Abstract
Current efforts toward achieving a cure for HIV are focused on developing strategies to eliminate latently infected CD4+ T cells, which represent the major barrier to virus eradication. Sensitive, precise, and practical assays that can reliably characterize and measure this HIV reservoir and can reliably measure the impact of a candidate treatment strategy are essential. PCR-based procedures for detecting integrated HIV DNA will overestimate the size of the reservoir by detecting replication-incompetent proviruses; however, viral outgrowth assays underestimate the size of the reservoir. Here, we describe the attributes and limitations of current procedures for measuring the HIV reservoir. Characterizing their relative merits will require rigorous evaluation of their performance characteristics (sensitivity, specificity, reproducibility, etc.) and their relationship to the results of clinical studies.
Introduction
Research focused on the eradication of HIV has highlighted the need for sensitive, precise, and practical assays to monitor the HIV reservoir. A latently infected cell contains a viral genome in a dormant state, from which virus replication can be induced. The latent HIV reservoir is comprised of those cells from which a replication-competent form of HIV can be induced in vivo. Latent proviruses are found in all CD4+ T cell subsets, but they are predominantly housed in the resting memory T cell subset (1–3). Approximately 1 in 105–108 CD4+ T cells is latently infected in most patients, with the rate of infection primarily depending on when antiretroviral therapy (ART) was started after infection and the individual’s HIV viral load set point (4). This set of cells may represent only 104–107 latently infected cells in the whole body, assuming a total of 1011 CD4+ T lymphocytes (5). These low frequencies highlight the many technical and practical challenges for measuring the HIV reservoir. The technical challenges require the assay to be (i) sensitive enough to detect small numbers in a background of a large number of cells, (ii) specific so that a rare event is a true event, and (iii) capable of distinguishing between replication-competent and -incompetent proviruses. Additionally, an assay must have the precision to permit a robust detection of a reduction in the HIV reservoir when assessing a candidate intervention for eradication. The practical challenges are to obtain enough cells to measure the rare events and to obtain enough rare events to measure a reduction. Because many assays are close to the limit of detection, the dynamic range that makes it possible to measure a reduction is usually quite small. Moreover, the coefficient of variation of assays almost always increases as they approach the limit of detection, making the interpretation more challenging. Another practical challenge is that only 1%–2% of the body’s lymphocytes are in the circulation (6). Over 98% of lymphocytes are in tissues that are more difficult to access; moreover, their distribution in those tissues is not mixed and homogeneous, as in the circulation. Finally, reports of assays in most publications fail to incorporate rigorous descriptions of performance characteristics such as sensitivity, specificity, or reproducibility, with the consequence that many assertions about the size or changes in the HIV reservoir may not be dependable.
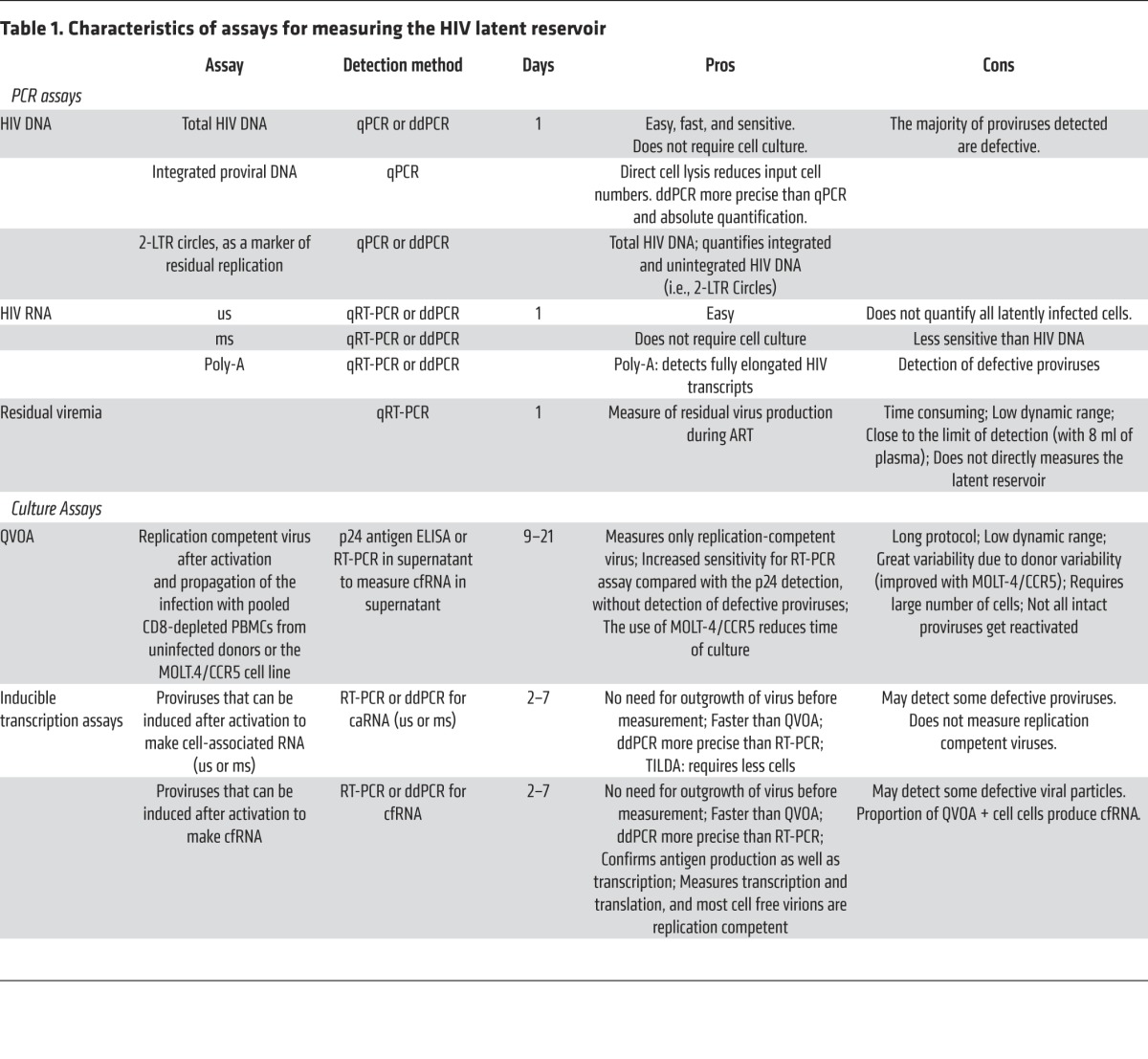
To date, there has been no convincing cellular marker that distinguishes the rare latently infected cell from uninfected cells. Measurements of the various forms of HIV-specific nucleic acids, which can be extracted or induced from latency and then detected, are typically the most sensitive. Despite this higher sensitivity, these assays fail to distinguish defective proviral genomes or other replication-incompetent forms of the virus. Terminal-dilution coculture assays (7–9) of replication-competent virus consistently detect only one infectious unit per hundreds of HIV proviral genomes (10). It is unclear how much of this low proportion is attributable to replication-incompetent proviruses and how much is attributable to suboptimal sensitivity of quantitative viral outgrowth assays (QVOA) for the detection of replication-competent, latently infected cells. The various assays that have been reported to date are summarized below, with attempts to characterize their applications, attributes, and limitations (Table 1).
