Abstract
The subventricular zone (SVZ) provides a specialized neurogenic microenvironment for proliferation and aggregation of basal progenitors (BPs). Our study reveals a mechanism for the aggregation of BPs within the SVZ required for their proliferation and generation of cortical layer neurons. The Autism-related IgCAM, MDGA1 is locally expressed in the BP cell membrane where it co-localizes and complexes with the gap junction protein Connexin43. To address MDGA1 function, we created a floxed allele of MDGA1 and deleted it from BPs. MDGA1 deletion results in reduced BP proliferation and size of the SVZ, with an aberrant population of BPs ectopically-positioned in the cortical plate. These defects are manifested in diminished production of cortical layer neurons and a significant reduction of the cortical layers. We conclude that MDGA1 functions to aggregate and maintain BPs within the SVZ providing the neurogenic niche required for their proliferation and generation of cortical layer neurons.
INTRODUCTION
The neocortex is the center for sensory perception, motor control and cognition, with a complex structuring that features six primary layers (Angevine and Sidman, 1961). Glutamatergic neurons comprise approximately 80% of cortical neurons and are generated within two distinct proliferative zones, the ventricular zone (VZ) and the subventricular zone (SVZ) (Bayer and Altman, 1991).
Early in cortical development, the stem-like neuroepithelial cells population of VZ progenitors undergoes symmetric divisions to expand the pool of cortical progenitors. Neuroepithelial cells subsequently differentiate into neurogenic radial glia (RG), which undergo asymmetric divisions to generate deep layer neurons of the cortical plate (CP), as well as replenish themselves. They also generate a distinct progenitor population, the basal progenitors (BPs) that leave the VZ and aggregate with one another to establish the SVZ, where they then generate upper and deeper cortical layer neurons (Molyneaux et al., 2007, Noctor et al., 2004, Sessa et al., 2008, Kowalczyk et al., 2009). The SVZ in mice begins to form laterally around E12.5 (Vasistha et al., 2015), but it is not fully developed until E13.5 (Kowalczyk et al., 2009, Tarabykin et al., 2001, Bayer and Altman, 1991). The SVZ undergoes a substantial increase in size and BPs number around E16.5, coincident with the peak of generation of upper layer neurons (Bayer and Altman, 1991, Molyneaux et al., 2007).
BPs of the SVZ are characterized by their expression of the T-box transcription factor, Tbr2, which is not expressed by RG or any other cortical cells (Englund et al., 2005, Arnold et al., 2008). Tbr2 is important for establishing the appropriate number of BPs, as shown by conditional deletion of Tbr2 that results in diminished number of BPs and the cortical neurons they produce (Sessa et al., 2008, Arnold et al., 2008). Little is known about the molecular mechanisms that aggregate BPs to form the SVZ and establish this progenitor niche required for the generation of cortical neurons (Javaherian and Kriegstein, 2009, Noctor et al., 2004, Molyneaux et al., 2007). Here we address this issue and show that the IgCAM, MDGA1, functions as a gap junction-associated protein to mediate adhesive interactions between BPs required to aggregate them within the SVZ and establish their proliferative state necessary for corticogenesis.
MDGA1 is an immunoglobulin (Ig) superfamily (IgSF) protein with a structure composed of six Ig domains, a fibronectin type III (FnIII) domain, a highly conserved MAM (meprin, A5 protein, receptor protein tyrosine phosphatase mu) domain, and a glycosylphosphatidylinositol (GPI) anchor domain (Litwack et al., 2004). The Ig and FnIII domains are characteristic of IgSF proteins that mediate cell adhesion, whereas the MAM motif is a specialized protein interaction domain. MDGA1 is exclusively associated with plasma membranes via its GPI anchor (Litwack et al., 2004) and is highly expressed in lipid rafts in carcinoma cell lines (Diaz-Lopez et al., 2005). MDGA2 is the only other known MDGA family member (Litwack et al., 2004). Membrane attachment via a GPI anchor makes MDGA proteins unique among all MAM containing proteins.
Expression of MDGA1 is restricted to the nervous system, with MDGA1 being robustly expressed in the upper cortical layers (Litwack et al., 2004, Takeuchi et al., 2007). MDGA1 has been shown with in vitro assays to enhance cell adhesion (Diaz-Lopez et al., 2010), a finding consistent with its domain structure and expression patterns, and supporting its proposed function as an IgCAM that has a role in adhesion-based mechanisms of neural development (Litwack et al., 2004, Takeuchi et al., 2007). In vitro studies also indicate that MDGA1 suppresses inhibitory synapse development through its selective association with Neuroligin2 (Lee et al., 2013, Pettem et al., 2013). Interest in MDGAs has been further enhanced by their identification as high susceptibility genes for several neurological disorders, with intronic single nucleotide polymorphisms and exonic deletions resulting in truncated and non-functional MDGA protein and a high correlation with Autism Spectrum Disorders, Bipolar Disorders and Schizophrenia (Kahler et al., 2008, Bucan et al., 2009, Li et al., 2011).
We show here that MDGA1 is expressed by Tbr2-positive (Tbr2+) BPs within the cortical SVZ and is locally concentrated in cell membrane domains at appositions between BPs. We find that MDGA1 co-localizes at these sites with the gap junction protein Connexin43 (Cx43) and, with biochemical assays, show that MDGA1 and Cx43 associate with one another. To address MDGA1 functions in corticogenesis, we created a floxed allele of MDGA1 and analyzed mice with conditional deletion of MDGA1 from BPs using a Nestin-Cre line. We show that MDGA1 is required to aggregate Tbr2+ BPs within the SVZ and to establish proper SVZ size and proliferation. Disrupting these functions of MDGA1 results in a reduced number and proliferation of BPs in the SVZ, a substantial population of Tbr2+ BPs ectopically-positioned in the CP, and a diminished production of cortical neurons. We conclude that MDGA1 expressed by BPs functions to aggregate and maintain them within the cortical SVZ through an interaction of MDGA1 with Cx43 and possibly other proteins, providing BPs with the specific SVZ niche required for their proliferation and generation of cortical neurons.
RESULTS
MDGA1 expression in the developing forebrain
MDGA1 is strongly expressed in the olfactory bulb (OB), rostral migratory stream (RMS), upper layer neurons and cortical niches (Figures S1, S2). In the OB, MDGA1 is expressed in the progenitor layer surrounding the olfactory ventricle and in the glomerular layer where it co-localizes with the gap junction protein Cx43 (Figure S1A-C’). In the RMS, MDGA1 co-localizes with subpopulations of Doublecortin (DCX)+ neurons and Glial Fibrillary Acidic Protein (GFAP)+ cells (Figure S1D-I).
In the cortex, MDGA1 is expressed in layers 2-3, and in layer 4 and 6a of the primary somatosensory area (Takeuchi et al., 2007) (Figure S2A-C). During cortical development, MDGA1 is expressed in upper layer neurons migrating from the SVZ and localizes to the cell surface at apposition sites between upper layer neurons (Figure S2D).
Several upper layer markers such as Svet1, Cux1 and Cux2 are also expressed by the SVZ (Tarabykin et al., 2001, Nieto et al., 2004). To determine whether BPs of the SVZ also express MDGA1, we use in situ hybridization (ISH) at key stages during the generation and formation of the upper layers.
The SVZ expands in thickness and cell density between E13.5 to P0 (Molyneaux et al., 2007). At E12.5, before SVZ emerges, MDGA1 starts to be observed in the VZ (Figure S2E), similarly to other genes such as Cux2 (Tarabykin et al., 2001, Nieto et al., 2004, Molyneaux et al., 2007). At E16.5, as the generation of upper layer neurons in the SVZ becomes prominent, MDGA1 is robustly expressed in the SVZ with some expression in the VZ (Figure S2A, A’). At P0, when the thickness of the SVZ and its cellular density peaks, MDGA1 remains strongly expressed in the SVZ (Figure S2B, B’). By P7, MDGA1 expression is no longer detected in the depleted germinal zones with the exception of the septo-callosal region (Figure S2C). Over this time frame, MDGA1 expression in the upper layers gradually increases in laminar thickness and density as upper layer neurons are added (Takeuchi et al., 2007).
In the embryonic VZ, MDGA1 expression describes a low-dorsomedial to high-lateral gradient (Figure 1A), suggesting a regional-dependent expression of MDGA1 in the neurogenic niches. Thus, MDGA1 expression in the SVZ/VZ has a 1) dorsomedial domain high in the SVZ (0.845±0.08) and low in the VZ (0.154±0.07) and related to the generation of upper layer neurons; and 2) a lateral domain high in the VZ (0.68±0.01; compared to SVZ (0.32±0.03)) and likely related to the generation of MDGA1+ neurons for the RMS (Figure 1A,S1). MDGA1+ cells in the lateral VZ co-localize with Nestin, which is consistent with MDGA1+ cells deriving from the Nestin progenitor lineage (Figure S2F-H).
