Abstract
Evolutionarily conserved receptor tyrosine kinase–like orphan receptor-1 and -2 (ROR1/2) are considered distinct receptors for Wnt5a and are implicated in noncanonical Wnt signaling in organogenesis and cancer metastasis. We found that Wnt5a enhanced proliferation and migration of chronic lymphocytic leukemia (CLL) cells and that these effects were blocked by the humanized anti-ROR1 mAb cirmtuzumab (UC-961). Treatment of CLL cells with Wnt5a induced ROR1 to oligomerize with ROR2 and recruit guanine exchange factors (GEFs), which activated Rac1 and RhoA; siRNA-mediated silencing of either ROR1 or ROR2 or treatment with UC-961 inhibited these effects. Using the ROR1-deficient CLL cell line MEC1, we demonstrated that ectopic ROR1 expression induced ROR1/ROR2 heterooligomers, which recruited GEFs, and enhanced proliferation, cytokine-directed migration, and engraftment potential of MEC1 cells in immune-deficient mice. Notably, treatment with UC-961 inhibited engraftment of ROR1+ leukemia cells in immune-competent ROR1-transgenic mice. Molecular analysis revealed that the extracellular Kringle domain is required for ROR1/ROR2 heterooligomerization and the cysteine-rich domain or intracellular proline-rich domain is required for Wnt5a-induced recruitment of GEFs to ROR1/ROR2. This study identifies an interaction between ROR1 and ROR2 that is required for Wnt5a signaling that promotes leukemia chemotaxis and proliferation.
Keywords: Hematology, Oncology
Introduction
The receptor tyrosine kinase–like orphan receptors ROR1 and ROR2 are evolutionarily conserved type I proteins (1–8). Studies on the developmental expression patterns of these proteins in Caenorhabditis elegans, Drosophila melanogaster, Xenopus laevis, and Mus musculus have shown striking conservation (9). ROR1 and ROR2 are expressed at the highest levels during the early stages of embryogenesis, being represented in most of the major systems in tissues derived from all 3 germ layers, but most prominently the neural crest. Notably, ROR1 expression is largely restricted to the neural mesenchyme (10, 11). Complete knockout of either ROR1 or ROR2, however, results in pervasive developmental abnormalities involving the heart, lungs, urogenital tracts, and other organs, suggesting that each potentially contributes broadly to organogenesis (12, 13).
Although low levels of ROR2 can be found on some adult tissues, postpartum expression of ROR1 is not apparent, except on a small subset of precursor B cells called hematogones (14). However, ROR1 can be found on the leukemia cells of patients with chronic lymphocytic leukemia (CLL) (15–17), and either ROR1 or ROR2 is expressed by neoplastic cells of a variety of different cancers (18, 19). Cancer-cell expression of ROR1 or ROR2 has been associated with enhanced cancer-cell migration, epithelial-messenchymal transition (EMT), increased associated risk for relapse and metastasis, and unfavorable prognosis (20, 21). More recently, ROR1 was identified on ovarian cancer stem cells, which have enhanced capacity for migration/spheroid formation in vitro and engraftment/metastasis in vivo (22).
ROR1 and ROR2 each may function as a receptor for Wnt5a (15, 23), which may induce noncanonical Wnt signaling, potentially leading to enhanced tumor-cell growth, directional migration, and/or tissue-cell polarity during organogenesis (18, 24–26). On the other hand, ROR2 also can repress transcription of Wnt target genes and modulate Wnt signaling by sequestering canonical Wnt ligands, thereby serving as a tumor suppressor in different cell contexts (27, 28). Although studies have shown that ROR1 or ROR2 can associate with other proteins to modify canonical Wnt signaling or induce noncanonical signaling (29, 30), ROR1 and ROR2 are each considered to function independently of one another. We examined primary CLL cells for ROR1-associated proteins and made the unexpected discovery that ROR1 formed heterooligomers with ROR2 in response to Wnt5a to recruit guanine exchange factors (GEFs) that activate Rho GTPases, which can enhance leukemia-cell chemotaxis and proliferation.
Results
Wnt5a enhances CLL proliferation and migration.
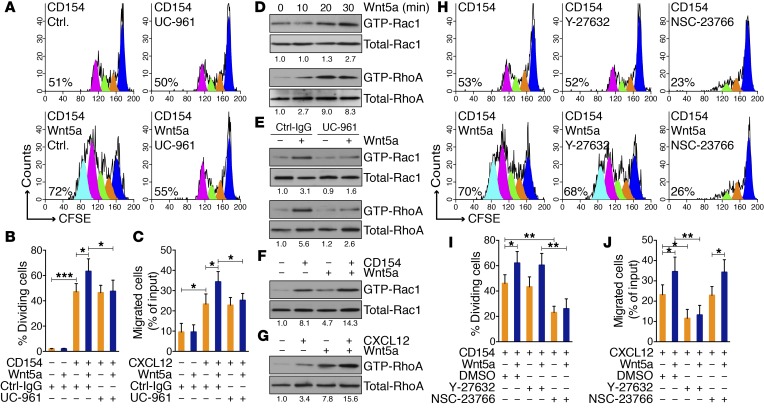
CLL cells can be induced to proliferate upon culture with cells expressing CD154 (HeLaCD154) in the presence of exogenous IL-4 and IL-10 (Figure 1, A and B). Addition of exogenous Wnt5a significantly enhanced the proportion of dividing cells and the number of cell divisions, as deduced from the fluorescence of cells labeled with CFSE; the enhanced proliferation induced by Wnt5a was inhibited by an anti-ROR1 mAb (UC-961) to levels comparable to those observed in cultures without Wnt5a (Figure 1, A and B). In contrast, CLL cells cocultured with HeLa cells were not induced to proliferate, even in the presence of IL-4/10 and/or Wnt5a (Supplemental Figure 1A; supplemental material available online with this article; doi:10.1172/JCI83535DS1).
Figure 1. Wnt5a can enhance CLL cell proliferation and migration.
(A) Fluorescence of CLL cells stained with CFSE and treated with CD154 with nonspecific IgG (Ctrl.) or UC-961 without (–) or with (+) Wnt5a. The percentage of dividing cells is indicated in each histogram. (B) Mean proportions of dividing CLL cells from each of 6 patients under conditions indicated at the bottom. (C) Mean proportions of CLL cells (n = 6) migrating in response to CXCL12 with Ctrl-IgG or UC-961, without (–) or with (+) Wnt5a, as indicated below. (D) Immunoblots of activated GTPase (top) or total GTPase (bottom) in parallel gels following treatment with Wnt5a for the times indicated on top (in minutes). Numbers below are the ratios of band densities of activated versus total GTPase normalized to that of untreated samples. (E) Immunoblots of activated or total GTPase in CLL cells treated with Ctrl-IgG or UC-961 without (–) or with (+) Wnt5a for 30 minutes. (F) Immunoblot of activated Rac1 in CLL cells treated with CD154 without (–) or with (+) Wnt5a for 30 minutes. (G) Immunoblot of activated or total RhoA in CLL cells treated with CXCL12 without (–) or with (+) Wnt5a for 30 minutes. (H) Fluorescence of CLL cells stained with CFSE and treated with CD154 without (–) or with (+) Wnt5a and without or with a Rac1 inhibitor (NSC-23766) or a RhoA inhibitor (Y-27632). (I) Mean proportions of CLL cells with diminished CFSE fluorescence from each of 6 patients in culture conditions indicated below. (J) Mean proportions of CLL cells (n = 6) that migrated in response to CXCL12 without (–) or with (+) Wnt5a and without or with NSC-23766 or Y-27632. Data are shown as mean ± SD. *P < 0.05; **P < 0.01; ***P < 0.001, as determined by 2-tailed Student’s t test.
We confirmed that exogenous Wnt5a also enhanced migration of CLL cells toward chemokines, e.g., CXCL12 (Figure 1C and ref. 31). The capacity of Wnt5a to enhance migration was inhibited by UC-961. However, exogenous Wnt5a without CXCL12 did not induce CLL-cell migration, and UC-961 did not inhibit the migration of CLL cells to CXCL12 without Wnt5a (Figure 1C).
Rho family proteins play important roles in regulating proliferation and/or migration (32), and Wnt5a has been reported to activate Rac1 and RhoA in other cell types (33, 34). We observed that Wnt5a induced activation of Rac1 and RhoA within 30 minutes (Figure 1D and Supplemental Figure 1B). Addition of UC-961 inhibited Wnt5a-induced activation of Rac1 and RhoA (Figure 1E and Supplemental Figure 1C). Coculture of CLL cells with HeLaCD154, but not HeLa, cells also induced activation of Rac1 (Figure 1F and Supplemental Figure 1D). Also, CXCL12 activated RhoA in CLL cells (Figure 1G, Supplemental Figure 1E, and ref. 35). In each case, exogenous Wnt5a enhanced the level of Rac1 or RhoA activated by CD154 or CXCL12, respectively (Figure 1, F and G). NSC-23766, an inhibitor of Rac1 GTPase, but not Y-27632, a selective inhibitor of p160ROCK, inhibited the proliferation induced by HeLaCD154 with or without exogenous Wnt5a. On the other hand, Y-27632, but not NSC-23766, inhibited chemotaxis to CXCL12 with or without exogenous Wnt5a, supporting the notion that activation of Rac1 or RhoA can promote CLL-cell proliferation or migration, respectively (Figure 1, H–J).
ROR1 oligomerizes with ROR2 in the context of Wnt5a.
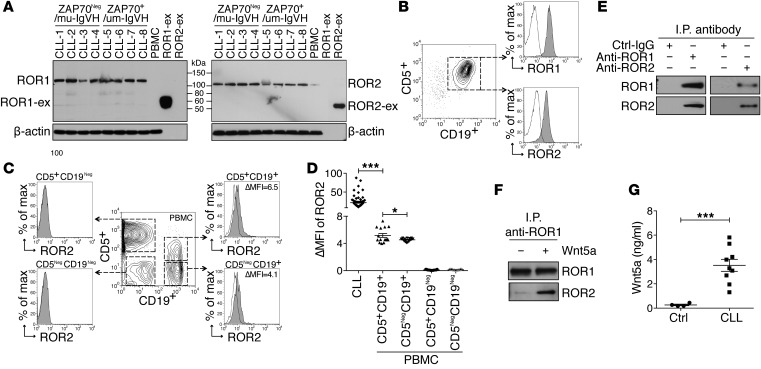
We performed mass spectrometry–based (MS-based) proteomic analysis on anti-ROR1 immune precipitates from CLL-cell lysates. Surprisingly, we detected ROR2 in addition to ROR1 (Supplemental Figure 2A). Detecting ROR2 was unexpected, as one group of investigators reported CLL cells specifically lacked expression of ROR2 (17). However, we detected ROR2 mRNA in isolated CLL cells (Supplemental Figure 2B) and both ROR1 and ROR2 in all samples examined by immunoblot analysis (Figure 2A). Surface expression of both proteins also was detected on CD5+CD19+ CLL cells via flow cytometry (Figure 2, B and D, and Supplemental Figure 2C).
