Abstract
During development transcription factor combinatorial codes define a large variety of morphologically and physiologically distinct neurons. Such a combinatorial code has been proposed for the differentiation of projection neurons of the somatic and visceral components of cranial nerves. It is possible that individual neuronal cell types are not specified by unique transcription factors, but rather emerge through the intersection of their expression domains. Brn3a, Brn3b and Brn3c, in combination with each other and/or transcription factors of other families, can define subgroups of Retinal Ganglion Cells (RGC), Spiral and Vestibular Ganglia, inner ear and vestibular hair cell neurons in the vestibuloacoustic system, and groups of somatosensory neurons in the Dorsal Root Ganglia (DRG). In the present study we investigated the expression and potential role of the Brn3b transcription factor in cranial nerves and associated nuclei of the brainstem. We report the dynamic expression of Brn3b in the somatosensory component of cranial nerves II, V, VII and VIII and visceromotor nuclei of nerves VII, IX, X, as well as other brainstem nuclei during different stages of development into adult stage. We find that genetically identified Brn3bKO RGC axons show correct but delayed pathfinding during the early stages of embryonic development. However loss of Brn3b does not affect the anatomy of the other cranial nerves normally expressing this transcription factor.
Keywords: Cranial Nerves, Transcription, Optic Nerve, Trigeminal Nerve, Facial Nerve, Glossopharyngeal Nerve, Vagus Nerve, Pou domain Brn3b
Introduction
In vertebrates, twelve pairs of cranial nerves provide general sensory, motor and visceral functions to the head, neck and inner organs of the thoracic and abdominal cavity, as well as some of the special senses such as olfaction, taste, vision, vestibular and auditory perception. The cell bodies of neurons building the cranial nerves are distributed in many nuclei spread from the midbrain to the rostral spinal cord, as well as in a multitude of ganglia and specialized structures such as the olfactory epithelium, retina and vestibuloacoustic complex. Their projections to or from the central nervous system are collected at the level of the cranial nerves that can be purely sensory (I, II), motor (III, IV, VI, XI, XII) or mixed (V, VII, VIII, IX, X) (Watson, et al., 2010). Amongst these, nerves II, V, VII, VIII, IX and X are of particular interest for this study, based on Brn3b expression in some or all of their components.
The optic nerve
The axon bundles of more than 20 retinal ganglion cell types (RGCs) leave the eye at the optic disc and carry just as many distinct channels of information through the optic nerve to the brain. After crossing the midline at the optic chiasm, RGC axons are included in either the main or the accessory optic tracts, targeting different retinorecipient nuclei of the brain based on the types of visual information they relay. At the chiasm, Brn3b negative, intrinsically photosensitive RGCs innervate the suprachiasmatic nucleus. Image forming information is relayed through the lateral geniculate nucleus of the thalamus to the primary visual cortex. Information for many other visual functions and reflexes necessary for light evoked behaviors, pupil constriction, coordination of head and eye motion, is projected to the pretectal area and the superior colliculus (SC) of the midbrain. Visual information required for vestibulo-ocular coordination and image stabilization is conveyed to several nuclei through a dedicated tract, constituting the Accessory Optic System (Pak, et al., 1987).
In mice the pioneering RGC axons reach the optic chiasm at E12.5 (Marcus, et al., 1995), and their most distally positioned target, the SC, at E15 (Edwards, et al., 1986). We previously analyzed the progression of Brn3bAP RGC axons through the neuraxis in late embryonic and postnatal stages of development (Badea et al., 2009a; McNeill et al., 2011), and found axon guidance defects at the level of the optic chiasm and optic tract in P9 Brn3bAP/KO mice (Badea et al., 2009a). Interestingly the observed defects, consisting of abnormal invasion of non-visual thalamic targets, seem to be transient since they were not observed in the adult Brn3bKO/KO animal. The reason for which 70% of RGCs are lost in adult Brn3bKO/KO mice (Gan et al., 1996; Erkman et al., 2000; Shi et al., 2013) is still poorly understood, but one of the potential mechanisms could be axon growth and path finding errors present as early as P0 (Erkman et al., 2000).
Trigeminal nerve
The trigeminal nerve (V) exits the brainstem on the ventrolateral aspect of the pons, corresponding developmentally to rhombomere 2 (r2), to join the trigeminal ganglion (TGG). The TGG contains the cell bodies of somatosensory neurons of this nerve and is derived during development from the dorsolateral placode and neural crest (Hamburger, 1961). The peripheral projections of TGG neurons are organized in three major branches: mandibular, maxillary and ophthalmic, which innervate the jaws, face, lips, eyes, and the specialized whisker system in rodents. The axons of TGG neurons enter the brainstem at pontine level and project rostrally to the principal trigeminal nucleus, situated in the pons, or caudally to the spinal trigeminal nucleus, which stretches in the adult rostrocaudally from the pons to the rostral spinal cord (rhombomeres r2 – r8) (Oury et al., 2006). The efferent, branchiomotor (BM) component of the nerve originates in the motor nucleus of the V (MoV) located in the pons, (rhombomeres r2 – r3) and innervates masticatory muscles (Gilland and Baker, 1993; Lumsden, 1990; Lumsden and Keynes, 1989).
A large collection of molecular markers are used to differentiate neuronal subtypes in dorsal root ganglia (DRG), which serve similar functions as the TGG for territories outside the head. Markers include neurofilament-200 (mechanoreceptors and proprioreceptors), parvalbumin (proprioceptors), calcitonin gene-related peptide (peptidergic nociceptors), neurotrophic receptors TrkA (nociceptors and mechanoreceptors), TrkB (mechanoreceptors), TrkC (mechanoreceptors and proprioceptors), cRet (Ret) (nociceptors and mechanoreceptors), transcription factors, Brn3a (mechanoreceptors, nociceptors, proprioceptors), Brn3b (mechanoreceptors) and Brn3c (peptidergic nociceptors) and Isolectin B4 (nonpeptidergic nociceptors) (Zylka et al., 2005, Komori et al., 2008, Luo et al., 2009, Badea et al., 2012).
Many of these markers have been successfully applied to the study of neuronal cell types in the TGG. A significant difference is the TrkC positive muscle spindle proprioceptor of the DRG (Mu et al., 1993; Klein et al., 1994). In the case of the TGG this class of proprioceptors is located in the mesencephalic trigeminal nucleus (MesV) (Alvarado-Mallart et al., 1975) and not in the ganglion. In the TGG TrkC labels mechanoreceptors (Funfschilling et al., 2004).
Facial nerve
The facial nerve (VII) exits the neuraxis at the junction between pons and medulla, as two distinguishable roots, the facial nerve proper and the nervus intermedius, in close apposition to the vestibulocochlear nerve (VIII). The motor component, facial nerve proper, controls the muscles for facial expression, upper neck, inner ear (Ashwell, 1982; Hinrichsen and Watson, 1984; Semba and Egger, 1986) and vibrissal movement in rodents (Henstrom et al., 2012; Semba and Egger, 1986). It originates in branchiomotor neurons (FBM), generated developmentally in r4 close to the midline (Lumsden, 1990; Lumsden and Keynes, 1989). During development FBM neurons migrate caudally through r5, followed by a ventrolateral migration that places them in their final adult positions in r6 (Auclair et al., 1996; Studer et al., 1996). As a result of this migration, the axons have a characteristic looping morphology or genu. The intermediate nerve carries a parasympathetic component, originating in visceromotor neurons (VM) of the superior salivatory nucleus (SSN), located in r5 (Auclair et al., 1996; Jacob and Guthrie, 2000). VM axons travel trough the intermediate nerve, passing through the geniculate ganglion (GG) and along the greater superficial petrosal (GSP) and chorda tympani (CT) branches to synapse onto parasympathetic neurons of the sphenopalatine, submandibular and sublingual ganglia, which innervate lacrimal and salivary glands respectively.
Developmentally the GG has dual origins, the distal ganglia composed of viscerosensory neurons is derived from the epibranchial placode, while the somatosensory neuron containing proximal part is derived from the neural crest (Ayer-Le Lievre and Le Douarin, 1982, D’Amico-Martel et al., 1983, Quina et al., 2012)
The sensory fibers of nerve VII originating from the GG project centrally through the nervus intermedius to the rostral part of the nucleus of the solitary tract at medullar levels (Spector and Glendinning, 2009). Their peripheral projections travel through the GSP, and CT to innervate the ear lobe, palate and taste buds of the anterior two thirds of the tongue.
Relatively few markers are known that separate the distinct general somatosensory and special viscerosensory cells of the GG. During embryonic development TrkB is expressed in a majority of GG neurons while TrkA, TrkC and cRet are present in fewer cells (Yamout et al., 2004). Viscerosensory neurons projecting in the CT and GSP and supplying taste innervation are known to express Phox2b but not Brn3a or Hmx1, while the general somatosensory neurons contributing to the auricular nerve are Brn3a and Hmx1 positive (Quina et al., 2012).
Vestibulocochlear nerve
The vestibulocochlear nerve (VIII) transmits information to and from the auditory and vestibular organs to the brain. The afferent branch of the nerve collects signals from inner ear and vestibular hair cells through projection sensory neurons with cell bodies located in the spiral and vestibular ganglia. The efferent component of the nerve is made up by a group of motor-like (Fritzsch, 1999; Tiveron et al., 2003) hindbrain neurons located in the vestibular efferent nucleus (VEN) and cochlear efferent nucleus (CEN) (White and Warr, 1983). These inner ear efferents (IEE) provide cholinergic input to receptors of the vestibular and cochlear apparatus and are believed to play a role in auditory pathway maturation (Glowatzki and Fuchs, 2000; Tritsch et al., 2007) and afferent response modulation of the vestibular (Highstein, 1991) and cochlear signals. VEN and CEN cells are born in r4 and bear the distinction of sending processes contralaterally across the floor plate (Simon and Lumsden, 1993) early in development, making them a topographic indicator of this rhombomere. Embryologically they share their origins with facial branchiomotor neurons (Fritzsch, 1996; Fritzsch, 1999).
Glossopharyngeal nerve
The glossopharyngeal nerve (IX) contains branchiomotor (BM), visceromotor (VM), viscerosensor (VS) general and special sensory fibers. BM fibers originate in the nucleus ambiguus (NA) located in the medulla and innervate the stylopharyngeus muscle that controls pharynx dilatation. VM fibers, from the inferior salivatory nucleus innervate the parasympathetic otic ganglion whose postganglionic fibers supply the parotid salivary glands (Chibuzo and Cummings, 1980; Satomi et al., 1979). Developmentally both the BM and VM cells originate within r6 (Simon and Lumsden, 1993). Sensory fibers relay taste information from the posterior part of the tongue (foliate papillae), somatosensory information from the back of the tongue, soft palate, tonsils, middle ear, pharynx, eustachian tubes, and viscerosensory information from carotid body and carotid sinus. General sensory and special sensory components of the nerve have their cell bodies in the petrosal ganglia (PG) derived from the second epibranchial placode (Ayer-Le Lievre and Le Douarin, 1982) and the more rostrally positioned neural crest derived superior ganglion (Narayanan and Narayanan 1980)
Vagus nerve
The vagus nerve (X) contains somatic and visceral afferents, as well as visceromotor and branchiomotor efferents. It supplies innervation to the pharyngeal – laryngeal muscles and all other organs of the abdomen and thorax. The cell bodies of the visceral sensory afferent are located in the nodose ganglion derived from the third epibranchial placode (Ayer-Le Lievre and Le Douarin, 1982) while the afferent somatosensory neurons are located in the neural crest derived jugular ganglion (Narayanan and Narayanan 1980).
They relay information from the gastrointestinal tract and other visceral structures to the caudal nucleus of the solitary tract, located in the medula (Altschuler et al., 1989). Visceral parasympathetic neurons of the dorsal motor nucleus of the vagus (dmnX) (located in the medulla) supply efferent fibers to the abdominal postganglionic neurons that synapse on virtually all internal organs. Branchiomotor vagal neurons are located in the NA of the medulla. These special visceral neurons innervate the striated (branchial) muscles of the pharynx, larynx and esophagus (Ross et al., 1985; Stuesse and Fish, 1984).
Combinatorial codes of transcription factors involved in cranial nerve development
Virtually all cranial nerve projection sensory neurons are placed outside the CNS, in ganglia or sensory epithelia of diverse origin. Efferent, somato- or viscero- motor neuron cell bodies are placed in the CNS, and are typically specified during the differentiation of the neural tube along the three cardinal body axes. Interestingly, a partially overlapping set of transcription factors including Lim homeobox, Pou4f and Phox2 family members is turned on in cranial nerve neurons, regardless of whether their cell bodies are in the brainstem nuclei or in the placode-derived ganglia. Visceral neurons express Phox2b, somatic sensory neurons Brn3a and somato-motor neurons Mnx1 and Lhx3/4 (Nomaksteinsky et. al. 2013).
Isl1 is expressed in the retina, cranial ganglia of nerves V, VII, IX, X (Liang et al., 2011), spinal cord motor neurons and DRGs (Ericson et al., 1992; Thor et al., 1991). In the central nervous system Isl1 can be detected in the ventral hindbrain and spinal motor neurons (MNs) (Ericson et al., 1992; Thor et al., 1991), dmnX, thalamic reticular nucleus, striatum, hypothalamus and all other brainstem motor neurons (Thor et al., 1991; Varela-Echavarria et al., 1996). Isl1 is required for proper motor neuron development in both the spinal cord and cranial nerves IX and XII (Liang et al., 2011). In addition, Isl1 is also necessary for TGG, DRG and retinal cell differentiation and survival (Elshatory et al., 2007; Pfaff et al., 1996) individually or in combination with Brn3a (Dykes et al., 2011) or Brn3b (Mu et al., 2008; Pan et al., 2008; Shi et al., 2013). Although Isl1 is expressed by viscerosensor (VS) and visceromotor (VM) neurons they can be distinguished by Phox2a and Phox2b expression.
Phox2b expression was described in all parasympathetic ganglia including ciliary, sphenopalatine, otic, submandibular and sublingual, and the sensory ganglia of the facial (GG), glossopharyngeal (PG), and vagal (NG) cranial nerves (Pattyn et al., 1997; Tiveron et al., 1996). Motor neurons expressing Phox2b include the BM nuclei of the V and VII nerves, NA, spinal accessory (XI) nucleus (Dubreuil et al., 2009; Pattyn et al., 1997), and the special motor – like neurons of the IEE and dmnX. In addition Phox2b is also expressed in the peritrigeminal neurons surrounding the MoV (Rose et al., 2009) and in a transient manner in the oculomotor and trochlear motoneurons (Pattyn et al., 1997)
Phox2b and to a lesser degree Phox2a play an essential role in the formation of the VII, IX and X placode-derived ganglia (Pattyn et al., 1999). Phox2b-mutant mice, fail to develop BM and VM neurons (Pattyn et al., 2000) while Phox2a knockouts fail to generate oculomotor and trochlear nuclei (Pattyn et al., 1997).
Brn3a, Brn3b and Brn3c constitute the POU4 domain subfamily of transcription factors (TF). Brn3s are expressed in retinal ganglion cells, inner ear hair cells, somatosensory, spiral and vestibular ganglia neurons (Badea and Nathans, 2011; Badea et al., 2012; Xiang et al., 1995), and required for the proper differentiation of several of these sensory neurons. Brn3a mutants die at birth. Affected structures include loss of cells in the TGG, as well as follicle-associated sensory nerve endings. Retinal phenotype includes an increased ratio of bistratified cellular morphologies (Badea et al., 2009a). Deletion of Brn3b leads to 70% loss of RGCs (Erkman et al., 2000; Xiang et al., 1998), with the remaining cells exhibiting a variety of axon guidance defects (Badea et al., 2009a; Erkman et al., 2000). Brn3c mutant animals lose their vestibular and auditory hair cells (Dykes et al., 2011; Xiang et al., 1997, 1995, 1998). Based on present data, the combinatorial expression pattern of Brn3 transcription factors plays an essential role in neuronal subtype specification for up to 15 types of RGCs and 11 types of DRG projecting neurons (Badea and Nathans, 2011; Badea et al., 2012). However the expression profile of Brn3s in the developing spinal cord hindbrain and midbrain widely varies between Brn3a and Brn3b (Turner et al 1994).
