Abstract
Deregulated protein and Ca2+ homeostasis underlie synaptic dysfunction and neurodegeneration in Huntington disease (HD); however, the factors that disrupt homeostasis are not fully understood. Here, we determined that expression of downstream regulatory element antagonist modulator (DREAM), a multifunctional Ca2+-binding protein, is reduced in murine in vivo and in vitro HD models and in HD patients. DREAM downregulation was observed early after birth and was associated with endogenous neuroprotection. In the R6/2 mouse HD model, induced DREAM haplodeficiency or blockade of DREAM activity by chronic administration of the drug repaglinide delayed onset of motor dysfunction, reduced striatal atrophy, and prolonged life span. DREAM-related neuroprotection was linked to an interaction between DREAM and the unfolded protein response (UPR) sensor activating transcription factor 6 (ATF6). Repaglinide blocked this interaction and enhanced ATF6 processing and nuclear accumulation of transcriptionally active ATF6, improving prosurvival UPR function in striatal neurons. Together, our results identify a role for DREAM silencing in the activation of ATF6 signaling, which promotes early neuroprotection in HD.
Introduction
Huntington’s disease (HD) is a progressive neurodegenerative disorder for which there is no cure, caused by the expansion of CAG triplets in the huntingtin (HTT) gene. As in other neurodegenerative diseases of genetic origin, symptoms in HD patients have a late onset. The reasons for this are not fully known, but might include early activation of endogenous neuroprotective mechanisms that are not well characterized (1). Despite the well-defined genetic cause, the cellular mechanisms that lead to disease onset and progression remain poorly understood. Several interconnected mechanisms that lead to altered protein and Ca2+ homeostasis have been proposed (2).
The unfolded protein response (UPR) is an ER process activated by accumulation of incorrectly folded proteins as well as in response to oxidative stress, energy depletion, and disruption of Ca2+ homeostasis (3). The UPR is thus a complex program that coordinates several signaling pathways. ER stress is sensed by 3 ER transmembrane receptors, activating transcription factor 6 (ATF6), inositol-requiring kinase 1 (IRE1), and double-stranded RNA-activated protein kinase–like endoplasmic reticulum kinase (PERK). In its initial phase, activation of the UPR is protective and initiates restoration of protein-folding homeostasis by increasing chaperone expression, reducing the protein translation rate, and increasing degradation of unfolded proteins. Persistent UPR activation leads to programmed cell death by mitochondria-mediated apoptosis or by autophagy (3).
Defective UPR has been implicated in a variety of neurodegenerative and metabolic disorders, including Alzheimer disease, Parkinson disease, HD, and prion diseases as well as diabetes and vascular diseases (4–6). In most cases, these pathologies are associated with chronic activation of PERK, which leads to a sustained increase in phospho-eIFα2 levels and the proapoptotic transcription factor CCAAT/enhancer-binding protein (CHOP) homologous protein (7–10). PERK inhibitors are suggested for treatment of these diseases (5, 11, 12). In the case of HD, ATF6 processing is reduced, with no change in phospho-eIF2, in HD mouse models and in HD patients (13).
ER stress is also induced by disrupted Ca2+ homeostasis, which recapitulates abnormal changes in various cell functions, including deficits in the mitochondrial respiratory chain and severe disruption of transcription regulation (2). Transcriptome analysis of the striatum from distinct HD models and from patients shows reduced expression of genes that encode regulatory proteins important for Ca2* homeostasis (14, 15). The precise molecular mechanisms that link abnormal Ca2+ homeostasis with HD pathogenesis are nonetheless incompletely understood.
The potassium channel–interacting protein (KChIP) subfamily of neuronal Ca2+ sensors (NCS) includes multifunctional Ca2+-binding proteins with specialized functions through specific protein-protein interactions and Ca2+-dependent DNA binding (16, 17). Downstream regulatory element antagonist modulator (DREAM), also known as KChIP3 or calsenilin (18, 19), is central to Ca2+ homeostasis. As a Ca2+-dependent transcriptional repressor, DREAM is a master regulator of activity-dependent gene expression (20) and controls genes important for Ca2+ homeostasis (21, 22). As an auxiliary protein in the plasma membrane, DREAM interacts with and regulates gating of Kv4 potassium channels (18), voltage-dependent Ca2+ channels (23, 24), and N-methyl-d-aspartate receptors (25, 26).
Here, we found that DREAM levels are reduced in mouse HD models shortly after birth and well before the appearance of locomotor dysfunction. The reduction in DREAM protein levels is a neuroprotective response, and chronic administration of repaglinide, a small molecule that binds DREAM in vitro, delayed disease onset, ameliorated locomotor symptoms, and prolonged life span in R6/2 mice. DREAM interacts with ATF6 in a Ca2+-dependent manner, which defines a mechanistic link between ER stress conditions and a role for DREAM in HD. Moreover, repaglinide competed with the DREAM-ATF6 interaction and increased nuclear accumulation of amino terminal ATF6 (Nt-ATF6) and ATF6-dependent transcription in striatal neurons in vivo. Our results suggest that an improved UPR after ATF6 derepression mediates the positive effect of DREAM silencing and repaglinide treatment in HD.
Results
Endogenous DREAM levels are reduced in murine HD models and in HD patients.
To study the functional implications of Ca2+ deregulation in HD, we focused on the potential role of DREAM, for which knockout and dominant active mouse models are available (27, 28). We analyzed DREAM levels in (a) R6/2 and R6/1 mice, 2 transgenic models of HD that develop disease symptoms shortly after birth or in adulthood, respectively (29); (b) the striatal STHdhQ111/111 cell line derived from knockin mice bearing 111 CAG repeats in the Htt gene (30); and (c) postmortem brain samples from HD patients.
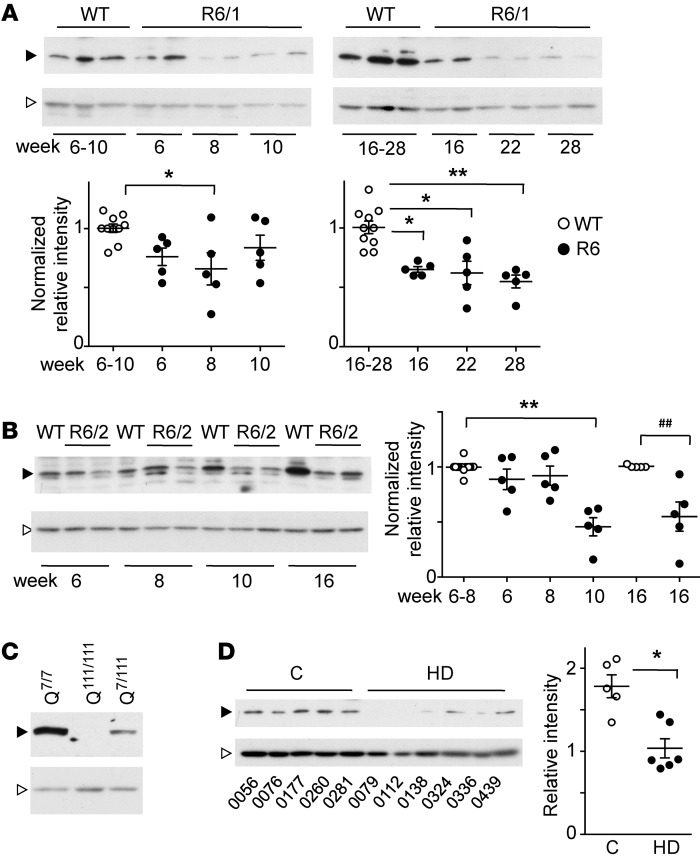
In R6 mice, DREAM expression was greatly reduced in the striatum (Figure 1, A and B) and in other brain areas including hippocampus and cortex (Supplemental Figure 1; supplemental material available online with this article; doi:10.1172/JCI82670DS1). In both animal models, this reduction was observed 8 to 9 weeks after birth, at a time when disease signs began to appear (R6/2) or were not yet apparent (R6/1). Reduced DREAM levels in the striatum were then maintained through the life span in both mouse models (Figure 1, A and B). DREAM protein was also reduced in heterozygous HdhQ111/7 relative to WT STHdhQ7/7 neurons and was virtually absent in homozygous STHdhQ111/111 cells (Figure 1C). Analysis of striatal samples from HD patients substantiated these observations; they showed a reduction in DREAM protein compared with that in age-matched controls (Figure 1D).
Figure 1. DREAM expression is reduced in murine in vivo and in vitro HD models and in HD patients.
(A and B) Western blot analysis of DREAM protein levels in striatum from R6/1 (A) and R6/2 mice (B) and corresponding WT littermates at the indicated weeks after birth. Five mice were analyzed in each group. Black arrowheads, DREAM immunoreactive band; white arrowheads, nonphosphorylated ERK1/2 loading control. After densitometric analysis, band intensity ratio vs. loading control was normalized to corresponding WT. Significant differences compared with WT were calculated using 1-way ANOVA (Kruskal-Wallis), followed by Dunnett’s multiple comparisons test or Mann-Whitney U test (##P = 0.0079) when comparing 2 groups (in B, 16 weeks). *P < 0.05; **P < 0.01. (C) Western blot analysis of DREAM levels in STHdhQ cells. DREAM-specific band intensity is reduced in heterozygous (Q7/111) compared with WT (Q7/7) cells and is below the detection limit in homozygous (Q111/111) cells. Black arrowhead, DREAM immunoreactive band; white arrowhead, α-tubulin loading control. (D) Western blot analysis of DREAM levels in striatal samples from controls (n = 5) and HD patients (n = 6). The corresponding brain bank code number (BCPA-) is shown below each lane. The DREAM-specific band (black arrowhead) was scanned, and intensity ratio vs. loading control (nonphosphorylated Erk1/2, white arrowhead) is shown (right). **P = 0.0019, Mann-Whitney U test. Black arrowheads (A–D) indicate the DREAM-specific band (Ab 670); white arrowheads show the loading control (ERK1/2 in A, B, and D; β-actin in C).
Reduced DREAM expression is a neuroprotective response in murine in vivo and in vitro HD models.