Table 1. Characteristics of assays for measuring the HIV latent reservoir.
PCR-based assays to measure the latent reservoir
The most sensitive, quickest, and easiest assays to measure the prevalence of HIV infected cells are PCR based (Figure 1A). These assays quantify total or integrated HIV DNA or HIV RNA transcripts.
Figure 1. Assays to measure the latent HIV reservoir.

(A) Blood plasma (or seminal plasma or cerebrospinal fluid) can be assayed for residual virion production with highly sensitive assays of HIV RNA that use large volumes of specimen, as described in the text. Latently infected cells from blood or from tissues that are more difficult to obtain can be examined for several different analytes. Extracted DNA can be assayed for total, integrated, or 2-LTR forms either by qPCR or ddPCR. (B) HIV RNA transcripts can be measured in resting cells either directly or after induction in the presence of antiretroviral drug to prevent cell-to-cell transmission. caRNA can be assayed for specific transcripts, and cfRNA can be measured for the production of virions. The induction of replication-competent provirus is assayed in the absence of antiretroviral drug and the addition of cells that permit the propagation HIV infection, as measured by the generation of p24 antigen or HIV RNA.
HIV DNA: Total, integrated, and 2-LTR circles.
The most common method for measuring HIV DNA is quantitative PCR (qPCR) for total DNA, which includes integrated and nonintegrated forms. Typically, qPCR is performed on DNA extracted from total peripheral blood mononuclear cells (PBMCs) or enriched CD4+ T cells using PCR primers and probes targeting conserved regions of the HIV genome (Pol or Gag) (10, 11). A standard curve is constructed with known copy numbers of proviral DNA (for example, using a plasmid standard such as pNL4-3), and values are normalized to the total cell number using a cellular gene (present as two copies per diploid cell) to calculate the number of infected cells. Several laboratories have improved this method in different ways. First, column-based DNA extraction protocols are inefficient and inconsistent. A direct cell lysis protocol using diverse detergents and/or proteinase K improves the efficiency of nucleic acid recovery; therefore, it is possible to reduce cell input to study cell populations with lower cell numbers accurately (1, 3, 12–15). Second, qPCR limits assay accuracy at low copy numbers by exponentially amplifying noise. Droplet digital PCR (ddPCR) has been proposed as an alternative to qPCR, with potentially improved accuracy and precision (16, 17).
Total HIV DNA includes stably integrated proviruses and extrachromosomal HIV DNA forms, which can be quantified separately. Alu PCR is the most common method to measure integrated HIV DNA (12, 18–21). With this protocol, one primer binds to the highly repetitive Alu elements in the human genome and the second targets the HIV Gag or long terminal repeat (LTR), after which nested PCR is used to enhance sensitivity using a second set of primers in the HIV LTR. A standard curve is required to quantify the levels of integrated HIV DNA, which can include a mixture of DNA from cells with different HIV integration sites at different distances from an Alu element. A correction factor can be used to account for proviruses that are too far from an Alu sequence to be detected. HIV-integrated DNA strongly correlated with total HIV DNA measured by ddPCR (10), indicating that most of the HIV DNA in ART-suppressed subjects is integrated. Integrated DNA modestly correlates with QVOA (10), and it might prove to be a more useful tool than total HIV DNA to estimate the size of an HIV reservoir when there is prolonged persistence or new accumulation of unintegrated DNA (i.e., 2-LTR circles). Integrated proviruses can also be measured using physical separation of high molecular weight (chromosomal) DNA (22), a linker-primer PCR assay (23), or inverse PCR (7).
Total HIV DNA PCR measurements can also detect other forms of unintegrated DNA, such as 1-LTR and 2-LTR circles, which are the result of failed integration. Although these forms are not considered part of the latent reservoir, 2-LTR circles have been used as a measure of recent infection during suppressive ART, especially during raltegravir intensification studies (24, 25), although in vitro evidence indicates that 2-LTR circles can persist for the life of the cell (26, 27). Several assays have been developed to study 2-LTR circles using primers flanking the 2-LTR circle junction. These assays have been performed in qPCR on PBMCs (24, 28) and CD4+ T cells (29), as well as ddPCR on PBMCs (10, 13, 25, 30, 31) and CD4+ T cells (14, 32, 33).
HIV cell-associated RNA in unstimulated cells.
HIV transcription results in more than 40 different viral RNA transcripts derived from alternative splicing of the primary transcript. Initially, multi-spliced (ms) RNA transcripts are produced, encoding the HIV regulatory proteins Tat, Rev, and Nef. There is then a shift toward production of unspliced (us) and single-spliced transcripts. The us-mRNA has two functions; it is translated into the structural precursor polyproteins for the Pol and Gag gene products, and it is incorporated into virions as genomic RNA. The different single-spliced mRNAs are translated into the envelope proteins, as well as other accessory proteins (Vif, Vpr, and Vpu), which will assemble into the budding virion. The vast majority of cells harboring latent HIV do not transcribe viral RNA; however, us- and msRNA have been detected in ART-treated subjects, even in cells that do not produce viral particles (34). usRNA is more abundant and therefore more easily detected; however, short HIV Gag RNA transcripts are more commonly produced (35). In contrast, the presence of msRNA reflects the ability of the cell to produce virus, but its presence does not imply productive replication per se (36). A temporal shift toward a higher msRNA/usRNA ratio in ART-treated subjects might be used as a biomarker of residual replication.
Cell-associated RNA (caRNA) can be measured from extracted RNA of total PBMCs or enriched CD4+ T cells. usRNA can be easily detected with a seminested or nested quantitative reverse transcription PCR (qRT-PCR) (36–39) or by ddPCR, using a set of primers and probe against Pol or Gag (17, 30, 33, 39). Alternatively, some investigators have adapted commercial quantitative tests for HIV-1 plasma viremia, such as Amplicor (Roche Diagnostics) (40, 41) or APTIMA HIV-1 Qualitative assay (Hologic) (42, 43) for the quantitation of caRNA. msRNA primers are designed to amplify a region containing the Tat/Rev (or Tat/Nef) exon-exon junction and can be quantified by nested qRT-PCR (36, 38, 39) or ddPCR (17, 39, 44). An assay to specifically quantify correctly terminated HIV-1 transcripts has been developed using a primer/probe against the polyadenylated tail present in mRNAs and HIV 3′-LTR, and it can be used in a qRT-PCR or ddPCR platform (17, 45, 46).
Plasma viremia, cell-free RNA.