Whereas Brn3a expression in cranial nerves has been widely studied, the precise expression pattern of Brn3b in the brainstem and cranial ganglia is poorly characterized, while Brn3c expression is limited to only a few neuron groups described above. In the present study we analyzed the detailed expression profile of Brn3b during development in the cranial nerve projecting neurons and other midbrain and hindbrain regions.
Materials and methods
Mouse lines
The following previously described mouse lines were used: (1) Conditional knock-in allele: Brn3bCKOAP (Badea et al., 2009a); (2) Drug inducible ubiquitous Knock- in Cre lines: ROSA26rtTA-CreERt (R26rtTA-CreERt, (Badea et al., 2009a)), ROSA26CreERt (R26CreERt, (Badea et al., 2003)), cRetCreERt knock-in line (Enomoto et al., 2001, Chi et al., 2009); (3) BAC transgenic Phox2b:Cre line (B6(Cg)-Tg(Phox2b-Cre)3Jke/J Stock Number: 016223) (Scott et al., 2011), Pax6αCre (Marquardt et al., 2001). (4) Knock-in allele cRetCFP/+ (Uesaka et al., 2008). To obtain sparse recombination, Brn3bCKOAP/CKOAP females were crossed to R26rtTA-CreERt/rtTA-CreERt; Brn3bKO/WT males.
Embryonic day 0.5 (E0.5) was set on the day a copulation plug was observed. Upon Cre mediated recombination, the endogenous Brn3b coding exons are deleted from the Brn3bCKOAP allele, and replaced with the Alkaline Phosphatase reporter (AP), which will now be under the control of the endogenous Brn3b promoter. The R26rtTA-CreERt line expresses in a constitutive ubiquitous manner an inactive tetracycline reverse transcriptional activator (rtTA) which can be induced by doxycyclin (Dox) and in turn activates transcription of a drug inducible form of Cre recombinase (CreER). CreER can then be activated by 4-hydroxytamoxiphen (4HT) that controls nuclear import and therefore activity of the Cre. To maintain a low level of Dox concentration in the pregnant females until the desired experimental time, we delivered a 0.2 mg Dox/g chow diet starting at E1.5 on two consecutive days, alternating with two days of regular diet, for 10 – 13 days. The CreER was then induced by intraperitoneal injection of 12.5–50 μg 4HT in sunflower seed oil 1–2 days prior to embryo collection. After 4HT administration the feeding cycle was halted. For full ubiquitous recombination Brn3bCKOAP/CKOAP females were crossed to R26CreERt/CreERt males and injected with 250 μg 4HT, 2 days before experiment. Phox2b:Cre × Brn3bCKOAP/CKOAP matings were used to restrict induction of the reporter to cell populations that had previously or simultaneously expressed Phox2b. In the case of cRetCreERt × Brn3bCKOAP sparse Cre induction was obtained by 2mg 4HT IP injection at 4 weeks of age. All adult animals were anesthetized with ketamine-xylazine and sacrificed by cervical dislocation. All mice used were of mixed C57Bl6/SV129 background. All mouse handling procedures used in this study were approved by the National Eye Institute Animal Care and Use Committee (ACUC) under protocols NEI 640, NEI 651 and NEI 652.
AP Histochemistry and Indirect Immunofluorescence staining
Collected embryos where fixed overnight in PBS containing 4% paraformaldehyde (PFA) at 4°C. Adult animals for AP staining were perfusion fixed by cardiac perfusion with 4% paraformaldehyde (PFA); if needed dissected brains were further immersion fixed in 4%PFA for an additional 2–4h.
Brains were sectioned on a vibratome at 150 – 250 μm thickness. After two washes in PBS, the tissues were heated in a water bath for 1–2 h at 65°C to inactivate the endogenous Alkaline Phosphatase (AP) activity. AP staining was performed in AP buffer (0.1mM Tris, 0.1M NaCl, 50mM MgCl2, pH 9.5), with 0.34 μg/ml nitroblue tetrazolium (NBT), and 0.175 μg/ml 5-bromo-4-chloro-3-indolyl-phosphate (BCIP) (Roche), for 1–12 h at room temperature with gentle agitation. After staining was complete, tissues were washed three times for 20 min in PBS, 0.1% Tween 20, and postfixed in PBS with 4% PFA overnight. Before imaging, samples were dehydrated through an ethanol series (50%–75%–85%–95%–100%) and then cleared with 2:1 benzyl benzoate (BB)/benzyl alcohol (BA). Immunostaining: embryos were immersion-fixed in PBS containing 2% paraformaldehyde, for either 2h (E12.5), or 4h (E15.5) 4°C. After fixation embryos were washed two times 20 minutes in PBS, then placed in 30% sucrose solution for overnight. Embryonic heads were placed in Optimal Cutting Temperature Compound (OCT Tissue-Tek-Sakura), and sectioned at 14 μm thickness on a cryostat. R26rtTA-CreERt/WT; Brn3bCKOAP/WT and R26rtTA-CreERt/WT; Brn3bCKOAP/WT embryos were embedded together in a single block. P14 animals for immunohistochemistry were fixed by cardiac perfusion with 2% PFA. Following fixation brains were removed, washed two times 20 minutes in PBS then cryoprotected overnight in 30% sucrose solution.
Images were collected on a Zeiss Discovery V8, and edited in ImageJ and Adobe Photoshop. Primary antibodies (see also table 1) :mouse monoclonal anti-Brn3a (Millipore, MAB1585, RRID:AB_94166), rabbit polyclonal anti-Brn3b (Xiang et al., 1993, 1995), rabbit polyclonal anti-Phox2b C-terminal 14-mer (Pattyn et al., 1997, RRID:AB_2313690). Commercial antibodies: goat anti-Isl1 (Neuromics #GT15051, RRID:AB_2126323), chicken anti-GFP (Abcam, #ab13970, RRID:AB_300798), goat anti-Parvalbumin (Swant, PVG-214, RRID:AB_10000345), chicken anti-NFH (Aves lab, #NF-H, RRID:AB_2313552), guinea pig anti-CGRP (Peninsula Laboratories T-5027, RRID:AB_2313552), goat anti-TrkA (R&D systems, AF1056, RRID:AB_2283049), goat anti-TrkB (R&D systems, AF1494, RRID:AB_2155264), goat anti-TrkC (R&D systems, AF1404, RRID:AB_2155412), goat anti-Chat (Chemicon, AB144P, RRID:AB 11214092)
Table 1.
Primary antibodies and dilutions used for immunohistochemistry.
| Antigen | Immunogen | Manufacturer, Species, type Catalogue number | Dilution |
|---|---|---|---|
| Brn3a | Human Brn3a aa186-224 – protein 10 fusion (pGEMEX) | Millipore, Mouse monoclonal, MAB1585 (RRID:AB_94166), clone 5A3.2 | 1:20 |
| Brn3b | Human Brn3b aa184-252 – protein 10 fusion (pGEMEX) | Rabbit polyclonal, previously described, see Xiang et al., 1995 | 1:20 |
| Phox2b | Mouse Phox2b aa301-314 | Rabbit polyclonal, described in Pattyn et al., 1997 (RRID:AB_2313690) | 1:500 |
| Isl1 | Recombinant human Islet-1 | Neuromics, Goat polyclonal, GT15051 (RRID:AB_2126323) | 1:500 |
| GFP | Recombinant GFP | Abcam, Chicken polyclonal, ab13970 (RRID:AB_300798) | 1:500 |
| TrkA | Mouse myeloma cell line NS0-derived recombinant rat TrkA. Ala33-Pro418 | R&D systems, Goat polyclonal, AF1056 (RRID:AB_2283049) | 1:40 |
| TrkB | Mouse myeloma cell line NS0-derived recombinant mouse TrkB. Cys32-His429 | R&D systems, Goat polyclonal, AF1494 (RRID:AB_2155264) | 1:40 |
| TrkC | Mouse myeloma cell line NS0-derived recombinant mouse TrkC. Cys32-Thr429 | R&D systems, Goat polyclonal, AF1404 (RRID:AB_2155412) | 1:40 |
| Parvalbumin | Rat muscle Parvalbumin | Swant, Goat polyclonal, PVG-213 (RRID:AB_10000345) | 1:400 |
| CGRP | Recombinant human CGRP | Peninsula Laboratories, Guinea pig polyclonal, T-5027 (RRID:AB_2314158) | 1:300 |
| NFH | Bovine NFH | Aves lab, Chicken polyclonal, #NF-H (RRID:AB_2313552) | 1:400 |
| Chat | Human placental choline acetyltransferase | Chemicon, Goat polyclonal, AB 144P (RRID:AB_11214092) | 1:100 |
Secondary antibodies: donkey antisera coupled to Alexa 488, 568, 555, and 647 dyes (Invitrogen/Life Technologies), IB4 (Invitrogen/Life Technologies).
In the case of combined immunostaining and AP histochemistry, to prevent masking of the fluorescent signal, heat inactivation and AP staining was performed only after completion of florescent imaging.
Antibody characterization
Brn3a
Mouse monoclonal anti-Brn3a (Millipore, MAB1585, RRID:AB_94166, clone 5A3.2) antibody was raised against amino acids 186–224 of human Brn3a fused to T7 gene 10 protein (Xiang et al., 1995). Based on manufacturer’s technical information the antibody shows no reactivity to Brn3a knockout mice (Xiang et al., 1996). On Western blot the antibody does not detect Brn3b or Brn3c. In rodent and monkey the antibody labels the nuclei of Brn3a positive retinal ganglion cells, and dorsal root ganglia neurons (Xiang et al., 1995, Badea et al., 2012)
Brn3b
A DNA sequence encoding Brn3b protein aa184–252 was cloned in the pGEMEX expression system. The resulting fusion protein was purified and used to immunize rabbits. Antibody specificity was shown by immunocytochemistry in 293S cells transfected with pCIS-Brn3b-cDNA expression vector, and by lack of staining in Brn3b knock-out tissues (Gan et al., 1996). On Western blot with cellular extract of pCIS-Brn3b-cDNA transfected 293S cells the antibody recognizes ~45kDa band corresponding to the molecular mass of Brn3b (Xiang et al., 1993, 1995).
Phox2b
Anti-Phox2b antibody (Pattyn et al., 1997 RRID:AB_2313690) was raised against the carboxy-terminal aa301–314 of mouse Phox2b. Specificity was shown by matching localizations of in situ hybridization and immunohistochemistry signal by Pattyn et al., 1997.
Isl1
Anti-Isl1 goat polyclonal antibody (Neuromics, GT15051, RRID:AB_2126323) was raised against recombinant human Islet-1. Specificity was tested by western blot by the manufacturer and in immunohistochemistry by colocalization with the mouse monoclonal anti-Isl1 DSHB, 39.4D5, RRID:AB_2314744 (Espana and Clotman 2012).
GFP
Anti-GFP antibody (Abcam, ab13970, RRID:AB_300798) was raised against full length GFP protein. Based on manufacturers technical information on Western blot the antibody recognizes 27–30 kDa band specific for GFP.
TrkA
Anti-TrkA antibody (R&D systems, AF1056, RRID:AB_2283049) was directed against mouse myeloma cell line NS0-derived recombinant rat TrkA Ala33-Pro418. The antibody shows approximately 5% cross reactivity with recombined human TrkA and less than 1% cross reactivity with recombined mouse TrkB and TrkC. On Western blot with rat striatum lysate the antibody recognizes a 140kDa band specific for TrkA. (manufacturers technical details). The labeling pattern in mouse TGG is in line with previous data, labeling nociceptive neurons (Matsumoto et al., 2012)
TrkB
Anti-TrkB antibody (R&D systems, AF1494, RRID:AB_2155264) produced against the extracellular domain (Cys32-His429) of recombined mouse TrkB. Based on manufacturers technical information the antibody detects mouse TrkB in direct ELISA and Western blot, and shows 25% cross reactivity with human TrkB, and less than 2% cross reactivity with recombined rat TrkA and TrkC. The antibody labels mechanoreceptors in the TGG as shown by Matsumoto et al., 2012.
TrkC
Anti-TrkC antibody (R&D systems, AF1404, RRID:AB_2155412) was raised in goat against the extracellular domain (Cys32-Thr429) of recombined mouse TrkC. The antibody has less than 2% cross reactivity with recominant TrkB, TrkA, as tested by ELISA and Western blotting with recombinant proteins. On Western blot with mouse DRG lysate it recognizes two bands, 100 kDa and 145 kDa predicted molecular weight of TrkC. (Kawamata et al., 2006, Komori et al., 2008). In DRG the antibody labels proprioceptive neurons (Komori et al., 2008, Hadjab et al., 2013).
Parvalbumin
Anti-Parvalbumin (Swant, PVG-214, RRID:AB_10000345) was raised in goat against rat muscle parvalbumin. On Western blots with mouse, rat, guinea pig, rabbit and macaque brain homogenate the antibody recognizes the 12-kDa band specific for parvalbumin (Schwaller et al. 1999, Glueckert et al. 2008).
CGRP
Anti-CGRP antibody raised in guinea pig (Peninsula Laboratories T-5027, RRID:AB_2313552) and was generated against the human α-CGRP with the following sequence ACDTATCVTHRLAGLLSRSGGVVKNNFVPTNVGSKAF* (Peleshok and Ribeiro-da-Silva 2011). Specificity and cross reactivity was tested by the manufacturer using radioimmunoassay against synthetic peptides. Antibody specificity and cross reactivity was analyzed by immunohistochemistry in Fundin et al., 1997; Peleshok and Ribeiro-da-Silva 2011.
NFH
Chicken IgY Anti-NFH antibody (Aves lab, #NF-H, RRID:AB_2313552) was raised against bovine NFH protein purified from spinal cords. Antibody is known to cross react with NFH protein from human, mouse and rat. Although Western blot data is not available, antibody specificity was shown by manufacturer and by Li and Ginty 2014
ChAT
Anti-Chat goat polyclonal antibody (Chemicon, AB144P, RRID:AB 11214092) was raised against human placental choline acetyltransferase. On Western blot with mouse brain lysate the antibody stains a single band of 68–70 kDa, apparent molecular weight for Chat (manufacturer’s technical data). Antibody specificity on tissue sections was shown by matching localizations of Chat:GFP signal with immunohistochemistry signal by Mesnage et al., 2011.
Cell counting
Trigeminal ganglia from four to five adult animals were embedded together in one OCT block. For each immunostaining cells were counted from 4–8 fields. Fields were chosen to cover peripheral and central regions of the TGG. In the case of the embryonic TGG, cells from 9 sections collected from 3 distinct embryos were analyzed. For the embryonic GG, cells from 6 sections collected from 3 distinct embryos were analyzed. In all experiments the number of counted cells were normalized to the total number of DAPI positive cells in the ganglion.
Results
Genetic strategies for revealing Brn3bAP cranial nerves
Given the previous findings demonstrating combinatorial expression of Brn3s in RGCs and DRGs, we decided to investigate Brn3b expression in cranial nerves. To more easily survey all cranial nerves, we focused on whole mount preparations and vibratome sections of E12.5 – E15.5 embryos, in which AP expression from the endogenous Brn3b locus (Brn3bAP) was induced using three complementary strategies (Figure 1).
Figure 1. Genetic-pharmacological and intersectional genetic strategies used to label Brn3b-positive cranial nerve neurons.