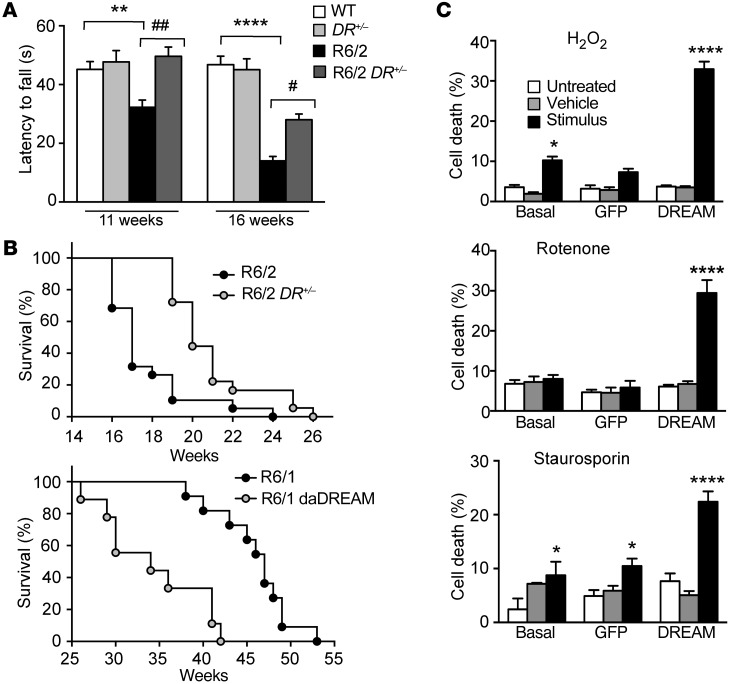
To determine the functional significance of the early reduction in DREAM expression in murine HD models, we modified endogenous DREAM levels in R6 mice crossed with DREAM–/– or transgenic daDREAM mice and analyzed the resulting phenotypes. Induced DREAM haplodeficiency in R6/2 DREAM+/– mice further reduced DREAM levels (Supplemental Figure 2A) and was associated with delayed appearance of motor coordination defects. In the rotarod test, early symptomatic 11-week-old R6/2 mice showed reduced latency to fall, whereas latency to fall in R6/2 DREAM+/– mice was comparable to that of WT littermates (Figure 2A). At 16 weeks of age, latency to fall was still longer in R6/2 DREAM+/– than in R6/2 mice, although it was lower in both genotypes than in WT controls (Figure 2A). We confirmed improved locomotion in R6/2 DREAM+/– mice using the footprint test (Supplemental Figure 2, B and C).
Figure 2. Reduced DREAM protein level is a neuroprotective response.
(A) Latency to fall in the rotarod test for mice (n = 24–30) of the indicated genotypes at 11 and 16 weeks of age. Significant differences between genotypes were found by 2-way ANOVA followed by Tukey’s test. **P < 0.01, ****P < 0.0001 vs. WT; #P < 0.05, ##P < 0.01 vs. R6/2. (B) Percentage survival (Kaplan-Meier) of R6/2 (n = 32) and R6/2 DREAM+/– (R6/2 DR+/–) (n = 43) (upper panel) or R6/1 (n = 11) and R6/1 daDREAM mice (n = 9) (lower panel). Curves were compared by log-rank (Mantel-Cox) test (P < 0.0001). (C) Cell death as a percentage of maximum LDH released for noninfected STHdhQ111/111 cells, cells infected with a GFP-expressing lentivirus, or cells infected with a DREAM-GFP–expressing lentivirus. For each group, cell death was compared for untreated cells vs. cells exposed to vehicle or to the indicated stimuli: H2O2 (10 μM), rotenone (100 nM), or staurosporine (5 nM). Data (mean ± SEM) are derived from 3 independent experiments in quadruplicate. Statistical significance was analyzed by 2-way ANOVA followed by Tukey’s test. *P < 0.05, ****P < 0.0001 vs. basal or vs. GFP groups.
Induced DREAM haplodeficiency in R6/2 DREAM+/– mice also led to a significant increase in life span compared with R6/2 mice, whereas DREAM overexpression in R6/1 daDREAM mice had the opposite effect and reduced life span compared with that of parental R6/1 mice (Figure 2B). A normalized gene expression profile in R6/2 DREAM+/– striatum (GEO GSE48104; Supplemental Tables 2 and 3) accompanied symptom amelioration in these mice (Supplemental Figure 3). DREAM haplodeficiency nevertheless did not modify progressive loss of body weight or the number of HTT-enriched inclusions in R6/2 striatal neurons (Supplemental Figure 4). These results suggest that early downregulation of DREAM expression in HD is a defense mechanism in R6/2 mice.
DREAM-related neuroprotection was also observed in a chemical model of HD based on administration of the mitochondrial toxin 3‑amino propionic acid (3-NPA) (ref. 31 and Supplemental Figure 5). Moreover, we reasoned that if reduced DREAM expression in STHdhQ111/111 cells is a neuroprotective mechanism, restoration of DREAM levels should sensitize STHdhQ111/111 cells to stress. We analyzed cell death in response to stress stimuli in STHdhQ111/111 cells after infection with a DREAM- or GFP-expressing lentivirus. Basal release of lactate dehydrogenase (LDH) did not differ between control and DREAM-overexpressing STHdhQ111/111 cells. Nonetheless, exposure to the mitochondrial toxins hydrogen peroxide (H2O2) (10 μM) or rotenone (100 nM) or to the more general toxin staurosporine (5 nM) elicited more LDH release in DREAM-infected STHdhQ111/111 cells than in naive or GFP-infected STHdhQ111/111 cells (Figure 2C).
As these results strongly suggested that DREAM silencing is part of an early neuroprotective response in HD, we explored DREAM potential as a therapeutic target.
Chronic administration of the DREAM-binding molecule repaglinide delays onset and progression of HD symptoms in R6/2 mice.
We hypothesized that small molecules able to bind and inhibit DREAM activity could be candidates for HD treatment. A literature search identified 2 reports of molecules that bind to NCS in vitro (32) or modulate formation of the KChIP-Kv4 potassium channel complex (33). The first study showed that repaglinide, a drug commonly used to stimulate insulin secretion, binds specifically to NCS and blocks NCS activity (32). In the second study, binding of a diaryl-urea derivative (CL-888) to KChIP1 disrupted KChIP‑1 activity on Kv4.3 channel function (33).
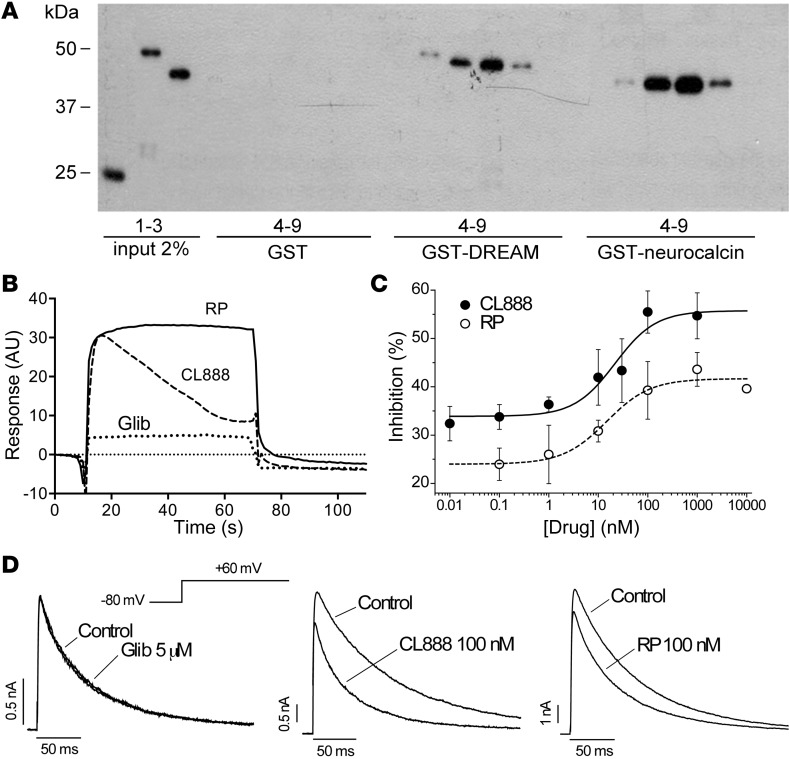
Since binding to recombinant DREAM was not evaluated directly in those studies, we used affinity pull-down and surface plasmon resonance experiments to test binding to DREAM. In both experiments, repaglinide or CL-888 binding to glutathione S-transferase–DREAM (GST-DREAM) was compared with binding to GST alone (negative control) and to GST-neurocalcin, an NCS (positive control) (32). In affinity pull-down, both repaglinide and CL‑888 bound GST-DREAM and GST-neurocalcin in a Ca2+-dependent manner (Figure 3A). Surface plasmon resonance confirmed these results and showed that repaglinide binds GST-DREAM with higher affinity than does CL-888 (Figure 3B).
Figure 3. Repaglinide binds to DREAM and blocks DREAM activity on potassium channel gating.
(A) Affinity pull-down assay of recombinant GST, GST-DREAM, or GST-neurocalcin (2% input in lanes 1–3). For each recombinant protein, the binding signal is shown to uncoupled Sepharose (lane 4), or to Sepharose coupled to (i) repaglinide with 4 mM EGTA (lane 5) or with 4 mM Ca2+ (lane 6) and (ii) other benzoic acid derivatives (lanes 7–9; cyanic acid, aspirin, and 3,4,5-trimethyoxybenzoic acid, respectively). A representative blot is shown. (B) Surface plasmon resonance analysis of the binding of repaglinide (RP), CL-888, and glibenclamide (Glib) (all at 2 μM) to immobilized GST-DREAM. A representative curve is shown. (C and D) Effects of repaglinide, CL-888, or glibenclamide on Kv4.3 channel gating. (C) Concentration curve for the inhibitory effect of repaglinide or CL-888. Experiments in CHO cells transfected with Kv4.3 and DREAM. Blockade was measured as the reduction in the amount of charge crossing the cell membrane (estimated from the integral of the current signal) during the 250 ms pulses to +60 mV. Lines represent the fit of the data to a Hill equation. Each point represents mean ± SEM of 3 to 9 experiments. (D) Representative Kv4.3+DREAM currents recorded on transiently transfected CHO cells. Current traces were obtained after depolarization to +60 mV from a holding potential of 80 mV. Currents were recorded in control conditions and after perfusion with 5 μM glibenclamide (left), 100 nM CL-888 (center), or 100 nM repaglinide (right).