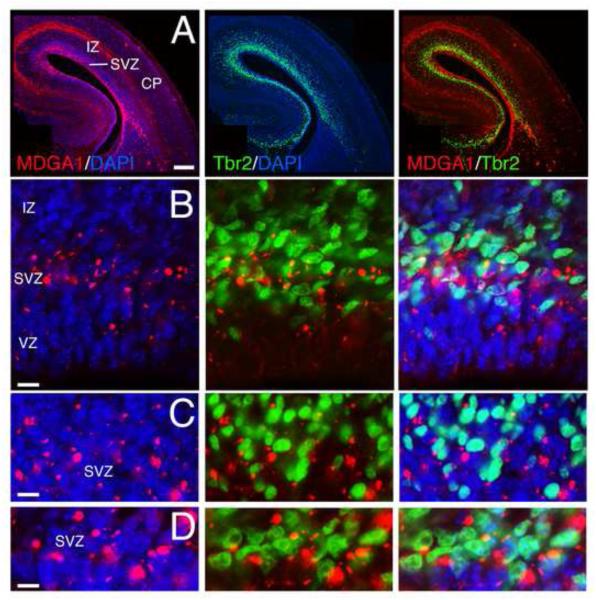
Figure 1. MDGA1 is expressed by basal progenitors of the subventricular zone.
Shown is immunolocalization of MDGA1 protein (red) and Tbr2 protein (green), a marker selective for basal progenitors of the subventricular zone (SVZ), on the same coronal cortical sections at E16.5. DAPI (blue) is shown as counterstaining. The C-D sets of photos are higher power views of SVZ cells from the A-B sets. Merged images of MDGA1 and Tbr2 are also shown. The majority of MDGA1+ cells in the SVZ (DM: 0.845±0.08; L: 0.32±0.01) co-localize with Tbr2 (tM: 0.54±0.06) and only a small fraction is negative for Tbr2. MDGA1 expression in the VZ describes a low-DM (0.154±0.07) to high-L (0.68±0.01) gradient. Abbreviations: CP: cortical plate; DM: dorsomedial; IZ: intermediate zone; L: lateral; VZ: ventricular zone. Scale bars: A (0.2 mm), B (100 μm) and C-D (50 μm).
To directly demonstrate that MDGA1 is expressed by BPs, we combined immunofluorescence for MDGA1 and the BP marker Tbr2 at E16.5 (Figure 1). Tbr2 is basically expressed in the SVZ labeling BPs, whereas MDGA1 is strongly expressed in the SVZ with some expression in the VZ (Figure 1B-D). Within the SVZ, the majority of cells that express Tbr2 also co-localize MDGA1 (Figure 1B-D). Within the VZ, MDGA1+ cells located in the top of the VZ closer to the SVZ co-localize Tbr2, whereas some MDGA1+ cells in the bottom of the VZ are negative for Tbr2 (Figure 1A-B). A quantitative analysis to determine the Mander’s co-localization coefficient (tM) using the thresholding algorithm of Costes (Costes et al., 2004, Mander, 1993) indicates a tM=0.54±0.06 (means±SEM) for MDGA1 and Tbr2 in SVZ/VZ. In the present study, we will focus on the role of MDGA1 in the SVZ and in the generation of MDGA1-expressing upper layer neurons.
MDGA1 is concentrated in cell membrane domains at appositions between BPs
MDGA1 is a 140 KDa protein exclusively associated with plasma membranes and anchored to them by a GPI linkage (Litwack et al., 2004). We confirmed these findings using a differential centrifugation assay in cortical tissue and a mouse neuroblastoma cell line (Figure S3A-C). Next we showed that MDGA1 is preferentially localized to the cell membrane by immunostaining 293 cells transfected with full-length mouse MDGA1 using rabbit polyclonal antibodies specific for MDGA1 (Figure 2A). This localized expression pattern resembles the labeling we describe in the cortical SVZ (Figures 1B-D, S2F-H) and in the upper layer neurons (Figure S2D), suggesting a likely physical interaction between apposing cells mediated by MDGA1.
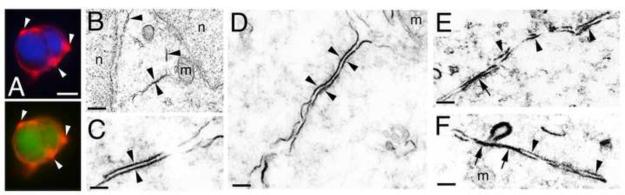
Figure 2. MDGA1 protein is localized in discrete cell membrane domains.
A: Immunolocalization of MDGA1 in vitro using 293 cells transfected with full-length MDGA1 and revealed using an antibody against MDGA1. MDGA1 is localized in discrete membrane domains (arrowheads). DAPI staining and GFP labeling confirming the transfections are also shown. B-F: Electron microscopy of ultrathin cortical sections at the level of the SVZ at E16.5. Arrowheads show MDGA1 immunolabeling of plasma membranes of adjacent BPs where an intermembrane space is observed. Arrows in E-F show MDGA1 expressed in tightly associated plasma membranes of adjacent BPs, in a gap junction-like manner. Abbreviations: m: mitochondria; n: nuclei of BP cell. Scale bars: A (10 μm), B (500 nm), C (100 nm), D (300 nm) and E-F (200 nm).
To obtain ultrastructural localization of MDGA1 in the SVZ, we used Electron Microscopy (EM) to analyze immunostained tissue from cortical levels where MDGA1 is mostly expressed in the SVZ. As described above, we find that MDGA1 is expressed in apposing cell membranes of adjacent BPs, with crisp expression focused on tightly apposed sections of BP cell membranes (Figure 2B-F). This localized MDGA1 expression is largely found in two distinct forms that relate to the separation between the apposing BP cell membranes: one where the apposing cell membranes are closely associated but a narrow space can be readily discriminated between them (Figure 2B-F), which resembles cell-cell contacts such as adherent junctions, and one where the apposing membranes come together in a very tight association that makes it difficult to distinguish a space between them, which resembles gap junctions (Figure 2E,F)(Brightman and Reese, 1969).
These findings show that within the SVZ, MDGA1 is preferentially localized at close appositions between BP cell membranes; a localization consistent with the domain structure of MDGA1 and is characteristic of many IgCAMs.
MDGA1 co-localizes and associates with gap junction proteins
Our data is consistent with MDGA1 being expressed in gap junctions, which serve important adhesive functions between neural cells (Elias et al., 2007). To address this, we performed double immunostaining at E16.5 for MDGA1 and Cx43, a gap junction protein preferentially expressed in the SVZ. We find that MDGA1 and Cx43 extensively co-localize in the BP membranes within the SVZ (Figure 3A,B). Most of the MDGA1 expression domains within the BP cell membranes also express Cx43, whereas some Cx43 expression domains are MDGA1-negative. The co-localization of MDGA1 and Cx43 within the SVZ persists to P0 (Figure S3D). We extended these in vivo observations using 293 cells transfected with Myc-tagged MDGA1 and double immunostained with antibodies specific for Myc-tag to identify MDGA1 and for Cx43. Both the transfected MDGA1 and endogenous Cx43 (Figure 3C) exhibited staining co-localized in the cell membrane in patterns similar to what we observed in vivo in the SVZ (Figure 3A,B).
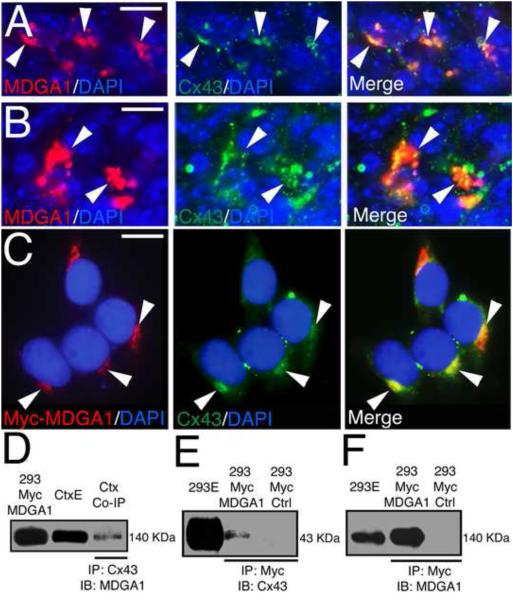
Figure 3. MDGA1 co-localizes and associates with the gap junction protein Connexin43.
