronic disease states, determining the mechanisms that contribute to the maintenance or loss of muscle mass is an important step in identifying potential therapeutic interventions. Using SNARK muscle–specific transgenic animals, we identified a role for SNARK in the maintenance of muscle mass with age and found that age-induced muscle loss in SDN mice coincided with accelerated age-induced adiposity, or sarcopenic obesity. Data from two independent mouse lines demonstrated that the severity of muscle loss in SDN mice was dependent on the amount of dominant-negative SNARK expressed in muscle, further strengthening the conclusion that SNARK is a critical regulator of muscle mass with age. Moreover, in an independent set of experiments using SNARK knockdown in cultured muscles, SNARK was shown to play an antiapoptotic role in myocytes following exposure to metabolic stress.
Remarkably, severe (50%) muscle atrophy in SDN mice occurred with the relatively mild physiological stress of aging to 1 year under chow diet conditions (Figure 4, C and D). Muscle loss does not typically occur in mice consuming a chow diet until greater than 2 years of age (31), a contention supported by the absence of age-induced muscle loss in control and SWT animals at 1 year. The precise mechanisms that result in age-induced loss of muscle mass are not known; however, there is evidence that dysregulated lipid metabolism, inflammation, or increased adiposity may be primary contributors (32–34). Aging to 1 year induced an increase in adiposity that may have contributed to age-induced muscle loss in SDN mice. However, muscle atrophy in SDN mice occurred with the relatively moderate increase in fat mass induced by aging and did not require supraphysiological experimental conditions designed to rapidly increase adiposity and induce large disruptions in lipid homeostasis (e.g., 60% high-fat diet or ob/ob background). These data demonstrate that SNARK may be necessary for protection of muscle mass with exposure to daily physiological and metabolic stressors, thus indicating the physiological relevance of these findings. This contention is supported by in vitro data demonstrating that a low dose (250 μM) of palmitate induced a 50% decrease in SNARK-knockdown cell number (Figure 2), while much larger doses of palmitate (>750 μM) are required to induce significant apoptosis in wild-type cells within 24 hours (35). SNARK expression was increased in response to muscle differentiation and palmitate treatment in cultured myocytes, high-fat feeding in mice, and aging in humans (Figure 1), thereby supporting a protective role for SNARK under conditions of physiological stress and in line with its classification as a stress-activated kinase (36, 37). This hypothesis is also supported by data in humans demonstrating an increase in skeletal muscle SNARK expression with obesity and in response to treatment with palmitate or TNF-α in human primary muscle culture (14). Future work should examine whether individuals prone to stress-induced muscle wasting have lower SNARK expression or activity in muscle and whether restoration of SNARK activity can prevent the muscle loss associated with metabolic stress or aging.
Increased myonuclear apoptosis in response to stressors, such as inflammation and metabolic dysfunction, is thought to contribute to the muscle loss that occurs with aging and disease (29). Loss of SNARK activation using a dominant-negative construct in vivo, or via reduction of SNARK expression in vitro, resulted in greatly increased apoptosis in response to physiological stressors (aging or palmitate exposure). In contrast, SNARK overexpression in vivo did not result in a change of muscle phenotype compared to littermate controls, indicating that endogenous SNARK was sufficient to protect against muscle apoptosis under our experimental conditions. Protection against stress-induced apoptosis in muscle represents a positive role for SNARK in the maintenance of health; however, the antiapoptotic role of SNARK may have negative implications in other cell types. For example, data in humans indicate that high SNARK expression is associated with impaired prognosis and survival of patients suffering from melanoma, while knockdown of SNARK expression can suppress melanoma tumor growth in mice (13, 38). Furthermore, SNARK was found to be upregulated by TNF-α and protective against death receptor CD95-mediated cell death in apoptosis-resistant tumor cells (39). In those experiments (39), knockdown of SNARK or overexpression of a kinase-dead SNARK mutant increased inflammation-mediated apoptosis in ACHN tumor cells, while SNARK overexpression increased cell survival. Thus, data from studies of cancer cell growth support a prosurvival role for SNARK. In contrast, one study has identified SNARK as having a proapoptotic role, concluding that SNARK heterozygous knockout mice were more susceptible to chemically induced colorectal tumorigenesis (15). These conflicting data indicate that the role of SNARK in cell survival may be tissue and context dependent.