Binding of CL-888 or repaglinide to DREAM counteracted the DREAM effect on A-type potassium currents, reducing peak current amplitude and accelerating inactivation kinetics (Figure 3, C and D). The IC50 values for this inhibition were 23.2 ± 13.9 and 15.8 ± 9.5 nM for CL-888 and repaglinide, respectively (Figure 3C). As also reported for CL-888 and KChIP1 (33), neither molecule altered recovery from inactivation kinetics or the voltage dependency of inactivation (Figure 3D) and both molecules blocked A-type current through Kv4.3 channels alone to a lesser extent than in the presence of DREAM (Supplemental Figure 6). Glibenclamide, used as a negative control, had no effect on A-type current amplitude or kinetics (Figure 3D). Repaglinide binding to recombinant DREAM, nonetheless, did not modify DREAM binding to downstream regulatory element (DRE) sites in vitro and did not alter basal expression of DREAM target genes (20) (Supplemental Figure 7); repaglinide binding to DREAM thus affects the protein-protein interaction with Kv4.3 potassium channels, but does not modify other activities of DREAM, i.e., its interaction with DNA.
Both repaglinide and glibenclamide are widely used in medical practice, which prompted us to explore their therapeutic potential in HD. We treated R6/2 mice with repaglinide (2 μg/ml), glibenclamide (2 μg/ml), or vehicle (DMSO, 0.2 μl/ml) in drinking water shortly after weaning and analyzed changes in HD phenotype.
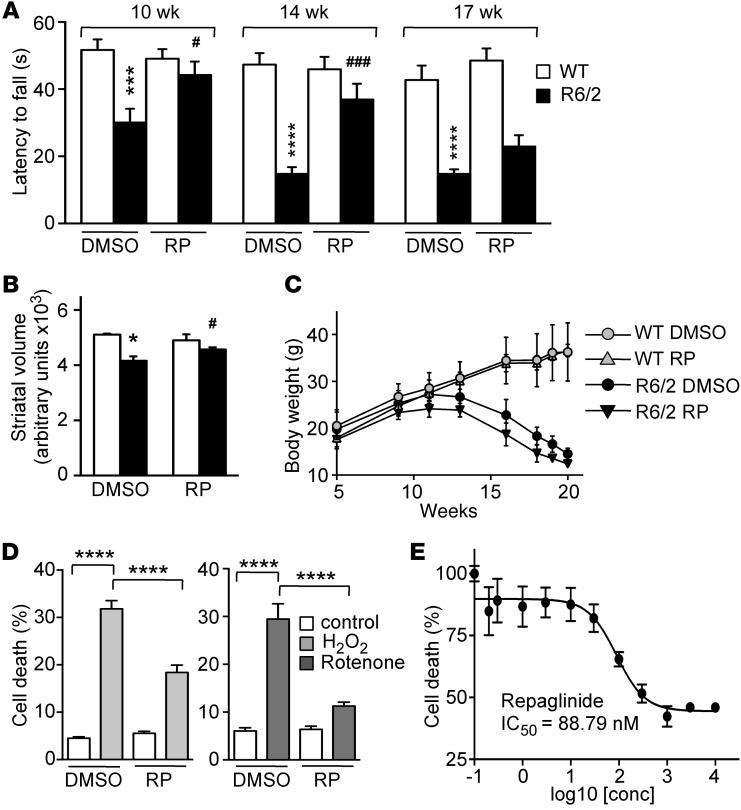
Repaglinide-treated R6/2 mice showed improved performance in the rotarod test. Latency was reduced in DMSO-treated R6/2 compared with WT mice at all ages tested, which was significant from 10 weeks of age (Figure 4A), the earliest time point analyzed. In repaglinide-treated R6/2 mice, however, reduction in latency to fall only became significant at 17 weeks of age (Figure 4A). We observed no effect on motor coordination in WT mice that received either DMSO or repaglinide in drinking water. Improved locomotion in treated R6/2 mice was paralleled by a reduction in striatal atrophy, measured as a reduction in striatal volume by NMR analysis (Figure 4B). Repaglinide did not reduce progressive body weight loss (Figure 4C) or the number of HTT-enriched nuclear inclusions in the striatum, which were similar in DMSO- and repaglinide-treated R6/2 mice (Supplemental Figure 8A). In these experiments, correct drug intake was monitored periodically by testing reduced postprandial increase in blood glucose levels after fasting (Supplemental Figure 8B). Chronic glibenclamide administration did not affect HD symptoms in R6/2 mice, in agreement with previous studies (34) and with its inability to interact with DREAM. These results support DREAM potential as a therapeutic target in HD and identify DREAM-binding molecules as prospective drugs to ameliorate HD symptoms.
Figure 4. Repaglinide ameliorates the HD phenotype.
(A) Latency to fall in the rotarod test of mice of the indicated genotypes and age, exposed chronically to vehicle (DMSO) or repaglinide. DMSO-treated R6/2 mice were significantly different from WT controls at all ages, and repaglinide-treated R6/2 differed from repaglinide-treated WT mice at all ages except at 17 weeks, when there was only a slight, nonsignificant improvement. ***P < 0.001, ****P < 0.0001 vs. WT; #P < 0.05, ###P < 0.001 vs. R6/2 (2-way ANOVA, Sidak’s post test; n = 13–17). (B) Nuclear magnetic resonance analysis of striatal volume in 18-week-old mice of indicated genotypes. *P < 0.0159, WT-DMSO vs. R6/2-DMSO; #P < 0.0303, R6/2-DMSO vs. R6/2-repaglinide (Mann Whitney U test; n = 4–7). (C) Body weight progression in male mice of the indicated genotypes exposed to DMSO or repaglinide. No significant differences (2-way ANOVA, Tukey’s multiple comparison test) were found within groups of untreated or repaglinide-treated mice (n = 10). (D) Cell death, as a percentage of maximum LDH released by DREAM-expressing STHdhQ111/111 cells. Cells were exposed to vehicle or repaglinide (100 nM) and stimulated with H2O2 (10 μM) or rotenone (100 nM). Data from 3 (rotenone) or 7 (H2O2) independent experiments in quadruplicate were analyzed by 2-way ANOVA followed by Tukey’s test. ****P < 0.0001. (E) Effect of repaglinide on LDH release from N2a neuroblastoma cells (n = 4) after H2O2 exposure (20 μM). A 4-parameter (variable slope) nonlinear curve fitting resulted in an IC50 of 88.79 ± 1.39 nM for repaglinide.
To further characterize the repaglinide effect on DREAM, we evaluated repaglinide treatment in STHdhQ111/111 cells sensitized to oxidative stress by DREAM overexpression. Exposure to repaglinide decreased H2O2- or rotenone-induced LDH release from sensitized STHdhQ111/111 cells (Figure 4D). Also, in these experiments, CL-888 (IC50 183 nM) was less potent than repaglinide (IC50 88.8 nM) (Figure 4E). Similar protection by repaglinide (IC50 30.7 nM) was obtained when disruption of mitochondrial transmembrane potential was used as an index of cell death after H2O2 exposure (Supplemental Figure 9).
Derepression of ATF6 transcription mediates DREAM-related neuroprotection in HD.
To define the mechanism of the neuroprotective effect of reduced DREAM function in HD, we focused on the interaction between GST-DREAM and the bZIP-containing transcription factor ATF6 (J.R. Naranjo, unpublished results), identified as a DREAM-interacting protein in nucleic acid–programmable protein arrays (NAPPA) (35). Since DREAM interacts with the bZIP protein cAMP responsive element binding protein (CREB) and this interaction abolishes CREB-dependent transactivation (36), we hypothesized that DREAM downregulation in HD neurons might derepress ATF6 and promote ATF6-dependent transcription as well as UPR-dependent survival.
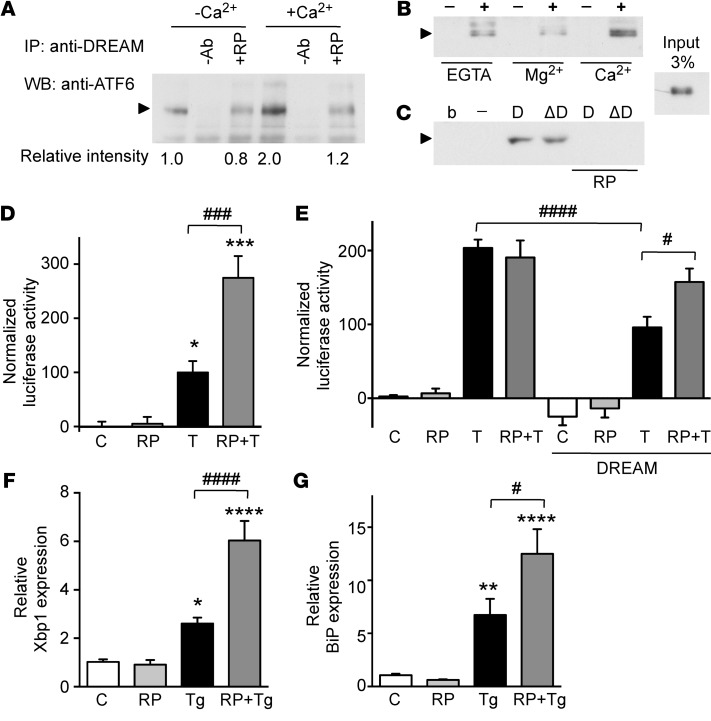
Coimmunoprecipitation experiments with STHdhQ7/7 cells confirmed interaction between endogenous DREAM and ATF6 (Figure 5A). This interaction was potentiated by Ca2+ and was reduced in the presence of repaglinide (Figure 5A). Pull-down assays corroborated the Ca2+ effect on the DREAM-ATF6 interaction and showed that Mg2+ binding to the second EF-hand in DREAM (37) did not increase the interaction with ATF6 (Figure 5B). Pull-down assays also showed that the N‑terminal part of DREAM (aa 1–90) and the N-terminal region of ATF6 (aa 1–376) interact in a repaglinide-dependent manner (Figure 5C). These results suggest that the DREAM-ATF6 interaction is involved in repaglinide-mediated neuroprotection.
Figure 5. Repaglinide blocks DREAM-ATF6 interaction and regulates derepression of ATF6 targets.