Despite the fact that ART can sustain suppression of virus replication below the level of detection of standard clinical assays for decades, more sensitive assays interrogating larger volumes of plasma can detect virion-associated HIV RNA in most subjects who are well suppressed on ART (47). In most subjects, this viremia is sustained for years at levels below 10 copies per ml plasma (48, 49). Evidence has been generated that at least some of these virions could be replication competent (50), but there is little evidence that this low-level viremia has undergone posttherapy evolution or has acquired drug resistance (51, 52), suggesting that there is viral production from persistently infected host cells or from the population of latently infected cells that is undergoing a steady state of activation and self-renewal. The detection of clonal populations among sequences in the same patient, even at different time points (53), provided the first evidence for a clonally proliferating infected cell population, which has subsequently been confirmed by the documentation of multiple clones of proviral integration sites (54, 55).
Although the source of this low-level viremia has not been clearly defined, its presence indicates a reservoir that must be purged to achieve eradication. The challenge is that measuring this reservoir requires a large volume of plasma and cumbersome procedures, and such measurements have a wide coefficient of variation (48, 56). As a measure of a latent reservoir, it also provides a very small dynamic range to assess the effectiveness of a candidate intervention. The assay as conducted involves the ultracentrifugation of 3–10 ml of plasma, followed by a qPCR assay with a standard control (48, 56). A second approach is under investigation that utilizes a highly sensitive, automated transcription-mediated amplification assay (TMA assay) to test 0.5-ml aliquots of plasma extract; quantitation is based on the proportion of aliquots providing a signal using Poissonian statistics (42).
Culture-based assays to measure HIV reservoir
Culture-based assays have been traditionally used to detect outgrowth of replication-competent virus. More recently, new RNA-based culture assays have been developed to quickly measure the inducible HIV RNA reservoir (Figure 1B).
Replication-competent proviruses.
The QVOA has been considered the gold-standard assay for determining the frequency of CD4+ T cells harboring replication-competent proviruses because replication-competent virus induced ex vivo almost certainly reflects failure to eradicate the virus in vivo. This assay consists of limiting dilutions of resting CD4+ T cells, which are activated using one of several methods, including phytohemagglutinin (PHA) in the presence of irradiated allogeneic PBMCs (8, 10, 57–60) and anti-CD3 and anti-CD28 antibodies (9). This generalized activation reverses latency and reinitiates the production of infectious HIV-1 from CD4+ T cells harboring replication-competent proviruses. Ex vivo viral propagation is mediated by PHA-stimulated CD4+ lymphoblasts obtained from uninfected donors (8, 10, 58–64) or by the 3×3 activation method of uninfected donor blasts (three donors, three activation conditions) (33, 65). After two or three weeks, viral outgrowth is assessed by an ELISA assay for HIV-1 p24 antigen or a PCR assay for HIV-1 RNA in the culture supernatant. Testing replicates in serial dilutions permits the quantification of latently infected cells because virus released from a single cell can spread through the culture and produce a detectable level of infection. The frequency of infected cells in the original population of CD4+ T cells, as well as a CI around this frequency, are then estimated based on Poisson statistics (66) and expressed as infectious units per million (IUPM) CD4+ T cells. Patients on long-term ART typically exhibit IUPM values between 0.1 and 1 — which is, on average, 300-fold less than HIV proviral DNA (8, 10, 61). It is important, especially for eradication studies, to explicitly report the 95% CI for a given IUPM value to allow comparison with infected cell frequency over time or among patients.
Currently, both QVOA and PCR-based methods are commonly used to assess the latent reservoir; however, there is little correlation between these two assays (10). These discrepancies can be attributed to the heterogeneous nature of the proviruses in CD4+ T cells. Only a small subset of proviruses can be induced to produce replication-competent virions. By sequence, many proviruses should be replication-competent, but these proviruses are not readily inducible in QVOA assays. Additionally, the proportion of sequence-competent but nonreplicating viruses is highly variable among individuals (67). Nevertheless, the majority of proviruses cannot produce replication-competent virus because these proviruses contain large internal deletions, G-to-A hypermutations introduced by apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 3G (APOBEC3G), or other inactivating mutations (67). Defective proviruses will not produce infectious virus that can be detected by QVOA, but these viruses can be detected by PCR-based assays. Observed differences may also be due to the lack of standardization and differences in QVOA protocols between laboratories (Table 2). For instance, different cell subsets can be used, such as resting or total CD4+ T cells, to measure the pool of cells harboring replication-competent virus. When using resting CD4+ T cells to exclude productively infected cells, CD69+, CD25+, or HLA-DR+CD4+ T cells are removed from cell culture; however, those markers not only identify truly activated CD4+ T cells, but also proliferating cells (HLA-DR+) and Tregs (CD25+). These cell subsets could also have a relevant contribution to HIV persistence. The activation protocol is also a main difference between the protocols used by different laboratories. Most protocols use PHA plus irradiated CD8-depleted PBMCs, causing high cell density in each well that is aggravated when feeder cells are added. In contrast, activation with anti-CD3/CD28 allows for a better control of cell concentration throughout the culture, which is an important factor in long protocols.
Table 2. Comparison of QVOAs and inducible RNA assays.

Despite the importance of the QVOA, this assay has several limitations. It is time consuming, labor intensive, and requires large blood volumes, two or three weeks of cell culture, and PBMCs from at least three uninfected blood donors per assay, making the assay relatively expensive. The pooled PBMCs from uninfected donors are required to propagate the HIV-1 released following the reversal of latency, resulting in high variability between experiments and limiting assay throughput. Cell lines such as MOLT-4/CCR5, which expresses high levels of CD4 and supports infection by both X4-tropic and R5-tropic HIV-1, can be used as target cells to amplify viruses that are released from activated, infected CD4+ T cells instead of uninfected donor cells, thereby providing greater uniformity for QVOA (68, 69). Despite reducing the variability between experiments by using a cell line, large blood volumes from the study subject and the other limitations of QVOA remain.
Not all replication-competent, latently infected cells are induced by the activation stimuli used in QVOA assays. These stimuli may not fully reproduce the complex activation conditions existing in vivo, particularly given the marked immunological heterogeneity of the latent reservoir cells (70). Moreover, the most efficient activation stimuli may vary by anatomic location or CD4+ T cell phenotype. Additionally, a single round of PHA stimulation is not enough to activate all intact proviruses (67). Thus, some latent viruses capable of generating infectious virions in vivo may not be recoverable in the QVOA.
QVOA also has a limited dynamic range because the levels of HIV provirus in most individuals on ART are close to the limit of detection, even with large blood draws; the 95% CI for individual determinations are usually ±0.7 log IUPM or 5-fold (63). These are additional challenges to observation of reductions in the reservoir during eradication strategies. Rosenbloom et al. have used a statistical approach for optimizing the QVOA experimental design to achieve any desired set of goals regarding sensitivity (determined by cell input) and precision (determined by number of replicates) (71).