Shown is co-localization of MDGA1 with Connexin43 (Cx43) in vivo in BPs of the SVZ (A-B) and in vitro in 293 cells transfected with Myc-MDGA1 (C), and the association of MDGA1 with Cx43 shown using co-immunoprecipitation (Co-IP) and immunoblotting (D-F). A-B: Immunofluorescence of MDGA1 (red) and Cx43 (green) on cortical sections at E16.5. Both MDGA1 and Cx43 show robust protein expression in the SVZ that is frequently co-localized in the plasma membrane of BPs in the SVZ. Arrowheads point to same cells in the respective series. C: 293 cells transfected with Myc-MDGA1 and immunostained for MDGA1 using a Myc antibody and for endogenous Cx43. Myc-MDGA1 (red) and the endogenous Cx43 (green) co-localize in discrete membrane domains. Arrowheads mark protein co-localization in the same domains in the series. Merged images are also shown. DAPI (blue) is used as counterstaining. D-F: Co-IP assays using protein extracts from WT cortex (E18, P7, D) or protein extracts of 293 cells transfected with Myc-MDGA1 (293 Myc MDGA1, E-F). D: Co-IP using WT cortical extracts (Ctx) and a Cx43 antibody, immunoblotted using an antibody specific for MDGA1 reveals a 140 KDa band as expected for MDGA1. E: Co-IP done with Myc antibody and immunoblot done with Cx43 antibody reveals a 43 KDa band as expected for Cx43. F: In a similar Co-IP, an immunoblot using an antibody specific for MDGA1 recognizes a 140 KDa band as expected for MDGA1. Abbreviations: 293E: whole extracts from 293 cells transfected with Myc-MDGA1; 293 Myc Ctrl: Myc negative control in 293 cells, no bands are labeled in the immunoblots; CtxE: whole cortex extract; IB: immunoblot; IP: immunoprecipitation. Scale bars: A-B (20 μm) and C (8 μm).
To address the potential for direct associations between MDGA1 and Cx43, we performed co-immunoprecipitation (Co-IP) assays using protein extracts from WT cortex (E18, P7, Figure 3D) and transfected 293 cells (Figure 3E,F). As a positive control, we showed that extracts from both 293 cells transfected with Myc-tagged full length MDGA1 and WT cortex blotted with an MDGA1 antibody labeled the expected 140 KDa band (Figure 3D). We performed the immunoprecipitation (IP) on these extracts with a Myc-specific antibody to pull down MDGA1, and performed the immunoblot with an antibody specific for Cx43 (Figure 3E) or for MDGA1 (Figure 3F). The IP using the Myc antibody pulled down both Cx43 and MDGA1, shown with the immunoblots using the Cx43 antibody labeling a 43 KDa band (Figure 3E) or using an MDGA1 antibody labeling a 140 KDa band (Figure 3F) as expected. As a negative control, we used 293 cells transfected with Myc-NKB; IPs done on extracts from these cells with the Myc antibody and immunoblots done with either the Cx43 or MDGA1 antibodies did not label any bands at the expected sizes (Figure 3E,F) eliminating the potential for false interactions. These associations were confirmed for endogenous MDGA1 and Cx43 by performing Co-IPs using cortical extracts (E18, P7), with the IP done using a Cx43 antibody and the immunoblot with an MDGA1 antibody that labeled the expected 140 KDa band (Figure 3D). These findings show that MDGA1 and Cx43 are co-expressed by BPs in the cortical SVZ, and co-localize to discrete BP cell membrane domains, suggesting that they form complexes within the cell membrane.
MDGA1 deletion from progenitors reduces SVZ size and BPs proliferation
We hypothesized that MDGA1 functions to mediate adhesion among BPs to aggregate them within the SVZ and thereby establish and maintain the SVZ. By “aggregation” we refer to a group of cells from the same cell type (e.g. BPs), which are tightly associated through cell-cell contacts (e.g. junctions). We predicted that MDGA1 function would also influence BPs proliferation. To address this, we generated a MDGA1 floxed allele (MDGA1fl) (Figure S4A, B) and used a Nestin-Cre line (Graus-Porta et al., 2001) to conditionally delete MDGA1 from cortical progenitors in the VZ, which generate BPs of the SVZ. The Nestin-Cre line produces robust recombination throughout the cortex by E11.5 and deletes floxed alleles from essentially all cortical progenitors (Chou et al., 2009). Offspring of mice homozygous for MDGA1 floxed alleles crossed to the Nestin-Cre line (cKO) are viable and have no MDGA1 transcripts detectable by ISH nor is MDGA1 protein evident in immunoblots (Figure S4C, D).
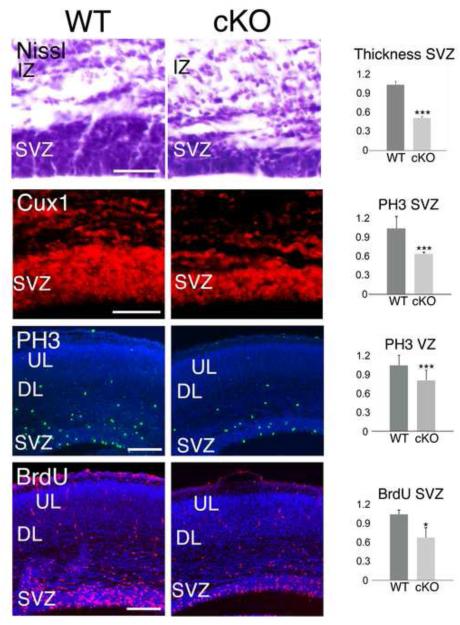
We first addressed the impact of the conditional deletion of MDGA1 on the integrity of the SVZ. Nissl staining and immunostaining for the SVZ marker Cux1 (Nieto et al., 2004) of E16.5 (Figure 4) and P0 (Figure S5A) cortical sections revealed that the SVZ is significantly reduced in the cKO at both E16.5 (Figure 4, 0.53±0.01, p= 0.005***) and P0 (Figure S5B, 0.682±0.03, p=0.003***) when compared to WT.
Figure 4. Conditional deletion of MDGA1 from cortical progenitors results in significant reduction of thickness and proliferation in the neurogenic niches.
Shown are coronal sections of E16.5 cortex from wild type (WT) and mice with a conditional deletion of MDGA1 using Nestin-Cre (cKO), demonstrating that both the thickness of the subventricular zone (SVZ, Nissl, Cux1) and proliferation in the SVZ (PH3, BrdU) are significantly reduced in the cKO. Nissl staining and immunofluorescence for the SVZ marker Cux1 shows a significant reduction in the thickness of the SVZ in the cKO versus WT (0.53±0.01, p= 0.005***, N=4). The number of PH3+ cells is also significantly reduced by 45% in the SVZ (0.559±0.03, p=0.0006***, N=4), and by 21% in the VZ (0.789±0.06, p=0.005***, N=4) in the cKO compared to WT. A 2-hour pulse of BrdU shows a reduction in BrdU incorporation by 35% in the SVZ of the cKO compared to WT (0.71±0.076, p=0.04*, N=4). Abbreviations: DL: deeper layers; IZ: intermediate zone; UL: upper layers; VZ: ventricular zone. Scale bars: Nissl and Cux1 panels (50 μm) and PH3 and BrdU panels (100 μm).
We next addressed the affect of the conditional deletion of MDGA1 on proliferation at E16.5, the peak of neurogenesis of upper layer neurons in the SVZ (Figure 4). To determine mitotic activity, we used immunostaining for Phospho-Histone 3 (PH3) and as a complementary approach, BrdU injections to label proliferating cells (Figure 4). At E16.5, the number of BPs in M-phase identified by PH3 labeling was significantly reduced in the SVZ of the cKO by 45% (Figure 4, 0.559±0.03, p=0.0006***) and in the VZ by 21% (Figure 4, 0.789±0.06, p=0.005***) respect to WT. In addition, a 2-hour pulse of BrdU also showed that the number of BrdU+ cells was significantly decreased in the SVZ of the cKO by 35% (Figure 4, 0.71±0.076, p=0.04*) compared to WT. The overall proliferative activity in the cKO is reduced; however, it is particularly significant in the SVZ with a reduction in proliferation proportional to its reduction in thickness. In the VZ of the cKO, only the reduction in proliferation is noticeable, and it is consistent with subpopulations of Tbr2+ BPs that are present in the VZ. Our findings indicate that MDGA1 expression by BPs is required to establish their appropriate numbers in the SVZ.