Figure 2. ROR1 couples with ROR2.
(A) Immunoblot analysis for ROR1 or ROR2 in lysates of CLL cells that were ZAP-70Neg and used mutated immunoglobulin heavy chain variable region genes (IGHV) (CLL 1–4) or were ZAP-70+ and used unmutated IGHV (CLL 5–8) or lysates of PBMCs from healthy adults. Purified recombinant extracellular ROR1 or ROR2 (ROR1-ex or ROR2-ex) was loaded onto separate lanes as controls. (B) Detection of ROR1 or ROR2 on CD5+CD19+ CLL cells by flow cytometry. (C) PBMCs of healthy adults were stained with anti–ROR2–Alexa Fluor 488, anti–CD19-PE, and anti–CD5-APC mAbs and analyzed by flow cytometry. The gating strategy is indicated in the center contour plot for each subgroup specified on the top of each histogram depicting the fluorescence of cells incubated with the anti-ROR2 mAb (gray histograms) versus an Alexa Fluor 488–conjugated Ctrl-IgG (white histograms). The ΔMFI for each of the CD19+ cell subsets is indicated in the top right. (D) ΔMFI for ROR2 of CLL samples (n = 80) or each of the gated lymphocyte subsets in PBMCs of healthy donors (n = 15). (E) Immunoblot analysis of anti-ROR1 or anti-ROR2 immune precipitates from lysates of freshly isolated CLL cells detecting the association of ROR1 with ROR2. (F) Immunoblot analysis of anti-ROR1 immune precipitates from lysates of CLL cells cultured in serum-free media and then treated without (−) or with (+) Wnt5a. (G) Wnt5a levels were assessed via ELISA in the plasma of CLL patients (n = 9) or age-matched healthy control subjects (n = 9). Data are shown as mean ± SEM. *P < 0.05; ***P < 0.001, as determined by 2-tailed Student’s t test.
We found that CD19+ blood B cells of healthy adults also expressed ROR2, including B cells that coexpressed CD5 (Figure 2C). We subtracted the mean fluorescence intensity (MFI) of cells stained with a fluorochrome-labeled, isotype-control mAb from the MFI of cells stained with anti-ROR2 to determine the ΔMFI. The mean ROR2 ΔMFI in CD5+CD19+ B cells of healthy subjects (5.1 ± 0.3; n = 15), was higher than that of CD5NegCD19+ B cells (4.5 ± 0.1), but still significantly lower than the mean ROR2 ΔMFI for CLL cells (21.8 ± 1.8, n = 80) (Figure 2D). We did not detect ROR2 on CD19Neg blood lymphocytes (Figure 2, C and D) or ROR1 on the mononuclear cells of healthy donors (Supplemental Figure 2C). Immunoblot analysis of anti-ROR1 or anti-ROR2 immune precipitates using CLL-cell lysates confirmed that ROR1 was coupled with ROR2 in CLL cells freshly isolated from patient blood samples (Figure 2E). However, when these CLL cells were cultured in media overnight, the association between ROR1 and ROR2 became less apparent, unless exogenous Wnt5a was added to the culture medium (Figure 2F). These data indicate that ROR1/ROR2 heterooligomers already were present on CLL cells in vivo. Such heterooligomers probably formed in response to endogenous Wnt5a, which we detected at high levels in the sera of patients with CLL relative to those of aged-matched control subjects (Figure 2G).
UC-961 disrupts Wnt5a-induced coupling of ROR1 with ROR2.
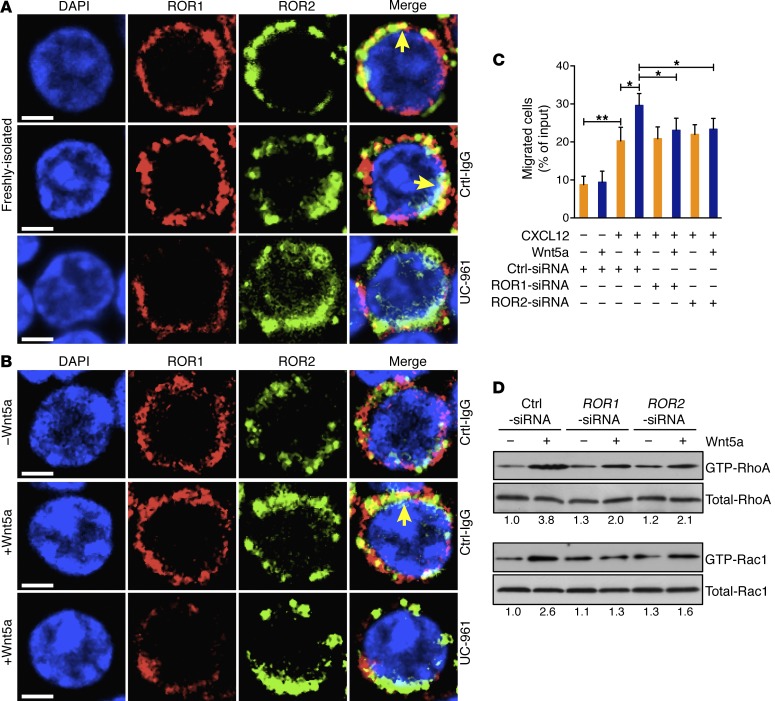
We performed fluorescence confocal microscopy, using a non-crossblocking mAb (4A5) specific for a ROR1 epitope distinct from that recognized by UC-961. This demonstrated that ROR1 colocalized with ROR2 in freshly isolated CLL cells (Figure 3A and Supplemental Figure 3A), but not with CD5 or CD19 (Supplemental Figure 3B). However, we detected little if any colocalization of ROR1 with ROR2 in CLL cells cultured in media, unless they were treated with exogenous Wnt5a (Figure 3B and Supplemental Figure 3C). Incubation of freshly isolated or Wnt5a-treated CLL cells with UC-961 apparently disrupted the ROR1/ROR2 heterooligomer, which otherwise was readily observed in freshly isolated or Wnt5a-treated CLL cells incubated with a nonspecific IgG (Ctrl-IgG) (Figure 3, A and B).
Figure 3. UC-961 inhibits Wnt5a-induced coupling of ROR1 with ROR2 and GTPase activation.
(A) Colocalization (yellow, with arrow) of ROR1 (red) with ROR2 (green) detected by confocal microscopy in freshly isolated CLL cells with or without Ctrl-IgG or UC-961, as indicated on the right margin of each row. Objective, ×100. Scale bars: 2 μm. (B) Confocal microscopy of serum-starved CLL cells stained for ROR1 and ROR2 after treatment with Ctrl-IgG or UC-961 without (–) or with (+) Wnt5a, presented as in panel A. Objective, ×100. Scale bars: 2 μm. (C) Mean proportions of CLL cells migrating toward CXCL12 without (–) or with (+) Wnt5a in samples (n = 6) transfected with control siRNA (Ctrl-siRNA) or siRNA specific for ROR1 or ROR2. Data are shown as mean ± SD. *P < 0.05; **P < 0.01, as determined by 2-tailed Student’s t test. (D) Activated RhoA or Rac1 was measured by Rho family protein activity pull-down assays on lysates of CLL cells transfected with Ctrl-siRNA or siRNA specific for ROR1 or ROR2 and cultured with or without Wnt5a. Whole-cell lysates were run on parallel gels to determine total RhoA or Rac1. Numbers below each lane are ratios of the band densities of activated versus total GTPase, normalized with respect to that of untreated samples.
Transfecting CLL cells with siRNA specific for ROR1 or ROR2, but not control siRNA, respectively, lowered expression of only ROR1 or ROR2 by immunoblot analysis or flow cytometry (Supplemental Figure 3, D and E). Silencing either ROR1 or ROR2 inhibited the capacity of Wnt5a to enhance CLL-cell migration to CXCL12 (Figure 3C) or to induce activation of RhoA or Rac1 (Figure 3D and Supplemental Figure 3F), indicating that optimal Wnt5a-induced signaling was dependent on the coexpression of both ROR1 and ROR2.
ROR1/ROR2 heterooligomers recruit GEFs.
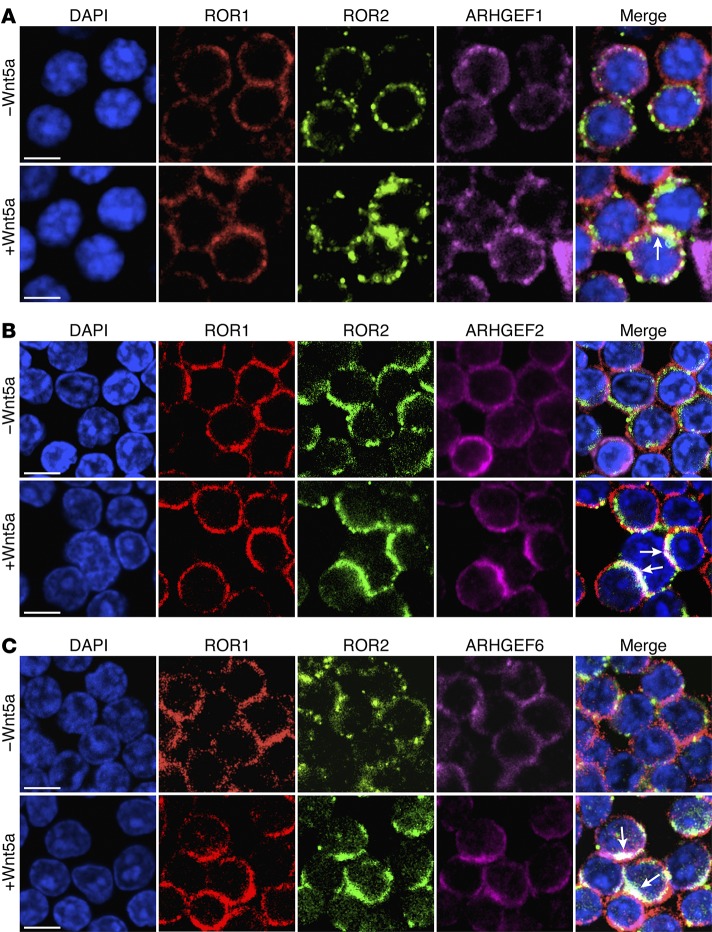
We performed MS analysis on anti-ROR1 immune precipitates generated from lysates of freshly isolated CLL cells. In addition to ROR1 and ROR2, we detected ARHGEF1, ARHGEF2, and ARHGEF6 in the anti-ROR1 immune precipitates (Supplemental Figure 4A). The association of each of these GEFs with ROR1 was confirmed by immunoblot analysis of the anti-ROR1 immune precipitates (Supplemental Figure 4B). Furthermore, the immune precipitates generated from lysates of freshly isolated CLL cells using mAbs specific for either ARHGEF1, ARHGEF2, or ARHGEF6 each contained ROR1 detectable by immunoblot analysis (Supplemental Figure 4B). Fluorescence confocal microscopy showed that ROR1 and ROR2 colocalized with ARHGEF1, ARHGEF2, or ARHGEF6 in cultured CLL cells that were treated with exogenous Wnt5a (Figure 4, A–C, and Supplemental Figure 4C).