Inducible HIV RNA transcription.
Several new assays that quantify the inducible HIV reservoir have been developed. These new assays reduce the contribution of most defective genomes, do not rely on an amplification step with virus propagation by coculture, and may have several advantages for measuring the HIV reservoir. First, many latency-reversing agents (LRAs) show a posttranscriptional block (72); therefore, the activity of these compounds requires measurement of HIV-inducible transcription ex vivo (73, 74). Additionally, because the assays that quantify the inducible HIV reservoir have a higher dynamic range than QVOA, such assays could make it easier to detect reductions in the reservoir ex vivo following effective in vivo interventions (75).
Different versions of assays to detect inducible HIV RNA transcription have been described. In general, CD4+ T cells are maximally activated as they are for QVOA, and then HIV RNA is directly measured from cell extracts (caRNA) (51, 69, 73–75) or cell supernatants (cell-free RNA [cfRNA]) (69, 73, 74). While caRNA measures transcriptionally competent provirus, it might also detect defective transcripts. cfRNA reflects the capacity to induce translation and form and release virions. In contrast to QVOA, various activation stimuli have been used in these assays, including anti-CD3/CD28 costimulation (17, 73), phorbol myristate acetate (PMA) and ionomycin (74–76), or PHA (51). Moreover, depending on the aim of the study, the format of the assay can be adapted easily. For example, a limiting dilution assay would be employed to measure the frequency of CD4+ T cells carrying inducible HIV provirus (17, 73, 76), while bulk activation of CD4+ T cells could be useful for a qualitative assay to test various concentrations of LRAs (74, 75), significantly reducing the cost of the assay.
For caRNA, ms- and usRNA transcripts can be measured by qRT-PCR (51, 56, 76) or ddPCR (17). After HIV transcription is induced, msRNA transcripts are generated, but as the replication cycle continues, the transcripts are increasingly exported as us- and single-spliced mRNA species. Interestingly, the majority of defective HIV genomes contain deletions in the Tat and Rev genes (67), indicating that transcripts from these genes are unlikely to be generated in cells harboring defective proviruses. Procopio et al. proposed the Tat/Rev-induced limiting dilution assay (TILDA) to measure caRNA. This assay uses small blood volumes (10 ml) and is relatively easy to perform without the expense and labor intensiveness of QVOA, making it an attractive option for assessing HIV persistence (76). However, similar to the QVOA, this assay relies on a single round of T cell activation (underestimating the HIV reservoir), and it can potentially measure transcripts derived from some defective proviruses, thereby overestimating the frequency of infected cells with replication-competent virus.
Despite the advantage of cfRNA over caRNA, the latter is more sensitive than the former (69, 73). Indeed, Cillo et al. showed that a mean of 1.5% of proviruses in CD4+ T cells produced detectable levels of cfRNA (virions) after full activation with anti-CD3/CD28 antibodies, while a mean of 7% of proviruses in CD4+ T cells were reactivated to produce us-caRNA (56). This study confirmed that the large majority of proviruses (98.5%) are not induced to produce virions with T cell activation. The approaches that will prove to have the best performance characteristics — and will correlate best with the actual HIV reservoir that needs to be reduced — remain important areas for future investigation.
Murine viral outgrowth assay
Transfer of infectious tissues between species can result in enhanced viral amplification and has been used historically to identify numerous pathogens using embryonated hen eggs or for the diagnosis of rabies virus (77, 78). Also, the adoptive transfer of PBMCs from ART-suppressed SIV-infected monkeys into uninfected monkeys is a sensitive method for detecting residual virus (79–81). Because humanized mice can be infected with HIV-1 (82), a new VOA was developed using mice to detect replication-competent virus: the murine VOA (83). In murine VOA, cells or tissues from HIV-1–infected subjects with undetectable plasma viral loads are transferred as xenografts into immunocompromised mice. Viral load is then monitored using qPCR. Virus production was detectable by murine VOA within 4 weeks in all subjects on long-term ART (83), consistent with the occurrence of viremia following interruption of ART in patients who initiated treatment during the chronic phase of HIV infection (84). Although murine VOA is not quantitative, it is more sensitive than the standard QVOA performed in parallel in the same subjects. The higher sensitivity of murine VOA compared with QVOA could be explained by a number of factors, including the ability to screen large numbers of host cells (up to 60 million per mouse) and xenogeneic human anti-mouse responses that lead to the activation of human CD4+ T cells. This method may have utility when ex vivo assays cannot detect latent infection in order to obviate an unnecessary treatment interruption; however, it is not readily amenable to quantitation.
Latency in cells other than CD4+ lymphocytes
The CD4+ lymphocyte is the cell documented to harbor the HIV reservoir. What is not known is whether additional cells are lurking in the background, obscured by this larger and proven reservoir. One other cell type, the macrophage and its related cell types like microglia, express the CD4 receptor and have been well documented to be host cells for HIV replication in vivo (85). In vitro and in vivo, HIV-infected macrophages can produce virus and survive for months: a much longer half-life than the productively infected lymphocyte. It has not been definitively determined whether HIV-infected macrophages in patients receiving effective ART produce virus with a prolonged but finite half-life or whether cells of this lineage include a reservoir of truly replication-competent, latently infected cells (85). Clinical studies of latency have focused on CD4+ lymphocytes from the blood and occasionally from tissues. The role of cells of the macrophage lineage represents not only an important biological question, but also an important practical challenge for measuring the latent reservoir. It will be necessary to investigate cells other than CD4+ lymphocytes and tissues other than blood, especially the CNS. Also of note, the problem of measuring DNA in CD4+ lymphocytes is further compounded in macrophages, which may contain viral DNA and protein as a result of their phagocytic function, rather than their role in replication (86).
The reservoir in tissues other than blood
The normal distribution of lymphocytes is 2% in the circulation and 98% in the lymphoid tissues. With HIV infection–induced immune activation, the distribution shifts to 1% and 99%, respectively (87). Therapy largely corrects this shift and results in redistribution of mostly CD45RO+ memory T cells from the lymphoid tissue back to the circulation (5, 88). The dense concentration of lymphoid cells promotes the cell-to-cell spread of HIV in anatomical compartments (89); consequently, the distribution of T cell subsets and the proportion of CD4+ lymphocytes that are latently infected may be higher in tissues than in the circulation (90, 91). Thus, measurements of the latent reservoir from blood represent a population that must be cleared to achieve a cure, but this population is a small and unrepresentative portion of the total reservoir.
Extracirculatory tissues are more difficult to access than blood and harbor latent infection that is not mixed and homogeneously distributed, as in blood. In addition to the large amount of lymphoid tissue in the body, two anatomic compartments — the CNS and the genital tract — have been shown to permit compartmental evolution and occasionally escape from the effective suppression of replication as measured in the blood (92–96). These two anatomic compartments differ from the rest of the body in being both pharmacologic and immunologic sanctuaries for HIV, and, in the case of the CNS, containing a large proportion of cells of the macrophage lineage. These differential selective pressures can account for independent evolution and replication during therapy. The practical issue for measuring the latent reservoir in these extracirculatory tissues is more attributable to the difficulty of obtaining these tissues than to assays different from the ones already described.