MDGA1 deletion results in ectopic Tbr2+ cells
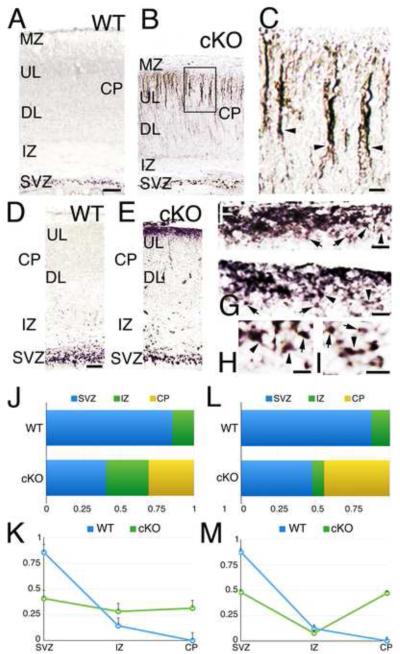
We hypothesized that the reduced number of BPs and proliferation in the cortical SVZ of the cKO is due to a proportion of BPs failing to aggregate within the SVZ after they are generated by RG and leave the VZ. To address this, we used Tbr2 to determine whether a subpopulation of BPs is found at ectopic positions within the developing cortex of cKO. In P0 WT, Tbr2+ cells were restricted to the SVZ (Figure 5A, SVZ: 0.877±0.03; IZ: 0.122±0.06; CP: 0). In P0 cKO, Tbr2+ cells were also present in the SVZ (0.472±0.01, p=0.0048***); but in addition, we found a sizeable number of ectopic Tbr2+ cells positioned in the upper CP (Figure 5B, IZ: 0.076±0.007, p=0.05; CP: 0.472±0, p=0.0095***). Contrary to the normal nuclear distribution of Tbr2 protein in BPs of the SVZ, we found that Tbr2 protein was concentrated in the cell cytoplasm and in radially aligned apical processes in these ectopic Tbr2+ cells (Figure 5B,C).
Figure 5. Tbr2-positive cells are ectopically located in the upper layers of the developing cortex in MDGA1 cKO mice.
A-I: Shown are coronal sections of the cortex from P0 (A-C) and E16.5 (D-I). WT and cKO immunostaining for Tbr2 reveals an ectopic population of Tbr2+ cells in the upper layers (UL) of the cKO cortex. A-C: At P0, Tbr2+ cells in WT cortex are limited to the SVZ (SVZ: 0.877±0.03; IZ: 0.122±0.06); whereas in cKO cortex, they are present in the SVZ (SVZ: 0.472±0.01, p=0.0048***; IZ: 0.076±0.007, p=0.05) and a substantial number of Tbr2+ cells are ectopically positioned in the upper layers of the cortical plate (CP, 0.472±0, p=0.0095***). Tbr2 protein is normally localized to the nucleus, as it is in basal progenitors of the SVZ in both WT and cKO cortex (A, B); but at P0 in the cKO (B, C), the ectopic Tbr2+ cells (arrowheads in C, which is a higher power of the boxed area in B) have aberrant Tbr2 protein localization to the apical process and a thin rim of cytoplasm surrounding the nucleus. D-I: At E16.5, Tbr2+ cells in WT cortex (D) are largely limited to the SVZ (SVZ: 0.856±0.02; IZ: 0.144±0.03). In cKO cortex (E), Tbr2+ cells are present in the SVZ (SVZ: 0.408±0.06, p=0.0008***; IZ: 0.282±0.05, p=0.1721), and a substantial number of Tbr2+ cells are ectopically positioned at the top of the CP (0.314±0, p=0.0029***). Tbr2 protein shows a variable localization (F-I), from being preferentially in the nucleus (arrows in F-I) to throughout the cell including multiple short processes extending from the cell body (arrowheads in F and G, and in the higher power panels H and I). J-M: quantification for Tbr2+ cells at E16.5 (J-K) and P0 (L-M) shows that the overall amount of Tbr2+ cells is not changed (P0: 1±0.01, p=0.8075; E16: 1±0.02, p=0.4051, J, L) but the laminar distribution is severely altered in the cKO (J-M). Tbr2+ cells in the SVZ are reduced in the cKO by 46% (E16.5) and by 55% (P0); whereas the CP, which does not express Tbr2 in WT, represents a 31% (E16.5) and a 46% (P0) of the Tbr2+ cells in the cKO. Abbreviation: DL: deeper layers; IZ: intermediate zone; MZ: marginal zone. Scale bars: A-B (100 μm), C (20 μm), D-E (100 μm) and F-I (20 μm).
A substantial number of ectopic Tbr2+ cells were observed in the cKO cortex at E16.5 (Figure 5E), but were not observed at any age in WT cortex (Figure 5A,D). In E16.5 WT and cKO, Tbr2+ cells were aggregated in the SVZ (WT: 0.856±0.02; cKO: 0.408±0.06, p=0.0008***) with a small proportion scattered in the intermediate zone (IZ, Figure 5D-E, WT: 0.144±0.03; cKO: 0.282±0.05, p=0.1721). However, in contrast to WT, the E16.5 cKO also had a high density of Tbr2+ cells ectopically positioned at the top of the CP (Figure 5E, WT: 0; cKO: 0.314±0, p=0.0029***). Tbr2 protein was localized to the nucleus in many of the ectopic Tbr2+ cells (Figure 5F-I), as it is in BPs normally positioned in the SVZ. In many others, Tbr2 protein was distributed throughout the cell body and the major processes extending from it (Figure 5F-I), revealing that the ectopic Tbr2+ cells have a multipolar-like morphology resembling that typical for Tbr2+ BPs in the SVZ of WT (Kowalczyk et al., 2009).
Our quantification analysis indicates that the overall amount of Tbr2+ cells is not changed between WT and cKO at P0 (1±0.01, p=0.8075) or at E16.5 (1±0.02, p=0.4051) (Figure 5J,L). However, the laminar distribution of the Tbr2+ cells is disrupted in the cortex of the cKO (Figure 5J-M): 1) in the SVZ, Tbr2+ cells are reduced by 46% at E16.5 and by 55% at P0, which is consistent with the reduction in thickness and proliferation observed in the SVZ (Figures 4, 5, S5A-B, S7); and 2) the CP does not express Tbr2 in WT, but Tbr2+ cells are represented by 31% at E16.5 and by 46% at P0 in the cKO, which is consistent with Tbr2+ BPs migrating out of the SVZ to occupy ectopic positions in the CP (Figures 5A-I, 6, S7).
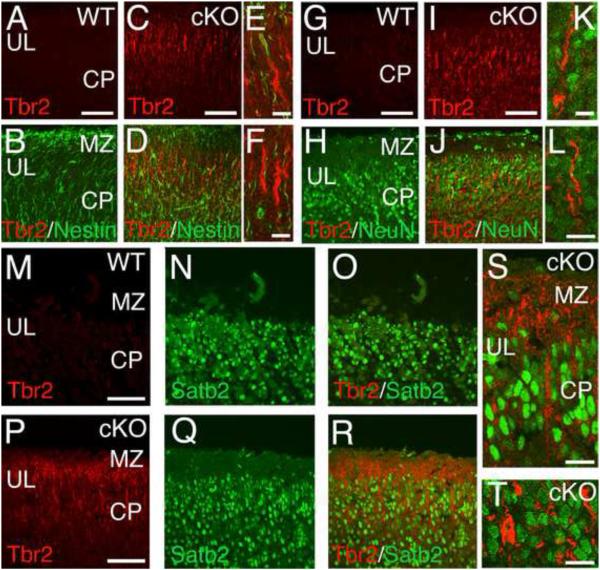
Figure 6. Ectopic Tbr2-positive cells in MDGA1 cKO cortex do not express neuronal or glial markers.
Shown are confocal images of sections from P0 WT and MDGA1 cKO cortex double immunostained with antibodies for Tbr2 (red), which in WT cortex is a marker specific for basal progenitors of the subventricular zone, and markers of radial glia (Nestin, green) and differentiated neurons (NeuN and Satb2, green), to determine possible cellular differentiation of the ectopic Tbr2+ cells found in the cortical plate (CP) of the cKO. The genotype and immunostaining for each panel is as indicated. Each panel shows only the upper layers (UL) of the CP and marginal zone (MZ). Ectopic Tbr2+ cells are found in the cKO cortex, but not in WT cortex; at this age (P0), they have a radial morphology and immunolocalization of Tbr2 protein (red) is confined to the apical process and cytoplasm of the cell body. The other markers (Nestin, NeuN and Satb2; green) are found in both WT and cKO cortex: in both, Nestin is localized to the radial processes of radial glia, which extend through the CP, whereas the transcription factors NeuN and Satb2 are localized to neuronal nuclei. Tbr2 immunostaining does not co-localize with immunostaining for any of the other three markers, as evident in the merged images from cKO cortex at low power (D) and high power (E,F) for Tbr2 and Nestin, at low power (J) and high power (K,L) for Tbr2 and NeuN, or at low power (R) and high power (S,T) for Tbr2 and Satb2. Scale bars: A-D (50 μm), E-F (20 μm), G-J (50 μm), K-L (20 μm), M-R (50 μm) and S-T (20 μm).