(A) Coimmunoprecipitation of endogenous ATF6 and DREAM proteins in STHdhQ7/7 cell lysates. Immunoprecipitated band (arrowhead) corresponds to full-length ATF6. Coimmunoprecipitation was performed in the absence or presence of Ca2+ (2 mM) and/or repaglinide (200 nM). Bottom, optical density of the ATF6 band from 3 independent immunoprecipitations is shown as mean relative intensity to the immunoprecipitate in the absence of Ca2+. (B) Pull-down using GST (–) and GST-DREAM1-256 (+) proteins as bait and in vitro–translated Nt-ATF61–376 as target in the presence of EGTA, Mg2+, or Ca2+ (2 mM). The experiment was repeated twice. (C) Pull-down in the presence of Ca2+ using GST (–), GST-DREAM1–256 (D), or N-terminal GST-DREAM1–90 (ΔD) proteins as bait and in vitro–translated Nt-ATF61–376 as target, alone or with repaglinide (200 nM). b, empty beads. The experiment was repeated twice. Input for B and C was 3%. (D) Luciferase reporter assays in STHdhQ7/7 cells transiently transfected with reporter plasmid p5×ATF6-GL3 and stimulated with tunicamycin (T) (200 ng/ml) alone or with repaglinide (100 nM). *P < 0.05, ***P < 0.001, vs. control; ###P < 0.001 for tunicamycin-stimulated cells alone or with repaglinide (1-way ANOVA, Bonferroni’s post-test). (E) Luciferase reporter assay in WT or DREAM-overexpressing HeLa cells stimulated as in C. ####P < 0.0001, tunicamycin-stimulated HeLa cells alone and with DREAM; #P < 0.05, tunicamycin-stimulated DREAM-expressing HeLa cells alone or with repaglinide (1-way ANOVA, Bonferroni’s post-test). (F and G) Real-time qPCR analysis of XBP1 and BIP mRNAs after stimulation (14 hours) with thapsigargin (Tg) (200 nM) of STHdhQ7/7 cells alone or with repaglinide (200 nM; 1 hour). Values are normalized relative to HPRT mRNA levels. The experiment was repeated 3 times in triplicate. Significant differences were determined as in D: **P < 0.01, ****P < 0.0001, vs. control; #P < 0.05, ####P < 0.0001 for thapsigargin-stimulated cells alone or with repaglinide (1-way ANOVA, Bonferroni’s post-test).
To study a derepression mechanism, we evaluated the repaglinide effect on ATF6-dependent transcription using a reporter plasmid containing 5 ATF6-binding sites (p5×ATF6-GL3) (38). Induction of ER stress with tunicamycin increased reporter activity in STHdhQ7/7 cells (Figure 5D). Repaglinide exposure slightly increased basal, but markedly increased tunicamycin-induced, luciferase activity (Figure 5D). To link the effect of repaglinide on ATF6 transactivation with DREAM-mediated derepression, we conducted similar reporter assays in HeLa cells, which do not express DREAM. Exposure of HeLa cells to tunicamycin greatly increased luciferase activity, which was unaffected by repaglinide (Figure 5E). DREAM overexpression in HeLa cells reduced basal and tunicamycin-induced luciferase activity, and repaglinide largely counteracted this DREAM effect on reporter activity (Figure 5E). To confirm a derepression mechanism, we analyzed directly the repaglinide effect on the expression of ATF6 target genes. Thapsigargin-induced ER stress in STHdhQ7/7 cells increased mRNA levels of the ATF6 downstream targets XBP1, BIP, CHOP, and calreticulin (Figure 5, F and G, and Supplemental Figure 10), as observed in other cells (39–42). STHdhQ7/7 cell exposure to repaglinide had no effect on basal conditions, but significantly increased thapsigargin-induced expression of ATF6 target genes (Figure 5, F and G, and Supplemental Figure 10).
Repaglinide activates ATF6 processing and redistribution in striatal neurons in R6/1 mice.
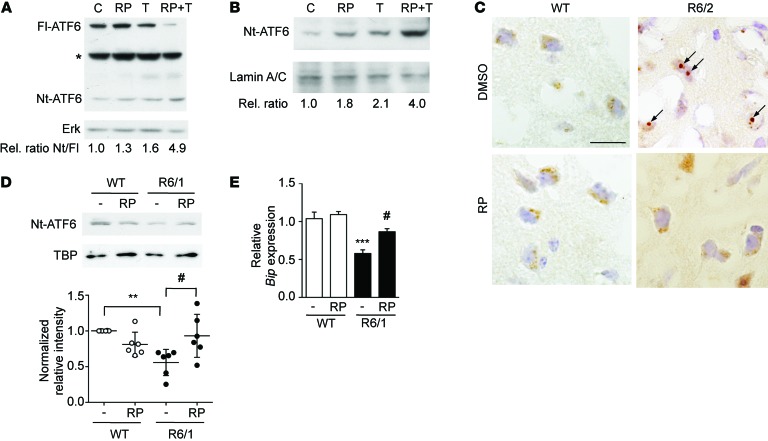
Increased ATF6 transcription in the presence of repaglinide might be linked to an increase in Nt-ATF6 processing and/or in DNA binding. Retardation assays nonetheless showed that DREAM-ATF6 interaction does not directly affect ATF6 binding to DNA in vitro (Supplemental Figure 11). Processing of ATF6 to the nuclear form involves protein-protein interactions and the sequential action of 2 proteases (3). We analyzed the effect of repaglinide on ATF6 processing in basal conditions and after tunicamycin stimulation. In whole-cell extracts of STHdhQ7/7 cells, basal Nt-ATF6 levels were low (Figure 6A), as reported for other cells (39, 43, 44). Repaglinide or tunicamycin exposure slightly increased Nt-ATF6 levels in STHdhQ7/7 cells; however, when both compounds were present, we observed a substantial increase in ATF6 processing, with a reduction in full-length ATF6 and a corresponding increase in Nt-ATF6 (Figure 6A). Analysis of nuclear extracts from STHdhQ7/7 cells exposed to repaglinide and tunicamycin showed a 4-fold increase in Nt-ATF6 compared with basal levels in untreated cells (Figure 6B and Supplemental Figure 12).
Figure 6. Repaglinide activates ATF6 processing in mouse striatal neurons.
(A) Western blot analysis of whole-cell extracts from STHdhQ7/7 cells stimulated with tunicamycin (200 ng/ml) and repaglinide (100 nM) alone or in combination. Migration of full-length ATF6 (Fl-ATF6) and the N-terminal ATF6 (Nt-ATF6) are shown. Asterisk marks a nonspecific band. Loading control with nonphosphorylated ERK is shown below. The relative ratio of Nt-ATF6 to Fl-ATF6 after scanning is shown (below). (B) Western blot analysis of nuclear extracts from STHdhQ7/7 cells stimulated as in A. After normalizing to the loading control (lamin A/C), optical density of Nt-ATF6 bands relative to control untreated cells is shown (below). (C) Immunohistochemistry for ATF6 in striatal neurons from WT and R6/2 mice receiving DMSO or repaglinide. A diffuse intracellular ATF6 labeling is observed in WT neurons. In striatal neurons from DMSO-treated R6/2 mice, ATF6 immunoreactivity appears as a dense intracellular aggregate (arrows). In repaglinide-treated R6/2 mice, ATF6 labeling has a diffuse distribution, similar to WT neurons. Scale bar: 20 μm. (D) Western blot analysis of nuclear extracts from striatum from WT and R6/1 mice receiving DMSO (–) or repaglinide. A representative autoradiogram is shown. After quantification of the autoradiograms, ratios vs. loading control TBP were normalized to DMSO-treated WT mice for each experiment. **P < 0.01, R6/1 vs. WT; #P < 0.05, R6/1 vs. repaglinide-treated R6/1 mice (1-way ANOVA [Kruskal-Wallis], followed by Dunn’s multiple comparisons test, n = 6 mice in each group). (E) Real-time qPCR analysis of Bip mRNA in the striatum from WT and R6/1 mice receiving DMSO (–) or repaglinide, as indicated. Values are normalized relative to Hprt mRNA levels. ***P < 0.001, DMSO-treated R6/1 vs. WT; #P < 0.05, R6/1 vs. RP-treated R6/1 mice (statistics as in D, n = 7 mice in each group).
We sought evidence of an effect in vivo after chronic administration of repaglinide to mice. In immunohistochemistry experiments, ATF6 immunoreactivity in striatal neurons from WT mice appeared as a diffuse staining (Figure 6C). This distribution was similar in WT mice receiving vehicle or repaglinide in the drinking water (Figure 6C). In R6/2 mice, however, ATF6 immunoreactivity appeared as a large and dense intracellular aggregate (Figure 6C), an aberrant distribution first reported in R6/1 striatal neurons (13). Administration of repaglinide to R6/2 mice reduced the size of ATF6 aggregates (Figure 6C) and restored a more diffuse cellular labeling, similar to WT neurons (Figure 6C). Western blot analysis of nuclear extracts from R6/1 mice showed reduced Nt-ATF6 levels in R6/1 compared with WT striatum (Figure 6D and Supplemental Figure 12), as described (13). Chronic administration of repaglinide did not affect nuclear Nt-ATF6 in WT mice, but significantly enhanced Nt-ATF6 levels in treated R6/1 mice (Figure 6D and Supplemental Figure 12). Repaglinide thus increased the levels of transcriptionally active Nt‑ATF6, which may contribute to enhanced ATF6-dependent transcription. Quantitative PCR (qPCR) analysis of mouse striatum showed decreased levels of Bip mRNA, an ATF6 target gene, in R6/1 compared with WT mice (Figure 6E). Administration of repaglinide to R6/1 mice partially reversed the decrease in BiP expression and restored levels of Bip mRNA that were not significantly different from WT values (Figure 6E).
These results indicate that DREAM regulates ATF6 processing and prosurvival UPR activity. Early neuroprotection in HD neurons is associated with a decrease in DREAM levels and can be potentiated by repaglinide administration.
Discussion
Most neurodegenerative diseases of genetic origin are slow-developing pathologies that manifest symptoms years after neuronal dysfunction has begun. A plausible reason for this delayed onset is early activation of endogenous neuroprotective mechanisms that temporarily maintain neuron function in a genetically unfavorable environment. We propose that reduction of DREAM activity, which leads to ATF6 derepression and improved prosurvival UPR, is one of these neuroprotective mechanisms. The contribution of other molecular mechanisms to the neuroprotective action following DREAM downregulation in HD cannot be excluded.
Neurodegenerative diseases are associated with accumulation of intracellular protein aggregates; the importance of defective UPR in these pathologies is increasingly recognized (5, 10, 12). Reduced ATF6 activity is reported in amyotrophic lateral sclerosis type VIII, a disease produced by a single mutation in vesicle-associated membrane protein–associated protein B (VAPB) that increases its ability to interact with ATF6 and block ATF6 transcription (45). Decreased ATF6 processing and nuclear accumulation of the transcriptionally active Nt-ATF6 fragment was also reported in the striatum of R6 mice and of HD patients (13). That study showed a decrease in active ATF6 in young mice well before neurological symptoms appeared, similar to our results for DREAM expression.