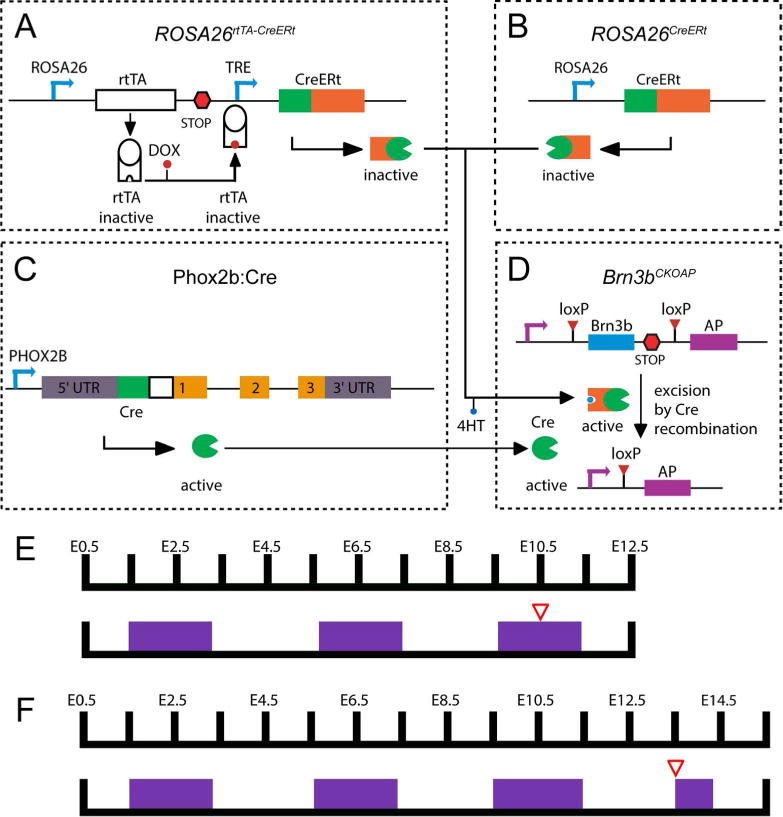
The Cre dependent Brn3b conditional knock-in AP reporter mouse line (A) was crossed to three alternative genetic drivers in order to obtain ubiquitous dense (B), neuronal subpopulation specific (C) or ubiquitous sparse (D) Cre activity. A, Conditional Knock-in reporter construct targeted at the Brn3b locus (Brn3bCKOAP). B, Near complete recombination was induced by 4HT induction of the CreERt activity from the R26CreERt knock-in allele. C, BAC transgenic line expressing constitutively active Cre in Phox2b positive neurons. D, Sparse random recombination was achieved by dual pharmacological control of a R26rtTA-CreERt knock-in construct. E, F Examples of dual pharmacological control regime used for Cre activity control in R26rtTA-CreERt; Brn3bCKOAP crosses (D × A). Plugged females were given regular feed alternating with 0.2 mg/g Dox chow (purple boxes) every two days, beginning with the day the plug was found (E0.5). 12.5 – 50 μg 4HT injected intraperitoneally two days prior to embryo harvesting (red triangle), at either E12.5 (E) or E15.5 (F).
Full Brn3b expression was induced in R26CreERt; Brn3bCKOAP embryos in which conditional recombination and Alkaline Phosphatase reporter (Figure 1A) induction was achieved by 4HT administration two days prior to embryo harvesting (Figure 1B). Cranial nerve neurons co-expressing Phox2b and Brn3b were visualized in Phox2b:Cre; Brn3bCKOAP embryos (Figure 1C). Sparse recombination was induced by modulating Doxycycline (DOX) and 4HT administration during early gestation of R26rtTA-CreERt; Brn3bCKOAP embryos (Figure 1D, E, F), in which Cre activity is under dual pharmacological control. Below we provide a survey of the expression patterns observed in these three genetic backgrounds, and then proceed to analyze in depth Brn3b distribution in each of the identified cranial nerves.
Brn3bAP is expressed in early stages of nerves II, V, VII, VIII, IX and X development
Brn3bAP expression in fully recombined embryos labels the entire neuraxis including spinal cord, brainstem, tectum and reaches into the dorsal thalamus by E12.5 (Figure 2 A1, A2). As early as E12.5 – E13, Brn3bAP is visible in the following cranial nerves: RGCs and the optic nerve (II); trigeminal (V) system including all three branches (ophthalmic – V1, maxillary – V2, mandibular – V3); facial nerve (VII) branches including the GSP, CT, main branch of the facial nerve, the vestibulocochlear (VIII) nerve; the glossopharyngeal (IX) nerve, its tympanic and pharyngeal branches, and the vagus nerve (X), (Figure 2 A1, A2). In Phox2b:Cre; Brn3bCKOAP E12.5 embryos, AP positivity is restricted to the nerves VII, IX and X, in keeps with the expected expression pattern of Phox2b, which excludes the TGG, tectum and visual system (Figure 2B1, B2). No staining was observed for cranial nerves I, III, IV, VI, XI and XII at any of the ages analyzed (E12.5 to E15.5).
Figure 2. Brn3bAP labeling in cranial nerves II, V, VII, VIII, IX and X during development.
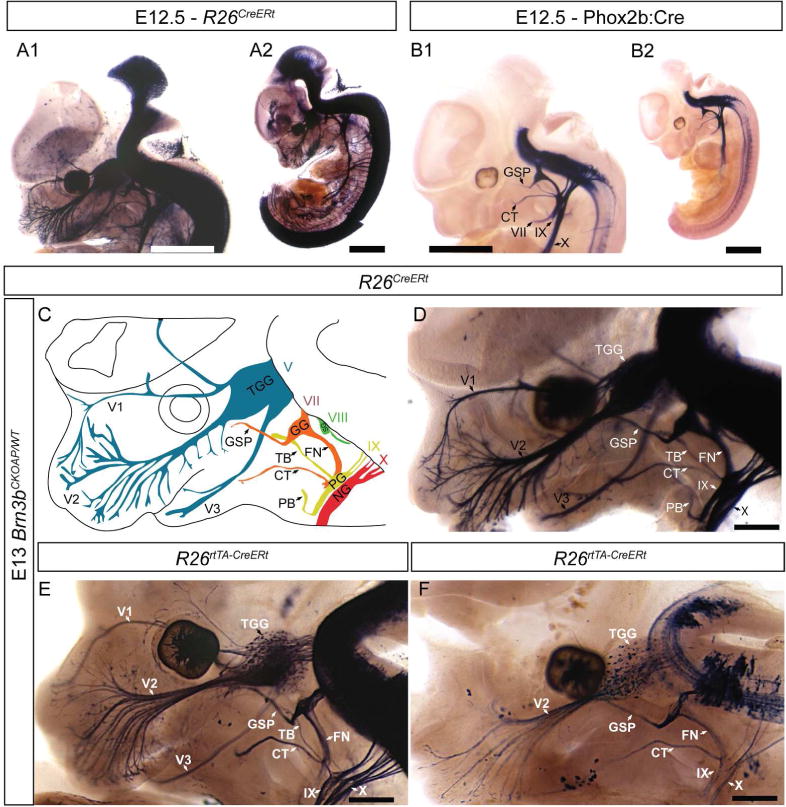
Full expression pattern of Brn3b determined by staining R26CreERt; Brn3bCKOAP embryos at E12.5 (A1, A2) and E13 (D). AP can be detected in the retina, spinal cord, dorsal root ganglia (DRG), brainstem, mesencephalon, trigeminal ganglion (TGG), trigeminal nerve (V), facial (VII), glossopharyngeal (IX) and vagal nerves (X) (schematized in C). B1, B2 Phox2b:Cre; Brn3bCKOAP E12.5 embryo shows AP positive signal in the cranial nerves VII, IX, X. C, Schematic of AP expressing nerves: 3 branches of Trigeminal nerve (V): ophthalmic – V1, maxillary V2 and mandibular V3. Facial nerve (VII) with Greater Superficial Petrosal (GSP), Chorda Tympani (CT), and main branch (FN). Vestibulocochlear nerve (VIII) Glossopharyngeal nerve (IX) with tympanic (TB) and pharyngeal (PB) branches. Vagus nerve (X). E,F R26rtTA-CreERt; Brn3bCKOAP embryos from pregnant females treated with 0.2mg/g doxycycline (DOX) and 50μg (E) or 17.5μg (F) of 4 hydroxytamoxifen (4HT). Different doses of 4HT – labeling off all or few nerve fibers in the developing cranial nerves (E – F). Scale bars: A1, A2, B1, B2=1mm, D – F=500μm.
In the spinal cord a set of Phox2b+, Brn3b+ interneurons can be seen, most likely due to read through transcription, or leaky expression of the Phox2b:Cre BAC transgenic line (Figure 2B1, B2). To determine the anatomic position of cranial nerve projecting neurons (Figure 2C, D) we used graded levels of sparse recombination (Figure 2E, F), this allows the visualization of individual Brn3bAP cell bodies and their projecting axons (Figure 2E, F).
Early stages of Brn3bAP RGC development
We examined individual Brn3bAP/KO and Brn3bAP/WT RGC axons during early stages of axon guidance within the retina, and through the optic nerve and tract, soon after the first RGCs are born (E12.5 – E15.5, Figure 3). At E12.5, cells with columnar shape, extending ventricular (apical) and vitreal (basal) end feet projections are visible, together with cells that have lost the ventricular attachment, and are sending axons into the early optic nerve (Figure 3A, B). In some instances, two cell bodies in close apposition are seen in one column, or as closely apposed columns (Figure 3B). The axonal process extension into the optic nerve fiber layer can coincide with retracting RGC ventricular processes (Figure 3B, panel 4). At this age, isolated Brn3bAP/WT RGC fibers have reached the chiasm, and can be seen climbing dorsally on the walls of the thalamus on the ipsilateral side (Figure 3C).
Figure 3. Brn3b expression in developing retinal ganglion cells.
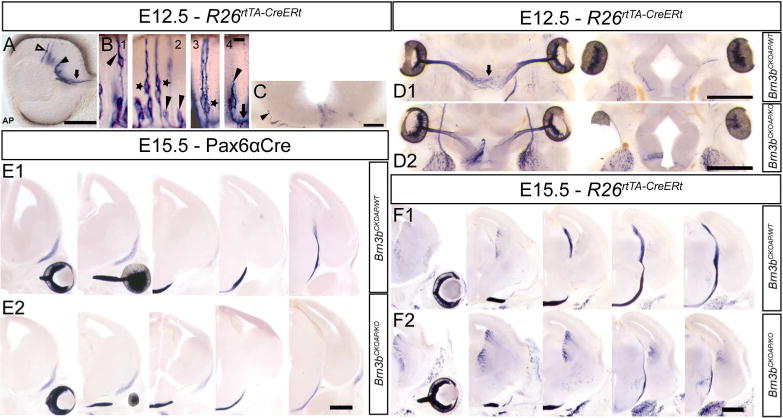
A, Optic cup section with sparsely labeled Brn3bAP RGCs at E12.5. RGCs at different developmental time points: columnar stage (hollow arrow), cells in the RGC layer with retracted ventricular end feet (black arrow head), cells projecting into the optic nerve (black arrow). B, Brn3bAP RGCs in intermediate stages of migration towards the GCL and ventricular (arrow head) process retraction (1). Note the presence of pairs of Brn3bAP RGCs (star) whose ventricular processes are cofasciculated (2) or closely apposed (3). End foot retraction (black arrowhead) and axon extension (black arrow) (4). C, Coronal section at the level of the optic chiasm at E12.5, ipsilateral Brn3bAP RGC axon (black arrow head) climbing on exterior wall of the thalamus. D1, D2, Serial horizontal sections at the level of the developing optic chiasm in R26rtTA-CreERt; Brn3bCKOAP/WT (D1) and R26rtTA-CreERt; Brn3bCKOAP/KO (D2) E12.5 littermates. E1, E2, Serial coronal sections at the level of the developing optic tract in Pax6α:Cre; Brn3bCKOAP/WT; (E1) and Pax6α:Cre; Brn3bCKOAP/KO (E2) E15.5 littermates. F1, F2, Serial coronal sections at the level of the developing optic tract in R26rtTA-CreERt; Brn3bCKOAP/WT (E1) and R26rtTA-CreERt; Brn3bCKOAP/KO; (E2) E15.5 littermates. Scale bars: A,C=100μm, B=10μm, D1,D2,E1–F2=500μm
Given the previously described RGC axon guidance defects at later developmental stages (Erkman et al., 2000) we have analyzed RGC axons at E12.5 (Figure 3D1,D2) and E15.5 (Figure 3E1,E2,F1,F2) in Brn3bAP/KO and Brn3bAP/WT animals. We do not find misguided axons in Brn3bAP/KO E12.5 (Figure 3D2 ) and E15.5 (Figure 3E2, F2) RGCs when compared with Brn3bAP/WT controls (Figure 3D1, E1, F1), however axon growth delays are observed as early as E12.5 at the level of the optic chiasm (Figure 3D2).
This axon growth delay was also evident at E15.5, when Brn3bAP/KO RGC just started climbing on the thalamic wall while Brn3bAP/WT littermate RGC axons have reached the level of the SC (Figure 3F1, F2). Similar axon growth delays were also observed, in Pax6α:Cre; Brn3bCKOAP/KO mice (Figure 3E1, E2). Thus, in the absence of Brn3b, Brn3bAP/KO RGC axons are able to correctly navigate the various developmental stages, intraretinally and through the optic nerve, chiasm and tract, however the apparent growth rate seems to be reduced.
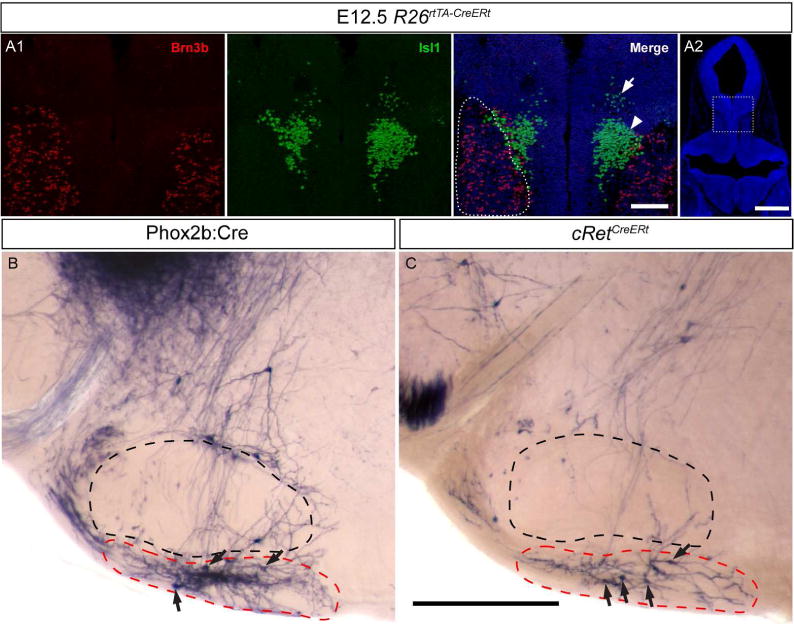
Brn3b is expressed in multiple stations of the trigeminal system
As previously illustrated in Figure 2A1, D all three peripheric branches of the trigeminal ganglion, ophthalmic, maxilar and mandibular, are positive for Brn3bAP, with clearly identifiable cell bodies, likely somatosensory neurons, in the TGG. To determine if the motor neurons projecting in the trigeminal nerve express Brn3b, we have analyzed Brn3bCKOAP;Phox2b:Cre E12.5 embryos. Since neither the branches nor the exit root of the trigeminal nerve appear AP positive in Brn3bCKOAP;Phox2b:Cre E12.5 whole mount preparations, Brn3b is unlikely to be expressed in the Phox2b+ motor component of the V (Figure 2B1, B2). By combined immunological and histochemical staining, we find that the Phox2b, Isl1 double positive MoV neurons are AP negative indicating lack of Brn3b expression. However the Isl1 negative peritrigeminal neurons (PT) expressing Phox2b (Rose et al., 2009) are AP and therefore Brn3b positive (Figure 4 A1, A2, B1, B2).