As a Ser/Thr protein kinase, it is likely that SNARK exerts its effect on myocyte survival through phosphorylation of substrate proteins. To date, the only identified substrate of SNARK is MYPT1, which is a regulatory subunit of the myosin phosphatase complex. Functionally, SNARK has been shown to regulate stress fiber formation (18) and cytoskeletal organization (40), both of which rely on myosin phosphatase regulation. Recent data show that, in addition to SNARK, the closely related AMPK-related kinase NUAK1 also phosphorylates MYPT1 (41). Interestingly, AMPK itself does not phosphorylate MYPT1 (41), indicating a distinct role for SNARK and NUAK1 in the regulation of cytoskeletal dynamics via the myosin phosphatase complex. NUAK1 regulates cell adhesion and cell cycle progression, with NUAK1 inhibitors acting to suppress cell proliferation, indicating that both SNARK and NUAK1 may be important regulators of cell survival (42). Rho kinase signaling has been identified as an important mediator of cell contractility, stress fiber formation, and survival signaling in many cell types, in part through its ability to regulate the myosin phosphatase complex via inhibitory phosphorylation of MYPT1 at T696 (16, 17). Increased myonuclear apoptosis in SDN mice and in SNARK-knockdown cells occurred in conjunction with severe blunting of MYPT1 phosphorylation at the Rho kinase site, T696. Thus, our data and data from other laboratories (18, 19, 42) demonstrate biochemical and functional interaction between the Rho kinase and SNARK signaling pathways, providing a potential mechanism by which SNARK exerts its prosurvival role in skeletal muscle. Indeed, exploitation of the strong antiapoptotic role of Rho kinase in many cell types has led to multiple successful preclinical trials of Rho kinase inhibitors as effective treatments in reducing tumor growth and metastasis (43). Our data suggest that SNARK is a positive regulator of Rho kinase signaling and provide evidence of a prosurvival role for SNARK/Rho kinase signaling in skeletal muscle.
The AMPK-related kinase family and AMPK in particular have been implicated in the etiology and treatment of multiple disease states and as mediators of numerous critical cellular processes. We now identify what we believe to be a novel physiological function for SNARK — a member of this family that has been considerably less well studied. Specifically, our investigation has identified a previously undiscovered role for SNARK as a regulator of skeletal muscle mass in vivo. In addition, we provide evidence that SNARK plays a protective role in myocyte survival under conditions of metabolic and physiological stress. Since increased stress-induced myonuclear apoptosis may be a contributor to muscle loss that occurs with several conditions and disease states, these data have broad implications for determining the etiology of muscle wasting and potential treatments for maintaining muscle mass.
Methods
Generation of SNARK transgenic mice.
Dominant-negative SNARK was generated by performing mutagenesis (T208A) on a full-length human SNARK cDNA template as previously described (11). A FLAG (DYKDDDDK) sequence was added to the N-termini of both wild-type and dominant-negative SNARK constructs using PCR, and the product was subcloned into a vector containing the MCK promoter. MCK-FLAG-SNARK constructs were linearized and purified, and mice were generated by pronuclear injection into fertilized oocytes from the C57BL/6 background. Founder mice overexpressing wild-type SNARK (SWT mice) or dominant-negative SNARK (SDN mice) were identified using PCR, and mouse lines were established by breeding transgenic founders to wild-type C57BL/6 mice mice. Two lines of SDN mice generated from independent founders that expressed lower levels (SDN-Low mice) and higher levels (SDN-High mice) of dominant-negative SNARK in muscle were studied to determine the “dose-response” effect of reduced SNARK activity in muscle; while data from two independent lines of mice with similar levels of wild-type SNARK overexpression were pooled (SWT mice) to determine the effect of increased SNARK activity in muscle. Young adult male (~18-week-old) and older (~58-week-old) male mice were studied to determine genotype-aging interactions. Body composition was measured in anesthetized mice using a Lunar PIXImus2 mouse densitometer.
Western blotting.