Figure 4. UC-961 inhibits Wnt5a-induced recruitment of GEFs.
(A) Colocalization (white, with arrow) of ARHGEF1 (purple) with ROR1 (red) and ROR2 (green) in CLL cells cultured without (–) or with (+) Wnt5a, as indicated on the left margin or each row. Objective, ×100. Scale bars: 5 μm. (B) Colocalization of ARHGEF2 (purple) with ROR1/ROR2 in CLL cells cultured without (–) or with (+) Wnt5a, as in panel A. Objective, ×100. Scale bars: 5 μm. (C) Colocalization of ARHGEF6 (purple) with ROR1/ROR2 in CLL cells cultured without (–) or with (+) Wnt5a, as in A. Objective, ×100. Scale bars: 5 μm.
However, treatment of the same cells with UC-961 inhibited the capacity of Wnt5a to induce recruitment of any one of these GEFs to either ROR1 or ROR2 (Supplemental Figure 5, A and B). Each of these GEFs was detected in CLL cells before and after 2 days of culture with CD154-bearing cells (Supplemental Figure 6A), which may induce expression of other GEFs, such as Tiam1 (35). We found that treatment of cultured CLL cells with Wnt5a increased the in vitro exchange activity for RhoA of immune precipitates generated with mAbs specific for either ARHGEF1 or ARHGEF2, but not ARHGEF6. Moreover, treatment of cultured CLL cells with Wnt5a increased the in vitro exchange activity for Rac1 using immune precipitates generated with mAbs specific for either ARHGEF2 or ARHGEF6, but not ARHGEF1 (Figure 5A and Supplemental Figure 6B). Treatment of the CLL cells with UC-961 inhibited the capacity of Wnt5a to induce activation of RhoA by immune precipitates generated from anti-ARHGEF1 or anti-ARHGEF2 or of Rac1 by immune precipitates of anti-ARHGEF2 or anti-ARHGEF6, respectively (Figure 5B).
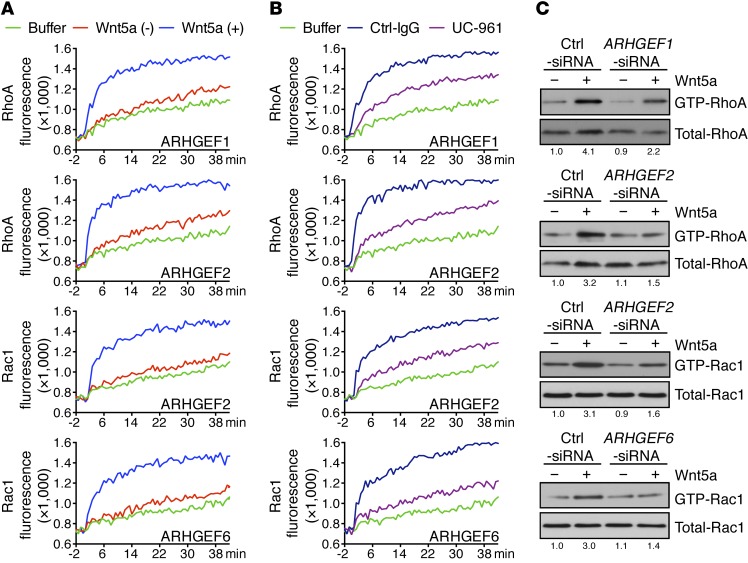
Figure 5. UC-961 inhibits Wnt5a-induced activation of RhoA and Rac1.
(A) In vitro exchange over time (in minutes) of RhoA (top 2 rows of graphs) or Rac1 (bottom 2 row of graphs) in immune precipitates using mAbs specific for ARHGEF1, ARHGEF2, or ARHGEF6 on lysates of CLL cells cultured with (blue lines) or without (red lines) Wnt5a, as indicated in the lower right of each graph. The green lines depict GTPase activation observed using buffer alone. (B) In vitro exchange assay on RhoA or Rac1 of immune precipitates from lysates of CLL cells cultured with UC-961 (purple lines) or Ctrl-IgG (blue lines), using mAbs specific for ARHGEF1, ARHGEF2, or ARHGEF6, as indicated in the bottom of each graph. The green lines depict GTPase activation using buffer alone. (C) Activation of RhoA or Rac1 following treatment without (–) or with (+) Wnt5a of CLL cells transfected with Ctrl-siRNA or siRNA specific for ARHGEF1, ARHGEF2, or ARHGEF6. Whole-cell lysates were run on parallel gels to determine total RhoA or Rac1. The number beneath each lane is the ratio of band densities for activated versus total GTPase normalized to that of untreated samples.
Wnt5a was less effective in activating RhoA in CLL cells transfected with siRNA specific for either ARHGEF1 or ARHGEF2 than in CLL cells transfected with control siRNA. On the other hand, Wnt5a was less effective in activating Rac1 in CLL cells transfected with siRNA specific for either ARHGEF2 or ARHGEF6 (Figure 5C and Supplemental Figure 6, C and D). However, CLL cells silenced for ARHGEF1 or ARHGEF6, respectively, did not have impaired Wnt5a-induced activation of Rac1 or RhoA (Supplemental Figure 6E), indicating that ARHGEF1 and ARHGEF2, or ARHGEF2 and ARHGEF6, were required for optimal Wnt5a-induced activation of RhoA or Rac1, respectively.
Structural domains of ROR1 required for Wnt5a-induced activation of RhoA and Rac1.
MEC1 cells were derived from human CLL cells and have been used as a cell model for this leukemia (36). However, we found that MEC1 cells lacked ROR1, but did express ROR2 (Supplemental Figure 7A). Stable transfection of MEC1 with vectors encoding full-length ROR1 or each of various truncated forms of ROR1 lacking distinct structural domains allowed us to generate MEC1 that expressed high levels of surface ROR1, which was detected via flow cytometry using the non-crossblocking anti-ROR1 mAb 4A5 (Supplemental Figure 7A). Expression of ROR1 or any one of the various truncated forms of ROR1 did not alter the expression levels of surface ROR2 (Supplemental Figure 7A).
Because MEC1 cells express high levels of Wnt5a (Supplemental Figure 7B and ref. 31), we assessed for ROR1/ROR2 heterooligomers on MEC1 cells made to express ROR1. We detected ROR1/ROR2 heterooligomers in each of the transfectants except for MEC1 transfected with ROR1 lacking the extracellular KNG domain (MEC1-ΔKNG) (Figure 6A and Supplemental Figure 7, C and D). Furthermore, fluorescence confocal microscopy demonstrated that ROR1 and ROR2 colocalized with ARHGEF1, ARHGEF2, and ARHGEF6 in MEC1-ROR1 cells (Figure 6B and Supplemental Figure 8A). However, we did not observe colocalization of ROR1 or ROR2 with any one of these GEFs in MEC1 cells or MEC1 cells that expressed any one of our truncated forms of ROR1 (Supplemental Figure 8, B and C, and Supplemental Figure 9, A and B). MEC1-ROR1 cells also had higher levels of activated Rac1 and RhoA than MEC1 (Figure 6C and Supplemental Figure 10A); such enhanced activation was inhibited by treatment with either UC-961 or neutralizing antibodies to Wnt5a (Figure 6D and Supplemental Figure 10B). Moreover, the MEC1 cells transfected with any one of the various truncated forms of ROR1 did not have levels of activated Rac1 or RhoA that were greater than those of the parental MEC1 cells.
Figure 6. Structural domains of ROR1 required for activation of RhoA or Rac1.
(A) Schematic depicts the structure of ROR1 or truncated forms of ROR1. Interaction of ROR1 with ROR2 was confirmed by immunoblot analysis of anti-ROR1 immune precipitates using lysates from nontransfected MEC1 (Ctrl.), MEC1-ROR1 (ROR1), or MEC1 cells transfected with each of the various truncated forms of ROR1. (B) Colocalization (white, with arrow) of ARHGEF1, ARHGEF2 or ARHGEF6 (purple) with ROR1 (red) and ROR2 (green) in MEC1-ROR1 cells. Objective, ×100. Scale bars: 2 μm. (C) Activated Rac1 or RhoA was measured in MEC1 (Ctrl.), MEC1-ROR1 (ROR1), or MEC1 cells expressing each of the truncated forms of ROR1. (D) Activated Rac1 or RhoA in MEC1 or MEC1-ROR1 cells treated with nonspecific IgG (Crtl-IgG), UC-961, or anti-Wnt5a. Whole-cell lysates were run on parallel gels to determine total RhoA or Rac1. The number beneath each lane is the ratio of band densities for activated versus total GTPase normalized to that of untreated samples.
Structural domains of ROR1 required for Wnt5a-enhanced CLL proliferation or migration.