The interaction between DREAM and ATF6 further suggests a link between these 2 early molecular events. Reduced DREAM levels derepress ATF6 activity and partially compensate for low ATF6 transcriptional activity in HD neurons. ATF6 target genes participate in the protein refolding and have antiapoptotic activity (3), which supports a neuroprotective mechanism related to the decrease in DREAM protein. Following UPR stimulation, ATF6 forms multiprotein complexes with XBP1 and NF-Y to activate transcription of target genes, for instance, to induce components of the ER-associated degradation pathway (ERAD) (41, 46). It is not known whether DREAM regulates Nt-ATF6 interaction with these nucleoproteins and thus modifies ATF6-dependent transcription. Ca2+ enhances DREAM-ATF6 interaction. Internal Ca2+ mobilization, e.g., with thapsigargin, activates ATF6 processing; the Ca2+-dependent interaction with DREAM suggests a feedback mechanism that terminates ATF6 activation in physiological conditions.
A defective UPR is a common feature in diabetes and neurodegenerative diseases. Diabetes is associated with several neurodegenerative diseases, including Parkinson disease, Alzheimer disease, and HD and might contribute to some of the neurological symptoms (47). R6/2 mice reproduced the diabetes phenotype (48), and abnormal glucose use is linked to progressive reduction in insulin release (34). These observations prompted several studies of hypoglycemic drugs for HD treatment. Glibenclamide and rosiglitazone, 2 oral hypoglycemic agents commonly used to treat diabetic patients, were tested in R6/2 mice. After chronic treatment, neither drug improved survival, weight loss, or motor coordination, although glibenclamide had an acute positive effect on glucose levels (34). These results supported previous studies in R6/2 mice, which showed that vaccination against murine HTT improves the diabetic phenotype, with no effect on body weight, progression of motor symptoms, or neuropathology (49). Here, we show that chronic administration of repaglinide, but not glibenclamide, delays onset and progression of disease symptoms in R6/2 mice. The effect of repaglinide administration or induced DREAM haplodeficiency on cognitive impairment in R6/2 mice was not analyzed in this study. Since changes in DREAM activity are associated with modified long-term potentiation (LTP) and long-term depression (LTD) (20, 26), future studies will analyze the potential effect of DREAM-binding molecules in cognition amelioration in R6/1 mice, a mouse model better suited for that purpose. At the molecular level, we show that repaglinide binding to DREAM does not affect its interaction with DNA, but blocks the DREAM-mediated increase in A‑type currents, reduces sensitization to oxidative stress, and disrupts the DREAM-ATF6 interaction to enhance ATF6 nuclear accumulation and expression of ATF6 target genes. Our in vivo results support the idea that repaglinide potentiates the UPR under ER stress, i.e., in R6/1 mice, and does not alter ATF6 processing in striatal neurons in WT mice. Identification of DREAM-binding molecules with greater affinity and/or selectivity might improve therapeutic effectiveness in delaying disease onset and ameliorating symptoms in HD patients.
Increased nuclear accumulation of ATF6 and expression of its target genes was described in ob/ob mice after chronic administration of the chemical chaperones 4-phenyl-butyric acid (4-PBA) (8) and taurine-ursodeoxycholic acid (TUDCA) (50). The mechanisms that link protein stabilization by chemical chaperones with ATF6 processing have not been identified; whether these compounds bind to DREAM or interfere with the DREAM/ATF6 interaction is also not known. Future studies will determine whether chemical chaperones modify onset and progression of disease symptoms in mouse HD models.
In conclusion, our results identify early DREAM downregulation in HD, which promotes neuroprotection via derepression of ATF6 signaling and activation of prosurvival UPR activity. We found that pharmacological inhibition of DREAM activity also induces UPR function, which opens a new avenue for therapeutic intervention in HD patients.
Methods
Mice and in vivo treatment.
R6/2 and R6/1 mice were originally from Jackson Laboratories. Our colonies were maintained by breeding R6 mice with CBA × C57BL/6 mice to obtain heterozygous mutants and WT offspring. Genotype and CAG-repeat length were determined by PCR-based amplification using genomic DNA extracted from tail biopsies. In R6/2 mice, the repeat length in the mutant transgene increased with successive crossings. Only R6/2 mice with fewer than 200 CAG repeats were used in these experiments. Homozygous DREAM knockout mice (27) or transgenic mice expressing dominant active mutant DREAM (21, 26), both in the CBA × C57BL/6 hybrid background, were crossed with R6/2 or R6/1 mice to generate R6/2 DREAM+/– and R6/1 daDREAM mice, respectively.
Repaglinide (2 μg/ml) or vehicle (DMSO, 0.2 μl/ml) was administered chronically in drinking water shortly after weaning. Four i.p. injections of 3-NPA (60, 60, 80, 80 mg/kg) were given, 1 every 12 hours, and motor skills on the rotarod were monitored 1 hour after each injection. For life span analysis, mice were monitored twice daily from 10 weeks of age until the end of the experiment. Death dates of R6/2 and R6/2 DREAM+/– mice were recorded; no natural deaths were observed for WT or DREAM+/– mice in this period.
Behavioral analysis.
Experiments were performed in adult R6/2 mice and WT littermates. Mice were initially housed 5 per cage in a temperature-controlled (21 ± 1°C) and humidity-controlled (65% ± 10%) room with a 12-hour light/12-hour dark cycle (lights on from 0800 to 2000 hours), with food and water ad libitum. All experiments took place during the light phase. All behavioral experiments were carried out in blind conditions.
The footprint test was used to compare the gait of R6/2 transgenic with that of WT control mice. To obtain footprints, mouse hind and forepaws were coated with blue and red nontoxic paints, respectively. Mice were then allowed to walk twice along a 40-cm long, 6-cm wide runway cylinder (with 6-cm high walls) into an enclosed box. A fresh sheet of white paper was placed on the runway floor for each run. Footprint patterns were analyzed using 2-step parameters. Hind and front-base width were each measured as the mean distance between left and right hind footprints and left and right front footprints, respectively. These values were determined by measuring the perpendicular distance of a given step to a line connecting its preceding and posterior steps. For each step parameter, 3 values were measured from each run, excluding footprints made at the beginning and end. The mean value of each set of 3 values was used in subsequent analyses.
The rotarod test was used to measure motor coordination and balance (Accelerating Model, Ugo Basile, Biological Research Apparatus). For basal rotarod performance, mice were tested on 2 consecutive days. On day 1 (training), each mouse was placed on the rotarod at a constant speed (4 rpm) for a maximum of 60 seconds. The procedure was repeated 3 times, with a rest period of 30 minutes between trials. On day 2 (experiment), mice received 1 training trial at constant speed (4 rpm) for a maximum of 60 seconds, followed by 3 test trials with acceleration from 4 to 40 rpm over a period of 60 seconds. The latency to fall off the rotarod within this time period was recorded. Any mice remaining on the apparatus after 60 seconds were removed and their time scored as 60 seconds. Data from the 3 test trials were averaged for each animal and used in statistical analyses.
Nuclear magnetic resonance.
Striatal volume was calculated by magnetic resonance imaging. Experiments were performed at the Nuclear Magnetic Resonance Centre (Universidad Complutense, Madrid, Spain) using a BIOSPEC BMT 47/40 (Bruker) operating at 4.7 T and equipped with a 12-cm actively shielded gradient system. Mice were anesthetized with oxygen/isofluorane and placed in prone position inside a cradle. The head was immobilized and placed beneath a 4-cm surface coil. Mice were monitored using a respiration sensor. Global shimmer was assessed, after which 3 gradient-echo scout images in axial, sagittal, and coronal directions were acquired (time to repetition/echo time = 100/3.2 ms, matrix = 128 × 128). A 3D fast spin-echo experiment with axial slice orientation was performed using the following acquisition parameters: time to repetition = 3,000 ms, effective echo time = 86.5 ms, number of averages = 2, field of view = 2.56 × 2.56 × 1.28 cm3, matrix size = 256 × 128 × 32. The reconstructed matrix size was 256 × 256 × 32. Total acquisition time was 27 minutes.
Human postmortem brain tissue.
Putamen samples from HD patients and age-matched controls were obtained from the Banco de Tejidos de la Fundacion CIEN (Madrid, Spain), following the ethical guidelines of the Declaration of Helsinki. Case information can be found in Supplemental Table 1.
Cells and cell viability.
STHdhQ knockin striatal neurons (30) were obtained from J.J. Lucas (Centro de Biología Molecular–CSIC, Madrid, Spain). N2a mouse neuroblastoma, HEK293, CHO, and HeLa cells were from ATCC. All cells were cultured in DMEM (with 10% FBS, penicillin/streptomycin, and Glutamax; all from Invitrogen). STHdhQ knockin striatal neurons were maintained at 32°C. Cells were treated with repaglinide at indicated concentrations 30 minutes before exposure to the mitochondrial toxins H2O2 (10 μM), rotenone (100 nM), or staurosporine (5 nM). Cell viability was assessed 3 hours after exposure to the toxin by measuring LDH release to medium using the Cytotoxicity Detection kit (Clontech) or by analyzing the disruption of mitochondrial transmembrane potential with the MitoCapture Mitochondrial Apoptosis Detection Fluorimetric Kit (BioVision). Cell cultures were routinely checked for mycoplasma infection.
Immunohistochemistry.
WT and R6/1 mice were fixed by transcardial perfusion with 4% paraformaldehyde (PFA) (pH 7.4) under deep pentobarbital anesthesia. The brains were dissected and post-fixed in PFA overnight, rinsed, and further incubated in 20% sucrose in 0.1 M PBS. Brains were sectioned on a cryo-microtome. For DREAM staining, sections were rinsed 3 times with PBS and incubated (overnight, room temperature) with polyclonal rabbit anti-DREAM (Ab1014, 1:20; ref. 28), followed by Cy3-conjugated secondary antibodies (1:200, Jackson Laboratories) (2 hours, room temperature). Samples were imaged and analyzed with a Nikon Eclipse microscope. For ATF6 staining, sections were incubated with rabbit polyclonal anti-ATF6 (H-280, Santa Cruz Biotechnology Inc.) and developed with Vectastain Elite ABC Kit (Dako).