Skeletal muscles and tissues were rapidly dissected, frozen in liquid N2, and homogenized using a TissueLyser (Qiagen) in a modified RIPA buffer containing 50 mM Tris-HCl (pH 7.5), 1 mM EDTA, 1 mM EGTA, 10% (v/v) glycerol, 1% (v/v) NP-40, 50 mM NaCl, 0.5% sodium deoxycholate, 50 mM NaF, 5 mM sodium pyrophosphate, 1 mM DTT, and protease inhibitor cocktail (Roche). For measurement of SNARK transgene tissue distribution, individual muscles or tissue aliquots were homogenized from 10-week-old SNARK transgenic mice or littermate controls. For measurement of catabolic and apoptotic markers, the TA, extensor digitorum longus, plantaris, and gastrocnemius muscles from 58-week-old mice were pulverized under liquid nitrogen, and an aliquot of mixed muscle was homogenized to ensure a representative sample of skeletal muscle was used for analysis. The protein content of muscle and tissue lysates was analyzed using a Bradford Assay (Bio-Rad), and samples containing equal amounts of protein were heated to 95°C for 5 minutes in Laemmli buffer. Samples were run on Criterion TGX 4-15% gradient gels (Bio-Rad) and transferred to nitrocellulose membranes. Membranes were blocked in 5% non-fat dry milk for 1 hour at room temperature and exposed to primary antibodies overnight at 4°C, followed by incubation with appropriate HRP-conjugated secondary antibodies and visualization on film with ECL. SNARK was detected using anti-SNARK sera provided by Cell Signaling Technology or anti-NUAK2 antibody (Abcam; ab107287). The following antibodies were used for the detection of phosphorylated and total protein levels: pMYPT1 T696 (Cell Signaling Technology 5163), MYPT1 (Cell Signaling Technology 8574), PP1C (Santa Cruz Biotechnology 7482), pJNK (Promega V7931), Caspase-3 (Cell Signaling Technology 9664), α-Tubulin (Cell Signaling Technology 2125), Caspase-7 (Cell Signaling Technology 8438), BAX (Cell Signaling Technology 2772), FLAG (Sigma-Aldrich F7425), PARP1 (Santa Cruz Biotechnology 56197), pCaMKII (Cell Signaling Technology 3362), CaMKII (Cell Signaling Technology 3361), NFAT (Santa Cruz Biotechnology 13033), RhoA (Cell Signaling Technology 2117), Beclin (Cell Signaling Technology 3495), ATG3 (Cell Signaling Technology 3415), ATG12 (Cell Signaling Technology 4180), LC3A/B (Cell Signaling Technology 4599/3868), P62 (Cell Signaling Technology 5114), pSMAD1/5 (Cell Signaling Technology 9516), SMAD1 (Cell Signaling Technology 9743), SMAD3 (Cell Signaling Technology 5339), SMAD3 (Cell Signaling Technology 9523), GAPDH (Cell Signaling Technology 2118), pAKT (Cell Signaling Technology 9271), pERK (Cell Signaling Technology 4370), pAMPK (Cell Signaling Technology 2531), and AMPKα (Cell Signaling Technology 5831).
Real-time PCR.
A portion of the TA muscle was collected from 52-week-old SNARK transgenic mice and littermate controls and stored for 24 hours in RNALater (Qiagen) at 4°C. RNA was extracted using Qiazol reagent (Qiagen) and purified using RNeasy columns (Qiagen). RNA samples underwent reverse transcription, and mRNA levels were measured using a Myogenesis and Myopathy PCR Array (Bio-Rad).
Human skeletal muscle samples.
Biopsies were taken from the vastus lateralis muscles of healthy young (age 22 ± 1 yr) and older (age 78 ± 5 yr) men at baseline after an overnight fast and stored in RNAlater (Ambion) for later analysis. Subject characteristics have been previously reported (33, 44).
In situ contraction.
In situ muscle contraction was performed as previously described (11). Briefly, the sciatic nerve was isolated from one leg and stimulated for 15 minutes, with the contralateral leg acting as a basal control. Following completion of the contraction protocol, the TA muscles from both legs were rapidly dissected and frozen in liquid N2. Muscles were weighed and processed for Western blotting.
C2C12 tissue culture.