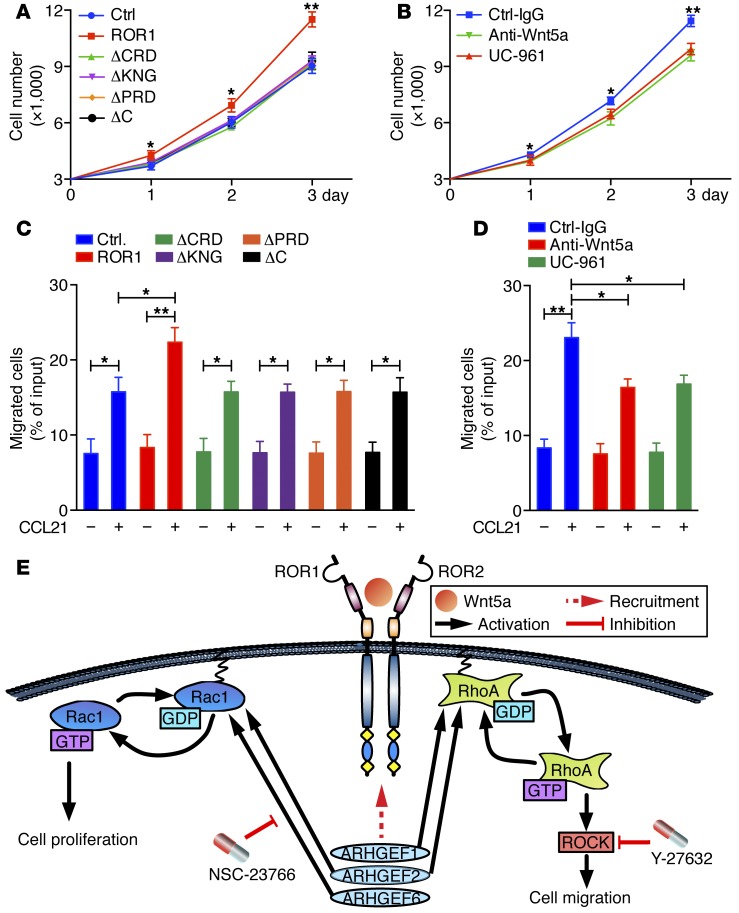
We examined the growth of MEC1 cells and of each of the MEC1-ROR1 transfectants. MEC1-ROR1 cells grew faster than parental MEC1 cells or MEC1 cells transfected with any one of the truncated forms of ROR1 (Figure 7A). Treatment with either neutralizing antibodies to Wnt5a or UC-961 inhibited the growth of MEC1-ROR1 cells, but not MEC1 cells (Figure 7B and Supplemental Figure 10C). Moreover, anti-Wnt5a mAbs also inhibited colocalization of ROR1 with ROR2 in MEC1-ROR1 cells, as assessed by confocal microscopy (Supplemental Figure 7, D and E). Furthermore, MEC1-ROR1 cells migrated significantly better in response to CCL21 than did MEC1 cells or MEC1 cells expressing any of the truncated forms of ROR1 (Figure 7C). Treatment with either neutralizing antibodies to Wnt5a or UC-961 was able to inhibit the migration of MEC1-ROR1 cells in response to CCL21 (Figure 7D). Collectively, these data support a model that Wnt5a induces ROR1 to couple with ROR2 and form ROR1/ROR2 heterooligomers, which recruit GEFs that in turn activate RhoA and Rac1 (Figure 7E).
Figure 7. Structural domains of ROR1 required for migration or proliferation.
(A) Mean numbers of MEC1 (blue circles), MEC1-ROR1 (red squares), or MEC1 cells expressing each of the truncated forms of ROR1 (colors indicated in the legend) in triplicate wells at the days indicated below the graph. (B) Mean numbers of MEC1-ROR1 cells cultured with Ctrl-IgG (blue squares), UC-961 (red triangles), or anti-Wnt5a (green inverted triangles) in triplicate wells at the times indicated below. (C) Bars indicate the mean proportions of MEC1 (blue), MEC1-ROR1 (red), or MEC1 cells transfected with each of the truncated forms of ROR1 (various colors, as indicated in the legend) migrating with (+) or without (–) CCL21, as indicated at the bottom. (D) Bars depict the mean proportions of MEC1-ROR1 cells migrating with (+) or without (–) CCL21 in the presence of Ctrl-IgG (blue bars), UC-961 (green bars), or anti-Wnt5a (red bars). Data in panels A–D are shown as mean ± SEM. *P < 0.05; **P < 0.01, as determined by 2-tailed Student’s t test. (E) Diagram of model for how Wnt5a can enhance proliferation and migration by inducing formation of a ROR1/ROR2 heterooligomer, which recruits GEFs that in turn activate Rac1 and RhoA.
UC-961 inhibits growth of MEC1-ROR1 cells in immune-deficient mice.
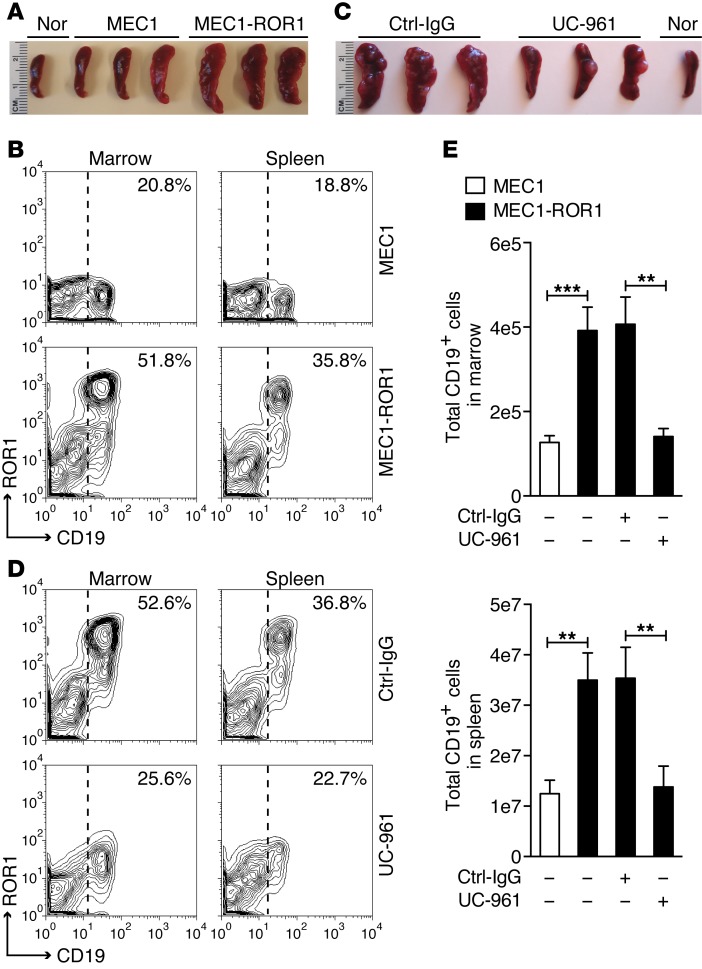
Prior studies indicated that MEC1 cells can engraft immune-deficient mice (37). Three weeks after i.v. infusion of equal numbers of MEC1 or MEC1-ROR1 cells, Rag2−/−γc−/− mice engrafted with MEC1-ROR1 cells had significantly greater splenic and marrow involvement with CD19+ human leukemia than did littermates infused with MEC1 (Figure 8, A and B). However, treatment with UC-961 significantly inhibited the growth of MEC1-ROR1, such that there was no longer a detectable difference in the leukemia-cell burden of mice engrafted with MEC1-ROR1 over that of mice engrafted with MEC1 cells (Figure 8, C–E). We also observed that the MEC1-ROR1 cells harvested from mice treated with UC-961 had lost or attenuated their expression of ROR1 (Figure 8D).
Figure 8. UC-961 inhibits engraftment of MEC1-ROR1.
(A) Representative spleens of Rag2−/−γc−/− mice 3 weeks after receiving an i.v. infusion of 1 × 106 MEC1 or MEC1-ROR1 cells. The spleen of an age-matched, nonengrafted Rag2−/−γc−/− mouse (Nor) is shown for comparison. (B) MEC1 or MEC1-ROR1 cells were collected from the marrow or spleens of mice engrafted 3 weeks earlier with MEC1 or MEC1-ROR1 cells. The fluorescence of cells stained with 4A5–Alexa Fluor 647 (ordinate) and anti-CD19-PE (abscissa) are shown in the contour plots. The percentages at the top right of each contour plot indicate the proportions of cells with fluorescence above the threshold indicated by the dotted line. (C) Representative spleens of Rag2−/−γc−/− mice 3 weeks after receiving an i.v. infusion of 1 × 106 MEC1-ROR1 cells and treatment with either Ctrl-IgG or UC-961. The spleen of an age-match nonengrafted Rag2−/−γc−/− mouse (Nor) is shown for comparison. (D) MEC1-ROR1 cells were collected from the marrow or spleens of mice engrafted 3 weeks earlier with MEC1-ROR1 and then treated with either Ctrl-IgG or UC-961, as indicated on the right. Cells were stained with 4A5–Alexa Fluor 647 and anti–CD19-PE to identify the MEC1-ROR1 cells, as in panel B. Percentages in the top right of each contour plot indicate the proportions of cells with fluorescence above the threshold indicated by the dotted line. (E) Bars indicate the average numbers of CD19+ human leukemia cells harvested from the marrow (left) or spleen (right) of mice engrafted 3 weeks earlier with MEC1 cells (white bars) or MEC1-ROR1 cells (black bars). Some groups of animals were treated with Ctrl-IgG or UC-961, as indicated at the bottom of each histogram. Data are shown as mean ± SD (n = 5). **P < 0.01; ***P < 0.001, as determined by 2-tailed Student’s t test.
UC-961 inhibited growth of ROR1×TCL1 leukemia cells in immune-competent mice.
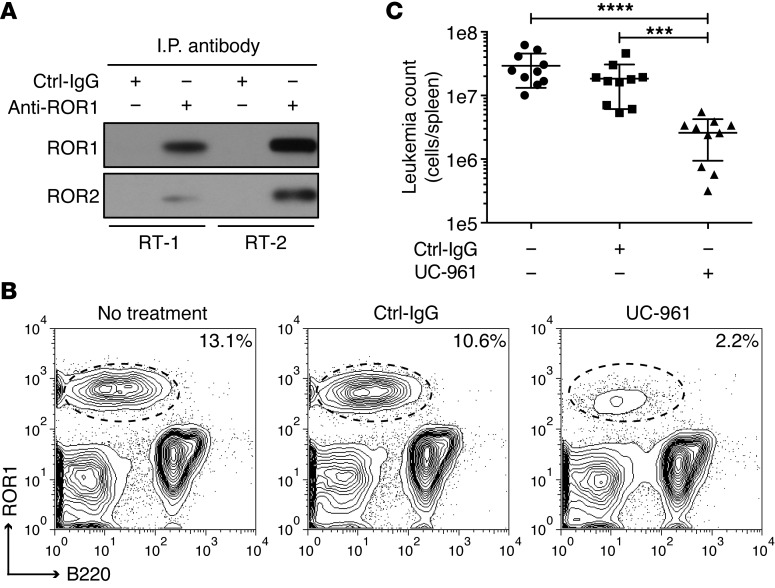
We examined the capacity of UC-961 to inhibit engraftment of CD5+B220loROR1+ B cell leukemia in immune-competent human ROR1 Tg mice (38). In contrast to leukemia cells that develop in TCL1-Tg mice, the leukemia cells that develop in double-Tg ROR1×TCL1 Tg mice express ROR1 (38). Co–immune-precipitation studies found that this human ROR1 also formed heterooligomers with mouse ROR2 in freshly isolated ROR1×TCL1 leukemia cells (Figure 9A), consistent with a model that proposes that the enhanced growth-promoting effect of ROR1 in this leukemia is mediated through ROR1:ROR2 heterooligomers, as in human CLL. To examine the activity of UC-961 against ROR1×TCL1 leukemia, we transferred 2 × 104 CD5+B220loROR1+ leukemia B cells to ROR1-Tg recipient mice and administered no treatment or weekly doses of 10 mg/kg of UC-961 or Ctrl-IgG. After 25 days, the proportion of CD5+B220loROR1+ leukemia cells in the spleen was determined by flow cytometric analysis, and the total number of CD5+B220loROR1+ B cells per spleen was enumerated. Compared with untreated or Ctrl-IgG–treated mice, UC-961–treated animals (n = 10) had significantly fewer CD5+B220loROR1+ B cells per spleen following weekly administration of anti-ROR1 mAbs (Figure 9, B and C).