We do not find ectopic Tbr2+ cells in the developing cortex of mice created by crossing MDGA1fl/fl mice to a Nex-Cre line (Figure S5C), which produces Cre-mediated recombination in newly generated cortical neurons prior to their migration (Goebbels et al., 2006). Our findings indicate that the ectopic Tbr2+ cells are aberrant BPs that failed to aggregate within the SVZ and migrate radially from the VZ to the top of the developing CP.
Ectopic Tbr2+ cells exhibit defective differentiation
To assess whether the ectopic Tbr2+ cells retain their proliferative state like Tbr2+ BPs that remain in their natural SVZ niche, we combined a 2-hour pulse of BrdU with PH3 immunostaining at E16.5 (Figure S6A). We found that a substantial number of SVZ cells were BrdU+ and a smaller number were PH3+ in both WT and cKO. However, both WT and cKO had very few BrdU+ cells and virtually no PH3+ cells superficial to the SVZ particularly at the top of the CP where the ectopic Tbr2+ cells are positioned in cKO (Figures 4, S6A). Thus, the distribution of proliferating cells is very similar in the cortex of E16.5 WT and cKO. These findings show that, unlike their Tbr2+ BPs counterparts in the SVZ, the ectopic Tbr2+ cells are in a non-proliferative state, consistent with recent reports that the SVZ provides a niche environment critical for BPs proliferation (Javaherian and Kriegstein, 2009, Ihrie and Alvarez-Buylla, 2011).
To determine whether the ectopic Tbr2+ cells have properties that define CP neurons and glia, we focused on P0 when the ectopic Tbr2+ cells are integrated into the upper layers of the CP and have acquired their radial morphology (Figure 6). Special attention was given to the Tbr2+ radial processes extended toward the pial surface by the ectopic Tbr2+ cells, which resemble the primary apical dendrite of cortical projection neurons and the radial processes of RG. We used confocal imaging co-immunostained with antibodies against Tbr2 combined with either the RG marker Nestin or the neuron-specific markers NeuN and Satb2. Robust Nestin immunolabeling of radial processes was observed in the CP of both WT (Figure 6A,B) and cKO (Figure 6C-F), but exhibited no overlap with the Tbr2 labeled radial processes of the ectopic Tbr2+ cells evident exclusively in the cKO (Figure 6C-F). We performed similar imaging using antibodies against Tbr2 and either NeuN, a pan-neuronal marker (Figure 6G-L), or Satb2, a cortical projection neurons marker (Figure 6M-T). Each neuronal marker produced robust immunolabeling in the CP of both WT (Figure 6G,H, M-O) and cKO (Figure 6I-L,P-T), but as with Nestin, we observed no co-localization between the Tbr2 immunolabeled cells and processes with neither NeuN (Figure 6I-L) or Satb2 (Figure 6P-T) in the cKO. Thus, the Tbr2+ cells in the CP of the cKO do not exhibit defining characteristics of either CP neurons or RG, consistent with their identification as ectopic BPs.
At early postnatal stages, we observed clusters of Caspase-3 immunolabeling in the upper layers of the cKO that are not present in WT, suggesting that the final fate of the ectopic Tbr2+ cells is presumably apoptosis (Figure S6B).
Cortical layers are diminished in MDGA1 cKO mice
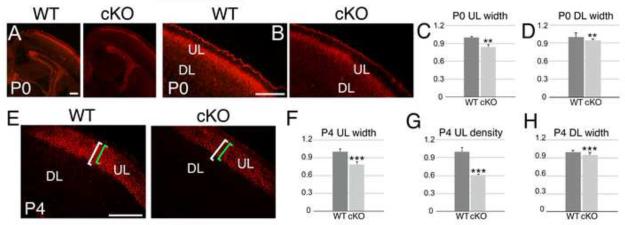
Tbr2+ BPs of the cortical SVZ contribute to all cortical layers, but their major contribution is to cell lineages that form upper layers (Sessa et al., 2008, Arnold et al., 2008, Vasistha et al., 2015). To address this, we used immunostaining for upper layer neurons marker Cux1 (Nieto et al., 2004). At P0 WT, Cux1+ upper layer neurons are evident, although they are still differentiating and a proportion of Cux1+ neurons are migrating to the upper layers (Figure 7A-B). In cKO, the density of Cux1+ upper layer neurons is substantially decreased and the upper layers width is significantly reduced by 16% compared to WT (Figure 7A-C, 0.830±0.02, p=0.021**). In adjacent Nissl sections, we measured the width of deeper layers and found a reduction by 6% (Figure 7D, 0.946±0.009, p=0.01**).
Figure 7. Thickness and neuronal density of cortical layers are significantly reduced in MDGA1 cKO mice.
Shown are coronal sections of the cortex of WT and cKO at P0 (A-B) and P4 (E) immunostained for the transcription factor Cux1 (A,B,E). Histograms quantifying the significant reduction in thickness (C: P0, 0.830±0.02, p=0.021**, N=4; F: P4, 0.781±0.05, p=0.0096***, N=4) and neuronal density (G: P4, 0.611±0.02, p=0.0013***, N=4) of the upper layers (UL), and quantification of the thickness (D: P0, 0.946±0.009, p=0.01**, N=4; H: P4, 0.943±0.01, p=0.006***, N=4) of the deeper layers (DL) are also shown. At P0, upper layers are reduced in thickness by 16% (C) and deeper layers by 6% (D). At P4, upper layers are reduced in thickness by 22% (F) and in cell density by 39% (G), while deeper layers are reduced in thickness by 6% (H). In E, brackets indicate comparisons of the thickness of Cux1-positive (+) upper layers in the cKO (green) and WT (white) cortex. At P4, in both WT and cKO, essentially all Cux1+ neurons are located in the upper layers with very few found in the deeper layers of the cortex, indicating that essentially all Cux1+ neurons complete this migration by P4. Scale bars in A (100μm), B (200 μm) and E (100 μm).
By P4, migration is essentially complete and virtually all Cux1+ neurons are localized to the upper layers with only a few scattered Cux1+ neurons still migrating (Figure 7E). The thickness and density of Cux1+ upper layers are markedly reduced in cKO compared to WT (Figure 7E-G). Quantitation shows that thickness and density are significantly reduced by 22% (Figure 7F, 0.781±0.05, p=0.0096***) and by 39% (Figure 7G, 0.611±0.02, 0.0013***), respectively. At P4, the thickness of deeper layers was reduced by 6% (Figure 7H, 0.943±0.01, p=0.006***). The significant reduction in upper layers in the cKO is consistent with a larger contribution of Tbr2+ BPs to upper layer neurons (Kowalczyk et al., 2009, Sessa et al., 2008, Vasistha et al., 2015). The similarity in the overall distribution of Cux1+ neurons between WT and cKO cortex indicates that the reduction in the upper layers in the cKO is not due to a migration defect. We conclude that the reduced BPs population and proliferation in the SVZ is manifested by both a reduced thickness and neuronal density in upper layers, with a minor reduction in deeper layers, and an overall loss of approximately half of the normal population of upper layer neurons in the cKO compared to WT.
DISCUSSION
We demonstrate a molecular mechanism required to properly establish and maintain the cortical SVZ, the consequences of deficiencies in this mechanism for the integrity of the SVZ and its proliferative capacity, the fate of BPs that normally populate the SVZ, and the production of cortical layer neurons. We present evidence that MDGA1 is a required component of this mechanism and mediates cellular interactions between BPs. We show that MDGA1 is expressed by cortical BPs, is localized to discrete membrane domains at appositions between BPs, co-localizes and associates with the gap junction protein Cx43 in the BP cell membranes. We propose that MDGA1 forms protein complexes with Cx43 and promotes adhesion among BPs to aggregate them in the SVZ, which is supported by our Co-IP assays and analyses of mice with a conditional deletion of MDGA1 from progenitors. We show that SVZ thickness and proliferation is significantly reduced in cKO, and we find an aberrant population of cells expressing Tbr2, a selective BP marker, ectopically positioned in the upper part of the developing CP. Concomitantly, we observe a substantial reduction in the number of cortical layer neurons produced by BPs of the SVZ.