C2C12 myoblasts (ATCC; passage 4–7) were used for all experiments and maintained in DMEM containing 10% FBS and 1% penicillin/streptomycin. Differentiation was induced by incubating the cells in DMEM containing 2% horse serum and 1% penicillin/streptomycin for 4 days. For experiments involving palmitate treatment, cells were incubated in DMEM containing 250 μM palmitate (from 1 mM stock in ethanol) conjugated to 2% fatty acid–free BSA or vehicle (ethanol/2% BSA) for 24 or 48 hours. SNARK-knockdown myoblasts were generated using siRNA against SNARK or a scrambled sequence obtained from Origene (SR30004, SR418439B) using the Lipofectamine RNAiMAX reverse transfection protocol (Life Technologies). Cell survival was assessed using the MTT viability assay. Briefly, cells were exposed to 500 μM MTT reagent for 3 hours, washed once with ice-cold PBS, and solubilized in DMSO. Absorbance was measured at 540 and 690 nm, and the relative number of viable cells was calculated for each well. Data represent the average of 3 independent experiments.
Muscle histology.
TA muscles from 58-week-old SNARK transgenic mice and littermate controls were frozen in isopentane cooled with liquid N2 and stored at –80°C until sectioning. Six-μm sections were cut using a cryostat microtome (Leica CM1850; Leica Microsystems) and affixed to slides. Sections were incubated overnight at 4°C in a humidified chamber with antibodies against Laminin (Sigma-Aldrich L9393) and myosin heavy chain type IIB (BF-F3, Developmental Studies Hybridoma Bank, University of Iowa, Iowa City, Iowa, USA). Subsequently, sections were incubated with appropriate conjugated secondary antibodies for 1 hour at 37°C (Alexa Fluor 488 and Alexa Fluor 568, Molecular Probes, Life Technologies). Slides were mounted with DAPI-containing mounting medium to stain myonuclei (Vectashield H-1500; Vector Laboratories Inc.). Histology image analysis for quantification of type I/IIA and type IIB muscle fiber CSA was performed across whole-section images by OracleBio Limited. Using Definiens Tissue Studio software, a customized algorithm was initially developed to automatically detect all individual fibers in the tissue section, as outlined by the laminin immunofluorescence image layer. Each detected fiber object was then automatically classified as type IIB (BF-F3 stained) or type I/IIA (unstained). Finally, each tissue slide was manually reviewed for exclusion of detected artefacts before the CSA (μm2) of each classified fiber was determined.
Statistics.
Analysis of changes in SNARK expression with exposure to metabolic stress or aging (Figure 1) and with SNARK knockdown (Figure 6, A and B) was performed using an unpaired t test. Analysis of genotype/age interactions in SNARK transgenic mice (Figure 3) as well as SNARK expression and cell survival (Figure 6, C and D) was performed using a 2-way ANOVA with Bonferroni post-hoc testing. Analysis of muscle phenotype (Figure 4) and protein expression/signaling within SNARK transgenic animals and littermate controls was performed with a 1-way ANOVA, followed by Bonferroni post-hoc testing. P values of less than 0.05 were considered significant.
Study approval.
All experiments were performed in accordance with a protocol approved by the Institutional Animal Care and Use Committee of Joslin Diabetes Center and with the Guide for the Care and Use of Laboratory Animals (8th ed. The National Academies Press. 2011.). The Tufts University Health Sciences Campus and Tufts Medical Center Institutional Review Board (IRB 8766) approved the collection of human tissues, and written informed consent was obtained from each participant.
Author contributions
SJL, LJG, RAF, and DAR designed the study. SJL, RAF, DAR, KS, HJK, MFH, and ALQ performed experiments. SJL, DAR, KS, MFH, and ALQ acquired data. SJL, DAR, and ALQ analyzed data. SJL and LJG wrote the manuscript.
Supplementary Material
Acknowledgments
This work was supported by NIH grants to L.J. Goodyear (AR42238 and DK068626) and the Joslin Diabetes Center Diabetes Research Center (P30 DK36836). H.J. Koh was supported by NIH grants (P30 DK036836 and P30DK040561). S.J. Lessard was supported by postdoctoral fellowships from the American Physiological Society (Physiological Genomics) and the Canadian Diabetes Association. This material is based upon the work supported by the US Department of Agriculture (USDA), under agreement 58-1950-0014.