Conclusions
The emerging interest in developing safe and affordable curative strategies to eliminate the need of lifelong therapy while improving the health of infected subjects and reducing the risk of viral transmission to uninfected individuals requires better assays to measure the latent reservoir. The actual latent reservoir is smaller than the proviral DNA population but larger than what is measured with the standard QVOA (10). The QVOA provides a minimal estimate of reservoir size; however, it does not successfully detect all the cells containing replication-competent virus after a single round of activation (67). In the context of eradication studies, this underestimation of the HIV reservoir is misleading if the absence of detection is interpreted as the detection of absence. The “Boston” patients (97) and the “Mississippi baby” (98) confirmed that the latent reservoir can persist below the limit of detection of current assays, allowing the infection to recur months to years later. As with remission after cancer chemotherapy, although no residual malignant disease can be detected, it can persist and recur years later.
Recently developed assays measuring induced RNA promise to be faster, more sensitive and precise, less demanding of blood volume, less expensive, and capatable of higher throughput than the standard QVOA; however, these assays must be thoroughly validated and standardized between laboratories. First, characterization of the performance of all assays is needed. Second, blinded assay comparisons are necessary to assess their relative performance characteristics with well-defined specimens. Such studies will provide important information regarding the most sensitive and specific assays that could be used in future eradication studies and would be useful in evaluating the qualities and features that would be necessary in a unique assay to measure the true size of the latent reservoir. Therefore, the creation of a consortium to specifically standardize the selected assay between laboratories would be fundamental, in terms of sample preparation, development of standard operating procedures (SOP), and data analysis.
Despite all the efforts in this area, other questions regarding the measurement of the latent reservoir remain open. Is it possible to reactivate all intact proviruses ex vivo? If not, what stimulation would maximally reactivate the latent reservoir? Similarly, the use of frozen samples would be the most convenient way to assess the decay of the latent reservoir in large eradication studies; however, is the quantification of the latent reservoir equally efficient in fresh and frozen samples? When studying eradication candidates, the shock-and-kill effect can be measured with inducible RNA-assays (HIV reactivation) and QVOA (decrease of infected cells), respectively. However, successful HIV cure strategies need to rely on host anti-HIV immune responses and parallel assessments of virological and immunological responses in order to accelerate the development of successful therapies for a functional cure. Despite all these concerns and unresolved questions, the validation of assays will be greatly facilitated by having an effective intervention on which these assays can be compared.
Acknowledgments
This work was supported by the Collaboratory for AIDS Research on Eradication (CARE; U19 AI096113), the UCSD Center for AIDS Research (AI306214), the Department of Veterans Affairs, and the James B. Pendleton Charitable Trust. M. Massanella is supported by a Beatriu de Pinos (AGAUR) postdoctoral fellowship.
Footnotes
Conflict of interest: D.D. Richman owns stock in Chimerix.
Reference information:J Clin Invest. 2016;126(2):464–472. doi:10.1172/JCI80567.
References
- 1.Chomont N, et al. HIV reservoir size and persistence are driven by T cell survival and homeostatic proliferation. Nat Med. 2009;15(8):893–900. doi: 10.1038/nm.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brenchley JM, et al. T-cell subsets that harbor human immunodeficiency virus (HIV) in vivo: implications for HIV pathogenesis. J Virol. 2004;78(3):1160–1168. doi: 10.1128/JVI.78.3.1160-1168.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Buzon MJ, et al. HIV-1 persistence in CD4+ T cells with stem cell-like properties. Nat Med. 2014;20(2):139–142. doi: 10.1038/nm.3445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Strain MC, et al. Effect of treatment, during primary infection, on establishment and clearance of cellular reservoirs of HIV-1. J Infect Dis. 2005;191(9):1410–1418. doi: 10.1086/428777. [DOI] [PubMed] [Google Scholar]
- 5.Zhang ZQ, et al. Kinetics of CD4+ T cell repopulation of lymphoid tissues after treatment of HIV-1 infection. Proc Natl Acad Sci U S A. 1998;95(3):1154–1159. doi: 10.1073/pnas.95.3.1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blum KS, Pabst R. Lymphocyte numbers and subsets in the human blood. Do they mirror the situation in all organs? Immunol Lett. 2007;108(1):45–51. doi: 10.1016/j.imlet.2006.10.009. [DOI] [PubMed] [Google Scholar]
- 7.Chun TW, et al. Quantification of latent tissue reservoirs and total body viral load in HIV-1 infection. Nature. 1997;387(6629):183–188. doi: 10.1038/387183a0. [DOI] [PubMed] [Google Scholar]
- 8.Finzi D, et al. Identification of a reservoir for HIV-1 in patients on highly active antiretroviral therapy. Science. 1997;278(5341):1295–1300. doi: 10.1126/science.278.5341.1295. [DOI] [PubMed] [Google Scholar]
- 9.Wong JK, et al. Recovery of replication-competent HIV despite prolonged suppression of plasma viremia. Science. 1997;278(5341):1291–1295. doi: 10.1126/science.278.5341.1291. [DOI] [PubMed] [Google Scholar]
- 10.Eriksson S, et al. Comparative analysis of measures of viral reservoirs in HIV-1 eradication studies. PLoS Pathog. 2013;9(2): doi: 10.1371/journal.ppat.1003174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rouzioux C, Mélard A, Avéttand-Fénoël V. Quantification of total HIV1-DNA in peripheral blood mononuclear cells. Methods Mol Biol. 2014;1087:261–270. doi: 10.1007/978-1-62703-670-2_21. [DOI] [PubMed] [Google Scholar]