Figure 4. Brn3b expression in peritrigeminal neurons.
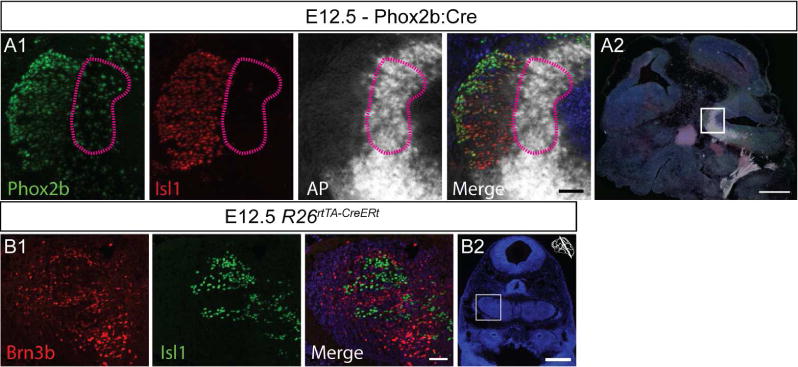
A1 A2, AP negative Phox2b and Isl1 positive MoV neurons at E12.5. AP indicating Phox2b and Brn3b coexpression is present in peritrigeminal neurons (purple lines).
B1 B2 Coronal section at the level of the MoV showing mutual exclusive expression of Brn3b positive neurons and Isl1 positive motoneurons. Scale bars: A1,B2=50μm, A2,B2=500μm
Peripheral endings of the trigeminal fibers can be seen as early as E13.5, distinguishing specialized whisker endings (deep vibrissal) from intervibrissal (Figure 5A, B1) as well as regular somatosensory fiber endings (Figure 5 B2). This indicates that Brn3b is expressed in at least some of the mechanoreceptors of the whisker system. Since previous work had documented expression of Brn3b in DRG mechanoreceptors, (Badea et al., 2012), we have analyzed the cell type distribution of Brn3b within the TGG, the principal somatosensory ganglion of the head.
Figure 5. Brn3b expression in embryonic trigeminal neurons.
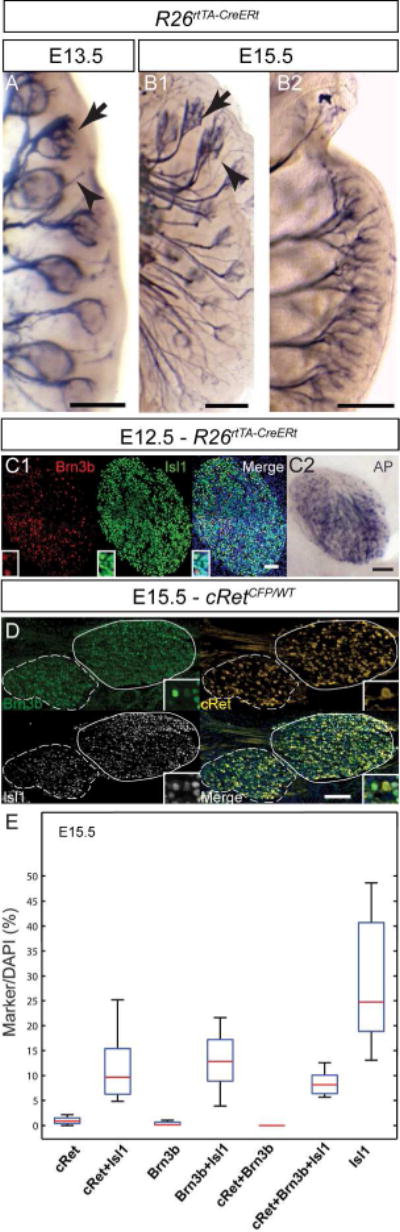
A, Brn3bAP positive deep vibrissal (arrow) and intervibrissal (arrow head) fibers in the mystacial pad are visible as early as E13.5. B1, Brn3bAP positive deep vibrissal (arrow) and intervibrissal (arrow head) as well as general sensory fibers (B2) innervating the lips of E15.5 embryo. C1, Extensive overlap of Brn3b and Isl1 in E12.5 embryos. C2, Brn3bAP positive TGG cells in ROSA26rtTACreERt; Brn3bCKOAP/WT E12.5 mice. D, Sagittal section of a E15.5 cRetCFP/WT maxillary (lines) and mandibular territories (doted lines) of TGG. Insets show combinatorial expression of Brn3b, Isl1, and cRet. E, Box whisker plot for cell populations of E15.5 TGG (D). Scale bars: A,B1,B2,C2,D=100μm, C1=50μm
Brn3b+ neurons can be detected in the TGG from E12.5, largely overlapping with Isl1 (Figure 5C1, C2). By E15.5, more than half of all TGG cells are Isl1 positive, either isolated or in combination with cRet or Brn3b. Essentially all cRet and/or Brn3b cells are also Isl1 positive, and Brn3b+ Isl1+, cRet+ Isl1+ and Brn3b+ cRet+ Isl1+ positive cells are represented in almost equal amounts, allowing the subdivision of the Brn3b+ TGG neuronal cell population in at least three distinct categories (Figure 5D, E). We next performed double immunofluorescence labeling with Brn3b in conjunction with TrkA, TrkB, TrkC, CGRP, IB4, NFH and Parv, a set of markers allowing the classification of major somatosensory populations in the adult DRG (Figures 6A, C). Like in the DRG, Brn3b did not co-localize with either TrkA, CGRP, or IB4, suggesting that TGG nociceptors do not express Brn3b.
Figure 6. Cell type distribution of Brn3b in adult trigeminal neurons.
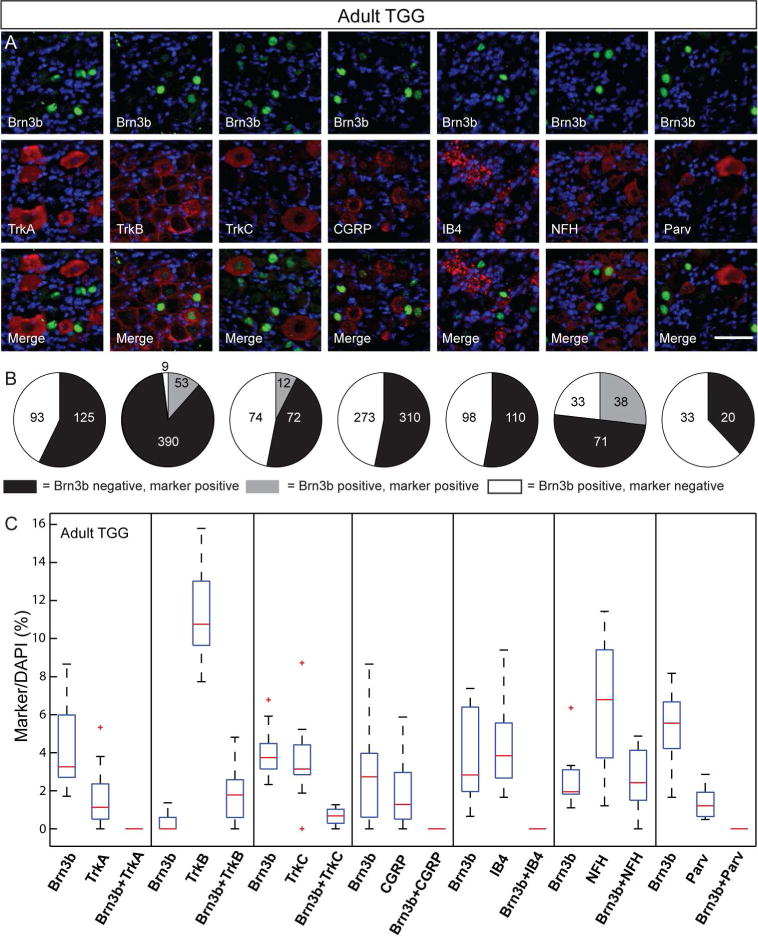
A, Double immunostaining for Brn3b (green) and indicated molecular markers (red) in the adult TGG. B Pie charts showing the number of TGG cells single and double positive for the various TGG markers and Brn3b stained in A. C, Box whisker plot for cell populations of the adult TGG neurons. Scale bars: A=50μm
NFH, TrkB and TrkC label sub-populations of Brn3b positive cells, indicating that TGG Brn3b+ neurons are at least in part mechanoreceptors. However Brn3b+ cells double labeled with either TrkB, TrkC or NFH (Figure 6A, B, C) do not account for all Brn3b+ TGG neurons, indicating additional cell types may be expressing the transcription factor.
Previous work (Funfschilling et al., 2004 and Hasegawa and Wang 2008) has further described combinatorial expression of TrkB, TrkC, CGRP and NFH in subpopulations of TGG mechanoreceptors of the mistacial pad follicle-sinus complexes (FSC). Specifically, CGRP is collocalizing with TrkC and/or TrkB in A beta lanceolate endings, while FSC A beta Merkel and/or spiny ending mechanoreceptors are NFH+ TrkB+/TrkC+ CGRP− (Funfschilling et al., 2004). Since Brn3b+ neurons are not positive for CGRP, we can infer that Brn3b is expressed in NFH+, CGRP−, TrkB+ and/or TrkC+ TGGs, which could represent FSC A beta Merkel and/or spiny ending mechanoreceptors. The lack of overlap with strong Parvalbumin staining would suggest that Brn3b+ mechanoreceptors do not include reticular or Ruffini endings of the vibrissal cavernous sinus (Rice et al., 1997).
Dynamic expression of Brn3b in the facial nerve
Several branches of the facial nerve are labeled by Brn3bAP (Figure 2A1, D). Brn3bAP labeling is transiently present in the branchiomotor branch of the facial nerve from E12.5 (Figure 2), and persists at least until E15.5 in GSP and CT (data not shown). Since the facial is a mixed nerve containing both branchio- and viscero-motor fibers with origins in the brainstem, and gustatory and sensory fibers originating in the geniculate ganglion, we used combined immunostaining and intersectional genetics approaches in order to identify the Brn3b positive facial nerve components.
AP staining of Phox2b:Cre; Brn3bCKOAP E11.5 embryos (Figure 7A), reveals labeling in two rostro-caudal Phox2b positive columns (Figure 7A) (Pattyn et al., 1997). Several efferent neuronal populations are defined by the intersection of Brn3b and Phox2b expression patterns: SSN, FBM and IEE neuron axons projecting in to cranial nerves VII and VIII are described here, while nuclei of the glossopharyngeal (IX) and vagal system, projecting through their respective nerves, will be addressed in subsequent sections. Individual cell bodies for several of these populations can be identified in equivalent R26rtTA-CreERt; Brn3bCKOAP preparations (Figure 7B).
Figure 7. Brn3b expression in the developing facial nerve (VII) system.
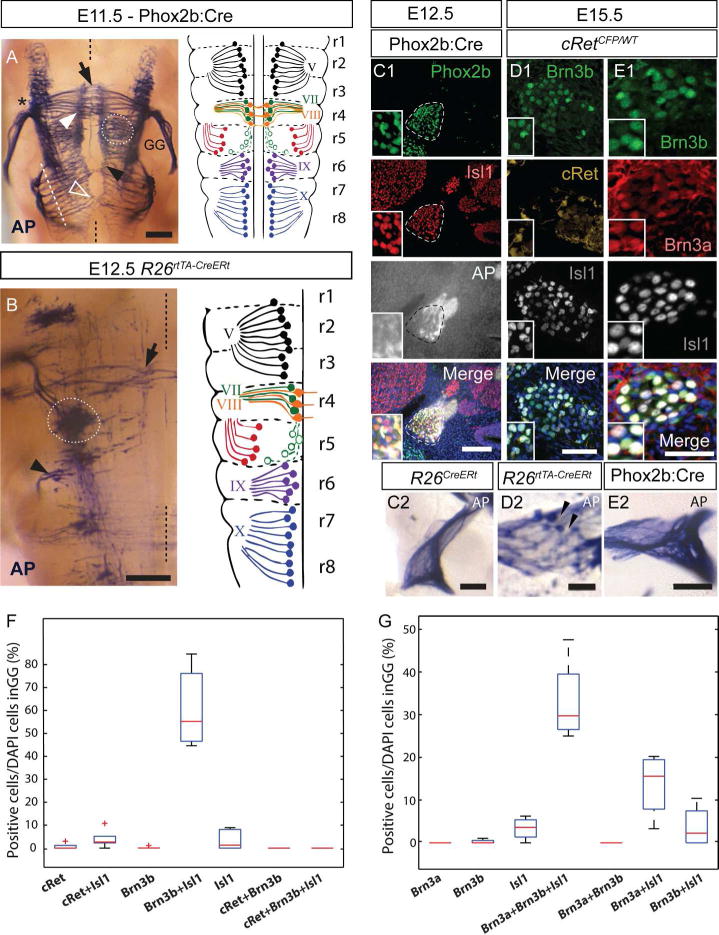
A, Left panel, Brn3bAP histochemistry of the E11.5 brainstem of Phox2b:Cre; Brn3bCKOAP embryo in whole mount dorsal perspective. AP Positive cell bodies include visceral motor neurons of the superior salivatory nucleus (SSN) (white stippled circle) and branchiomotor neurons (white arrow head). Inner ear efferent fibers (black arrow) are crossing the midline (black stippled line). Exit point of the nerves VII and VIII are indicated by black asterisk. Cell bodies of the glossopharyngeal (black arrow head) and vagal (white hollow arrow) nuclei exiting the neuraxis through multiple rootlets, arranged in a continuous fashion (white stippled line). Right panel, schematic of the rhombomeric distribution of neuronal cell bodies, axonal processes and exit points of cranial nerves V, VII, VIII, IX and X, as they map onto the rhombomeres (adapted from Lumsden and Krumlauf, 1996). B, Whole mount Brn3bAP histochemistry same perspective as A, in an E12.5 R26rtTA-CreER; Brn3bCKOAP embryo. Left side of the animal, with the midline indicated by a black stippled line. Isolated cell bodies or small groups of the nuclei indicated in A and their axonal projections are visible. Black arrow indicates inner ear efferent neuron axons extending contralaterally across the midline. Superior salivary nucleus (white stippled circle). Group of cells with axons projecting in the glosspharyngeal nerve (black arrow head). C1, Combined Brn3bAP histochemistry and IIF of the GG (stippled lines) in the Phox2b:Cre; Brn3bCKOAP E12.5 embryo, showing extensive Phox2b and Isl1 double positive neurons in the GG. C2 Coronal section at the level of the GG of the R26CreERt; Brn3bCKOAP E12.5 mouse, with no visible AP positive cell bodies. D1,E1 Immunostaining of the GG in cRetCFP/WT E15.5 embryos. D2, Coronal section at the level of the GG of the R26rtTA-CreERt; Brn3bCKOAP E15.5 embryo, showing Brn3bAP positive cells (black arrowheads). E2 AP positive fibers crossing the GG of Phox2b:Cre; Brn3bCKOAP E15.5 embryo with no visible AP positive cell bodies. F box whisker plots for cell populations in D1. G box whisker plots for cell populations in E1. Scale bars: A = 250μm, B = 200μm, C1,C2, D1, D2, E2 = 100μm, E1=50μm
IEE and FBM: at the level of rhombomere r4, a medially placed column can be seen, projecting ipsilateral fibers towards the exit root of the facial, as well as contralaterally across the midline (Figure 7A, black arrow), in a fashion characteristic for the IEE neurons (Fritzsch, 1996; Fritzsch and Nichols, 1993; Simon and Lumsden, 1993), whose differentiation is Phox2b dependent (Tiveron et al., 2003). Intermingled with the IEE neurons are the FBM neurons (Lumsden, 1990; Lumsden and Keynes, 1989), that transiently express Brn3b, and send their fibers into the main branch of the VII around E11.5 (Figure 7A, black asterisk). Immunohistochemistry confirms Brn3b and Isl1 co-expression in the r4 postmitotic precursor cell pool that gives rise to the IEE and FBM (Figure 8A1, A2), and lack of Brn3b staining in Isl1 positive migrating FBM cells (Studer 1996) at the level of r5–r6 (Figure 8B1, B2).