Mutant huntingtin immunoreactive inclusions (anti-HTT, MAB5374, Millipore) were counted in 5-μm cryostat sections of a coronal midstriatal level of mouse brain, and each animal was assessed in a single section. A 0.15-mm2 rectangular area (0.5 × 0.3 mm) was defined at 1 striatum and was divided in 2 separate subareas comprising 20 sections each. Positive inclusions in 10 square sections in each subarea were counted. A final average count was obtained by calculating the average counts of both 20-section subareas. Final data are expressed as positive inclusions per mm2.
Western blot.
STHdhQ cells were lysed in lysis buffer (50 mM Tris HCl, pH 7.5, 150 mM NaCl, 1 mM EDTA, 1% Triton X-100, 0.5% deoxycholate, 0.1% SDS) supplemented with a protease inhibitor cocktail (Roche); cell pellets were incubated on ice in lysis buffer for 45 minutes, with occasional vortexing. Nuclear extracts from STHdhQ7/7 cells and mouse striatal tissue were prepared using NE-PER Nuclear and Cytoplasmic Extraction Reagents (Pierce). To prepare total striatal extracts, mouse and human striatal tissues were homogenized on ice in RIPA buffer (9806, Cell Signaling Technology) supplemented with 1 mM PMSF. Extracts were cleared by centrifugation (14,000 g, 20 minutes). Samples (20–30 μg) were analyzed by SDS-PAGE and immunoblot. PVDF membranes were incubated overnight at 4°C with the following specific antibodies: anti-DREAM (Ab730, ref. 51, or 1B1, ref. 36), ATF6α (H-280, Santa Cruz Biotechnology, Inc.), and as controls, with ERK1/2 (MK12, Millipore), α-tubulin (DM1A, Millipore), PI3K p85b (52), TBP (MAB3658, Millipore), CREB (06-863, Millipore), or lamin A/C (MAB636, ThermoFisher). Secondary antibodies used were HRP-conjugated donkey anti-rabbit or anti-mouse IgG (Jackson ImmunoResearch; 1 hour, room temperature), and signals were detected with ECL (SuperSignal West Femto, Thermo Scientific or ECL Select, GE Healthcare). Band intensity was quantified with QuantityOne software (Bio-Rad).
Coimmunoprecipitation.
DREAM and ATF6 were coimmunoprecipitated from whole-cell extracts (300 μg) from STHdhQ7/7 cells, incubated with 0.5 μg affinity-purified rabbit anti-DREAM Ab1014 (28), with addition of calcium (2 mM) or repaglinide (400 nM) to the whole-cell extract where appropriate. Samples were immunoblotted with rabbit anti-ATF6α (H-280, Santa Cruz Biotechnology Inc.) and proteins visualized with HRP-conjugated secondary antibody (Jackson ImmunoResearch) followed by ECL (ECL Select). Blots were quantified using Quantity One software (Bio-Rad).
The interaction between DREAM and ATF6 was determined using NAPPA (Protoarray v5.0, Life Technologies) (J.R. Naranjo, data not shown). Arrays were hybridized with recombinant GST-DREAM or GST alone and developed using affinity-purified rabbit anti-DREAM (Ab1014; ref. 28). Arrays were scanned using an Axon GenePix 4100A microarray reader with the GenePixPro 6.0 program and results analyzed with Prospector software (Invitrogen). Z-scores were calculated to assess statistical significance according to the following formula: z = (x – μ)/σ where x is the raw value, μ the population mean, and σ the SD for the population. The Z-score for the DREAM-ATF6 interaction was 5.6, above the cut-off value for statistical significance (Z‑score ≥ 2).
Electrophoretic mobility shift analysis.
Band-shift assays with Nt-ATF6 (38) or DREAM (53) were performed as reported for respective recombinant protein.
Luciferase reporter assays.
STHdhQ7/7 or HeLa cells (4 × 105 cells/p60 mm plate) were transfected 3 hours after seeding using JetPEI (Polyplus Transfections), with 3 μg reporter plasmid p5×ATF6-GL3 (Addgene) per plate of STHdhQ7/7 cells. For HeLa cells, 0.5 μg reporter plasmid and 2.5 μg empty pcDNA3 or DREAM expression vector were used per plate. At 20 hours after transfection, cells were exposed to repaglinide or vehicle 1 hour before tunicamycin stimulation and harvested 14 hours later. Luciferase reporter assays were as described (21).
Affinity pull-down assay.
Molecules were coupled to EAH Sepharose (Pierce) resin according to the manufacturer’s protocol. Coupled resin samples and noncoupled control resin were blocked with 5% BSA in 20 mM Tris HCl, pH 7.5, and washed once with Tris buffer. Equimolar amounts of recombinant GST-DREAM or GST protein in 1 ml binding buffer (20 mM Tris HCl, pH 7.5, 150 mM NaCl, 4 mM CaCl2 or EGTA, Complete Mini EDTA-free protease inhibitors [Roche], 0.1% BSA) were added to 100 μl resin samples and incubated overnight at 4°C on a rotating wheel. Samples were washed 3 times with wash buffer (20 mM Tris HCl, pH 7.5, 500 mM NaCl, 4 mM CaCl2 or EGTA, Complete Mini EDTA-free protease inhibitors, 0.1% BSA) and twice with the same buffer without BSA. Proteins were eluted from the resin with 2× SDS sample buffer. Samples were analyzed by Western blot using anti–GST-HRP (Sigma-Aldrich, A-7340, 1:2000).
GST-protein pull-down.
Equimolar amounts of GST, full-length GST-DREAM (aa 1–256), and amino terminal GST-DREAM (aa 1–90) recombinant proteins were bound to glutathione Sepharose beads (GEH) in binding buffer (PBS, 5 mM DTT, 1 mM EDTA, Complete Mini EDTA-free protease inhibitors [Roche]) for 2 hours with rotation at 4°C. After extensive washing with binding buffer, the beads were blocked with 5% BSA overnight at 4°C, washed with binding buffer, and reconstituted in pull-down buffer (20 mM Tris, pH 8.0, 150 mM NaCl, 10% glycerol, 0.1% NP40, 5 mM DTT, Complete Mini EDTA-free protease inhibitors [Roche] supplemented with 2 mM CaCl2 or MgCl2 or EGTA). In vitro translated (TNT Quick Coupled Transcription/Translation, Promega) Nt-ATF6 (aa 1–376) was added to the beads, as well as repaglinide (400 nM) as indicated, and incubated (1 hour, with rotation at 4°C). After extensive washing with pull-down buffer, bound protein was eluted with 2× SDS sample buffer and Western blot developed with an ATF6 antibody (H-280, Santa Cruz Biotechnology Inc.).
Surface plasmon resonance.
The Biacore X100 was used to perform interaction analyses at 20°C. GST-DREAM or GST-neurocalcin were immobilized on a CM5 sensor chip consisting of a carboxymethyl-modified dextran polymer linked to a gold-covered glass support. GST-DREAM and GST-neurocalcin were diluted to 100 μg/ml in acetate buffer, pH 4, and immobilized to the sensor chip by amine coupling. The surface was activated by injecting a solution of 0.2 M N-ethyl-N-dimethylaminopropyl-carbodiimide (EDC) and 50 mM N-hydroxysuccinimide (NHS) for 7 minutes. GST-DREAM or GST-neurocalcin was injected for 7 minutes and the surface was blocked by injecting 1 M ethanolamine at pH 8.5 for 7 minutes. The final immobilization level was 6,900 resonance units (RU) for GST-DREAM and 7,500 RU for GST-neurocalcin. Immobilization was followed by several washes with DMSO running buffer (50 mM Tris, pH 7.5, 50 mM NaCl, 2 mM CaCl2 with 2% DMSO and 0.05% Tween X100) to equilibrate the protein surface. Unmodified dextran was used as a reference surface. Compounds were injected over the reference and protein flow cells for 1 minute at a flow rate of 90 μl/min. Each cycle consisted of a 1-minute waiting period to monitor baseline stability, a 1-minute injection of compound, a 1-minute undisturbed dissociation phase, and a wash of the flow system with a 1:1 mixture of DMSO and running buffer.
Electrophysiology.
Electrophysiological experiments were performed in CHO cells transiently transfected with cDNA encoding Kv4.3 or Kv4.3 plus DREAM (KChIP3) and reporter plasmid CD8. Cells were cultured at 37°C in Iscove’s medium supplemented with 10% FBS, 1% l‑glutamine, and 1% penicillin-streptomycin in 5% CO2. Transient transfection was performed with Fugene 6 (Roche). Before experimental use, cells were incubated with polystyrene microbeads precoated with anti-CD8 antibody (Dynabeads M450; Dynal).
Potassium currents were recorded at room temperature (20°C–22°C) using the whole-cell patch-clamp technique with an Axopatch 200B patch-clamp amplifier (Molecular Devices). Currents were filtered at 2 kHz (4-pole Bessel filter) and sampled at 4 kHz. Micropipettes were pulled from borosilicate glass capillary tubes (Narishige GD-1) on a programmable horizontal puller (Sutter Instrument Co.) and heat polished with a microforge (Narishige). Micropipette resistance was 1.8-3 MΩ. Capacitance and series resistance compensation were optimized, with 80% compensation of the effective access resistance usually obtained. The intracellular pipette-filling solution contained 80 mM K-aspartate, 42 mM KCl, 3 mM phosphocreatine, 10 mM KH2PO4, 3 mM MgATP, 5 mM HEPES-K, and 5 mM EGTA and was adjusted to pH 7.25 with KOH. The bath solution contained 136 mM NaCl, 4 mM KCl, 1.8 mM CaCl2, 1 mM MgCl2, 10 mM HEPES-Na, and 10 mM glucose and was adjusted to pH 7.40 with NaOH.
Real-time qPCR.
RNA was isolated from tissues or cell suspensions using TRIzol, treated with DNase (Ambion), and reverse transcribed using hexamer primer and Moloney murine leukemia virus reverse transcriptase. To confirm the absence of genomic DNA, each sample was processed in parallel without reverse transcriptase. Real-time qPCR for endogenous DREAM was performed as described (28). Assays from Applied Biosystems were used for the DREAM target genes c-fos (Mm00487425_m1), Junb (Mm00492781_m1), and Mef2c (Mm0122839_m1) and for the ATF6 target genes calreticulin (Mm00482936_m1), Xbp1 (Mm00457359_m1), and Chop (Mm00492097_m1). For Bip, the primers 5′-ACTTGGAATGACCCTTCGGTG-3′ and 5′-TGCTTGTCGCTGGGCATC-3′ as well as SYBR technology were used. ATF6 target genes were quantified by the normalized expression method using Hprt as a reference with primers 5′-TTGGATACAGGCCAGACTTTGTT-3′ and 5′-CTGAAGTACTCATTATAGTCAAGGGCATA-3′ and the MGB probe 5′-TTGAAATTCCAGACAAGTTT-3′.