Figure 9. UC-961 inhibits engraftment of ROR1×TCL1 leukemia.
(A) Immunoblot analysis of anti-ROR1 immune precipitates from lysates of ROR1×TCL1 leukemia cells detecting the association of ROR1 with ROR2. RT-1, leukemia that developed in ROR1×TCL1 mouse no. 1; RT-2, ROR1×TCL1 leukemia that developed in ROR1×TCL1 mouse no. 2. (B) Anti-ROR1 mAb UC-961 inhibits engraftment of CD5+B220loROR1+ leukemia cells in ROR1-Tg mice. ROR1-Tg mice were engrafted with 2 × 104 CD5+B220loROR1+ B cells and then given i.v. injections of 10 mg/kg of mAb on days 1, 3, 7, 14, and 21. Contour plots depict the fluorescence of splenic lymphocytes harvested on day 25 after adoptive transfer from representative mice (n = 10) that received no treatment (left panel), nonspecific human IgG (center panel), or UC-961 mAb (right panel), as assessed by light scatter after staining the cells with fluorochrome-conjugated mAb specific for B220 (abscissa) or human ROR1 (ordinate), using an anti-ROR1 mAb that did not crossreact with UC-961. The percentages in the top right of each contour plot indicate the proportion of CD5+B220loROR1+ leukemia cells in blood mononuclear cells. (C) Total number of CD5+B220loROR1+ leukemia cells in spleens of recipient ROR1 Tg mice 25 days after adoptive transfer of 2 × 104 CD5+B220loROR1+ B cells that received weekly injections of 10 mg/kg of control human IgG or UC-961 mAb, as determined by flow cytometric analysis and cell count. Each symbol represents the number of leukemia cells found in individual mice. Data are shown as mean ± SD for each group of animals (n = 10). ***P < 0.001; ****P < 0.0001, compared with the untreated or Ctrl-IgG–treated group, respectively, as calculated using Student’s t test.
Discussion
We found that CLL cells express ROR1/ROR2 heterooligomers. Such heterooligomers apparently formed in response to Wnt5a, which we found present at relatively high levels in the plasma of patients with CLL. When cultured in media lacking Wnt5a, the ROR1/ROR2 heterooligomer dissociated, but was reformed by treatment with exogenous Wnt5a, which induced recruitment of GEFs and the activation of RhoA and Rac1. Activation of RhoA was associated with enhanced chemokine-induced migration, whereas activation of Rac1 was associated with enhanced leukemia-cell proliferation in response to membrane-bound CD154. Inhibiting expression of either ROR1 or ROR2 impaired the capacity of Wnt5a to enhance chemotaxis, as did treatment with UC-961. The interaction between ROR2 and ROR1 to achieve optimal noncanonical Wnt5a signaling also was observed in MEC1, a CLL-cell line that we found expressed Wnt5a and ROR2, but did not express ROR1. Transfection of MEC1 to express ROR1 resulted in formation of ROR1/ROR2 heterooligomers, which subsequently recruited GEFs, resulting in heightened activation of RhoA and Rac1, enhanced cellular chemokine-directed migration, and enhanced growth relative to that of parental MEC1 cells; such changes were inhibited by treatment with UC-961 or neutralizing antibodies to Wnt5a. Collectively, our studies indicate that neither ROR1 nor ROR2 was sufficient for optimal Wnt5a-induced activation of RhoA or Rac1.
We found the KNG domain of ROR1 was required for it to oligomerize with ROR2. KNG domains contain intradomain disulfide bridges, which define polypeptide loops that often are involved in protein-protein interactions (39). As such, the KNG domain of ROR1 may interact with the KNG of ROR2 to form heterooligomers in response of Wnt5a (e.g., Figure 6A). Although the failure of the ΔKNG-ROR1 to oligomerize with ROR2 may be secondary to steric constraints introduced by truncation of the ROR1 extracellular domain, ΔCRD-ROR1 (where CRD indicates cysteine-rich domain), lacking the larger extracellular CRD, formed heterooligomers with ROR2, apparently in response to Wnt5a. As the CRD is the putative binding site for Wnt proteins (40, 41), it is conceivable that Wnt5a binding to ROR2 may allow it to oligomerize with ROR1. Alternatively, the CRD may have residues that ordinarily inhibit the heterooligomerization of ROR1 and ROR2 unless the CRD is bound to Wnt5a. In any case, neither ΔCRD-ROR1 nor any one of the other truncated forms of ROR1 used in this study allowed for recruitment of GEFs to ROR1/ROR2 heterooligomers or enhanced activation of RhoA and Rac1 in transfected MEC1 cells. Because each of the truncated forms of ROR1, except for ΔKNG-ROR1, allowed for ROR1 to oligomerize with ROR2, the formation of ROR1/ROR2 heterooligomers was not sufficient to trigger optimal Wnt5a-induced activation of RhoA or Rac1.
Our data demonstrate that the Wnt5a-induced ROR1-ROR2 heterooligomer can recruit, anchor, and activate GEFs. The intracellular domains of ROR1 appear necessary for such docking, including the proline-rich domain (PRD), which contains several potential SH3-binding sites. ARHGEF6 has an SH3 domain that may permit docking of this GEF to the PRD of ROR1, possibly in a manner similar to the way in which it docks with PAK1 (42). However, ARHGEF1 and ARHGEF2 do not have SH3 domains (43), and most likely, they require an adapter protein or proteins to dock with the intracellular domains of the ROR1/ROR2 heterooligomer. Furthermore, it is possible that GEFs other than ARHGEF1, ARHGEF2, or ARHGEF6 also contribute to ROR1 signaling. For example, culture of CLL cells with CD154-bearing cells can induce expression of other GEFs, such as Tiam1, which does not appear to be expressed at high levels by nonstimulated CLL cells (35). Conceivably, the particular GEFs recruited by ROR1/ROR2 may be idiosyncratic to the type of cell or its stage of activation/differentiation. Furthermore, additional studies are required to define the other proteins recruited to the Wnt5a-induced ROR1/ROR2 complex that ordinarily are not bound to either ROR1 or ROR2 alone in the absence of Wnt5a.
A model proposing that ROR1 oligomerizes with ROR2 for Wnt5a signaling has support from data on the developmental expression and the implied function of these orphan receptors. The spatial and temporal expression of ROR1 and ROR2 mostly overlap in face, limbs, heart, and lungs during mouse embryogenesis (44). In the absence of either ROR1 or ROR2, mice have impaired development of multiple organs (45). Furthermore, ROR1 and ROR2 colocalize in the mouse central neurons and function in Wnt5a-activated signaling pathways, leading to synapse formation in the mammalian central nervous system (46).
However, in different cell contexts, ROR2 might provide for signaling independently of ROR1 and vice versa. This is implied from studies on mice made deficient in expression of either ROR1 or ROR2 or both orphan receptors; only mice made deficient in both ROR1 and ROR2 had developmental defects that entirely mimicked those of animals made deficient for expression of Wnt5a (26). More recent studies also have shown that signaling via Wnt5a and ROR1/2 was necessary for embryo implantation into the uterus (47). Knockout of Wnt5a or ROR1 and ROR2 precluded embryo implantation, whereas knockout of either ROR1 or ROR2 alone was not sufficient to manifest this functional defect. These studies imply that expression of only one type of orphan receptor potentially might compensate for loss of the other. Perhaps relevant in this regard are studies demonstrating that Wnt5a induces homooligomerization and activation of ROR2 in skeletal morphogenesis to promote osteoblast differentiation and bone formation (48). However, if such homooligomerization also occurs in CLL cells, then it does not appear sufficient to recruit GEF or activate RhoA/Rac1, at least not as efficiently as the ROR1/ROR2 heterooligomer.
ROR1 or ROR2 also might oligomerize with other surface proteins, such as Frizzled (Fzd) receptors, which then could compensate for loss of signaling via a ROR1/ROR2 heterooligomer. Furthermore, the type of signaling provided by ROR1 or ROR2 could be influenced by the relative expression levels of these orphan receptors with other surface proteins and Fzd receptors that might otherwise complex with LRP5/6 in response to canonical Wnt factors, such as Wnt3a. Through such a mechanism, Wnt5a inhibits the capacity of Wnt factors, such as Wnt3a, to induce canonical Wnt signaling (27, 33, 49–51). Furthermore, studies have demonstrated that isolated overexpression of ROR2 in 293 cells actually enhanced the capacity of Wnt5a to repress expression of β-catenin target genes (28). As such, the capacity of Wnt5a to act as tumor suppressor versus tumor promoter may depend upon the relative expression of ROR2, ROR1, LPR5/6, and various Fzd receptors, which either bind Wnt5a or interact to influence the response to this or other Wnt factors.
ROR2 appears more widely expressed on postpartum tissues than ROR1, which appears confined to a rare subpopulation of precursor B cells, called hematogones (14). In this study, we found ROR2 on normal CD5 B cells, which are the presumed normal counterpart to CLL (52). Acquisition of ROR1 by such cells may be an important step in leukemogenesis, allowing the neoplastic B cells to activate RhoA and Rac1 in response to Wnt5a, which we found at relatively high levels in the plasma of patients with CLL. This may help explain earlier observations that CLL cells have higher levels of activated RhoA, Rac1, and activated ARHGEF2 than normal B cells of healthy adults (35).