Figure 8. Brn3b expression in the developing facial nucleus.
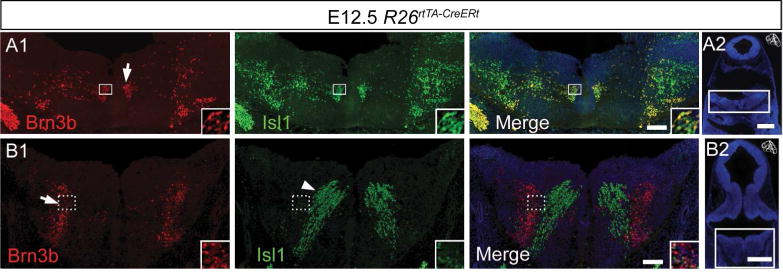
A1, A2 Coronal section at the level of genu of the facial nerve (arrow) containing Brn3b (red) and Isl1 (green) positive cells. B1, B2, Coronal section at the level of the migrating stream of facial branchiomotor (Isl1 positive, Brn3b negative, white arrowhead). Scale bars: A1, B1 = 100μm, A2 B2 = 500μm
Geniculate ganglion: The major afferent fibers of the facial nerve are provided by gustatory (Phox2b+) and somatosensory neurons (Brn3a+) present in the GG (Huang et al., 2001). During early stages (E11.5 and E12.5), no Brn3bAP cell bodies can be detected in the GGs of either Phox2b:Cre; Brn3bCKOAP (Figure 7A, C1), or fully recombined R26CreERt; Brn3bCKOAP (Figure 7C2) embryos. However the ganglion is wrapped in AP positive fibers likely originating from the brainstem. By E15.5 Brn3b is expressed in the neural crest-derived proximal region of the GG (Figure 7D1), exhibiting a partially overlapping pattern with Brn3a (Figure 7E1, G) however no overlap with cRet (Figure 7D1, F). Brn3b expression in the E15.5 GG was also confirmed by the presence of AP positive cell bodies in sparsely recombined R26rtTA-CreERt; Brn3bCKOAP embryos (Figure 7D2). The lack of Brn3bAP positive cells in the GG of Phox2b:Cre; Brn3bCKOAP E15.5 embryos, infers that Brn3b, like Brn3a, is not co-expressed with Phox2b in the GG (Figure 7E2, (Quina et al., 2012)).
Visceromotor fibers: Brn3bAP labeling can be detected in the CT and GSP of E12.5 Phox2b:Cre; Brn3bCKOAP (Figure 2B1) animals. At this stage of development, neurons sending fibers in these branches include the somatosensory and/or gustatory neurons of the GG and superior salivatory nucleus (SSN) neurons located in r5. As described above, no Brn3b+ or Brn3bAP positive neurons are present in the GG at E12.5. However, Brn3bAP cell bodies positioned laterally, away from the floor plate, in r5 can be observed in wholemount Phox2b:Cre; Brn3bCKOAP E11.5 and R26rtTA-CreERt; Brn3bCKOAP E12.5 embryos (Figure 7A, B stippled circle), and Brn3b protein can be detected by immunostaining at the same position (E12.5, Figure 8B1), in a column lateral to the intensely Isl1+ migratory stream. The anatomic location of the cell bodies together with the CT and GSP projections of their fibers suggest to us that Brn3bAP cells in r5 represent the lateral neuron population believed to give rise to the superior salivatory nucleus (SSN) (Figure 7A stippled circle) as described by McKay et al., 1997, Auclair et al., 1996, Coppola et al., 2010). It should be noted that some of these cells are also weakly Isl1+ (Figure 8B1 inset, Mikaels et al., 2000).
To summarize, Brn3b+ cRet− Phox2b− cells can be found at E15.5 but not E11.5–12.5 in the GG. These cells fall in at least two distinct categories based on presence or absence of Brn3a, and may represent somatosensory innervation to parts of the face. In addition Brn3b is expressed in IEE and potentially in the SSN nuclei, as well as the postmitotic facial branchiomotor neurons located in r4.
Afferent and Efferent innervation of the vestibuloacoustic system is Brn3b positive
Afferent fibers of the vestibulo-acoustic system originate in the spiral ganglia of the inner ear and vestibular ganglion, while efferent components originate in several nuclei in the brainstem. Brn3b expression has been reported in the adult and embryonic spiral and vestibular ganglia (Huang et al., 2001, Badea et al., 2012, Deng et al., 2014). In the sparsely labeled E15.5 R26rtTA-CreERt; Brn3bCKOAP embryo we can observe afferent projecting fibers originating from the spiral ganglia neurons (Figure 9A), and dendritic processes penetrating into the cochlear epithelium interdigitating with the developing inner ear hair cells. In contrast, in Phox2b:Cre; Brn3bCKOAP the IEE fibers originating from the cochlear efferent nucleus (see section on facial nerve) are selectively labeled, due to the lack of Phox2b expression in the spiral ganglia (Figure 9B). At E15.5 the IEEs seem to innervate specifically the spiral ganglion, and avoid the territory where future inner ear hair cells develop.
Figure 9. Brn3b expression in the afferent and efferent projecting cells of the developing cochlear nerve.
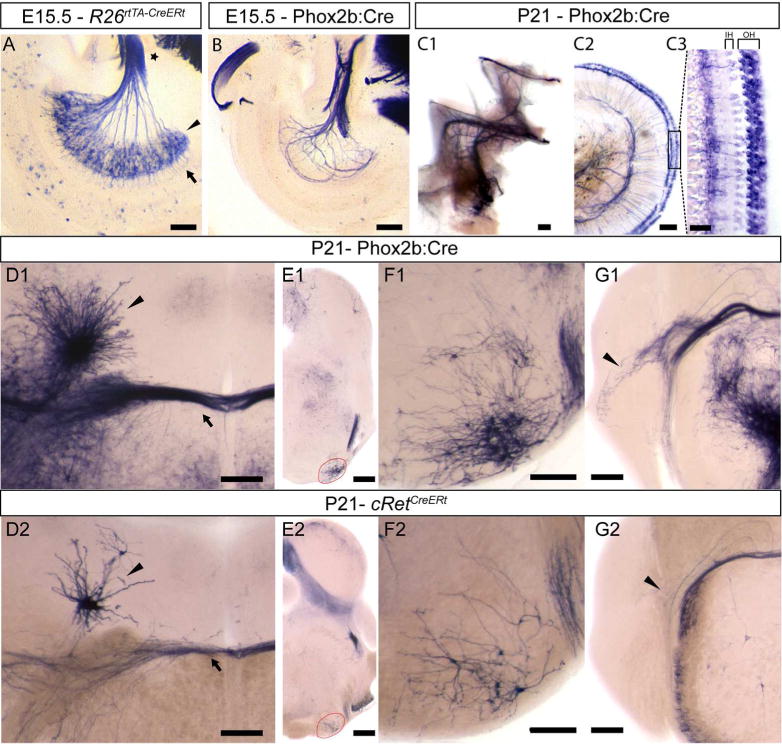
A, Spiral ganglion neuron cell bodies (black arrowhead), axons forming the cochlear nerve (star) and dendritic processes innervating the cochlea (arrow) in a R26rtTA-CreERt; Brn3bCKOAP E15.5 embryo. B, Efferent axon innervations of the cochlea, in a Phox2b:Cre; Brn3bCKOAP E15.5 embryo and adult (C1,C2). C3 Efferent innervation of the three rows of outer hair cells. D1–G2 coronal sections through the adult pons and brainstem. D1, D2 Dense and sparse labeling of Eve (nucleus of origin of efferents of the vestibular nerve) (arrow head) and OCB (Olivocochlear Bundle) (arrow) in adult Phox2b:Cre; Brn3bCKOAP (D1) and cRetCreERt; Brn3bCKOAP (D2) animals. E1, F1 Brn3b/Phox2b and E2, F2 Brn3b/cRet double positive neurons of the ventral nucleus of the trapezoid body (VNTB), at the level of the superior olivary complex (SOC) (red circle). G1, G2 Phox2b+ Brn3b+ positive (G1) and cRet+ Brn3b+ positive (G2) VNTB projections to the cochlear nucleus (black arrow head). Scale bars: A,B,C1,C2 = 100μm, C3=25μm, D1,D2,F1,F2,G1,G2,=200μm, E1,E2=500μm
In the adult cochlea of Phox2b:Cre; Brn3bCKOAP (Figure 9C1) only the outer hair cell innervating efferent fibers are AP positive (Figure 9 C2, C3) indicating that not all inner ear projecting brainstem nuclei are Brn3b, Phox2b double positive.
The vestibular and cochlear efferent projecting nuclei are positioned in distinct anatomical territories. Due to their developmental migration pattern, the vestibular efferent neurons are spread in areas adjacent to the facial nerve inflection point (genu): 1) nucleus of origin of vestibular efferents (Eve) placed dorso-laterally to the facial genu (White and War 1983, Schwarz et al., 1986, Paxinos et al.,2012); 2) a group of cells placed dorso-medially to the genu (Schwarz et al., 1986); 3) the ventro-medially placed caudal pontine reticular nucleus (Schwarz et al., 1986, Wang et al., 2013a). Brn3bAP cell bodies can only be seen in Eve cells in both adult Phox2b:Cre; Brn3bCKOAP (Figure 9D1) and cRetCreERt; Brn3bCKOAP (Figure 9D2) injected with 4HT in adult stage. Thus, adult Eve neurons express Phox2b, cRet and Brn3b (Kang 2007, (Figure 9 D1, D2)). Morphologically the cells bodies of the Eve are tightly grouped, with dendritic branches radiating isotropically, with the axonal projections joining the olivocochlear bundle (OCB) (Figure 9 D1, D2 arrow).
The brainstem efferent nuclei projecting to the cochlea (olivocochlear, acoustic efferents) are positioned in the lateral and medial superior olive (LSO and MSO) of the superior olivary complex (SOC) and the ventral nucleus of the trapezoid body (VNTB). Cells of the VNTB form the medial olivocochlear bundle (MOC) which projects to the cochlear nucleus (Faye-Lund, 1986; Warr and Beck 1996) and the outer hair cells of the cochlea (Thompson and Schofield 2000). Brn3bAP cell bodies with characteristic morphologies located in the MOC of the VNTB (Figure 9E1, F1, E2, F2), and Brn3bAP positive fibers projecting to the cochlear nucleus (Figure 9G1, G2) and outer hair cells are visible in both Phox2b:Cre; Brn3bCKOAP (Figure 9C1, C2, C3) and cRetCreERt; Brn3bCKOAP (Figure 9E2, F2, G2) animals (Brown and Levine 2008). Thus, the VNTB expresses Brn3b in conjunction with Phox2b (Kang et al., 2007) and/or cRet.
Brn3b expression of Eve and VNTB neurons was confirmed by immunofluorescence on P14 brain sections with anti-Brn3b and anti-Chat, a known marker for Eve (Figure 10A1 arrow, A2) and VNTB (Figure 10B1,B2) neurons (Leijon and Magnusson 2014). Note the Chat positive neurons of the abducens nucleus do not express Brn3b (Figure 10A1 arrow head).
Figure 10. Brn3b expression in the Eve and VNTB of P14 animals.
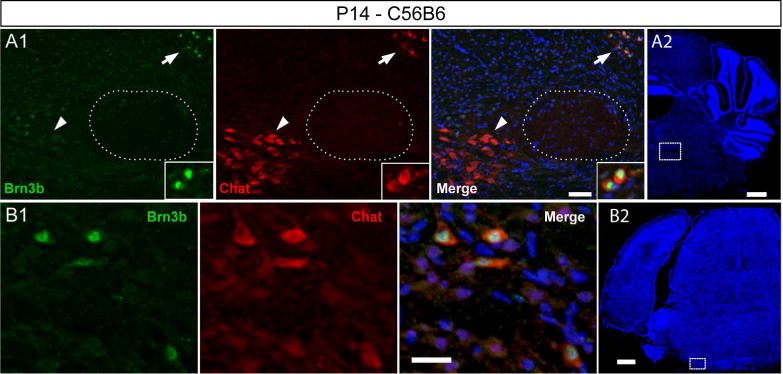
A1 A2 Double immunostaining at the level of the genu (stippled circle) in P14 animals. Two Chat positive nuclei visible, Brn3b positive Eve (white arrow), and Brn3b negative nucleus abducens (white arrow head). B1, B2 Brn3b Chat double positive cells in the VNTB. Scale bars: A1,B1=25μm, A2,B2=500μm
Brn3b is expressed in central nuclei but not peripheral ganglia of the IX and X nerves
Both nerves IX and X are Brn3bAP positive in R26CreER; Brn3bCKOAP and Phox2b:Cre; whole-mount preparations of E11.5 – E13.5 embryos (Figures 2, Figure 7A, B). Phox2b:Cre; Brn3bCKOAP embryos reveal a dorsal column of cell bodies in r6 – r8, in continuation with the FBM, and projecting their axons into the continuous exit roots of the glossopharyngeal and vagus nerve (Figure 11A). To determine if the AP signal originates from neurons located in the brainstem or the cranial ganglia of nerves IX/X we have analyzed fully recombined R26CreER; Brn3bCKOAP (Figure 11B1, B2) and sparsely recombined R26rtTA-CreERt; Brn3bCKOAP (Figure 11C) E12.5 embryos. No AP positive cell bodies were observed in the N/P complex of E12.5 R26CreER; Brn3bCKOAP animals (Figure 11B1), however intense AP signal was evident at the level of dmnX (Figure 11B2), and individual cell bodies can be detected in the sparsely recombined dmnX (Figure 11C). In addition, no AP positive cell bodies were observed among the Isl1 and Phox2b positive cells of the N/P complex of Phox2b:Cre; Brn3bCKOAP animals (Figure 11D) at E12.5. Similarly at E15.5 we observed AP positive cells in dmnX but not the N/P ganglia in Phox2b:Cre; Brn3bCKOAP (Figure 11E1, E2) and R26rtTA-CreERt; Brn3bCKOAP (Figure 11F1, F2) animals. Brn3b was not detected by immunostaining in E15.5 N/P ganglia (Figure 11G).
Figure 11. Brn3b expression in the developing glossopharyngeal and vagal systems.