Gene-expression microarray.
Whole striatum RNA from WT and transgenic mice was prepared using TRIzol (Invitrogen) and the RNAeasy Mini Kit (QIAGEN). RNA was quantified and quality was controlled with a 2100 Bioanalyzer (Agilent Technologies). cDNA was synthesized from 4 μg total RNA using 1-cycle target labeling and control reagents (Affymetrix) to produce biotin-labeled cRNA. The cRNA preparation (15 μg) was fragmented (94°C, 35 minutes) into 35–200 bases in length. Labeled cRNAs were hybridized to Affymetrix oligonucleotide arrays (GeneChip Mouse Genome 430 2.0 Array). Each sample was added to a hybridization solution containing 100 mM 2-(N-morpholino) ethanesulfonic acid, 1 M Na+, and 20 mM EDTA with 0.01% Tween-20 to a final cRNA concentration of 0.05 μg/ml and hybridized (16 hours, 45°C). Each microarray was washed, stained with streptavidin-phycoerythrin in a Fluidics Station 450 (Affymetrix), and scanned at 1.56 μm resolution in a GeneChip Scanner 3000 7G System (Affymetrix).
Microarray data analysis.
Four biological replicates were hybridized independently for each transcriptomic comparison. GeneChip intensities were background corrected, normalized, and summarized by the RMA method (54) using the Affy package (55) from Bioconductor. After normalization, GeneChip probeSetIDs sharing the same gene symbol and gene title, according to the annotations provided by Affymetrix, were merged and their expression values were averaged. For each comparison, a moderated t test was applied to identify differentially expressed genes as implemented in the Limma package (56) from Bioconductor. Raw P values were adjusted using the false discovery rate (FDR) method (57). Genes with a FDR of less than 0.005 were included in the list of induced (fold change > 0) or repressed (fold change < 0) candidates. The FIESTA viewer was used to select these numerical filters and the candidate genes in each comparison (http://bioinfogp.cnb.csic.es/tools/FIESTA/index.php). The Venny tool was used to create Venn diagrams with all combinations of up- and downregulated genes in the 2 comparisons, R6/2 DREAM+/– versus WT and R6/2 versus WT (http://bioinfogp.cnb.csic.es/tools/venny/index.html). GeneCodis (58) was used for functional enrichment analysis of relevant genes, where Molecular Function terms of the generic GO slim set were considered (Mundodi and Ireland; http://www.geneontology.org/GO.slims.shtml).
Statistics.
All data values are shown as mean ± SEM. Differences were considered significant at P < 0.05. Comparisons between groups were analyzed with 1- or 2-way ANOVA when appropriate, followed by multiple comparisons using the indicated post tests. In the case of small sample size or non-Gaussian distribution, nonparametric tests were used. Two-group comparisons were performed with unpaired 2-tailed Student’s t test. Animal experiments were randomized. Sample size was not predetermined by statistical method. Prism GraphPad Software 6.0 was used to plot graphs and for statistical analysis.
Study approval.
Behavioral tests and animal care were conducted in accordance with standard ethical guidelines (European Communities Directive 86/609 EEC; NIH 1995) and approved by the CNB-CSIC ethical committee.
Author contributions
JRN conceived the study, designed and performed experiments, analyzed data, and wrote the paper. HZ performed and analyzed biochemical experiments and immunochemical staining experiments. DV performed and analyzed biochemical and behavioral experiments and edited the manuscript. PG performed lentiviral infections, cell and tissue biochemical experiments, and real-time qPCR. XMD did mouse genotyping and behavioral experiments. JMO carried out cell death assays. EH performed biochemical analysis. JCO designed and analyzed microarray data. MDA performed immunological staining experiments and quantification of Htt aggregates. AP performed and analyzed electrophysiological recordings. PC performed surface plasmon resonance experiments and chemical synthesis of CL-888. TG contributed to electrophysiological recordings. ADLC contributed to electrophysiological recordings. CV designed, analyzed, and supervised electrophysiological experiments and contributed to manuscript writing. JCV performed protein array experiments. AR provided human postmortem striatal samples and supervised immunological staining experiments. MGR designed and supervised surface plasmon resonance experiments and contributed to manuscript writing. JYL designed and supervised biochemical experiments and immunochemical staining experiments and edited the manuscript. BM designed, supervised, and performed biochemical experiments, analyzed data, and wrote the paper.
Supplementary Material
Acknowledgments
We thank José J. Lucas for critical reading of the manuscript, Ignaci Sahún and Mara Dierssen for advice on footprint analysis, and Catherine Mark for editorial assistance. A. De la Cruz holds a RECAVA contract, A. Prieto and P. Cercós hold FPI fellowships, and T. González holds a Ramón y Cajal contract. J. Casado-Vela holds a JAE-DOC (CSIC) from the Spanish Ministerio de Economía y Competitividad (MINECO), cofunded by the European Social Fund. This work was funded by the Instituto de Salud Carlos III/CIBERNED (to J.R. Naranjo, B. Mellström, and A. Rábano), FISS-RIC RD12/0042/0019 (to C. Valenzuela), Madrid regional government/Neurodegmodels (to J.R. Naranjo), MINECO grants SAF2010-21784 and SAF2014-53412-R (to J.R. Naranjo), SAF2012-32209 (to M. Gutierrez-Rodriguez), SAF2010-14916 and SAF2013-45800-R (to C. Valenzuela), and a grant from the Swedish Research Council (J.Y. Li).
Footnotes
Hongyu Zhang’s present address is: University of Bordeaux, Interdisciplinary Institute for Neuroscience, CNRS UMR 5297, Bordeaux, France.
Conflict of interest: The authors have declared that no conflict of interest exists.
Reference information:J Clin Invest. 2016;126(2):627–638. doi:10.1172/JCI82670.
References
- 1.Kreiner G, et al. A neuroprotective phase precedes striatal degeneration upon nucleolar stress. Cell Death Differ. 2013;20(11):1455–1464. doi: 10.1038/cdd.2013.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blackstone C. Huntington’s disease: from disease mechanisms to therapies. Drug Discov Today. 2014;19(7):949–950. doi: 10.1016/j.drudis.2014.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chakrabarti A, Chen AW, Varner JD. A review of the mammalian unfolded protein response. Biotechnol Bioeng. 2011;108(12):2777–2793. doi: 10.1002/bit.23282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lee J, Ozcan U. Unfolded protein response signaling and metabolic diseases. J Biol Chem. 2014;289(3):1203–1211. doi: 10.1074/jbc.R113.534743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Halliday M, Mallucci GR. Modulating the unfolded protein response to prevent neurodegeneration enhance memory. Neuropathol Appl Neurobiol. 2014;41(4):414–427. doi: 10.1111/nan.12211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hetz C, Mollereau B. Disturbance of endoplasmic reticulum proteostasis in neurodegenerative diseases. Nat Rev Neurosci. 2014;15(4):233–249. doi: 10.1038/nrn3689. [DOI] [PubMed] [Google Scholar]
- 7.Chang RC, Wong AK, Ng HK, Hugon J. Phosphorylation of eukaryotic initiation factor-2alpha (eIF2α) is associated with neuronal degeneration in Alzheimer’s disease. Neuroreport. 2002;13(18):2429–2432. doi: 10.1097/00001756-200212200-00011. [DOI] [PubMed] [Google Scholar]
- 8.Ozcan U, et al. Chemical chaperones reduce ER stress and restore glucose homeostasis in a mouse model of type 2 diabetes. Science. 2006;313(5790):1137–1140. doi: 10.1126/science.1128294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.O’Connor T, et al. Phosphorylation of the translation initiation factor eIF2α increases BACE1 levels and promotes amyloidogenesis. Neuron. 2008;60(6):988–1009. doi: 10.1016/j.neuron.2008.10.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ma T, et al. Suppression of eIF2alpha kinases alleviates Alzheimer’s disease-related plasticity and memory deficits. Nat Neurosci. 2013;16(9):1299–1305. doi: 10.1038/nn.3486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sidrauski C, et al. Pharmacological brake-release of mRNA translation enhances cognitive memory. Elife. 2014;2: doi: 10.7554/eLife.00498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Moreno JA, et al. Oral treatment targeting the unfolded protein response prevents neurodegeneration and clinical disease in prion-infected mice. Sci Transl Med. 2014;5(206): doi: 10.1126/scitranslmed.3006767. [DOI] [PubMed] [Google Scholar]
- 13.Fernandez-Fernandez MR, Ferrer I, Lucas JJ. Impaired ATF6α processing, decreased Rheb and neuronal cell cycle re-entry in Huntington’s disease. Neurobiol Dis. 2011;41(1):23–32. doi: 10.1016/j.nbd.2010.08.014. [DOI] [PubMed] [Google Scholar]
- 14.Luthi-Carter R, et al. Decreased expression of striatal signaling genes in a mouse model of Huntington’s disease. Hum Mol Genet. 2000;9(9):1259–1271. doi: 10.1093/hmg/9.9.1259. [DOI] [PubMed] [Google Scholar]
- 15.Hodges A, et al. Regional and cellular gene expression changes in human Huntington’s disease brain. Hum Mol Genet. 2006;15(6):965–977. doi: 10.1093/hmg/ddl013. [DOI] [PubMed] [Google Scholar]
- 16.Carrion AM, Link WA, Ledo F, Mellstrom B, Naranjo JR. DREAM is a Ca2+-regulated transcriptional repressor. Nature. 1999;398(6722):80–84. doi: 10.1038/18044. [DOI] [PubMed] [Google Scholar]
- 17.Rivas M, Aurrekoetxea K, Mellstrom B, Naranjo JR. Redox signaling regulates transcriptional activity of the Ca2+-dependent repressor DREAM. Antioxid Redox Signal. 2011;14(7):1237–1243. doi: 10.1089/ars.2010.3385. [DOI] [PubMed] [Google Scholar]