We found that treatment of CLL cells with the anti-ROR1 mAb UC-961 disrupted the coupling of ROR1 with ROR2 and interfere with activation of Rho GTPases in response to Wnt5a, thereby abrogating the capacity of Wnt5a to enhance chemokine-induced migration or CD154-induced proliferation. Similarly, UC-961 eliminated the advantages that MEC1-ROR1 cells had over parental MEC1 cells in proliferation or chemokine-induced migration in vitro. Prior studies found that treatment with UC-961 of immune-deficient mice bearing primary ovarian cancer patient–derived xenografts caused depletion and senescence of ovarian cancer stem cells, which express high levels of ROR1 (22), suggesting a role for such noncanonical Wnt signaling in maintaining cancer stem cell survival and/or renewal. Because of the restricted expression of ROR1, mAbs such as UC-961 may selectively block the activation of Rho GTPases in ROR1-expressing cancers in response to Wnt5a, which also can be found at high levels in other cancers (53, 54). Collectively, our studies imply that UC-961 can block ROR1-dependent signaling, providing additional rationale for the clinical evaluation of this antibody in patients with CLL or other cancers that are complemented by ROR1-dependent, noncanonical Wnt5a signaling.
Methods
CLL specimens.
Blood samples were collected from CLL patients at the Moores Cancer Center. Peripheral blood mononuclear cells (PBMCs) were isolated by density centrifugation with Ficoll-Paque PLUS (GE Healthcare Life Sciences) and suspended in 90% FBS (Omega Scientific) and 10% DMSO (Sigma-Aldrich) for viable storage in liquid nitrogen. Samples with more than 95% CD19+CD5+ CLL cells were used without further purification throughout this study. Plasma samples were collected by spinning blood samples for 10 minutes at 187 g and stored at –20°C.
Cell proliferation assay.
Primary CLL cell proliferation assay was performed as described (55). CLL cells were labeled by CFSE (Life Technologies) and plated at 1.5 × 106/well/ml in a 24-well tray on a layer of irradiated HeLaCD154 cells (80 Gy) at a CLL/HeLaCD154 cell ratio of 15:1 in complete RPMI-1640 medium supplemented with 5 ng/ml of recombinant human IL-4 (R&D Systems) and 15 ng/ml of recombinant human IL-10 (R&D Systems). Wnt5a (200 ng/ml, R&D Systems) or UC-961 (10 µg/ml) was added, as indicated in the text. CFSE-labeled CLL cells were analyzed by flow cytometry; Modfit LT software (version 3.0, Verity Software House) was used for analysis of cell proliferation.
MEC1 cell proliferation assay was also performed as described (18). To monitor for differences in cell proliferation, MEC1, MEC1-ROR1, or MEC1 transfected with truncated forms of ROR1 was plated in 96-well plates at 5 × 103 cells/well in complete RPMI-1640 medium. MEC1-ROR1 cells were plated in 96-well plates at 5 × 103 cells/well in complete RPMI-1640 medium in the absence or presence of Ctrl-IgG, anti-Wnt5a, or UC-961 mAbs. A total of 10 μl CCK-8 (Dojindo Molecular Technologies) was added to each well after 3 days, and the cells were cultured for an additional 3 hours. The absorbance at 450 nm was measured to calculate the numbers of viable cells in each well.
Chemotaxis assay.
The chemotaxis assay across 5.0 μm pore size polycarbonate membranes was performed as described (56). A total of 5 × 105 cells were serum starved for 12 hours and then treated with or without Wnt5a (400 ng/ml) for 30 minutes and seeded in the upper compartment of Transwell inserts. Cells were incubated for 2 hours in serum-free medium at 37°C and 5% CO2, and the migration toward chemokine (CXCL12, 100 ng/ml or CCL21, 100 ng/ml) was analyzed by flow cytometry. The percentage of migrating cells was calculated as the number of migrated cells in response to chemokine divided by the total number of input cells.
RhoA and Rac1 activation assay.
RhoA and Rac1 activation assay reagents were purchased from Cytoskeleton and used per the manufacturer’s instructions. Briefly, GTP-bound active RhoA or Rac1 was pulled down with Rhotekin-RBD or PAK-PBD beads, respectively, and then subjected to immunoblot analysis. Immunoblots of whole-cell lysates were used to assess for total RhoA or Rac1. The integrated optical density (IOD) of bands was evaluated by densitometry and analyzed using Gel-Pro Analyzer 4.0 software (Media Cybernetics).
Immunoblot and immunoprecipitation analysis.
Western blot analysis was performed as described (15). Equal amounts of total protein from each sample were fractionated by SDS-PAGE and blotted onto polyvinylidene difluoride membrane. Western blot analysis was performed using primary mAbs specific for ROR1 (catalog 4102), ARHGEF1 (catalog 3669), or ARHGEF6 (catalog 4515), which were detected using secondary antibodies conjugated with horseradish peroxidase (Cell Signaling Technology). mAbs specific for ROR2 (ab92397) or ARHGEF2 (ab90783) were purchased from Abcam. Anti–β-actin antibody (sc-47778) was obtained from Santa Cruz Biotechnology Inc., and anti-Wnt5a antibody (MAB645) was from R&D Systems. Anti-mouse ROR2 antibody (catalog 14700) was obtained from Cell Signaling Technology. We controlled the specificity of each of the mAbs for ROR1 or ROR2 using the extracellular domains of ROR1 (ROR1-ex) or ROR2 (ROR2-ex) (Supplemental Figure 2D). Immunoprecipitation analysis was performed as described (38). Cells were lysed in a buffer containing 50 mM Tris-HCl (pH 7.6), 150 mM NaCl, 0.1% SDS, 0.1% sodium deoxycholate, 1% Triton X-100, and cOmplete Protease Inhibitor Cocktail (Roche Applied Sciences). The lysates were cleared by centrifugation at 16,000 g for 15 minutes. Immune precipitates were generated using the antibody of interest and protein G/A agarose beads, followed by immunoblot or MS analysis, as described. Antibodies for immune precipitation were as follows: the anti-ROR1 antibody was generated in our laboratory; the anti-ROR2 antibody (sc-374174) was obtained from Santa Cruz Biotechnology; the anti-ARHGEF1 antibody (catalog 3669) was purchased from Cell Signaling Technology; the anti-ARHGEF2 (ab90783) and anti-ARHGEF6 (ab91562) antibodies were obtained from Abcam.
MS analysis.
Bound proteins were digested by trypsin (Roche) directly on beads for the MS analysis. Beads were washed by 1 ml of 50 mM HEPES buffer (pH 7.2) 3 times and then were suspended in 100 μl Hepes buffer (50 mM; pH 7.2). Cystines were reduced and alkylated using 1 mM Tris (2-carboxyethyl) phosphine (Fisher) at 95°C for 5 minutes and then 2.5 mM iodoacetamide (Fisher) at 37 °C in darkness for 15 minutes. Proteins were digested with 0.5 μg trypsin (trypsin/protein [w/w] ratio = 1:50) overnight. Supernatant containing digested peptides was collected and passed through a 0.22-μM filter via centrifugation. MS methods were as described (57). Briefly, digested peptides were separated by online 2D-nanoLC and detected by LTQ linear ion trap mass spectrometers. Each sample took 22.5 hours to analyze and about 200,000 MS/MS spectra were collected for each run. Raw data were extracted and searched using Spectrum Mill (Agilent, v3.03) database search software against the NCBI refseq database limited to human taxonomy (version 44).
Flow cytometry analysis.
Flow cytometry analysis was performed as described (15). The following antibodies were used to stain cells at 4°C for 20 minutes: anti-ROR1 mAb (4A5) conjugated with Alexa Fluor 647 (4A5–Alexa Fluor 647) was generated in our laboratory; anti-ROR2 mAb (clone 231509) conjugated with Alexa Fluor 488 (anti-ROR2–Alexa Fluor 488) was obtained from R&D Systems; phycoerythrin-conjugated (PE-conjugated) anti-CD19 (anti-CD19–PE, catalog 561741) and allophycocyanin-conjugated (APC-conjugated) anti-CD5 (anti–CD5-APC, catalog 555355) antibodies were from BD Biosciences. The specificity of anti-ROR2 mAb was confirmed by flow cytometry analysis of stained CHO cells transfected to express human ROR2 (Supplemental Figure 2E). The stained cells were washed twice with FACS buffer (PBS, pH 7.4, 3% FBS) and examined by 4-color, multiparameter flow cytometry using a dual-laser FACSCalibur (BD Biosciences). Data were analyzed using FlowJo software (TreeStar). We subtracted the MFI of cells stained with a fluorochrome-labeled, isotype-control mAb from the MFI of the same cells stained with anti-ROR1 or anti-ROR2 to determine the specific increase in MFI (ΔMFI).
Reverse-transcription PCR.
Total RNA was extracted using TRIzol (Life Technologies). A 10-μg volume of total RNA was incubated with 10 U RNase-free DNase I (Life Technologies) at 37°C for 30 minutes. RNA was further purified with an RNeasy Mini Kit (QIAGEN). The purified total RNA (2 μg) was reverse transcribed with 200 U Superscript III Reverse Transcriptase (Life Technologies). Taq 2× Master Mix (NEB) was used for PCR per the manufacturer’s protocol.
Plasma Wnt5a ELISA.
An ELISA kit (MyBioSource) was used to measure Wnt5a levels in plasma samples from 9 patients with CLL and 9 healthy individuals per the manufacturer’s instruction.
Confocal microscopy imaging.
Cells were stained with anti-ROR1 mAb (4A5–Alexa Fluor 647) and anti-ROR2–Alexa Fluor 488 mAbs at 4°C for 20 minutes, washed twice with FACS buffer, and then fixed with 4% paraformaldehyde for 20 minutes at 4°C. Cells were washed twice with PBS and permeabilized with 0.3% Triton X-100 in PBS at 4°C for 10 minutes. After the cells were washed twice with PBS, they were blocked with 5% BSA in PBS at 4°C for 30 minutes. Anti-ARHGEF1 antibody (Cell Signaling Technology), anti-ARHGEF2 antibody (Abcam), or anti-ARHGEF6 antibody (Cell Signaling Technology) was added in blocking buffer and incubated overnight at 4°C. Cells were washed twice with PBS, and Alexa Fluor 594–conjugated goat anti-rabbit secondary antibody was added and incubated at room temperature for 60 minutes. Cells were washed twice with PBS, and DAPI was added and incubated at room temperature for 10 minutes. The stained cells were washed and attached to slides via Cytospin. Confocal images were collected on a Nikon A1R confocal microscope using a ×100 objective with an NA of 1.4. The laser lines for the different fluorophores were as follows: 405 nm for DAPI, 488 nm for Alexa Fluor 488, 561 nm for Alexa Fluor 594, and 647 nm for Alexa Fluor 647. Control antibodies, anti-CD5–Alexa Fluor 647, and anti-CD19–Alexa Fluor 488 were from BD Biosciences. The percentage of the area that had colocalized signals was analyzed by ImageJ colocalization plugin (http://imagej.nih.gov/ij/). Colocalized regions were measured in pixels and related to the total cell area.