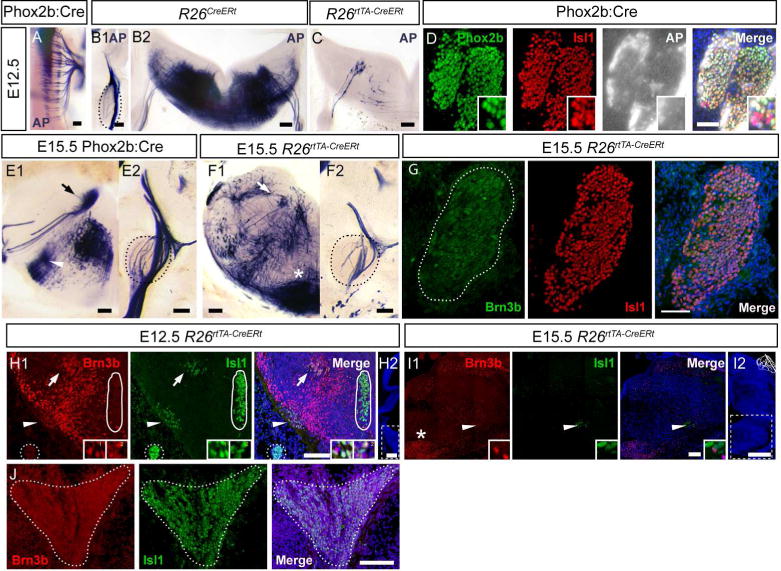
A, Dorsal view of the brainstem of a E12.5 Phox2b:Cre; Brn3bCKOAP embryo showing Brn3bAP positive cell bodies at the level of the dorsal motor nucleus of the vagus. B1, B2, AP histochemistry of coronal sections through R26CreERt; Brn3bCKOAP E12.5 embryos. B1, Brn3bAP positive fibers wrapping around the N/P ganglia complex. B2, Full Brn3bAP histochemistry of brainstem at the level of the dmnX. C, Hemisection through a sparsely labeled R26rtTA-CreERt; Brn3bCKOAP E12.5 embryo, showing a few dmnX cell bodies and their projections into the Xth nerve. D, Immunostaining for Phox2b and Isl1 combined with histochemistry for Brn3bAP, in sections of Nodose and Petrose Ganglia (N/P) of E12.5 Phox2b:Cre; Brn3bCKOAP embryo. E1, Brn3bAP histochemistry of brainstem sections in E15.5 Phox2b:Cre; Brn3bCKOAP embryos. Note the precise isolation of the dmnX (arrow) and NA (arrowhead), from several other Brn3bAP positive nuclei. E2, N/P ganglia from the same animal as in E1, wrapped in Brn3bAP fibers, but devoid of AP positive nuclei. F1, F2, Brn3bAP histochemistry in E15.5 R26rtTA-CreERt; Brn3bCKOAP embryos labeled at intermediate sparse levels, showing AP positivity in the dmnX (arrow), and lack of AP positive cell bodies in the N/P (black stippled lines). G Immunostaining of N/P ganglia shows Isl1 but no Brn3b positivity at E15.5. H1,H2 Coexpression of Brn3b (red) and Isl1 (green) in the dmnX (white arrow) and nucleus ambiguus (NA, white arrowhead). Insets in bottom right corner show high magnification of Brn3b and Isl1 co-expression in NA (1) and dmnX (2). I1,I2, Expression of Brn3b in the NA at E15.5. H2,I2 Stippled lines show the regions in H1 and I1. Asterisk in F1 and I1 marks E15.5 expression of Brn3b (I1) and Brn3bAP (F1) in the inferior olive. J Sagittal section at the level of superior and jugular ganglia (stippled line) of E12.5 embryo show now Brn3b expression. Scale bars: A,B1,B2,C,E1,E2,F1,F2,G,H1,I1,J=100μm,D=50μm, H2,I2=500μm.
The dmnX (Figure 11H1 arrow) and NA (Figure 11H1 arrow head) can be identified in double immunostaining for Brn3b and Isl1 at E12.5 (Figure 11H1). Note that in the same section the Isl1+ hypoglossal nucleus (Figure 11H1 circle) does not express Brn3b. As shown in coronal sections (Figure 11 H1) and sagittal sections (Figure 11J) at E12.5 the Isl1 positive superior and jugular ganglia do not express Brn3b in detectable levels by our antibody.
Similarly to Brn3a (McEvilly et al., 1996) at E15.5 Brn3b is expressed in cells of the NA. (Figure 11I1). Interestingly, Brn3b colocalizes with Isl1 in the NA at E12.5 but not at E15.5.
In the adult brainstem, the dorsal vagal complex consists of the dmnX, the nucleus of the solitary tract and the Area Postrema (Gao et al., 2009). In Phox2b:Cre; Brn3bCKOAP adult mice, the AP signal in the dmnX is visibly reduced, while the nucleus of the solitary tract and the Area Postrema show intense AP staining (Figure 12A1). AP signal was also present in the anatomic position of the NA in both Phox2b:Cre; Brn3bCKOAP (Figure 12A2) and cRetCreERt; Brn3bCKOAP animals (Figure 12B).
Figure 12. Brn3b expression in the area postrema of the postnatal mouse.
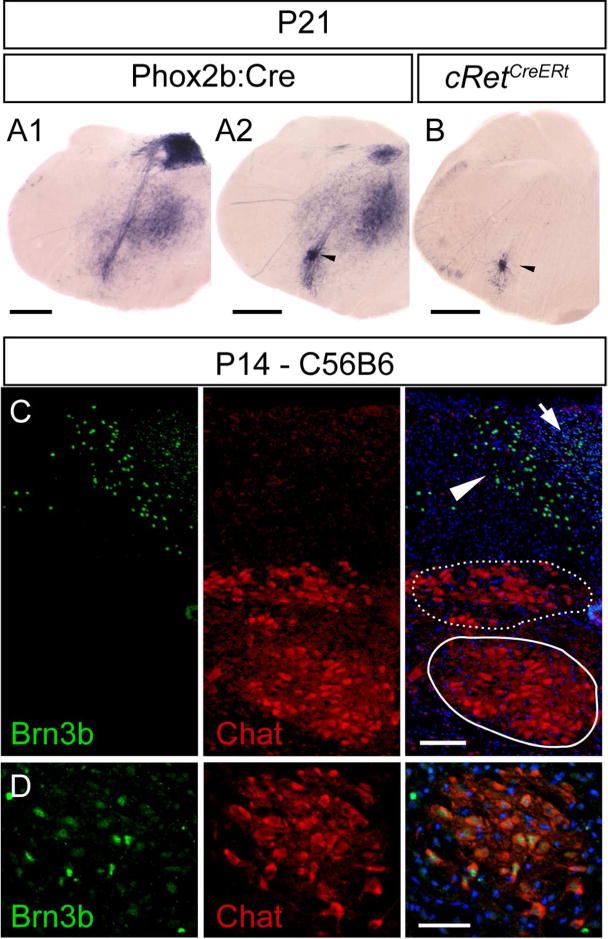
A1, Brn3AP positive areas include central stations of the vagus nerve (AP, NTS, and dmnX), in an adult Phox2b:Cre; Brn3bCKOAP animal. A2 B, AP signal in the anatomic position of the NA (black arrow head) in Phox2b:Cre; Brn3bCKOAP (A2) and cRetCreERt; Brn3bCKOAP (B) mouse. C, D Immunostaining analysis did not reveal any Brn3b expression in the Chat positive dmnX (Figure 12C stippled circle) or hypoglossal nuclei (12C circle). Expression was limited to the Area postrema (arrow), subdividions of the nucleus of the solitary tract (C arrow head) and NA (D). Scale bars: A1,A2,B=500μm, C,D=50μm.
In P14 mice, Brn3b protein is absent from the Chat positive dmnX (Figure 12C stippled circle) or hypoglossal nuclei (Figure 12C circle), but can be detected in subdivisions of the NTS (Figure 12C, arrowhead), Area Postrema (Figure 12C arrow) and NA (Figure 12D). It is worth highlighting that not all Chat positive cells of the NA are also Brn3b+ thus suggesting heterogeneity of the NA (Figure 12D).
The presented data indicates that similarly to FBM neurons Brn3b is transiently expressed in the dmnX during early development while it is constantly present in the NA. In addition the observation that the Brn3b positive cells of the NA are also cRet positive, indicates a complex combinatorial expression signature for these neurons.
Other Brainstem nuclei
Whole mount preparations of R26CreERt; Brn3bCKOAP and Phox2b:Cre; Brn3bCKOAP embryos at E12.5 – E15.5 (Figure 2 and data not shown) showed no AP staining in cranial nerves III, IV, VI and XII. In addition the central nuclei of these nerves were negative for both Brn3b protein and AP (abducens – VI – Figure 10A1 arrow head; oculomotor – III and trochlear – IV – Figure 13A1; hypoglossal – XII – Figure 11H1 circle). However Brn3b positive cells were identified latero-ventral to the trochlear nucleus (Figure 13A1, A2).
Figure 13. Other Brainstem nuclei.
A1 A2, Isl1 positive oculomotor (arrow) and trochlear (arrow head) nuclei do not express Brn3b. Brn3b positive cells in proximity of to the trochlear nucleus (stippled line). B, C AP positive cell bodies (black arrows) ventral to the facial nucleus (black stippled lines) in the retrotrapezoid area (red lines) indicating Phox2b/Brn3b (B) and Brn3b/cRet (C) coexpression. Scale bar A1=100μm, A2, B, C = 500μm
As previously described, transient inferior olive expression of Brn3bAP (Figure 11F1 star) and Brn3b (Figure 11I1 star), was observed at E15.5. No Brn3bAP positive cells remain in the inferior olives of adult Phox2b:Cre; Brn3bCKOAP, cRetCreERt; Brn3bCKOAP, (Figure 12 A1, A2, B) and R26CreERt; Brn3bCKOAP (data not shown) mice.
The Phox2b positive retrotrapezoid nucleus (RTN) of the rostral ventrolateral brainstem contains CO2 sensitive neurons that regulate breathing amplitude and frequency (Wang et al., 2013b, Bochorishvili et al., 2012, Marina et al., 2010, Kang et al., 2007). At the anatomical position of the RTN, AP labeled cell bodies are present both in Phox2b:Cre; Brn3bCKOAP (Figure 13B) and cRetCreERt; Brn3bCKOAP (Figure 13C) adult mice. Since recombination, and hence AP expression was induced in the cRetCreERt; Brn3bCKOAP mice at the adult stage, it is unlikely that the staining is derived from previous transient Brn3b expression. However we were not able to detect Brn3b expression by immunostaining, either due to low levels of Brn3b protein undetectable by our antibody, or potential misexpression of the Brn3bAP allele in these cells. In addition, at E15.5 and adult we find intense Brn3bAP (Phox2b:Cre; CKOAPBrn3b) staining but not Brn3b+ cell bodies in a position consistent with the medullary reticular nucleus, ventral part (MdV, Paxinos) (Figure 11E1, 11F1, 12A1). We interpret this staining as fibers of passage originating in a population of spinal cord interneurons that do express Brn3b during early stages of development.
Differences between Brn3bCKOAP/KO and Brn3bCKOAP/WT
We have analyzed 92 R26rtTA-CreERt; Brn3bCKOAP/KO and 87 R26rtTA-CreERt ; Brn3bCKOAP/WT embryos of various ages, including E12.5, E13.5, E14.5 and E15.5. Labeling efficiency was similar in the two genotypes, 33.6% of Brn3bCKOAP/KO animals showed sparse labeling while 21.7% dense labeling and 44.5% no labeling. In the case of Brn3bCKOAP/WT 33.3% animals showed sparse labeling while 18.4% dense labeling and 48.3% no labeling.
In spite of delayed axon projections of RGC, anatomical survey of other AP expressing cranial nerves showed no obvious defects in the R26rtTA-CreERt; Brn3bCKOAP/KO animals when compared to their R26rtTA-CreERt; Brn3bCKOAP/WT littermates. Since sparse labeling could result in different structures being differentially labeled in different experimental animals, we scored the presence or absence of all Brn3b positive nuclei over the total sets of R26rtTA-CreERt; Brn3bCKOAP/KO mice and R26rtTA-CreERt; Brn3bCKOAP/WT littermates, and find that all nuclei are represented at comparable levels in the investigated animals (Figure 14, 15, 16 17).
Figure 14.
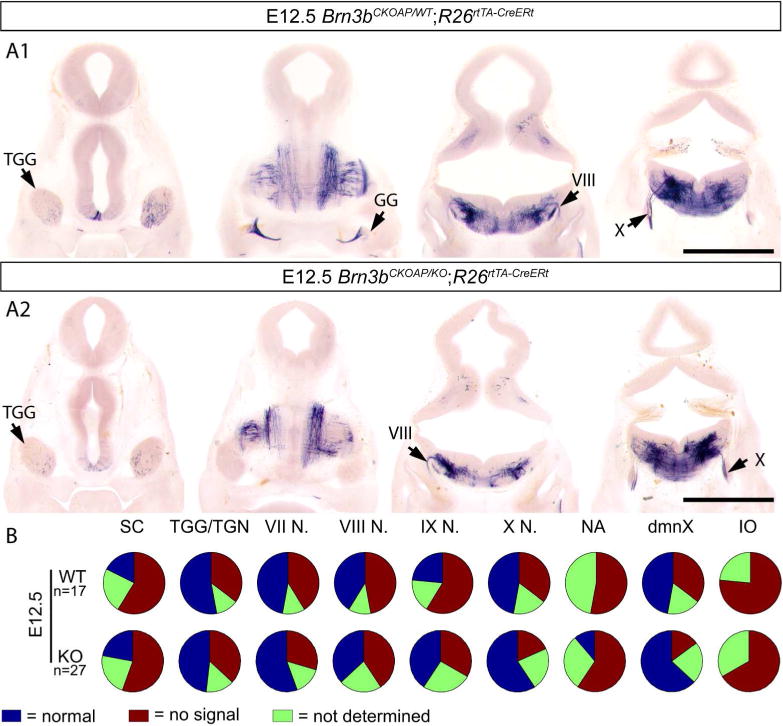
Analysis of Brn3bAP positive structures in E12.5 R26rtTA-CreERt; Brn3bCKOAP/WT (A1) R26rtTA-CreERt; Brn3bCKOAP/KO (A2). B, n = the total number of analyzed embryos per condition. Scale bars A1, A2= 1mm
Figure 15.
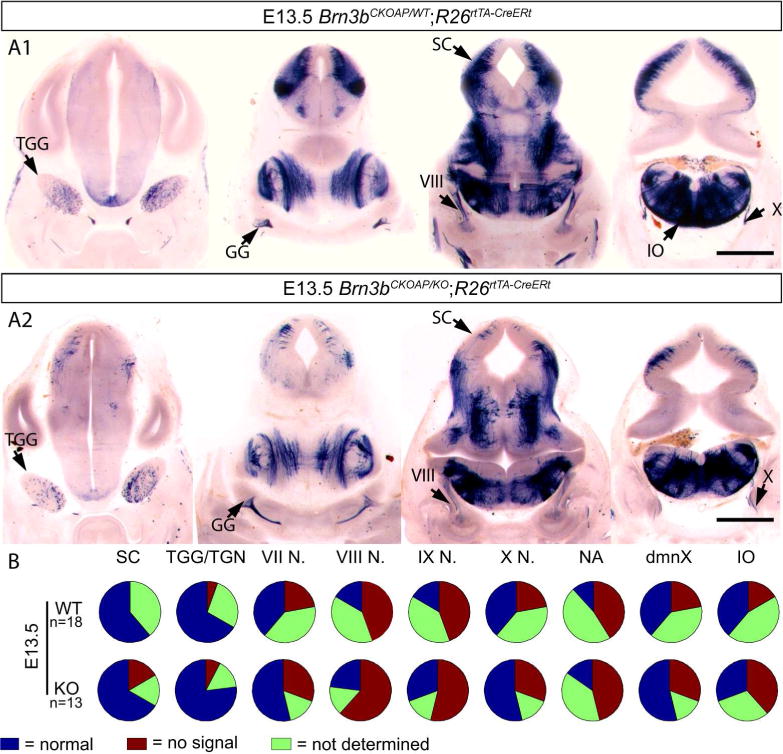
Analysis of Brn3bAP positive structures in E13.5 R26rtTA-CreERt; Brn3bCKOAP/WT (A1) R26rtTA-CreERt; Brn3bCKOAP/KO (A2). B, n = the total number of analyzed embryos per condition. Scale bars A1, A2= 1mm
Figure 16.