- 18.An WF, et al. Modulation of A-type potassium channels by a family of calcium sensors. Nature. 2000;403(6769):553–556. doi: 10.1038/35000592. [DOI] [PubMed] [Google Scholar]
- 19.Buxbaum JD, et al. Calsenilin: a calcium-binding protein that interacts with the presenilins and regulates the levels of a presenilin fragment. Nat Med. 1998;4(10):1177–1181. doi: 10.1038/2673. [DOI] [PubMed] [Google Scholar]
- 20.Mellstrom B, et al. DREAM controls the on/off switch of specific activity-dependent transcription pathways. Mol Cell Biol. 2014;34(5):877–887. doi: 10.1128/MCB.00360-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gomez-Villafuertes R, et al. Downstream regulatory element antagonist modulator regulates Ca2+ homeostasis and viability in cerebellar neurons. J Neurosci. 2005;25(47):10822–10830. doi: 10.1523/JNEUROSCI.3912-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Naranjo JR, Mellstrom B. Ca2+-dependent transcriptional control of Ca2+ homeostasis. J Biol Chem. 2012;287(38):31674–31680. doi: 10.1074/jbc.R112.384982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thomsen MB, Wang C, Ozgen N, Wang HG, Rosen MR, Pitt GS. Accessory subunit KChIP2 modulates the cardiac L-type calcium current. Circ Res. 2009;104(12):1382–1389. doi: 10.1161/CIRCRESAHA.109.196972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Anderson D, et al. Regulation of neuronal activity by Cav3-Kv4 channel signaling complexes. Nat Neurosci. 2010;13(3):333–337. doi: 10.1038/nn.2493. [DOI] [PubMed] [Google Scholar]
- 25.Zhang Y, et al. The DREAM protein negatively regulates the NMDA receptor through interaction with the NR1 subunit. J Neurosci. 2010;30(22):7575–7586. doi: 10.1523/JNEUROSCI.1312-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wu LJ, et al. DREAM (Downstream Regulatory Element Antagonist Modulator) contributes to synaptic depression and contextual fear memory. Mol Brain. 2010;3: doi: 10.1186/1756-6606-3-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cheng HY, et al. DREAM is a critical transcriptional repressor for pain modulation. Cell. 2002;108(1):31–43. doi: 10.1016/S0092-8674(01)00629-8. [DOI] [PubMed] [Google Scholar]
- 28.Savignac M, Pintado B, Gutierrez-Adan A, Palczewska M, Mellstrom B, Naranjo JR. Transcriptional repressor DREAM regulates T-lymphocyte proliferation and cytokine gene expression. EMBO J. 2005;24(20):3555–3564. doi: 10.1038/sj.emboj.7600810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mangiarini L, et al. Exon 1 of the HD gene with an expanded CAG repeat is sufficient to cause a progressive neurological phenotype in transgenic mice. Cell. 1996;87(3):493–506. doi: 10.1016/S0092-8674(00)81369-0. [DOI] [PubMed] [Google Scholar]
- 30.Trettel F, et al. Dominant phenotypes produced by the HD mutation in STHdh(Q111) striatal cells. Hum Mol Genet. 2000;9(19):2799–2809. doi: 10.1093/hmg/9.19.2799. [DOI] [PubMed] [Google Scholar]
- 31.Beal MF, et al. Neurochemical and histologic characterization of striatal excitotoxic lesions produced by the mitochondrial toxin 3-nitropropionic acid. J Neurosci. 1993;13(10):4181–4192. doi: 10.1523/JNEUROSCI.13-10-04181.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Okada M, Takezawa D, Tachibanaki S, Kawamura S, Tokumitsu H, Kobayashi R. Neuronal calcium sensor proteins are direct targets of the insulinotropic agent repaglinide. Biochem J. 2003;375(pt 1):87–97. doi: 10.1042/BJ20030376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bowlby MR, et al. Identification and characterization of small molecule modulators of KChIP/Kv4 function. Bioorg Med Chem. 2005;13(22):6112–6119. doi: 10.1016/j.bmc.2005.06.042. [DOI] [PubMed] [Google Scholar]
- 34.Hunt MJ, Morton AJ. Atypical diabetes associated with inclusion formation in the R6/2 mouse model of Huntington’s disease is not improved by treatment with hypoglycaemic agents. Exp Brain Res. 2005;166(2):220–229. doi: 10.1007/s00221-005-2357-z. [DOI] [PubMed] [Google Scholar]
- 35.Ramachandran N, et al. Next-generation high-density self-assembling functional protein arrays. Nat Methods. 2008;5(6):535–538. doi: 10.1038/nmeth.1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ledo F, Kremer L, Mellstrom B, Naranjo JR. Ca2+-dependent block of CREB-CBP transcription by repressor DREAM. EMBO J. 2002;21(17):4583–4592. doi: 10.1093/emboj/cdf440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Osawa M, Dace A, Tong KI, Valiveti A, Ikura M, Ames JB. Mg2+ and Ca2+ differentially regulate DNA binding and dimerization of DREAM. J Biol Chem. 2005;280(18):18008–18014. doi: 10.1074/jbc.M500338200. [DOI] [PubMed] [Google Scholar]
- 38.Wang Y, Shen J, Arenzana N, Tirasophon W, Kaufman RJ, Prywes R. Activation of ATF6 and an ATF6 DNA binding site by the endoplasmic reticulum stress response. J Biol Chem. 2000;275(35):27013–27020. doi: 10.1074/jbc.M003322200. [DOI] [PubMed] [Google Scholar]
- 39.Yoshida H, Matsui T, Yamamoto A, Okada T, Mori K. XBP1 mRNA is induced by ATF6 and spliced by IRE1 in response to ER stress to produce a highly active transcription factor. Cell. 2001;107(7):881–891. doi: 10.1016/S0092-8674(01)00611-0. [DOI] [PubMed] [Google Scholar]
- 40.Yoshida H, Matsui T, Hosokawa N, Kaufman RJ, Nagata K, Mori K. A time-dependent phase shift in the mammalian unfolded protein response. Dev Cell. 2003;4(2):265–271. doi: 10.1016/S1534-5807(03)00022-4. [DOI] [PubMed] [Google Scholar]
- 41.Yamamoto K, et al. Transcriptional induction of mammalian ER quality control proteins is mediated by single or combined action of ATF6alpha and XBP1. Dev Cell. 2007;13(3):365–376. doi: 10.1016/j.devcel.2007.07.018. [DOI] [PubMed] [Google Scholar]
- 42.Carnemolla A, et al. Rrs1 is involved in endoplasmic reticulum stress response in Huntington disease. J Biol Chem. 2009;284(27):18167–18173. doi: 10.1074/jbc.M109.018325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Haze K, Yoshida H, Yanagi H, Yura T, Mori K. Mammalian transcription factor ATF6 is synthesized as a transmembrane protein and activated by proteolysis in response to endoplasmic reticulum stress. Mol Biol Cell. 1999;10(11):3787–3799. doi: 10.1091/mbc.10.11.3787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yoshida H, Haze K, Yanagi H, Yura T, Mori K. Identification of the cis-acting endoplasmic reticulum stress response element responsible for transcriptional induction of mammalian glucose-regulated proteins. Involvement of basic leucine zipper transcription factors. J Biol Chem. 1998;273(50):33741–33749. doi: 10.1074/jbc.273.50.33741. [DOI] [PubMed] [Google Scholar]
- 45.Gkogkas C, et al. VAPB interacts with and modulates the activity of ATF6. Hum Mol Genet. 2008;17(11):1517–1526. doi: 10.1093/hmg/ddn040. [DOI] [PubMed] [Google Scholar]
- 46.Yoshida H, et al. ATF6 activated by proteolysis binds in the presence of NF-Y (CBF) directly to the cis-acting element responsible for the mammalian unfolded protein response. Mol Cell Biol. 2000;20(18):6755–6767. doi: 10.1128/MCB.20.18.6755-6767.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Craft S, Watson GS. Insulin and neurodegenerative disease: shared and specific mechanisms. Lancet Neurol. 2004;3(3):169–178. doi: 10.1016/S1474-4422(04)00681-7. [DOI] [PubMed] [Google Scholar]
- 48.Bjorkqvist M, et al. The R6/2 transgenic mouse model of Huntington’s disease develops diabetes due to deficient beta-cell mass and exocytosis. Hum Mol Genet. 2005;14(5):565–574. doi: 10.1093/hmg/ddi053. [DOI] [PubMed] [Google Scholar]
- 49.Miller TW, Shirley TL, Wolfgang WJ, Kang X, Messer A. DNA vaccination against mutant huntingtin ameliorates the HDR6/2 diabetic phenotype. Mol Ther. 2003;7(5 pt 1):572–579. doi: 10.1016/s1525-0016(03)00063-7. [DOI] [PubMed] [Google Scholar]
- 50.Engin F, et al. Restoration of the unfolded protein response in pancreatic beta cells protects mice against type 1 diabetes. Sci Transl Med. 2013;5(211): doi: 10.1126/scitranslmed.3006534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Savignac M, et al. Increased B cell proliferation and reduced Ig production in DREAM transgenic mice. J Immunol. 2010;185(12):7527–7536. doi: 10.4049/jimmunol.1000152. [DOI] [PubMed] [Google Scholar]
- 52.Cortes I, et al. p85beta phosphoinositide 3-kinase subunit regulates tumor progression. Proc Natl Acad Sci U S A. 2012;109(28):11318–11323. doi: 10.1073/pnas.1118138109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Link WA, et al. Day-night changes in downstream regulatory element antagonist modulator/potassium channel interacting protein activity contribute to circadian gene expression in pineal gland. J Neurosci. 2004;24(23):5346–5355. doi: 10.1523/JNEUROSCI.1460-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Irizarry RA, Bolstad BM, Collin F, Cope LM, Hobbs B, Speed TP. Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res. 2003;31(4): doi: 10.1093/nar/gng015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gautier L, Cope L, Bolstad BM, Irizarry RA. affy — analysis of Affymetrix GeneChip data at the probe level. Bioinformatics. 2004;20(3):307–315. doi: 10.1093/bioinformatics/btg405. [DOI] [PubMed] [Google Scholar]
- 56. Smyth GK. limma: Linear models For microarray data. In: Gentleman R, Carey VJ, Huber W, Irizarry RA, Dudoit S, eds. Bioinformatics and Computational Biology Solutions Using R and Bioconductor. New York, New York, USA: Springer; 2005:397–420. [Google Scholar]
- 57.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Statist Soc B. 1995;57(1):289–300. [Google Scholar]
- 58.Tabas-Madrid D, Nogales-Cadenas R, Pascual-Montano A. GeneCodis3: a non-redundant and modular enrichment analysis tool for functional genomics. Nucleic Acids Res. 2012;40(Web Server issue):W478–W483. doi: 10.1093/nar/gks402. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.