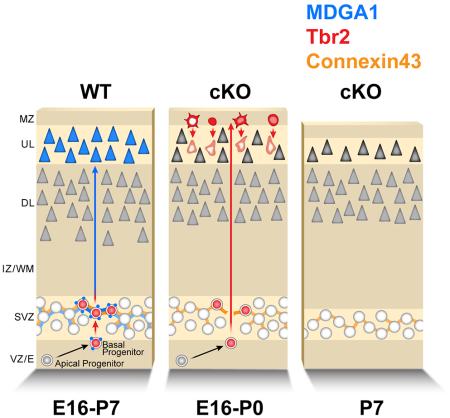
Our findings indicate that MDGA1 functions as a gap junction-associated protein and interacts with other membrane-associated proteins expressed by BPs, such as Cx43, to mediate the adhesion required for proper aggregation of newly generated BPs within the SVZ, a mechanism necessary for BP proliferation and their generation of the appropriate population of cortical layer neurons. When BPs do not express MDGA1, the adhesion among BPs is diminished and a significant proportion of BPs exhibit an aberrant migration to the top of the CP, along the radial migratory path and following the “inside-out” gradient, with the most recently generated cells migrating to the top of the developing CP (Angevine and Sidman, 1961). As the remaining cortical layer neurons are generated, they in turn migrate past the ectopic BPs to the top of the CP, differentiate and in the process displace the ectopic Tbr2+ BPs to the upper layers of the CP. Coincident with this displacement process and the normal laminar differentiation of the CP, the morphology of the ectopic Tbr2+ BPs transitions from their initial multipolar morphology, similar to that of BPs in the SVZ, to their radial morphology, similar to that exhibited by differentiating CP neurons. The ultimate consequence of the conditional deletion of MDGA1 is a significant reduction in the generation of cortical layer neurons, mainly upper layer neurons (Figure S7).
MDGA1 adhesive mechanisms to aggregate BPs in the SVZ
IgCAMs interact both homophilically and heterophilically to mediate adhesion (Brummendorf and Rathjen, 1996). However, a recent study only showed heterophilic binding of MDGA1 to either MDGA1-expressing neurons or axons (Fujimura et al., 2006). Thus, MDGA1 likely mediates adhesion between BPs through heterophilic binding. One possibility is “in trans” binding of MDGA1 to other adhesion proteins of apposing BPs or to components of the extracellular matrix (ECM). Consistent with that, MDGA1 enhances heterophilic cell adhesion to non-MDGA1 expressing cells, as well as to the ECM (Diaz-Lopez et al., 2010). Another possibility is an “in cis” association of MDGA1 with other membrane proteins (Lee et al., 2013, Pettem et al., 2013) to form complexes that generate adhesion between BPs.
MDGA1 and Cx43 are largely expressed by BPs in the neocortical SVZ and both co-localize in discrete membrane domains at appositions between cells. Cx43-mediated adhesion between migrating cortical neurons and RG fibers is due to Cx43 forming gap junction hemichannels in the membrane that interact heterophilically with other proteins in adjacent membranes to provide adhesive contacts (Elias et al., 2007). Because MDGA1 is an extracellular protein anchored to the cell membrane by a GPI-linkage, the association of MDGA1 with Cx43 is likely through the extracellular domains of Cx43.
Conditional deletion of Cx43 from progenitors using a Nestin-Cre line results in a reduction in thickness in upper layers (Cina et al., 2009). This is consistent with MDGA1 cooperating with Cx43 to influence the aggregation of BPs and proliferation in the cortical SVZ. We suggest that Cx43 hemi-junctions form adhesive contacts among BPs, and that MDGA1 facilitates or enhances this gap junction adhesion by “in cis” or “in trans” heterophilic binding to Cx43. Other GPI-anchored proteins have been shown to participate in junction formation between cells of the vertebrate nervous system, including Contactin in myelinated peripheral nerves (Boyle et al., 2001) and Nogo66 in the cerebellum (Liu et al., 2005).
Characteristics of ectopic BPs and contributing mechanisms
BPs are generated within the VZ by RG, and migrate superficially from the VZ to aggregate on top of it to form the SVZ (Molyneaux et al., 2007). BPs undergo a single terminal division in the SVZ to generate two postmitotic neurons; a small proportion of BPs generate additional BPs prior to their terminal division (Miyata et al., 2004, Noctor et al., 2004, Molyneaux et al., 2007). Coincident with their terminal division, their progeny fated to become neurons down-regulate Tbr2, a process required for activation of their neuronal differentiation program (Englund et al., 2005, Alcamo et al., 2008, Tsui et al., 2013). Consistent with their failure to down-regulate Tbr2, the ectopic Tbr2+ cells in cKO do not express markers for cortical neurons, nor for other components of the CP and remain in an undifferentiated state to presumably undergo apoptosis. Our findings indicate that the ectopic Tbr2+ cells are aberrant BPs, and that their defective undifferentiated state is due to their ectopic position and failure to execute their normal sequence of proliferation and differentiation that occurs in their SVZ niche, particularly the absence of their normal terminal division that leads to down-regulation of Tbr2 and the engagement of a neuronal differentiation program. Analyses of mice with an early conditional deletion of Tbr2 show that the proliferation and number of BPs in the SVZ is reduced by ~ 25% (Arnold et al., 2008, Sessa et al., 2008). It is likely that the aberrant cellular localization of Tbr2 in the ectopic Tbr2+ BPs in the cKO cortex might affect Tbr2 function and regulation, and contributes to the abnormal characteristics of the ectopic Tbr2+ BPs.
Several studies have shown that the cortical SVZ provides a neurogenic niche for BPs, and contains neurotransmitters, growth factors, morphogens, ECM, vasculature microenvironment, as well as cell-cell interactions that function in a non-cell autonomous fashion to regulate progenitor fate and proliferation (Javaherian and Kriegstein, 2009, Ihrie and Alvarez-Buylla, 2011, Stubbs et al., 2009). The deletion of the centrosome protein Sas-4 from progenitors leads to ectopic populations of Pax6+ RG and their Tbr2+ BP progeny in the CP that are eliminated by cell death (Insolera et al., 2014), similarly to the data we report here in cKO. Our complementary findings reinforce the importance of the molecular and genetic mechanisms that operate in the neurogenic niches to properly establish the fate of cortical progenitors.
Consequence of reduced SVZ for cortical layers
The significant reduction in BPs and proliferation in the SVZ of the cKO results in a reduction of the number of cortical layer neurons, which is particularly significant for upper layer neurons. Interestingly, our findings in the cKO show that upper layer neurons, which normally express MDGA1 (Takeuchi et al., 2007), are able to migrate to their appropriate CP positions, with Cux1+ neurons normally positioned within the cortex and their overall distribution is similar to WT. This differs from our previous finding that acute down-regulation of MDGA1 in migrating upper layer neurons results in their aberrant migration with few of them reaching the upper layers (Takeuchi and O'Leary, 2006). Our findings in the cKO are consistent with a recent study of MDGA1 null mice that finds only a transient slowing in the migration of a small subset of upper layer neurons that ultimately reach their appropriate laminar destinations (Ishikawa et al., 2011).
None-the-less, inconsistencies between the effects of acute RNA knockdowns and gene knockouts on cortical neuronal migration, with knockdowns leading to substantial defects not replicated by knockouts, have been reported for components of the DCX signaling pathway (Bai et al., 2003). The complete deficiency of the protein at earlier stages may induce compensatory mechanisms that reduce the severity of the phenotype. In our cKO, the significant reduction in upper layer neurons remains evident after their migration is complete, with no evidence of ectopic upper layer neurons indicative of a migration defect, confirming that it is due to the corresponding reduction in the population of BPs and their proliferation in the SVZ rather than a migration defect.
In the present study, we describe an MDGA1-dependent mechanism that serves to aggregate newly generated BPs and form the SVZ, a prominent proliferative zone responsible for producing cortical layer neurons. From an evolutionary perspective, it would be interesting to determine if genes such as MDGA1 and Cux2 might be related with the expansion of the SVZ and subsequently increase in upper layers size in the mammalian cortex. MDGA1 also functions in synapse formation, repressing the formation of inhibitory synapses through selective binding to Neuroligin2 (Lee et al., 2013, Pettem et al., 2013). Based on its expression patterns in brain and spinal cord (Litwack et al., 2004, Takeuchi et al., 2007), it is likely MDGA1 will be found to have other critical functions in neural development and plasticity, and will likely underlie the neurological disorders that are tightly linked to defined mutations in human MDGA genes, including Autism Spectrum Disorders, Schizophrenia and Bipolar Disorders (Kahler et al., 2008, Bucan et al., 2009, Li et al., 2011).