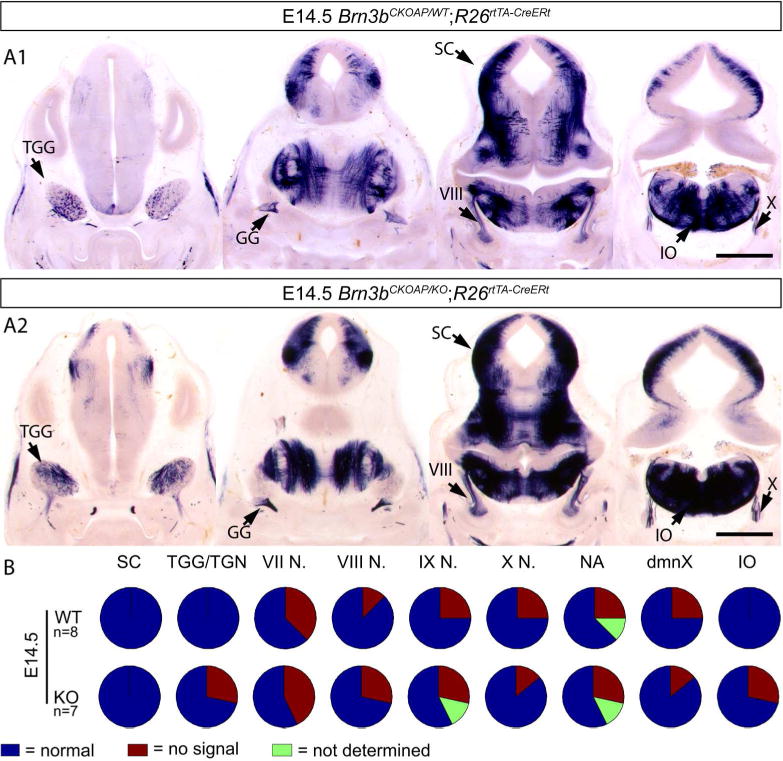
Analysis of Brn3bAP positive structures in E14.5 R26rtTA-CreERt; Brn3bCKOAP/WT (A1) R26rtTA-CreERt; Brn3bCKOAP/KO (A2). B, n = the total number of analyzed embryos per condition. Scale bars A1, A2= 1mm
Figure 17.
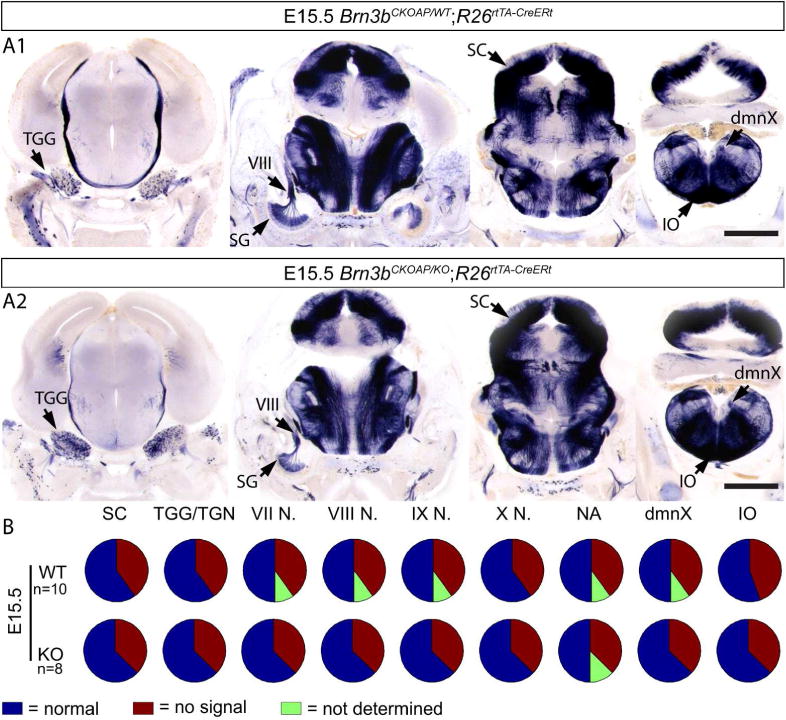
Analysis of Brn3bAP positive structures in E15.5 R26rtTA-CreERt; Brn3bCKOAP/WT (A1) R26rtTA-CreERt; Brn3bCKOAP/KO (A2). B, n = the total number of analyzed embryos per condition. Scale bars A1, A2= 1mm
We should mention that, as can be seen in Figures 2A, 14, 15, 16 and 17 and previously reported, Brn3b is also expressed in the superior colliculus, where it persists until the adult age (Badea and Nathans, unpublished). We have confirmed this pattern of expression by immunostaining (data not shown), but find no evidence for SC defects in Brn3bKO mice.
Discussion
Previous work had pointed at Brn3 transcription factors as potential regulators of neuronal cell type diversity in specific classes of first order projection neurons of the visual, auditory and somatosensory pathway. The expression and role of Brn3a in the development cranial nerves (optic, trigeminal, facial, vestibuloacoustic), has previously been addressed. We report here that Brn3b is dynamically expressed in several ganglionic or intra-CNS stations of nerves II, V, VII, IX and X, and several other nuclei of the brainstem (summarized in Table 2). Using the partial overlap of expression between Brn3b, Isl1 and Phox2b, as well as anatomic location of Brn3b+ cell bodies and projection patterns of axonal and dendritic arbors, we demonstrate Brn3b expression in sensory and visceral but not somatomotor components of these nerves. We bring further refinement to the described Brn3b phenotype in RGCs, but do not observe any abnormalities in the other nuclei or ganglia expressing the transcription factor.
Table 2.
Summary of Brn3b expression in different neuronal structures as determined by AP staining or immunohistochemistry. Observed Brn3b expression (+), not determined (ND).
| E11.5 | E12.5 | E13.5 | E14.5 | E15.5 | P14 | Adult | |
|---|---|---|---|---|---|---|---|
| RGC | + | + | + | + | + | + | + |
| TGG | ND | + | + | + | + | + | + |
| PT | ND | + | ND | ND | ND | ND | ND |
| GG – viscerosensory | − | − | ND | ND | − | ND | ND |
| GG – somatosensory | − | − | ND | ND | + | ND | ND |
| SG | ND | + | + | + | + | + | + |
| N/P | − | − | − | − | − | ND | ND |
| S/J | ND | − | ND | ND | ND | ND | ND |
| dmnX | + | + | + | + | + | − | − |
| FBM | + | − | − | − | − | − | − |
| VNTB | ND | ND | ND | ND | ND | + | + |
| Eve | ND | ND | ND | ND | ND | + | + |
| IO | ND | + | + | + | + | − | − |
| SSN | + | + | ND | ND | ND | ND | ND |
| IEE | + | + | ND | + | + | ND | ND |
| NA | ND | + | ND | ND | + | + | + |
Axon guidance in RGCs lacking Brn3b
Previously published work revealed three major types of axon guidance defects in RGCs lacking Brn3b. DiI injections in the retina revealed intra-ocular axon guidance defects at E14.5 while genetic labeling of individual RGCs showed abnormal branching in adult Brn3bAP/KO axons. Along the optic path, DiI tracing showed invasion of the contra-lateral optic nerve and increased ipsilateral projections through the optic chiasm at P0, and stalling at the transition from the pretectal area to the superior colliculus around P1. Genetically labeled Brn3bAP/KO RGC axons exhibit transient misrouting and invasion of other thalamic areas in early (P6 – P9) postnatal development, while the LGN and SC have substantial amounts of innervation (Badea et al 2009). Finally, Brn3bKO RGC axons have major growth defects in retinal explant cultures. We find that Brn3bAP/KO RGC exhibit delayed axon growth, evidenced as early as E12.5 and persisting through E15.5. However RGC axons formed correctly within the retina, and tracked to the optic nerve chiasm and tract. It therefore appears that the initial steps of RGC axon navigation are robust to loss of Brn3b and its transcriptional targets. Therefore it may be that the transient postnatal axon misrouting defects are secondary to the failure of establishing productive synapses in the target tissues (sprouting), rather than a bona fide axon guidance defect.
Cell type distribution of Brn3b in the TGG is similar to that of DRG projecting neurons
Whereas DRG neurons, including mechanoreceptors are entirely derived from neural crest precursors, the TGG has dual origin, with neural crest derived cells giving birth to small nocicepors and glial cells, while the other sensory modality neurons are derived from placode precursors. Previously we reported selective Brn3b expression in DRG mechanoreceptors beginning with postnatal stages of development. In the trigeminal ganglion, Brn3b is expressed as early as E12.5 and by E15.5, at least 4 distinct neuronal populations can be separated: (1) Isl1+, (2) Isl1+, Brn3b+, (3) Isl1+, cRet+, and (4) Isl1+, cRet+, Brn3b+. Since, in the E15.5 DRG, the neurotrophin receptor cRet is expressed in subpopulations of mechanoreceptors and nonpeptidergic nociceptors, populations (2) and (4) may represent two distinct subsets of mechanoreceptors, whereas population (3) might be comprised of nociceptors. In the adult, Brn3b+ TGG neurons are TrkB+, TrkC+, NFH+, consistent with Brn3b expression in subpopulations of mechanoreceptors, including some of the elements of the special somatosensory sense associated with the vibrissae. Hence Brn3b cell type distribution is similar for both DRG and TGG.
Brn3s expression in the projection neurons of the special senses
We report here that Brn3b is absent from the gustatory component of the GG of the VII, and not expressed at all in the nodose/petrose ganglion of the IX/X. Since Brn3a is also expressed in the GG in a pattern mutually exclusive with Phox2b, it appears that Brn3 factors do not participate in taste projection neuron specification (D’Autreaux et al., 2011; Huang et al., 2001; Quina et al., 2012), but rather are restricted to somatosensory neurons innervating the head. Since no Brn3 expression has been reported in olfactory neurons (Badea and Nathans, and Sajgo and Badea, data not shown), it appears that POU4 TFs are not associated with sensory neurons of the chemical senses, unlike the situation in drosophila where Acj6, the POU4 orthologue is expressed in the olfactory receptor neurons (ORN) (Komiyama et al 2004, Bai and Carlson 2010). Thus the combinatorial code of Brn3s in the mammalian system seems to be restricted to general somatosensation, vision and vestibuloacoustic senses. However, more targeted experiments are needed to determine whether Brn3s or other POU domain transcription factors are expressed and/or play any role in the development or specification of chemical sense neurons.
Dynamic expression of Brn3b in the inner ear efferent neurons and FBM precursors
Previously Brn3b expression was documented in E12.5 statoacoustic ganglion, and its adult derivatives the vestibular and spiral ganglia (Badea et al., 2012, Deng et al., 2014). By genetic intersection strategies we now demonstrate expression of Brn3b in the Phox2b positive IEE population. The complex migration pattern of the FBM is mirrored in the dynamic expression of Isl1, Phox2b and Brn3b in these neurons. While Phox2b marks the entire population as early as E9 in the adult it’s expression is restricted to the IEE efferent origin nuclei, Eve and VNTB (Song et al., 2006, Kang et al., 2007). In contrast Isl1 marks this population and all its derivatives from its origin in r4 at E10.5 until their final location and in to adult age (Thor et al 1991, Song et al., 2006). We find that Brn3b expression is limited to the FBMs in r4, (E11.5–E12.5), while the migrating FBMs, which will generate the nucleus of the VII and provide motor innervation to the face, are Isl1+, Phox2b+, Brn3b−. The early co-expression of Brn3b and Phox2b in both cell populations is consistent with the hypothesis that IIEs are evolutionarily derived from FBMs (Fritzsch, 1999). Interestingly, In the case of the Brn3a−, Phox2b+ nodose/petrose viscerosensory neurons, ablation of Phox2b results in a fate change towards somatosensory neurons and misexpression of Brn3a (D’Autreaux et al. 2011). Thus combinatorial expression of Phox2b and Brn3s could help determine specific cell fates, visceral, somatomotor or somatosensory in a context dependent manner.
In the adult, the molecular distinction between Brn3b+, Phox2b− afferent and Brn3b+ Phox2b+ efferent projecting neurons of the auditory and vestibular systems is preserved. Adult inductions of Brn3bCKOAP recombination using a cRetCreERt label the Eve and VNTB, confirming co-expression of cRet and Brn3b in the adult. Given the complex pattern of overlap between cRet and Brn3b in RGCs, TGG, GG, Eve and VNTB, it would be interesting to investigate potential links between transcriptional regulation and neurotrophic signaling in cell type specification of these populations. The partially overlapping expression profiles of the Brn3s, cRet, and Phox2b loci, in conjunction with the existence of a large variety of genetic tools, offers the opportunity to selectively label or ablate specific circuit elements.
Role of Brn3 transcription factors in cranial nerve development and function
The relative importance of the three POU 4 family members (Brn3a, Brn3b or Brn3c) in distinct sensory compartments is suggested by their spatiotemporal expression profile. Although all three Brn3s are expressed in RGCs, the most striking phenotype results from ablation of the earliest expressed family member, Brn3b. Ablation of Brn3a, whose expression is delayed by at least one day compared to Brn3b results in subtle morphological changes and only modestly affects RGC numbers. The importance of the temporal aspect of Brn3 expression is further supported by the apparent rescue of RGC numbers in mice expressing the Brn3a cDNA instead of Brn3b at the Brn3b locus, and hence at an earlier time point (Pan et al., 2005). In contrast, during TGG, DRG and SAG development, Brn3a expression precedes that of Brn3b and/or Brn3c and Brn3a ablation leads to down regulation of Brn3b and/or Brn3c and major cell loss (Huang et al., 1999, Dykes et al., 2010, Zou et al., 2012, Badea et al., 2012). Even though we report no major phenotypes associated with loss of Brn3b in other cranial nerves it is possible that more subtle, cell type specific changes are present in adult Brn3bKO mice.
It has been proposed that transcription factors of the POU domain 4 (Brn3) family are somatosensory markers and/or determinants as part of a larger transcriptional combinatorial code including sensory, motor and visceral projection neurons, conserved from gastropods to vertebrates (Nomaksteinsky et al., 2013). However we now report that the POU domain transcription factor Brn3b is also expressed in visceromotor (dmnX, SSN, NA) and other brainstem nuclei (Eve, VNTB).
In addition to elucidating the mechanisms of cell type specification, such studies could also guide genetic circuit manipulation aimed at understanding the functions of these diverse cell populations, using strategies analogous to the ones employed in the visual system. In many cases, the anatomic – molecular correlate afforded by the conditional knock-in reporters could help in the identification of the cell populations involved, by virtue of their characteristic peripheric or central innervation territories, already well described in classical anatomic studies.
Combinatorial gene expression can be further used to study specific cell types within the nervous system. This approach can possibly isolate unique cell types within individual brain nuclei making it possible to study their cellular morphologies, connections and physiology. Thus using combinatorial expression it would be possible to create a unique genetic identifier for all neuronal cell types.
Acknowledgments
The Authors acknowledge Hideki Enomoto, Anna Vysochan, and Wenqin Luo for Ret-CFP and embryos and cRetCre-ERt, Jean Francois Brunet for gift of Phox2b antibody, Lars von Buchholtz and Silvia Arber for helpful comments on the manuscript, and Nadia Parmhans for help with genetic line crosses and genotyping.