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Reference information:J Clin Invest. 2016;126(2):560–570. doi:10.1172/JCI79197.
References
- 1.Fielding RA, et al. Sarcopenia: an undiagnosed condition in older adults. J Am Med Dir Assoc. 2011;12(4):249–256. doi: 10.1016/j.jamda.2011.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.McLean RR, et al. Criteria for clinically relevant weakness and low lean mass and their longitudinal association with incident mobility impairment and mortality: the foundation for the National Institutes of Health (FNIH) sarcopenia project. A Biol Sci Med Sci. 2014;69(5):576–583. doi: 10.1093/gerona/glu012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang C, Bai L. Sarcopenia in the elderly: basic and clinical issues. Geriatr Gerontol Int. 2012;12(3):388–396. doi: 10.1111/j.1447-0594.2012.00851.x. [DOI] [PubMed] [Google Scholar]
- 4.Wannamethee SG, Shaper AG, Lennon L, Whincup PH. Decreased muscle mass and increased central adiposity are independently related to mortality in older men. Am J Clin Nutr. 2007;86(5):1339–1346. doi: 10.1093/ajcn/86.5.1339. [DOI] [PubMed] [Google Scholar]
- 5.Stenholm S, Harris TB, Rantanen T, Visser M, Kritchevsky SB, Ferrucci L. Sarcopenic obesity: definition, cause and consequences. Curr Opin Clin Nutr Metab Care. 2008;11(6):693–700. doi: 10.1097/MCO.0b013e328312c37d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Anker SD, et al. Wasting as independent risk factor for mortality in chronic heart failure. Lancet. 1997;349(9058):1050–1053. doi: 10.1016/S0140-6736(96)07015-8. [DOI] [PubMed] [Google Scholar]
- 7.Kim TN, et al. Prevalence and determinant factors of sarcopenia in patients with type 2 diabetes: the Korean Sarcopenic Obesity Study (KSOS) Diabetes Care. 2010;33(7):1497–1499. doi: 10.2337/dc09-2310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Martin L, et al. Cancer cachexia in the age of obesity: skeletal muscle depletion is a powerful prognostic factor, independent of body mass index. J Clin Oncol. 2013;31(12):1539–1547. doi: 10.1200/JCO.2012.45.2722. [DOI] [PubMed] [Google Scholar]
- 9.Hardie DG, Ross FA, Hawley SA. AMPK: a nutrient and energy sensor that maintains energy homeostasis. Rev Mol Cell Biol. 2012;13(4):251–262. doi: 10.1038/nrm3311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jessen N, Sundelin EI, Møller AB. AMP kinase in exercise adaptation of skeletal muscle. Drug Discov Today. 2014;19(7):999–1002. doi: 10.1016/j.drudis.2014.03.009. [DOI] [PubMed] [Google Scholar]
- 11.Koh HJ, et al. Sucrose nonfermenting AMPK-related kinase (SNARK) mediates contraction-stimulated glucose transport in mouse skeletal muscle. Proc Natl Acad Sci U S A. 2010;107(35):15541–15546. doi: 10.1073/pnas.1008131107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sun X, Gao L, Chien HY, Li WC, Zhao J. The regulation and function of the NUAK family. J Mol Endocrinol. 2013;51(2):R15–R22. doi: 10.1530/JME-13-0063. [DOI] [PubMed] [Google Scholar]
- 13.Namiki T, et al. AMP kinase-related kinase NUAK2 affects tumor growth, migration, and clinical outcome of human melanoma. S Proc Natl Acad Sci U S A. 2011;108(16):6597–6602. doi: 10.1073/pnas.1007694108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rune A, Osler ME, Fritz T, Zierath JR. Regulation of skeletal muscle sucrose, non-fermenting 1/AMP-activated protein kinase-related kinase (SNARK) by metabolic stress and diabetes. Diabetologia. 2009;52(10):2182–2189. doi: 10.1007/s00125-009-1465-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tsuchihara K, et al. Susceptibility of Snark-deficient mice to azoxymethane-induced colorectal tumorigenesis and the formation of aberrant crypt foci. Cancer Sci. 2008;99(4):677–682. doi: 10.1111/j.1349-7006.2008.00734.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Street CA, Bryan BA. Rho kinase proteins — pleiotropic modulators of cell survival and apoptosis. Anticancer Res. 2011;31(11):3645–3657. [PMC free article] [PubMed] [Google Scholar]