- 12.Vandergeeten C, et al. Cross-clade ultrasensitive PCR-based assays to measure HIV persistence in large-cohort studies. J Virol. 2014;88(21):12385–12396. doi: 10.1128/JVI.00609-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Oliveira MF, et al. Comparative analysis of cell-associated HIV DNA levels in cerebrospinal fluid and peripheral blood by droplet digital PCR. PLoS One. 2015;10(10): doi: 10.1371/journal.pone.0139510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Martínez-Bonet M, et al. Establishment and replenishment of the viral reservoir in perinatally HIV-1-infected children initiating very early antiretroviral therapy. Clin Infect Dis. 2015;61(7):1169–1178. doi: 10.1093/cid/civ456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Christopherson C, et al. PCR-Based assay to quantify human immunodeficiency virus type 1 DNA in peripheral blood mononuclear cells. J Clin Microbiol. 2000;38(2):630–634. doi: 10.1128/jcm.38.2.630-634.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Strain MC, et al. Highly Precise measurement of HIV DNA by droplet digital PCR. PLoS One. 2013;8(4): doi: 10.1371/journal.pone.0055943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. doi: 10.21769/bioprotoc.1492. Massanella M, Gianella S, Lada SM, Richman DD, Strain MC. Quantification of Total and 2-LTR (Long terminal repeat) HIV DNA, HIV RNA and Herpesvirus DNA in PBMCs. Bio-protocol. 2015;5(11):e1492. http://www.bio-protocol.org/e1492. [DOI] [PMC free article] [PubMed]
- 18.O’Doherty U, Swiggard WJ, Jeyakumar D, McGain D, Malim MH. A sensitive, quantitative assay for human immunodeficiency virus type 1 integration. J Virol. 2002;76(21):10942–10950. doi: 10.1128/JVI.76.21.10942-10950.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mexas AM, et al. Concurrent measures of total and integrated HIV DNA monitor reservoirs and ongoing replication in eradication trials. AIDS. 2012;26(18):2295–2306. doi: 10.1097/QAD.0b013e32835a5c2f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liszewski MK, Yu JJ, O’Doherty U. Detecting HIV-1 integration by repetitive-sampling Alu-gag PCR. Methods. 2009;47(4):254–260. doi: 10.1016/j.ymeth.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brady T, et al. Quantitation of HIV DNA integration: effects of differential integration site distributions on Alu-PCR assays. J Virol Methods. 2013;189(1):53–57. doi: 10.1016/j.jviromet.2013.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Koelsch KK, et al. Dynamics of total, linear nonintegrated, and integrated HIV-1 DNA in vivo and in vitro. J Infect Dis. 2008;197(3):411–419. doi: 10.1086/525283. [DOI] [PubMed] [Google Scholar]
- 23.Vandegraaff N, Kumar R, Burrell CJ, Li P. Kinetics of human immunodeficiency virus type 1 (HIV) DNA integration in acutely infected cells as determined using a novel assay for detection of integrated HIV DNA. J Virol. 2001;75(22):11253–11260. doi: 10.1128/JVI.75.22.11253-11260.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Buzon MJ, et al. HIV-1 replication and immune dynamics are affected by raltegravir intensification of HAART-suppressed subjects. Nat Med. 2010;16(4):460–465. doi: 10.1038/nm.2111. [DOI] [PubMed] [Google Scholar]
- 25.Hatano H, et al. Increase in 2-long terminal repeat circles and decrease in D-dimer after raltegravir intensification in patients with treated HIV infection: a randomized, placebo-controlled trial. J Infect Dis. 2013;208(9):1436–1442. doi: 10.1093/infdis/jit453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pierson TC, et al. Intrinsic stability of episomal circles formed during human immunodeficiency virus type 1 replication. J Virol. 2002;76(8):4138–4144. doi: 10.1128/JVI.76.8.4138-4144.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bushman F. Measuring covert HIV replication during HAART: the abundance of 2-LTR circles is not a reliable marker. AIDS. 2003;17(5):749–750. doi: 10.1097/00002030-200303280-00014. [DOI] [PubMed] [Google Scholar]
- 28.Zhu W, et al. Rapid Turnover of 2-LTR HIV-1 DNA during early stage of highly active antiretroviral therapy. PLoS One. 2011;6(6): doi: 10.1371/journal.pone.0021081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pace MJ, et al. Directly infected resting CD4+ T cells can produce HIV Gag without spreading infection in a model of HIV latency. PLoS Pathog. 2012;8(7): doi: 10.1371/journal.ppat.1002818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gianella S, et al. Cytomegalovirus replication in semen is associated with higher levels of proviral HIV DNA and CD4+ T cell activation during antiretroviral treatment. J Virol. 2014;88(14):7818–7827. doi: 10.1128/JVI.00831-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bosman KJ, et al. Comparison of digital PCR platforms and semi-nested qPCR as a tool to determine the size of the HIV reservoir. Sci Rep. 2015;5: doi: 10.1038/srep13811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Soriano-Sarabia N, et al. Peripheral Vγ9Vδ2 T cells are a novel reservoir of latent HIV Infection. PLoS Pathog. 2015;11(10): doi: 10.1371/journal.ppat.1005201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Puertas MC, et al. Effect of lithium on HIV-1 expression and proviral reservoir size in the CD4+ T cells of antiretroviral therapy suppressed patients. AIDS. 2014;28(14):2157–2159. doi: 10.1097/QAD.0000000000000374. [DOI] [PubMed] [Google Scholar]
- 34.Fischer M, et al. Attenuated and nonproductive viral transcription in the lymphatic tissue of HIV-1-infected patients receiving potent antiretroviral therapy. J Infect Dis. 2004;189(2):273–285. doi: 10.1086/380797. [DOI] [PubMed] [Google Scholar]
- 35.Lassen KG, Ramyar KX, Bailey JR, Zhou Y, Siliciano RF. Nuclear retention of multiply spliced HIV-1 RNA in resting CD4+ T cells. PLoS Pathog. 2006;2(7): doi: 10.1371/journal.ppat.0020068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pasternak AO, et al. Highly sensitive methods based on seminested real-time reverse transcription-PCR for quantitation of human immunodeficiency virus type 1 unspliced and multiply spliced RNA and proviral DNA. J Clin Microbiol. 2008;46(7):2206–2211. doi: 10.1128/JCM.00055-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pasternak AO, et al. Cellular levels of HIV unspliced RNA from patients on combination antiretroviral therapy with undetectable plasma viremia predict the therapy outcome. PLoS One. 2009;4(12): doi: 10.1371/journal.pone.0008490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schmid A, et al. Profound depletion of HIV-1 transcription in patients initiating antiretroviral therapy during acute infection. PLoS One. 2010;5(10): doi: 10.1371/journal.pone.0013310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kiselinova M, Pasternak AO, De Spiegelaere W, Vogelaers D, Berkhout B, Vandekerckhove L. Comparison of droplet digital PCR and seminested real-time PCR for quantification of cell-associated HIV-1 RNA. PLoS One. 2014;9(1): doi: 10.1371/journal.pone.0085999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yerly S, Rutschmann OT, Opravil M, Marchal F, Hirschel B, Perrin L. Cell-associated HIV-1 RNA in blood as indicator of virus load in lymph nodes. The Swiss HIV Cohort Study. J Infect Dis. 1999;180(3):850–853. doi: 10.1086/314932. [DOI] [PubMed] [Google Scholar]