Nucleofection of siRNA and plasmids.
Human B Cell Nucleofector Kit for siRNA or plasmid transfection was from Lonza. B-CLL cells or MEC1 cells (5 × 106) were suspended in 100 μl Nucleofector Solution with siRNA (Life Technologies) or plasmids and transfected with the Nucleofector II device (program U-015). The transfected cells were cultured in 12-well plates in complete medium for 48 hours and then subjected to immunoblot analysis and assays. Endofree Plasmid Maxi Kits (QIAGEN) were used to purify plasmids for transfection. G418 (1.5 mg/ml) was used for selection of stable MEC1 transfectants, which then were examined via flow cytometry.
RhoGEF nucleotide exchange activity assay.
RhoGEF exchange assay kit was from Cytoskeleton and was used per the manufacturer’s instructions. For in vitro guanine nucleotide exchange activity on Rac1 or RhoA, ARHGEF1, ARHGEF2, or ARHGEF6 was immunoprecipitated from CLL cells that previously had been treated with or without Wnt5a (R&D Systems), Ctrl-IgG, or UC-961. Reactions were measured in a Tecan Spectrofluor plus fluorimeter (λex = 360 nm, λem = 460 nm). ARHGEF1, ARHGEF2, or ARHGEF6 was added after 120 seconds. Readings were taken at 20°C every 30 seconds for a total reaction time of 44 minutes. The exchange curve was generated by exporting raw data to Microsoft Excel and analyzing the data using GraphPad Prism 6.0.
Adoptive transfer and UC-961 administration.
We designed 2 strategies to address the role of ROR1 in the progression of CLL and the antileukemia activity of UC-961 in immune-deficient mice. First, we injected 1 × 106 MEC1 versus MEC1-ROR1 cells into Rag2−/−γc−/− mice i.v. and then sacrificed the mice for necropsy 3 weeks later. Second, we injected 1 × 106 MEC1-ROR1 cells into Rag2−/−γc−/− mice and treated the engrafted mice with Ctrl-IgG (Life Technologies) or UC-961 at 10 mg/kg every 3 days until they were sacrificed for necropsy 3 weeks later.
Also, we evaluated the antileukemia activity of UC-961 in immune-competent mice. ROR1×TCL1 leukemia B cells (CD5+B220loROR1+) were isolated from the spleen, enriched via density gradient centrifugation, suspended in sterile PBS, and injected i.v. into ROR1-Tg recipient mice at 2 × 104 cells per animal. Samples used for transplantation were verified by flow cytometry to be more than 90% leukemia B cells. Recipient mice received either no treatment or weekly i.v. injections of 10 mg/kg of UC-961 or Ctrl-IgG (rituximab) beginning on day 1. All mice were sacrificed on day 25, and single-cell suspensions of splenocytes were purged of red blood cells by hypotonic lysis in ammonium-chloride-potassium (ACK) lysis solution, washed, suspended in 2% (w/v) BSA (Sigma-Aldrich) in PBS (pH = 7.4), and stained for surface expression of CD3 (17A2), CD5 (53-7.3), B220 (RA3-6B2), and ROR1 (4A5) using optimized concentrations of fluorochrome-conjugated mAbs. Cells were examined by 4-color, multiparameter flow cytometry using dual-laser FACSCalibur (BD), and the data were analyzed using FlowJo software (TreeStar). The total number of leukemia cells per spleen was calculated by determining the percentage of CD5+B220loROR1+cells of total lymphocytes by flow cytometry and multiplying this number by the total spleen cell count.
Statistics.
Data are presented as mean ± SEM or mean ± SD, as indicated, for data sets that satisfied conditions for a normal distribution, as determined by the Kolmogorov-Smirnov test. Differences between 2 groups were determined by unpaired 2-tailed Student’s t test. Differences between multiple groups were determined by 1-way ANOVA with post-hoc Tukey HSD test. All P values of less than 0.05 were considered significant. Analysis for significance was performed with GraphPad Prism 6.0 (GraphPad Software Inc.).
Study approval.
Blood samples were collected from CLL patients at the Moores Cancer Center who satisfied diagnostic and immunophenotypic criteria for common B cell CLL and who provided written, informed consent, in compliance with the Declaration of Helsinki and the Institutional Review Board (IRB) of the UCD (IRB approval number 080918). All experiments with mice were carried out in accordance with Guide for the Care and Use of Laboratory Animals (8th ed. The National Academies Press. 2011), and the University of California San Diego Institutional Animal Care and Use Committee (IACUC) and Animal Care Program approved the study protocol.
Author contributions
JY and TJK conceived the project. JY, LC, GFW, BC, ZS, RW, LZ, and SZ performed the experiments. JY, GFW, ZS, SPB, and TJK analyzed the data. JY and TJK wrote the manuscript.
Supplementary Material
Acknowledgments
We thank Laura Rassenti, Jessie Fecteau, Esther Avery, Zhuhong Zhang, and Gang Cheng for technical assistance; Kersi Pestonjamasp for fluorescence confocal microscopy analysis; the Moores Cancer Center Microscopy Core Facility for technical help; and Andrew Greaves for the graphic drawing. We thank the California Institute for Regenerative Medicine (CIRM) (grant DR3-06924) for supporting us in generating anti-ROR1 mAbs and UC-961. This work was supported by the UCSD Foundation Blood Cancer Research Fund (BCRF), a SPORE grant (7005-14) from the Leukemia and Lymphoma Society, and a PO1 grant (5P01CA081534-14) from the NIH for the CLL Research Consortium.
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Reference information:J Clin Invest. 2016;126(2):585–598. doi:10.1172/JCI83535.
References
- 1.Masiakowski P, Carroll RD. A novel family of cell surface receptors with tyrosine kinase-like domain. J Biol Chem. 1992;267(36):26181–26190. [PubMed] [Google Scholar]
- 2.Wilson C, Goberdhan DC, Steller H. Dror, a potential neurotrophic receptor gene, encodes a Drosophila homolog of the vertebrate Ror family of Trk-related receptor tyrosine kinases. Proc Natl Acad Sci U S A. 1993;90(15):7109–7113. doi: 10.1073/pnas.90.15.7109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Forrester WC, Dell M, Perens E, Garriga G. A C. elegans Ror receptor tyrosine kinase regulates cell motility and asymmetric cell division. Nature. 1999;400(6747):881–885. doi: 10.1038/23722. [DOI] [PubMed] [Google Scholar]
- 4.Koga M, Take-uchi M, Tameishi T, Ohshima Y. Control of DAF-7 TGF-(alpha) expression and neuronal process development by a receptor tyrosine kinase KIN-8 in Caenorhabditis elegans. Development. 1999;126(23):5387–5398. doi: 10.1242/dev.126.23.5387. [DOI] [PubMed] [Google Scholar]
- 5.McKay SE, et al. Aplysia ror forms clusters on the surface of identified neuroendocrine cells. Mol Cell Neurosci. 2001;17(5):821–841. doi: 10.1006/mcne.2001.0977. [DOI] [PubMed] [Google Scholar]
- 6.Hikasa H, Shibata M, Hiratani I, Taira M. The Xenopus receptor tyrosine kinase Xror2 modulates morphogenetic movements of the axial mesoderm and neuroectoderm via Wnt signaling. Development. 2002;129(22):5227–5239. doi: 10.1242/dev.129.22.5227. [DOI] [PubMed] [Google Scholar]
- 7.Rodriguez-Niedenfuhr M, Prols F, Christ B. Expression and regulation of ROR-1 during early avian limb development. Anat Embryol (Berl) 2004;207(6):495–502. doi: 10.1007/s00429-004-0381-6. [DOI] [PubMed] [Google Scholar]
- 8.Katoh M, Katoh M. Comparative genomics on ROR1 and ROR2 orthologs. Oncol Rep. 2005;14(5):1381–1384. doi: 10.3892/or.14.5.1381. [DOI] [PubMed] [Google Scholar]
- 9.Minami Y, Oishi I, Endo M, Nishita M. Ror-family receptor tyrosine kinases in noncanonical Wnt signaling: their implications in developmental morphogenesis and human diseases. Dev Dyn. 2010;239(1):1–15. doi: 10.1002/dvdy.21991. [DOI] [PubMed] [Google Scholar]
- 10.Oishi I, et al. Spatio-temporally regulated expression of receptor tyrosine kinases, mRor1, mRor2, during mouse development: implications in development and function of the nervous system. Genes Cells. 1999;4(1):41–56. doi: 10.1046/j.1365-2443.1999.00234.x. [DOI] [PubMed] [Google Scholar]
- 11.Al-Shawi R, Ashton SV, Underwood C, Simons JP. Expression of the Ror1 and Ror2 receptor tyrosine kinase genes during mouse development. Dev Genes Evol. 2001;211(4):161–171. doi: 10.1007/s004270100140. [DOI] [PubMed] [Google Scholar]
- 12.Nomi M, et al. Loss of mRor1 enhances the heart and skeletal abnormalities in mRor2-deficient mice: redundant and pleiotropic functions of mRor1 and mRor2 receptor tyrosine kinases. Mol Cell Biol. 2001;21(24):8329–8335. doi: 10.1128/MCB.21.24.8329-8335.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lyashenko N, Weissenbock M, Sharir A, Erben RG, Minami Y, Hartmann C. Mice lacking the orphan receptor ror1 have distinct skeletal abnormalities and are growth retarded. Dev Dyn. 2010;239(8):2266–2277. doi: 10.1002/dvdy.22362. [DOI] [PubMed] [Google Scholar]
- 14.Broome HE, Rassenti LZ, Wang HY, Meyer LM, Kipps TJ. ROR1 is expressed on hematogones (non-neoplastic human B-lymphocyte precursors) and a minority of precursor-B acute lymphoblastic leukemia. Leuk Res. 2011;35(10):1390–1394. doi: 10.1016/j.leukres.2011.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fukuda T, et al. Antisera induced by infusions of autologous Ad-CD154-leukemia B cells identify ROR1 as an oncofetal antigen and receptor for Wnt5a. Proc Natl Acad Sci U S A. 2008;105(8):3047–3052. doi: 10.1073/pnas.0712148105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Daneshmanesh AH, et al. Ror1, a cell surface receptor tyrosine kinase is expressed in chronic lymphocytic leukemia and may serve as a putative target for therapy. Int J Cancer. 2008;123(5):1190–1195. doi: 10.1002/ijc.23587. [DOI] [PubMed] [Google Scholar]