- 17.Feng J, et al. Inhibitory phosphorylation site for Rho-associated kinase on smooth muscle myosin phosphatase. J Biol Chem. 1999;274(52):37385–37390. doi: 10.1074/jbc.274.52.37385. [DOI] [PubMed] [Google Scholar]
- 18.Vallenius T, Vaahtomeri K, Kovac B, Osiceanu AM, Viljanen M, Makela TP. An association between NUAK2 and MRIP reveals a novel mechanism for regulation of actin stress fibers. J Cell Sci. 2011;124(pt 3):384–393. doi: 10.1242/jcs.072660. [DOI] [PubMed] [Google Scholar]
- 19.Yamamoto H, et al. Identification of a novel substrate for TNFalpha-induced kinase NUAK2. Biochem Biophys Res Commun. 2008;365(3):541–547. doi: 10.1016/j.bbrc.2007.11.013. [DOI] [PubMed] [Google Scholar]
- 20.Horsley V, Friday BB, Matteson S, Kegley KM, Gephart J, Pavlath GK. Regulation of the growth of multinucleated muscle cells by an NFATC2-dependent pathway. J Cell Biol. 2001;153(2):329–338. doi: 10.1083/jcb.153.2.329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pu WT, Ma Q, Izumo S. NFAT transcription factors are critical survival factors that inhibit cardiomyocyte apoptosis during phenylephrine stimulation in vitro. Circ Res. 2003;92(7):725–731. doi: 10.1161/01.RES.0000069211.82346.46. [DOI] [PubMed] [Google Scholar]
- 22.Lexell J, Taylor CC, Sjostrom M. What is the cause of the ageing atrophy? Total number, size and proportion of different fiber types studied in whole vastus lateralis muscle from 15- to 83-year-old men. J Neurol Sci. 1988;84(2–3):275–294. doi: 10.1016/0022-510x(88)90132-3. [DOI] [PubMed] [Google Scholar]
- 23.Lexell J, Henriksson-Larsen K, Winblad B, Sjostrom M. Distribution of different fiber types in human skeletal muscles: effects of aging studied in whole muscle cross sections. Muscle Nerve. 1983;6(8):588–595. doi: 10.1002/mus.880060809. [DOI] [PubMed] [Google Scholar]
- 24.Sartori R, Gregorevic P, Sandri M. TGFβ and BMP signaling in skeletal muscle: potential significance for muscle-related disease. Trends Endocrinol Metab. 2014;25(9):464–471. doi: 10.1016/j.tem.2014.06.002. [DOI] [PubMed] [Google Scholar]
- 25.Paul PK, et al. Targeted ablation of TRAF6 inhibits skeletal muscle wasting in mice. J Cell Biol. 2010;191(7):1395–1411. doi: 10.1083/jcb.201006098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Paul PK, Kumar A. TRAF6 coordinates the activation of autophagy and ubiquitin-proteasome systems in atrophying skeletal muscle. Autophagy. 2011;7(5):555–556. doi: 10.4161/auto.7.5.15102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sartori R, et al. BMP signaling controls muscle mass. Nat Genet. 2013;45(11):1309–1318. doi: 10.1038/ng.2772. [DOI] [PubMed] [Google Scholar]
- 28.Sandri M. Protein breakdown in muscle wasting: role of autophagy-lysosome and ubiquitin-proteasome. Cell Biol. 2013;45(10):2121–2129. doi: 10.1016/j.biocel.2013.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Marzetti E, Calvani R, Bernabei R, Leeuwenburgh C. Apoptosis in skeletal myocytes: a potential target for interventions against sarcopenia and physical frailty - a mini-review. Gerontology. 2012;58(2):99–106. doi: 10.1159/000330064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chaitanya GV, Steven AJ, Babu PP. PARP-1 cleavage fragments: signatures of cell-death proteases in neurodegeneration. Cell Commun Signal. 2010;8: doi: 10.1186/1478-811X-8-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shavlakadze T, McGeachie J, Grounds MD. Delayed but excellent myogenic stem cell response of regenerating geriatric skeletal muscles in mice. Biogerontology. 2010;11(3):363–376. doi: 10.1007/s10522-009-9260-0. [DOI] [PubMed] [Google Scholar]