EXPERIMENTAL PROCEDURES
Animals
Analyses were done blind to genotype. For in vivo analyses, we used C57/B6 mice, including MDGA1fl/fl mice created for this study, which were used to generate a conditional knockout of MDGA1 by crossing to Nestin-Cre mice (MDGA1fl/fl;NestinCre/Cre, or MDGA1fl/fl;Nestin+/Cre)(Graus-Porta et al., 2001) or Nex-Cre mice (MDGA1fl/fl;NexCre/Cre, or MDGA1fl/fl;Nex+/Cre)(Goebbels et al., 2006). The wild type mice used in this study were MDGA1fl/fl and MDGA1fl/+ mice, both negative for Cre.
Antibodies
The following primary antibodies were used: mouse anti-Satb2 (1:250, Abcam), rabbit anti-Doublecortin (1:500, Cell Signaling), rabbit-anti GFAP (1:500, Abcam), rabbit anti-Tbr2 (1:250. Abcam), mouse anti-NeuN (1:500, Millipore), rat anti-Nestin (1:500, BD Pharmingen), rabbit anti-PH3 (1:250, Millipore), rat anti-BrdU (1:500, Abcam), rabbit anti-Cux1 (1:300, Santa Cruz), goat anti-MDGA1 (1:750, Santa Cruz), rabbit anti-MDGA1 (1:750, Millipore), rabbit anti-Connexin43 (1:400, kindly provided by T. Hunter) and mouse anti-Myc antibody (1:500, kindly provided by T. Hunter).
Immunohistochemistry
For immunostaining, 10-20 μm thick sections (cryostat and paraffin) were developed for DAB (Di-Amino-Benzidine) colorimetric reactions or for immunofluorescence following standard protocols (Chou et al., 2009).
In situ hybridization
An antisense RNA probe for MDGA1 was labeled using a DIG-RNA labeling kit (Roche). Standard methods for in situ hybridization on 20 μm cryostat sections and 10 μm paraffin sections were used (Chou et al., 2009).
Additional Experimental Procedures can be found in the Supplemental Information.
Supplementary Material
ACKNOWLEDGMENTS
We thank T. Hurtado, J. Simon, B. Higgins and H. Gutierrez for technical assistance. Animal work was done according to animal use protocols approved by the IACUC of the Salk Institute. This work was funded by NIH grants R01 NS31558, R01 MH086147, P30 NS072031 and the Vincent J. Coates Chair in Molecular Neurobiology to D.D.M.O’L.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
AUTHOR’S CONTRIBUTIONS
C.G.P-G. and D.D.M.O’L. designed the study, targeting vectors for generating the MDGA1 floxed allele, analyzed findings, prepared figures and wrote the paper. C.G.P-G. performed the primary experiments.
REFERENCES
- ALCAMO EA, CHIRIVELLA L, DAUTZENBERG M, DOBREVA G, FARINAS I, GROSSCHEDL R, MCCONNELL SK. Satb2 regulates callosal projection neuron identity in the developing cerebral cortex. Neuron. 2008;57:364–77. doi: 10.1016/j.neuron.2007.12.012. [DOI] [PubMed] [Google Scholar]
- ANGEVINE JB, JR., SIDMAN RL. Autoradiographic study of cell migration during histogenesis of cerebral cortex in the mouse. Nature. 1961;192:766–8. doi: 10.1038/192766b0. [DOI] [PubMed] [Google Scholar]
- ARNOLD SJ, HUANG GJ, CHEUNG AF, ERA T, NISHIKAWA S, BIKOFF EK, MOLNAR Z, ROBERTSON EJ, GROSZER M. The T-box transcription factor Eomes/Tbr2 regulates neurogenesis in the cortical subventricular zone. Genes Dev. 2008;22:2479–84. doi: 10.1101/gad.475408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BAI J, RAMOS RL, ACKMAN JB, THOMAS AM, LEE RV, LOTURCO JJ. RNAi reveals doublecortin is required for radial migration in rat neocortex. Nat Neurosci. 2003;6:1277–83. doi: 10.1038/nn1153. [DOI] [PubMed] [Google Scholar]
- BAYER SA, ALTMAN J. Neocortical Development. Raven Press; New York: 1991. [Google Scholar]
- BOYLE ME, BERGLUND EO, MURAI KK, WEBER L, PELES E, RANSCHT B. Contactin orchestrates assembly of the septate-like junctions at the paranode in myelinated peripheral nerve. Neuron. 2001;30:385–97. doi: 10.1016/s0896-6273(01)00296-3. [DOI] [PubMed] [Google Scholar]
- BRIGHTMAN MW, REESE TS. Junctions between intimately apposed cell membranes in the vertebrate brain. Journal of Cell Biology. 1969;40:648–677. doi: 10.1083/jcb.40.3.648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BRUMMENDORF T, RATHJEN FG. Structure/function relationships of axon-associated adhesion receptors of the immunoglobulin superfamily. Curr Opin Neurobiol. 1996;6:584–93. doi: 10.1016/s0959-4388(96)80089-4. [DOI] [PubMed] [Google Scholar]
- BUCAN M, ABRAHAMS BS, WANG K, GLESSNER JT, HERMAN EI, SONNENBLICK LI, ALVAREZ RETUERTO AI, IMIELINSKI M, HADLEY D, BRADFIELD JP, KIM C, GIDAYA NB, LINDQUIST I, HUTMAN T, SIGMAN M, KUSTANOVICH V, LAJONCHERE CM, SINGLETON A, KIM J, WASSINK TH, MCMAHON WM, OWLEY T, SWEENEY JA, COON H, NURNBERGER JI, LI M, CANTOR RM, MINSHEW NJ, SUTCLIFFE JS, COOK EH, DAWSON G, BUXBAUM JD, GRANT SF, SCHELLENBERG GD, GESCHWIND DH, HAKONARSON H. Genome-wide analyses of exonic copy number variants in a family-based study point to novel autism susceptibility genes. PLoS Genet. 2009;5:e1000536. doi: 10.1371/journal.pgen.1000536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CHOU SJ, PEREZ-GARCIA CG, KROLL TT, O'LEARY DD. Lhx2 specifies regional fate in Emx1 lineage of telencephalic progenitors generating cerebral cortex. Nat Neurosci. 2009;12:1381–9. doi: 10.1038/nn.2427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CINA C, MAASS K, THEIS M, WILLECKE K, BECHBERGER JF, NAUS CC. Involvement of the cytoplasmic C-terminal domain of connexin43 in neuronal migration. J Neurosci. 2009;29:2009–21. doi: 10.1523/JNEUROSCI.5025-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- COSTES SV, DAELEMANS D, CHO EH, DOBBIN Z, PAVLAKIS G, LOCKETT S. Automatic and quantitative measurement of protein-protein colocalization in live cells. Biophys J. 2004;86:3993–4003. doi: 10.1529/biophysj.103.038422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DIAZ-LOPEZ A, INIESTA P, MORAN A, ORTEGA P, FERNANDEZ-MARCELO T, SANCHEZ-PERNAUTE A, TORRES AJ, BENITO M, DE JUAN C. Expression of Human MDGA1 Increases Cell Motility and Cell-Cell Adhesion and Reduces Adhesion to Extracellular Matrix Proteins in MDCK Cells. Cancer Microenviron. 2010;4:23–32. doi: 10.1007/s12307-010-0055-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DIAZ-LOPEZ A, RIVAS C, INIESTA P, MORAN A, GARCIA-ARANDA C, MEGIAS D, SANCHEZ-PERNAUTE A, TORRES A, DIAZ-RUBIO E, BENITO M, DE JUAN C. Characterization of MDGA1, a novel human glycosylphosphatidylinositol-anchored protein localized in lipid rafts. Exp Cell Res. 2005;307:91–9. doi: 10.1016/j.yexcr.2005.02.016. [DOI] [PubMed] [Google Scholar]
- ELIAS LA, WANG DD, KRIEGSTEIN AR. Gap junction adhesion is necessary for radial migration in the neocortex. Nature. 2007;448:901–7. doi: 10.1038/nature06063. [DOI] [PubMed] [Google Scholar]
- ENGLUND C, FINK A, LAU C, PHAM D, DAZA RA, BULFONE A, KOWALCZYK T, HEVNER RF. Pax6, Tbr2, and Tbr1 are expressed sequentially by radial glia, intermediate progenitor cells, and postmitotic neurons in developing neocortex. J Neurosci. 2005;25:247–51. doi: 10.1523/JNEUROSCI.2899-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FUJIMURA Y, IWASHITA M, MATSUZAKI F, YAMAMOTO T. MDGA1, an IgSF molecule containing a MAM domain, heterophilically associates with axon- and muscle-associated binding partners through distinct structural domains. Brain Res. 2006;1101:12–9. doi: 10.1016/j.brainres.2006.05.030. [DOI] [PubMed] [Google Scholar]