- 17.Baskar S, et al. Unique cell surface expression of receptor tyrosine kinase ROR1 in human B-cell chronic lymphocytic leukemia. Clin Cancer Res. 2008;14(2):396–404. doi: 10.1158/1078-0432.CCR-07-1823. [DOI] [PubMed] [Google Scholar]
- 18.Zhang S, et al. The onco-embryonic antigen ROR1 is expressed by a variety of human cancers. Am J Pathol. 2012;181(6):1903–1910. doi: 10.1016/j.ajpath.2012.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Debebe Z, Rathmell WK. Ror2 as a therapeutic target in cancer. Pharmacol Ther. 2015;150:143–148. doi: 10.1016/j.pharmthera.2015.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cui B, et al. Targeting ROR1 inhibits epithelial-mesenchymal transition and metastasis. Cancer Res. 2013;73(12):3649–3660. doi: 10.1158/0008-5472.CAN-12-3832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li X, et al. Activation of Wnt5a-Ror2 signaling associated with epithelial-to-mesenchymal transition of tubular epithelial cells during renal fibrosis. Genes Cells. 2013;18(7):608–619. doi: 10.1111/gtc.12064. [DOI] [PubMed] [Google Scholar]
- 22.Zhang S, et al. Ovarian cancer stem cells express ROR1, which can be targeted for anti-cancer-stem-cell therapy. Proc Natl Acad Sci U S A. 2014;111(48):17266–17271. doi: 10.1073/pnas.1419599111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Oishi I, et al. The receptor tyrosine kinase Ror2 is involved in non-canonical Wnt5a/JNK signalling pathway. Genes Cells. 2003;8(7):645–654. doi: 10.1046/j.1365-2443.2003.00662.x. [DOI] [PubMed] [Google Scholar]
- 24.Green JL, Inoue T, Sternberg PW. Opposing Wnt pathways orient cell polarity during organogenesis. Cell. 2008;134(4):646–656. doi: 10.1016/j.cell.2008.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.He F, et al. Wnt5a regulates directional cell migration and cell proliferation via Ror2-mediated noncanonical pathway in mammalian palate development. Development. 2008;135(23):3871–3879. doi: 10.1242/dev.025767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ho HY, et al. Wnt5a-Ror-dishevelled signaling constitutes a core developmental pathway that controls tissue morphogenesis. Proc Natl Acad Sci U S A. 2012;109(11):4044–4051. doi: 10.1073/pnas.1200421109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ford CE, Qian Ma SS, Quadir A, Ward RL. The dual role of the novel Wnt receptor tyrosine kinase, ROR2, in human carcinogenesis. Int J Cancer. 2013;133(4):779–787. doi: 10.1002/ijc.27984. [DOI] [PubMed] [Google Scholar]
- 28.Mikels AJ, Nusse R. Purified Wnt5a protein activates or inhibits β-catenin-TCF signaling depending on receptor context. PLoS Biol. 2006;4(4): doi: 10.1371/journal.pbio.0040115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gentile A, Lazzari L, Benvenuti S, Trusolino L, Comoglio PM. Ror1 is a pseudokinase that is crucial for Met-driven tumorigenesis. Cancer Res. 2011;71(8):3132–3141. doi: 10.1158/0008-5472.CAN-10-2662. [DOI] [PubMed] [Google Scholar]
- 30.Yamaguchi T, et al. NKX2-1/TITF1/TTF-1-Induced ROR1 is required to sustain EGFR survival signaling in lung adenocarcinoma. Cancer Cell. 2012;21(3):348–361. doi: 10.1016/j.ccr.2012.02.008. [DOI] [PubMed] [Google Scholar]
- 31.Kaucka M, et al. The planar cell polarity pathway drives pathogenesis of chronic lymphocytic leukemia by the regulation of B-lymphocyte migration. Cancer Res. 2013;73(5):1491–1501. doi: 10.1158/0008-5472.CAN-12-1752. [DOI] [PubMed] [Google Scholar]
- 32.Etienne-Manneville S, Hall A. Rho GTPases in cell biology. Nature. 2002;420(6916):629–635. doi: 10.1038/nature01148. [DOI] [PubMed] [Google Scholar]
- 33.Nishita M, et al. Ror2/Frizzled complex mediates Wnt5a-induced AP-1 activation by regulating Dishevelled polymerization. Mol Cell Biol. 2010;30(14):3610–3619. doi: 10.1128/MCB.00177-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee JG, Heur M. Interleukin-1β-induced Wnt5a enhances human corneal endothelial cell migration through regulation of Cdc42 and RhoA. Mol Cell Biol. 2014;34(18):3535–3545. doi: 10.1128/MCB.01572-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hofbauer SW, et al. Tiam1/Rac1 signals contribute to the proliferation and chemoresistance, but not motility, of chronic lymphocytic leukemia cells. Blood. 2014;123(14):2181–2188. doi: 10.1182/blood-2013-08-523563. [DOI] [PubMed] [Google Scholar]
- 36.Stacchini A, et al. MEC1 and MEC2: two new cell lines derived from B-chronic lymphocytic leukaemia in prolymphocytoid transformation. Leuk Res. 1999;23(2):127–136. doi: 10.1016/S0145-2126(98)00154-4. [DOI] [PubMed] [Google Scholar]
- 37.Bertilaccio MT, et al. A novel Rag2–/–γc–/–-xenograft model of human CLL. Blood. 2010;115(8):1605–1609. doi: 10.1182/blood-2009-05-223586. [DOI] [PubMed] [Google Scholar]
- 38.Widhopf GF, et al. ROR1 can interact with TCL1 and enhance leukemogenesis in Emu-TCL1 transgenic mice. Proc Natl Acad Sci U S A. 2014;111(2):793–798. doi: 10.1073/pnas.1308374111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Graversen JH, Sigurskjold BW, Thogersen HC, Etzerodt M. Tetranectin-binding site on plasminogen kringle 4 involves the lysine-binding pocket and at least one additional amino acid residue. Biochemistry. 2000;39(25):7414–7419. doi: 10.1021/bi000155j. [DOI] [PubMed] [Google Scholar]
- 40.Roszmusz E, Patthy A, Trexler M, Patthy L. Localization of disulfide bonds in the frizzled module of Ror1 receptor tyrosine kinase. J Biol Chem. 2001;276(21):18485–18490. doi: 10.1074/jbc.M100100200. [DOI] [PubMed] [Google Scholar]
- 41.Green JL, Kuntz SG, Sternberg PW. Ror receptor tyrosine kinases: orphans no more. Trends Cell Biol. 2008;18(11):536–544. doi: 10.1016/j.tcb.2008.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Manser E, et al. PAK kinases are directly coupled to the PIX family of nucleotide exchange factors. Mol Cell. 1998;1(2):183–192. doi: 10.1016/S1097-2765(00)80019-2. [DOI] [PubMed] [Google Scholar]
- 43.Rossman KL, Der CJ, Sondek J. GEF means go: turning on RHO GTPases with guanine nucleotide-exchange factors. Nat Rev Mol Cell Biol. 2005;6(2):167–180. doi: 10.1038/nrm1587. [DOI] [PubMed] [Google Scholar]
- 44.Matsuda T, et al. Expression of the receptor tyrosine kinase genes, Ror1 and Ror2, during mouse development. Mech Dev. 2001;105(1–2):153–156. doi: 10.1016/s0925-4773(01)00383-5. [DOI] [PubMed] [Google Scholar]
- 45.van Amerongen R, Berns A. Knockout mouse models to study Wnt signal transduction. Trends Genet. 2006;22(12):678–689. doi: 10.1016/j.tig.2006.10.001. [DOI] [PubMed] [Google Scholar]
- 46.Paganoni S, Bernstein J, Ferreira A. Ror1-Ror2 complexes modulate synapse formation in hippocampal neurons. Neuroscience. 2010;165(4):1261–1274. doi: 10.1016/j.neuroscience.2009.11.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cha J, et al. Appropriate crypt formation in the uterus for embryo homing and implantation requires Wnt5a-ROR signaling. Cell Rep. 2014;8(2):382–392. doi: 10.1016/j.celrep.2014.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liu Y, Rubin B, Bodine PV, Billiard J. Wnt5a induces homodimerization and activation of Ror2 receptor tyrosine kinase. J Cell Biochem. 2008;105(2):497–502. doi: 10.1002/jcb.21848. [DOI] [PubMed] [Google Scholar]
- 49.Grumolato L, et al. Canonical and noncanonical Wnts use a common mechanism to activate completely unrelated coreceptors. Genes Dev. 2010;24(22):2517–2530. doi: 10.1101/gad.1957710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Petrova IM, Malessy MJ, Verhaagen J, Fradkin LG, Noordermeer JN. Wnt signaling through the Ror receptor in the nervous system. Mol Neurobiol. 2014;49(1):303–315. doi: 10.1007/s12035-013-8520-9. [DOI] [PubMed] [Google Scholar]
- 51.Sato A, Yamamoto H, Sakane H, Koyama H, Kikuchi A. Wnt5a regulates distinct signalling pathways by binding to Frizzled2. EMBO J. 2010;29(1):41–54. doi: 10.1038/emboj.2009.322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Seifert M, et al. Cellular origin and pathophysiology of chronic lymphocytic leukemia. J Exp Med. 2012;209(12):2183–2198. doi: 10.1084/jem.20120833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bo H, et al. Upregulation of Wnt5a promotes epithelial-to-mesenchymal transition and metastasis of pancreatic cancer cells. BMC Cancer. 2013;13: doi: 10.1186/1471-2407-13-496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Weeraratna AT, et al. Wnt5a signaling directly affects cell motility and invasion of metastatic melanoma. Cancer Cell. 2002;1(3):279–288. doi: 10.1016/S1535-6108(02)00045-4. [DOI] [PubMed] [Google Scholar]
- 55.Fecteau JF, et al. Lenalidomide inhibits the proliferation of CLL cells via a cereblon/p21(WAF1/Cip1)-dependent mechanism independent of functional p53. Blood. 2014;124(10):1637–1644. doi: 10.1182/blood-2014-03-559591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Burger JA, Burger M, Kipps TJ. Chronic lymphocytic leukemia B cells express functional CXCR4 chemokine receptors that mediate spontaneous migration beneath bone marrow stromal cells. Blood. 1999;94(11):3658–3667. [PubMed] [Google Scholar]
- 57.Qiao H, et al. Processing and subcellular trafficking of ER-tethered EIN2 control response to ethylene gas. Science. 2012;338(6105):390–393. doi: 10.1126/science.1225974. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.