- 32.Rivas DA, Morris EP, Fielding RA. Lipogenic regulators are elevated with age and chronic overload in rat skeletal muscle. Acta Physiol (Oxf) 2011;202(4):691–701. doi: 10.1111/j.1748-1716.2011.02289.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rivas DA, et al. Increased ceramide content and NFκB signaling may contribute to the attenuation of anabolic signaling after resistance exercise in aged males. J Appl Physiol (1985) 2012;113(11):1727–1736. doi: 10.1152/japplphysiol.00412.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rivas DA, et al. Diminished skeletal muscle microRNA expression with aging is associated with attenuated muscle plasticity and inhibition of IGF-1 signaling. FASEB J. 2014;28(9):4133–4147. doi: 10.1096/fj.14-254490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Taheripak G, Bakhtiyari S, Rajabibazl M, Pasalar P, Meshkani R. Protein tyrosine phosphatase 1B inhibition ameliorates palmitate-induced mitochondrial dysfunction and apoptosis in skeletal muscle cells. Free Radic Biol Med. 2013;65:1435–1446. doi: 10.1016/j.freeradbiomed.2013.09.019. [DOI] [PubMed] [Google Scholar]
- 36.Lefebvre DL, et al. Identification and characterization of a novel sucrose-non-fermenting protein kinase/AMP-activated protein kinase-related protein kinase, SNARK. Biochem J. 2001;355(pt 2):297–305. doi: 10.1042/0264-6021:3550297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lefebvre DL, Rosen CF. Regulation of SNARK activity in response to cellular stresses. Biochim Biophys Acta. 2005;1724(1–2):71–85. doi: 10.1016/j.bbagen.2005.03.015. [DOI] [PubMed] [Google Scholar]
- 38.Namiki T, Coelho SG, Hearing VJ. NUAK2: an emerging acral melanoma oncogene. Oncotarget. 2011;2(9):695–704. doi: 10.18632/oncotarget.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Legembre P, Schickel R, Barnhart BC, Peter ME. Identification of SNF1/AMP kinase-related kinase as an NF-κB-regulated anti-apoptotic kinase involved in CD95-induced motility and invasiveness. J Biol Chem. 2004;279(45):46742–46747. doi: 10.1074/jbc.M404334200. [DOI] [PubMed] [Google Scholar]
- 40.Hoppe PE, Chau J, Flanagan KA, Reedy AR, Schriefer LA. Caenorhabditis elegans unc-82 encodes a serine/threonine kinase important for myosin filament organization in muscle during growth. Genetics. 2010;184(1):79–90. doi: 10.1534/genetics.109.110189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zagorska A, et al. New roles for the LKB1-NUAK pathway in controlling myosin phosphatase complexes and cell adhesion. Sci Signal. 2010;3(115): doi: 10.1126/scisignal.2000616. [DOI] [PubMed] [Google Scholar]
- 42.Banerjee S, Zagórska A, Deak M, Campbell DG, Prescott AR, Alessi DR. Interplay between Polo kinase, LKB1-activated NUAK1 kinase, PP1βMYPT1 phosphatase complex and the SCFβTrCP E3 ubiquitin ligase. Biochem J. 2014;461(2):233–245. doi: 10.1042/BJ20140408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fritz G, Kaina B. Rho GTPases: promising cellular targets for novel anticancer drugs. Curr Cancer Drug Targets. 2006;6(1):1–14. [PubMed] [Google Scholar]
- 44.Chalé A, Cloutier GJ, Hau C, Phillips EM, Dallal GE, Fielding RA. Efficacy of whey protein supplementation on resistance exercise-induced changes in lean mass, muscle strength, and physical function in mobility-limited older adults. J Gerontol A Biol Sci Med Sci. 2013;68(6):682–690. doi: 10.1093/gerona/gls221. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.