- GOEBBELS S, BORMUTH I, BODE U, HERMANSON O, SCHWAB MH, NAVE KA. Genetic targeting of principal neurons in neocortex and hippocampus of NEX-Cre mice. Genesis. 2006;44:611–21. doi: 10.1002/dvg.20256. [DOI] [PubMed] [Google Scholar]
- GRAUS-PORTA D, BLAESS S, SENFTEN M, LITTLEWOOD-EVANS A, DAMSKY C, HUANG Z, ORBAN P, KLEIN R, SCHITTNY JC, MULLER U. Beta1-class integrins regulate the development of laminae and folia in the cerebral and cerebellar cortex. Neuron. 2001;31:367–79. doi: 10.1016/s0896-6273(01)00374-9. [DOI] [PubMed] [Google Scholar]
- IHRIE RA, ALVAREZ-BUYLLA A. Lake-front property: a unique germinal niche by the lateral ventricles of the adult brain. Neuron. 2011;70:674–86. doi: 10.1016/j.neuron.2011.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- INSOLERA R, BAZZI H, SHAO W, ANDERSON KV, SHI SH. Cortical neurogenesis in the absence of centrioles. Nat Neurosci. 2014;17:1528–35. doi: 10.1038/nn.3831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ISHIKAWA T, GOTOH N, MURAYAMA C, ABE T, IWASHITA M, MATSUZAKI F, SUZUKI T, YAMAMOTO T. IgSF molecule MDGA1 is involved in radial migration and positioning of a subset of cortical upper-layer neurons. Dev Dyn. 2011;240:96–107. doi: 10.1002/dvdy.22496. [DOI] [PubMed] [Google Scholar]
- JAVAHERIAN A, KRIEGSTEIN A. A stem cell niche for intermediate progenitor cells of the embryonic cortex. Cereb Cortex. 2009;19(Suppl 1):i70–7. doi: 10.1093/cercor/bhp029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KAHLER AK, DJUROVIC S, KULLE B, JONSSON EG, AGARTZ I, HALL H, OPJORDSMOEN S, JAKOBSEN KD, HANSEN T, MELLE I, WERGE T, STEEN VM, ANDREASSEN OA. Association analysis of schizophrenia on 18 genes involved in neuronal migration: MDGA1 as a new susceptibility gene. Am J Med Genet B Neuropsychiatr Genet. 2008;147B:1089–100. doi: 10.1002/ajmg.b.30726. [DOI] [PubMed] [Google Scholar]
- KOWALCZYK T, PONTIOUS A, ENGLUND C, DAZA RA, BEDOGNI F, HODGE R, ATTARDO A, BELL C, HUTTNER WB, HEVNER RF. Intermediate neuronal progenitors (basal progenitors) produce pyramidal-projection neurons for all layers of cerebral cortex. Cereb Cortex. 2009;19:2439–50. doi: 10.1093/cercor/bhn260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LEE K, KIM Y, LEE SJ, QIANG Y, LEE D, LEE HW, KIM H, JE HS, SUDHOF TC, KO J. MDGAs interact selectively with neuroligin-2 but not other neuroligins to regulate inhibitory synapse development. Proc Natl Acad Sci U S A. 2013;110:336–41. doi: 10.1073/pnas.1219987110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LI J, LIU J, FENG G, LI T, ZHAO Q, LI Y, HU Z, ZHENG L, ZENG Z, HE L, WANG T, SHI Y. The MDGA1 gene confers risk to schizophrenia and bipolar disorder. Schizophr Res. 2011;125:194–200. doi: 10.1016/j.schres.2010.11.002. [DOI] [PubMed] [Google Scholar]
- LITWACK ED, BABEY R, BUSER R, GESEMANN M, O'LEARY DD. Identification and characterization of two novel brain-derived immunoglobulin superfamily members with a unique structural organization. Mol Cell Neurosci. 2004;25:263–74. doi: 10.1016/j.mcn.2003.10.016. [DOI] [PubMed] [Google Scholar]
- LIU X, LIU YY, JIN WL, LIU HL, JU G. Nogo-66 receptor at cerebellar cortical glia gap junctions in the rat. Neurosignals. 2005;14:96–101. doi: 10.1159/000086291. [DOI] [PubMed] [Google Scholar]
- MANDER EEM, VERBEEK FJ, ATEN JA. Measurement of co-localization of objects in dual-color confocal images. Journal of Microscopy. 1993;169:375–382. doi: 10.1111/j.1365-2818.1993.tb03313.x. [DOI] [PubMed] [Google Scholar]
- MIYATA T, KAWAGUCHI A, SAITO K, KAWANO M, MUTO T, OGAWA M. Asymmetric production of surface-dividing and non-surface-dividing cortical progenitor cells. Development. 2004;131:3133–45. doi: 10.1242/dev.01173. [DOI] [PubMed] [Google Scholar]
- MOLYNEAUX BJ, ARLOTTA P, MENEZES JR, MACKLIS JD. Neuronal subtype specification in the cerebral cortex. Nat Rev Neurosci. 2007;8:427–37. doi: 10.1038/nrn2151. [DOI] [PubMed] [Google Scholar]
- NIETO M, MONUKI ES, TANG H, IMITOLA J, HAUBST N, KHOURY SJ, CUNNINGHAM J, GOTZ M, WALSH CA. Expression of Cux-1 and Cux-2 in the subventricular zone and upper layers II-IV of the cerebral cortex. J Comp Neurol. 2004;479:168–80. doi: 10.1002/cne.20322. [DOI] [PubMed] [Google Scholar]
- NOCTOR SC, MARTINEZ-CERDENO V, IVIC L, KRIEGSTEIN AR. Cortical neurons arise in symmetric and asymmetric division zones and migrate through specific phases. Nat Neurosci. 2004;7:136–44. doi: 10.1038/nn1172. [DOI] [PubMed] [Google Scholar]
- PETTEM KL, YOKOMAKU D, TAKAHASHI H, GE Y, CRAIG AM. Interaction between autism-linked MDGAs and neuroligins suppresses inhibitory synapse development. J Cell Biol. 2013;200:321–36. doi: 10.1083/jcb.201206028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SESSA A, MAO CA, HADJANTONAKIS AK, KLEIN WH, BROCCOLI V. Tbr2 directs conversion of radial glia into basal precursors and guides neuronal amplification by indirect neurogenesis in the developing neocortex. Neuron. 2008;60:56–69. doi: 10.1016/j.neuron.2008.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- STUBBS D, DEPROTO J, NIE K, ENGLUND C, MAHMUD I, HEVNER R, MOLNAR Z. Neurovascular congruence during cerebral cortical development. Cereb Cortex. 2009;19(Suppl 1):i32–41. doi: 10.1093/cercor/bhp040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TAKEUCHI A, HAMASAKI T, LITWACK ED, O'LEARY DD. Novel IgCAM, MDGA1, expressed in unique cortical area- and layer-specific patterns and transiently by distinct forebrain populations of Cajal-Retzius neurons. Cereb Cortex. 2007;17:1531–41. doi: 10.1093/cercor/bhl064. [DOI] [PubMed] [Google Scholar]
- TAKEUCHI A, O'LEARY DD. Radial migration of superficial layer cortical neurons controlled by novel Ig cell adhesion molecule MDGA1. J Neurosci. 2006;26:4460–4. doi: 10.1523/JNEUROSCI.4935-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TARABYKIN V, STOYKOVA A, USMAN N, GRUSS P. Cortical upper layer neurons derive from the subventricular zone as indicated by Svet1 gene expression. Development. 2001;128:1983–93. doi: 10.1242/dev.128.11.1983. [DOI] [PubMed] [Google Scholar]
- TSUI D, VESSEY JP, TOMITA H, KAPLAN DR, MILLER FD. FoxP2 Regulates Neurogenesis during Embryonic Cortical Development. J Neurosci. 2013;33:244–58. doi: 10.1523/JNEUROSCI.1665-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VASISTHA NA, GARCIA-MORENO F, ARORA S, CHEUNG AF, ARNOLD SJ, ROBERTSON EJ, MOLNAR Z. Cortical and Clonal Contribution of Tbr2 Expressing Progenitors in the Developing Mouse Brain. Cereb Cortex. 2015;25:3290–302. doi: 10.1093/cercor/bhu125. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.