- 41.Fischer M, et al. Highly sensitive methods for quantitation of human immunodeficiency virus type 1 RNA from plasma, cells, and tissues. J Clin Microbiol. 1999;37(5):1260–1264. doi: 10.1128/jcm.37.5.1260-1264.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nugent CT, et al. Detection of HIV-1 in alternative specimen types using the APTIMA HIV-1 RNA Qualitative Assay. J Virol Methods. 2009;159(1):10–14. doi: 10.1016/j.jviromet.2009.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hatano H, et al. A randomized, controlled trial of raltegravir intensification in antiretroviral-treated, HIV-infected patients with a suboptimal CD4+ T cell response. J Infect Dis. 2011;203(7):960–968. doi: 10.1093/infdis/jiq138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gianella S, et al. Cytomegalovirus DNA in semen and blood is associated with higher levels of proviral HIV DNA. J Infect Dis. 2013;207(6):898–902. doi: 10.1093/infdis/jis777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bullen CK, Laird GM, Durand CM, Siliciano JD, Siliciano RF. New ex vivo approaches distinguish effective and ineffective single agents for reversing HIV-1 latency in vivo. Nat Med. 2014;20(4):425–429. doi: 10.1038/nm.3489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shan L, et al. A novel PCR assay for quantification of HIV-1 RNA. J Virol. 2013;87(11):6521–6525. doi: 10.1128/JVI.00006-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dornadula G, et al. Residual HIV-1 RNA in blood plasma of patients taking suppressive highly active antiretroviral therapy. JAMA. 1999;282(17):1627–1632. doi: 10.1001/jama.282.17.1627. [DOI] [PubMed] [Google Scholar]
- 48.Palmer S, et al. New real-time reverse transcriptase-initiated PCR assay with single-copy sensitivity for human immunodeficiency virus type 1 RNA in plasma. J Clin Microbiol. 2003;41(10):4531–4536. doi: 10.1128/JCM.41.10.4531-4536.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Palmer S, et al. Low-level viremia persists for at least 7 years in patients on suppressive antiretroviral therapy. Proc Natl Acad Sci U S A. 2008;105(10):3879–3884. doi: 10.1073/pnas.0800050105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sahu GK, Sarria JC, Cloyd MW. Recovery of replication-competent residual HIV-1 from plasma of a patient receiving prolonged, suppressive highly active antiretroviral therapy. J Virol. 2010;84(16):8348–8352. doi: 10.1128/JVI.00362-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hermankova M, et al. Analysis of human immunodeficiency virus type 1 gene expression in latently infected resting CD4+ T lymphocytes in vivo. J Virol. 2003;77(13):7383–7392. doi: 10.1128/JVI.77.13.7383-7392.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kieffer TL, et al. Genotypic analysis of HIV-1 drug resistance at the limit of detection: virus production without evolution in treated adults with undetectable HIV loads. J Infect Dis. 2004;189(8):1452–1465. doi: 10.1086/382488. [DOI] [PubMed] [Google Scholar]
- 53.Bailey JR, et al. Residual human immunodeficiency virus type 1 viremia in some patients on antiretroviral therapy is dominated by a small number of invariant clones rarely found in circulating CD4+ T cells. J Virol. 2006;80(13):6441–6457. doi: 10.1128/JVI.00591-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wagner TA, et al. HIV latency. Proliferation of cells with HIV integrated into cancer genes contributes to persistent infection. Science. 2014;345(6196):570–573. doi: 10.1126/science.1256304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Maldarelli F, et al. Specific HIV integration sites are linked to clonal expansion and persistence of infected cells. Science. 2014;345(6193):179–183. doi: 10.1126/science.1254194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cillo AR, et al. Improved single-copy assays for quantification of persistent HIV-1 viremia in patients on suppressive antiretroviral therapy. J Clin Microbiol. 2014;52(11):3944–3951. doi: 10.1128/JCM.02060-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chun TW, et al. Presence of an inducible HIV-1 latent reservoir during highly active antiretroviral therapy. Proc Natl Acad Sci U S A. 1997;94(24):13193–13197. doi: 10.1073/pnas.94.24.13193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ylisastigui L, Archin NM, Lehrman G, Bosch RJ, Margolis DM. Coaxing HIV-1 from resting CD4 T cells: histone deacetylase inhibition allows latent viral expression. AIDS. 2004;18(8):1101–1108. doi: 10.1097/00002030-200405210-00003. [DOI] [PubMed] [Google Scholar]
- 59.Archin NM, et al. Valproic acid without intensified antiviral therapy has limited impact on persistent HIV infection of resting CD4+ T cells. AIDS. 2008;22(10):1131–1135. doi: 10.1097/QAD.0b013e3282fd6df4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lehrman G, et al. Depletion of latent HIV-1 infection in vivo: a proof-of-concept study. Lancet. 2005;366(9485):549–555. doi: 10.1016/S0140-6736(05)67098-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Siliciano JD, et al. Long-term follow-up studies confirm the stability of the latent reservoir for HIV-1 in resting CD4+ T cells. Nat Med. 2003;9(6):727–728. doi: 10.1038/nm880. [DOI] [PubMed] [Google Scholar]
- 62.Finzi D, et al. Latent infection of CD4+ T cells provides a mechanism for lifelong persistence of HIV-1, even in patients on effective combination therapy. Nat Med. 1999;5(5):512–517. doi: 10.1038/8394. [DOI] [PubMed] [Google Scholar]
- 63.Siliciano JD, Siliciano RF. Enhanced culture assay for detection and quantitation of latently infected, resting CD4+ T-cells carrying replication-competent virus in HIV-1-infected individuals. Methods Mol Biol. 2005;304:3–15. doi: 10.1385/1-59259-907-9:003. [DOI] [PubMed] [Google Scholar]
- 64.Soriano-Sarabia N, et al. Quantitation of replication-competent HIV-1 in populations of resting CD4+ T cells. J Virol. 2014;88(24):14070–14077. doi: 10.1128/JVI.01900-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Morón-López S, et al. Short-term treatment with IFNα diminishes expression of HIV-1 and reduces CD4+ T-cell activation in HIV/HCV-coinfected patients on antiretroviral therapy. J Infect Dis. doi: 10.1093/infdis/jiv5. [published online ahead of print: November 1, 2015]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rosenbloom DI, et al. Designing and interpreting limiting dilution assays: general principles and applications to the latent reservoir for Human Immunodeficiency Virus-1. Open Forum Infect Dis. 2015;2(4):ofv123. doi: 10.1093/ofid/ofv123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ho YC, et al. Replication-competent noninduced proviruses in the latent reservoir increase barrier to HIV-1 cure. Cell. 2013;155(3):540–551. doi: 10.1016/j.cell.2013.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Laird GM, et al. Rapid quantification of the latent reservoir for HIV-1 using a viral outgrowth assay. PLoS Pathog. 2013;9(5): doi: 10.1371/journal.ppat.1003398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Richman DD. Detection and Quantification of HIV Reservoirs: Measuring the latent reservoir. Presented at: Keystone Symposia. Mechanisms of HIV Persistence: Implications for a Cure 2015; April 26–May 1, 2015; Boston, Massachusetts, USA. [Google Scholar]
- 70.Weinberger AD, Weinberger LS. Stochastic fate selection in HIV-infected patients. Cell. 2013;155(3):497–499. doi: 10.1016/j.cell.2013.09.039. [DOI] [PubMed] [Google Scholar]
- 71.Rosenbloom DIS, Elliott O, Hill AL, Henrich TJ, Siliciano JL, Siliciano JF. Designing and interpreting limiting dilution assays: general principles and applications to the latent reservoir for Human Immunodeficiency Virus-1. Open Forum Infect Dis. 2015;2(4) doi: 10.1093/ofid/ofv123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Mohammadi P, et al. Dynamics of HIV latency and reactivation in a primary CD4+ T cell model. PLoS Pathog. 2014;10(5): doi: 10.1371/journal.ppat.1004156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Cillo AR, et al. Quantification of HIV-1 latency reversal in resting CD4+ T cells from patients on suppressive antiretroviral therapy. Proc Natl Acad Sci U S A. 2014;111(19):7078–7083. doi: 10.1073/pnas.1402873111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Laird GM, et al. Ex vivo analysis identifies effective HIV-1 latency-reversing drug combinations. J Clin Invest. 2015;125(5):1901–1912. doi: 10.1172/JCI80142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Elliott JH, et al. Activation of HIV transcription with short-course vorinostat in HIV-infected patients on suppressive antiretroviral therapy. PLoS Pathog. 2014;10(11): doi: 10.1371/journal.ppat.1004473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Procopio FA, et al. A novel assay to measure the magnitude of the inducible viral reservoir in HIV-infected individuals. EBioMedicine. 2015;2(8):872–881. doi: 10.1016/j.ebiom.2015.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Webster WA, Casey GA, Charlton KM. The mouse inoculation test in rabies diagnosis: early diagnosis in mice during the incubation period. Can J Comp Med. 1976;40(3):322–325. [PMC free article] [PubMed] [Google Scholar]
- 78.Burnet FM. The use of the developing egg in virus research. Med Res Council Spec Rep Ser. 1936: [Google Scholar]
- 79. Okoye AA, et al. Early Treatment in Acute SIV Infection Limits the Size and Distribution of the Viral Reservoir. Presented at: Conference on Retroviruses and Opportunistic Infections 2014; March 3–6, 2014; Boston, Massachusetts, USA. Abstract 136LB. [Google Scholar]
- 80.Hansen SG, et al. Immune clearance of highly pathogenic SIV infection. Nature. 2013;502(7469):100–104. doi: 10.1038/nature12519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lifson JD, et al. Evaluation of the safety, immunogenicity, and protective efficacy of whole inactivated simian immunodeficiency virus (SIV) vaccines with conformationally and functionally intact envelope glycoproteins. AIDS Res Hum Retroviruses. 2004;20(7):772–787. doi: 10.1089/0889222041524661. [DOI] [PubMed] [Google Scholar]
- 82.Denton PW, García JV. Humanized mouse models of HIV infection. AIDS Rev. 2011;13(3):135–148. [PMC free article] [PubMed] [Google Scholar]
- 83.Metcalf Pate KA, et al. A murine viral outgrowth assay to detect residual HIV type 1 in patients with undetectable viral loads. J Infect Dis. 2015;212(9):1387–1396. doi: 10.1093/infdis/jiv230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wit FW, et al. Safety of long-term interruption of successful antiretroviral therapy: the ATHENA cohort study. AIDS. 2005;19(3):345–348. [PubMed] [Google Scholar]
- 85.Campbell JH, Hearps AC, Martin GE, Williams KC, Crowe SM. The importance of monocytes and macrophages in HIV pathogenesis, treatment, and cure. AIDS. 2014;28(15):2175–2187. doi: 10.1097/QAD.0000000000000408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Calantone N, et al. Tissue myeloid cells in SIV-infected primates acquire viral DNA through phagocytosis of infected T cells. Immunity. 2014;41(3):493–502. doi: 10.1016/j.immuni.2014.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Haase AT. Population biology of HIV-1 infection: viral and CD4+ T cell demographics and dynamics in lymphatic tissues. Annu Rev Immunol. 1999;17:625–656. doi: 10.1146/annurev.immunol.17.1.625. [DOI] [PubMed] [Google Scholar]
- 88.Pakker NG, et al. Biphasic kinetics of peripheral blood T cells after triple combination therapy in HIV-1 infection: a composite of redistribution and proliferation. Nat Med. 1998;4(2):208–214. doi: 10.1038/nm0298-208. [DOI] [PubMed] [Google Scholar]
- 89.Costiniuk CT, Jenabian MA. Cell-to-cell transfer of HIV infection: implications for HIV viral persistence. J Gen Virol. 2014;95(pt 11):2346–2355. doi: 10.1099/vir.0.069641-0. [DOI] [PubMed] [Google Scholar]
- 90.Sathaliyawala T, et al. Distribution and compartmentalization of human circulating and tissue-resident memory T cell subsets. Immunity. 2013;38(1):187–197. doi: 10.1016/j.immuni.2012.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Yukl SA, et al. The distribution of HIV DNA and RNA in cell subsets differs in gut and blood of HIV-positive patients on ART: implications for viral persistence. J Infect Dis. 2013;208(8):1212–1220. doi: 10.1093/infdis/jit308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Cu-Uvin S, et al. Genital tract HIV-1 RNA shedding among women with below detectable plasma viral load. AIDS. 2010;24(16):2489–2497. doi: 10.1097/QAD.0b013e32833e5043. [DOI] [PubMed] [Google Scholar]
- 93.Lambert-Niclot S, et al. Detection of HIV-1 RNA in seminal plasma samples from treated patients with undetectable HIV-1 RNA in blood plasma on a 2002-2011 survey. AIDS. 2012;26(8):971–975. doi: 10.1097/QAD.0b013e328352ae09. [DOI] [PubMed] [Google Scholar]
- 94.Bednar MM, et al. Compartmentalization, viral evolution, and viral latency of HIV in the CNS. Curr HIV/AIDS Rep. 2015;12(2):262–271. doi: 10.1007/s11904-015-0265-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Hellmuth J, Valcour V, Spudich S. CNS reservoirs for HIV: implications for eradication. J Virus Erad. 2015;1(2):67–71. [PMC free article] [PubMed] [Google Scholar]
- 96.Pillai SK, et al. Semen-specific genetic characteristics of human immunodeficiency virus type 1 env. J Virol. 2005;79(3):1734–1742. doi: 10.1128/JVI.79.3.1734-1742.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Henrich TJ, et al. Antiretroviral-free HIV-1 remission and viral rebound after allogeneic stem cell transplantation: report of 2 cases. Ann Intern Med. 2014;161(5):319–327. doi: 10.7326/M14-1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Luzuriaga K, et al. Viremic relapse after HIV-1 remission in a perinatally infected child. N Engl J Med. 2015;372(8):786–788. doi: 10.1056/NEJMc1413931. [DOI] [PMC free article] [PubMed] [Google Scholar]