This work was supported by the National Eye Institute Intramural Funding to TC Badea
Abbreviations
- II
Optic nerve
- III
Oculomotor nerve
- IV
Trochlear nerve
- V
Trigeminal nerve
- VI
Abducens nerve
- VII
Facial nerve
- VIII
Vestibulocochlear nerve
- IX
Glossopharyngeal nerve
- X
Vagus nerve
- XI
Accessory nerve
- XII
Hypoglossal nerve
- r
Rhombomere
- RGC
Retinal ganglion cell
- TGG
Trigeminal ganglia
- V1
Ophthalmic nerve of the trigeminal
- V2
Maxillary nerve of the trigeminal
- V3
Mandibular nerve if the trigeminal
- MoV
Motor nucleus of the trigeminal
- MesV
Mesencephalic trigeminal nucleus
- GG
Geniculate ganglion
- CT
Chorda tympani nerve
- GSP
Greater superficial petrosal nerve
- FBM
Facial branchiomotor nucleus
- FSC
Mistacial pad follicle-sinus complexes
- SG
Spiral ganglion
- S/J
Superior-jugular ganglion
- VEN
Vestibular efferent nucleus
- CEN
Cochlear efferent nucleus
- NG
Nodose ganglion
- PG
Petrose ganglion
- NA
Nucleus ambiguus
- dmnX
Dorsal motor nucleus of the vagus
- IO
Inferior olive
- DRG
Dorsal root ganglion
- IEE
Inner ear efferent
- SC
Superior colliculus
- BM
Branchiomotor Neurons
- VM
Visceromotor neurons
- VS
Viscerosensor neurons
- SSN
Superior salivatory nucleus
- Eve
Nucleus of origin of vestibular efferents
- OCB
Olivocochlear bundle
- MOC
Medial olivocochlear bundle
- LSO
Lateral superior olive
- MSO
Medial superior olive
- SOC
Superior olivary complex
- VNTB
Ventral nucleus of the trapezoid body
- RTN
Retrotrapezoid nucleus
Footnotes
No known conflicts of interest.
Author contributions: Study concept and design: S.S. O.P. T.C.B. Performed Experiments: S.S. S.A. Acquisition of data: S.S. Statistical analysis: S.S. Analysis and interpretation of data: S.S., T.C.B. Drafting and Revising the manuscript: S.S. O.P. T.C.B.
References
- Altschuler SM, Bao X, Bieger D, Hopkins DA, Miselis RR. Viscerotopic representation of the upper alimentary tract in the rat: sensory ganglia and nuclei of the solitary and spinal trigeminal tracts. Journal of Comparative Neurology. 1989;283(2):248–268. doi: 10.1002/cne.902830207. [DOI] [PubMed] [Google Scholar]
- Alvarado-Mallart MR, Batini C, Buisseret-Delmas C, Corvisier J. Trigeminal representations of the masticatory and extraocular proprioceptors as revealed by horseradish peroxidase retrograde transport. Experimental brain research. 1975;23(2):167–179. doi: 10.1007/BF00235459. [DOI] [PubMed] [Google Scholar]
- Ashwell KW. The adult mouse facial nerve nucleus: morphology and musculotopic organization. Journal of anatomy. 1982;135(Pt 3):531–531. [PMC free article] [PubMed] [Google Scholar]
- Auclair F, Valdés N, Marchand R. Rhombomere-specific origin of branchial and visceral motoneurons of the facial nerve in the rat embryo. Journal of Comparative Neurology. 1996;369(3):451–461. doi: 10.1002/(SICI)1096-9861(19960603)369:3<451::AID-CNE9>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- Ayer-Le Lievre, CS Le, Douarin NM. The early development of cranial sensory ganglia and the potentialities of their component cells studied in quail-chick chimeras. Developmental biology. 1982;94(2):291–310. doi: 10.1016/0012-1606(82)90349-9. [DOI] [PubMed] [Google Scholar]
- Badea TC, Cahill H, Ecker J, Hattar S, Nathans J. Distinct roles of transcription factors brn3a and brn3b in controlling the development, morphology, and function of retinal ganglion cells. Neuron. 2009;61(6):852–864. doi: 10.1016/j.neuron.2009.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badea TC, Hua ZL, Smallwood PM, Williams J, Rotolo T, Ye X, Nathans J. New mouse lines for the analysis of neuronal morphology using CreER (T)/loxP-directed sparse labeling. PLoS One. 2009;4(11):e7859–e7859. doi: 10.1371/journal.pone.0007859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badea TC, Nathans J. Morphologies of mouse retinal ganglion cells expressing transcription factors Brn3a, Brn3b, and Brn3c: analysis of wild type and mutant cells using genetically-directed sparse labeling. Vision research. 2011;51(2):269–279. doi: 10.1016/j.visres.2010.08.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badea TC, Wang Y, Nathans J. A noninvasive genetic/pharmacologic strategy for visualizing cell morphology and clonal relationships in the mouse. J Neurosci. 2003;23(6):2314–2322. doi: 10.1523/JNEUROSCI.23-06-02314.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badea TC, Williams J, Smallwood P, Shi M, Motajo O, Nathans J. Combinatorial expression of Brn3 transcription factors in somatosensory neurons: genetic and morphologic analysis. The Journal of Neuroscience. 2012;32(3):995–1007. doi: 10.1523/JNEUROSCI.4755-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai L, Carlson JR. Distinct functions of acj6 splice forms in odor receptor gene choice. The Journal of Neuroscience. 2010;30(14):5028–5036. doi: 10.1523/JNEUROSCI.6292-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bochorishvili G, Stornetta RL, Coates MB, Guyenet PG. Pre-Botzinger complex receives glutamatergic innervation from galaninergic and other retrotrapezoid nucleus neurons. Journal of Comparative Neurology. 2012;520(5):1047–1061. doi: 10.1002/cne.22769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown MC, Levine JL. Dendrites of medial olivocochlear neurons in mouse. Neuroscience. 2008;154(1):147–159. doi: 10.1016/j.neuroscience.2007.12.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi X, Michos O, Shakya R, Riccio P, Enomoto H, Licht JD, Asai N, Takahashi M, Ohgami N, Kato M, et al. Ret-dependent cell rearrangements in the Wolffian duct epithelium initiate ureteric bud morphogenesis. Developmental cell. 2009;17(2):199–209. doi: 10.1016/j.devcel.2009.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chibuzo GA, Cummings JF. Motor and sensory centers for the innervation of mandibular and sublingual salivary glands: a horseradish peroxidase study in the dog. Brain research. 1980;189(2):301–313. doi: 10.1016/0006-8993(80)90092-x. [DOI] [PubMed] [Google Scholar]
- Coppola E, Rallu M, Richard J, Dufour S, Riethmacher D, Guillemot F, Goridis C, Brunet JF. Epibranchial ganglia orchestrate the development of the cranial neurogenic crest. Proceedings of the National Academy of Sciences. 2010;107(5):2066–2071. doi: 10.1073/pnas.0910213107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Amico-Martel A, Noden DM. Contributions of placodal and neural crest cells to avian cranial peripheral ganglia. American Journal of Anatomy. 1983;166(4):445–468. doi: 10.1002/aja.1001660406. [DOI] [PubMed] [Google Scholar]
- D’Autreaux F, Coppola E, Hirsch M-R, Birchmeier C, Brunet JF. Homeoprotein Phox2b commands a somatic-to-visceral switch in cranial sensory pathways. Proc Natl Acad Sci U S A. 2011;108(50):20018–20023. doi: 10.1073/pnas.1110416108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis BM, Fundin BT, Albers KM, Goodness TP, Cronk KM, Rice FL. Overexpression of nerve growth factor in skin causes preferential increases among innervation to specific sensory targets. Journal of Comparative Neurology. 1997;387(4):489–506. doi: 10.1002/(sici)1096-9861(19971103)387:4<489::aid-cne2>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- Deng M, Yang H, Xie X, Liang G, Gan L. Comparative expression analysis of POU4F1, POU4F2 and ISL1 in developing mouse cochleovestibular ganglion neurons. Gene Expression Patterns. 2014;15(1):31–37. doi: 10.1016/j.gep.2014.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubreuil V, Barhanin J, Goridis C, Brunet JF. Breathing with phox2b Philosophical Transactions of the Royal Society B. Biological Sciences. 2009;364(1529):2477–2483. doi: 10.1098/rstb.2009.0085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dykes IM, Tempest L, Lee S-I, Turner EE. Brn3a and Islet1 act epistatically to regulate the gene expression program of sensory differentiation. The journal of Neuroscience. 2011;31(27):9789–9799. doi: 10.1523/JNEUROSCI.0901-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards MA, Schneider GE, Caviness VS. Development of the crossed retinocollicular projection in the mouse. Journal of Comparative Neurology. 1986;248(3):410–421. doi: 10.1002/cne.902480309. [DOI] [PubMed] [Google Scholar]
- Elshatory Y, Deng M, Xie X, Gan L. Expression of the LIM-homeodomain protein Isl1 in the developing and mature mouse retina. Journal of Comparative Neurology. 2007;503(1):182–197. doi: 10.1002/cne.21390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enomoto H, Crawford PA, Gorodinsky A, Heuckeroth RO, Johnson EM, Milbrandt J. RET signaling is essential for migration, axonal growth and axon guidance of developing sympathetic neurons. Development. 2001;128(20):3963–3974. doi: 10.1242/dev.128.20.3963. [DOI] [PubMed] [Google Scholar]
- Ericson J, Thor S, Edlund T, Jessell TM, Yamada T. Early stages of motor neuron differentiation revealed by expression of homeobox gene Islet-1. Science. 1992;256(5063):1555–1560. doi: 10.1126/science.1350865. [DOI] [PubMed] [Google Scholar]
- Erkman L, Yates PA, McLaughlin T, McEvilly RJ, Whisenhunt T, O’Connell SM, Krones AI, Kirby MA, Rapaport DH, Bermingham JR, et al. A POU domain transcription factor–dependent program regulates axon pathfinding in the vertebrate visual system. Neuron. 2000;28(3):779–792. doi: 10.1016/s0896-6273(00)00153-7. [DOI] [PubMed] [Google Scholar]
- Espana A, Clotman F. Onecut factors control development of the Locus Coeruleus and of the mesencephalic trigeminal nucleus. Molecular and Cellular Neuroscience. 2012;50(1):93–102. doi: 10.1016/j.mcn.2012.04.002. [DOI] [PubMed] [Google Scholar]
- Faye-Lund H. Projection from the inferior colliculus to the superior olivary complex in the albino rat. Anatomy and embryology. 1986;175(1):35–52. doi: 10.1007/BF00315454. [DOI] [PubMed] [Google Scholar]
- Fritzsch B. Development of the Labyrinthine Efferent Systema. Annals of the New York Academy of Sciences. 1996;781(1):21–33. doi: 10.1111/j.1749-6632.1996.tb15690.x. [DOI] [PubMed] [Google Scholar]
- Fritzsch B. The Efferent Auditory System: Basic Science and Clinical Applications. Singular Publishing Group, Inc; 1999. Ontogenetic and evolutionary evidence for the motoneuron nature of vestibular and cochlear efferents; p. 31. [Google Scholar]
- Fritzsch B, Nichols DH. DiI reveals a prenatal arrival of efferents at the differentiating otocyst of mice. Hear Res. 1993;65:1–2. 51–60. doi: 10.1016/0378-5955(93)90200-k. [DOI] [PubMed] [Google Scholar]
- Fundin BT, Silos-Santiago I, Ernfors P, Fagan AM, Aldskogius H, DeChiara TM, Phillips HS, Barbacid M, Yancopoulos GD, Rice FL. Differential dependency of cutaneous mechanoreceptors on neurotrophins, trk receptors, and P75 LNGFR. Developmental biology. 1997;190(1):94–116. doi: 10.1006/dbio.1997.8658. [DOI] [PubMed] [Google Scholar]
- Funfschilling U, Ng Y-G, Zang K, Miyazaki J-I, Reichardt LF, Rice FL. TrkC kinase expression in distinct subsets of cutaneous trigeminal innervation and nonneuronal cells. Journal of Comparative Neurology. 2004;480(4):392–414. doi: 10.1002/cne.20359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan L, Xiang M, Zhou L, Wagner DS, Klein WH, Nathans J. POU domain factor Brn-3b is required for the development of a large set of retinal ganglion cells. Proceedings of the National Academy of Sciences. 1996;93(9):3920–3925. doi: 10.1073/pnas.93.9.3920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao H, Glatzer NR, Williams KW, Derbenev AV, Liu D, Smith BN. Morphological and electrophysiological features of motor neurons and putative interneurons in the dorsal vagal complex of rats and mice. Brain research. 2009;1291:40–52. doi: 10.1016/j.brainres.2009.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilland E, Baker R. Conservation of neuroepithelial and mesodermal segments in the embryonic vertebrate head. Cells Tissues Organs. 1993;148:2–3. 110–123. doi: 10.1159/000147530. [DOI] [PubMed] [Google Scholar]
- Glowatzki E, Fuchs PA. Cholinergic synaptic inhibition of inner hair cells in the neonatal mammalian cochlea. Science Signaling. 2000;288(5475):2366–2366. doi: 10.1126/science.288.5475.2366. [DOI] [PubMed] [Google Scholar]
- Glueckert R, Bitsche M, Miller JM, Zhu Y, Prieskorn DM, Altschuler RA, Schrott-Fischer A. Deafferentiation-associated changes in afferent and efferent processes in the guinea pig cochlea and afferent regeneration with chronic intrascalar brain-derived neurotrophic factor and acidic fibroblast growth factor. Journal of Comparative Neurology. 2008;507(4):1602–1621. doi: 10.1002/cne.21619. [DOI] [PubMed] [Google Scholar]
- Hadjab S, Franck MCM, Wang Y, Sterzenbach U, Sharma A, Ernfors P, Lallemend F. A Local Source of FGF Initiates Development of the Unmyelinated Lineage of Sensory Neurons. The Journal of Neuroscience. 2013;33(45):17656–17666. doi: 10.1523/JNEUROSCI.1090-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamburger V. Experimental analysis of the dual origin of the trigeminal ganglion in the chick embryo. Journal of Experimental Zoology. 1961;148(2):91–123. doi: 10.1002/jez.1401480202. [DOI] [PubMed] [Google Scholar]
- Hasegawa H, Wang F. Visualizing mechanosensory endings of TrkC-expressing neurons in HS3ST-2-hPLAP mice. Journal of Comparative Neurology. 2008;511(4):543–556. doi: 10.1002/cne.21862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henstrom D, Hadlock T, Lindsay R, Knox CJ, Malo J, Vakharia KT, Heaton JT. The convergence of facial nerve branches providing whisker pad motor supply in rats: Implications for facial reanimation study. Muscle & nerve. 2012;45(5):692–697. doi: 10.1002/mus.23232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Highstein SM. The central nervous system efferent control of the organs of balance and equilibrium. Neuroscience research. 1991;12(1):13–30. doi: 10.1016/0168-0102(91)90096-h. [DOI] [PubMed] [Google Scholar]
- Hinrichsen CFL, Watson CD. The facial nucleus of the rat: representation of facial muscles revealed by retrograde transport of horseradish peroxidase. The Anatomical Record. 1984;209(3):407–415. doi: 10.1002/ar.1092090321. [DOI] [PubMed] [Google Scholar]
- Huang EJ, Liu W, Fritzsch B, Bianchi LM, Reichardt LF, Xiang M. Brn3a is a transcriptional regulator of soma size, target field innervation and axon pathfinding of inner ear sensory neurons. Development. 2001;128(13):2421–2432. doi: 10.1242/dev.128.13.2421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang EJ, Zang K, Schmidt A, Saulys A, Xiang M, Reichardt LF. POU domain factor Brn-3a controls the differentiation and survival of trigeminal neurons by regulating Trk receptor expression. Development. 1999;126(13):2869–2882. doi: 10.1242/dev.126.13.2869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob J, Guthrie S. Facial visceral motor neurons display specific rhombomere origin and axon pathfinding behavior in the chick. The Journal of Neuroscience. 2000;20(20):7664–7671. doi: 10.1523/JNEUROSCI.20-20-07664.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang BJ, Chang DA, Mackay DD, West GH, Moreira TS, Takakura AC, Gwilt JM, Guyenet PG, Stornetta RL. Central nervous system distribution of the transcription factor Phox2b in the adult rat. Journal of Comparative Neurology. 2007;503(5):627–641. doi: 10.1002/cne.21409. [DOI] [PubMed] [Google Scholar]
- Kawamata T, Ninomiya T, Toriyabe M, Yamamoto J, Niiyama Y, Omote K, Namiki A. Immunohistochemical analysis of acid-sensing ion channel 2 expression in rat dorsal root ganglion and effects of axotomy. Neuroscience. 2006;143(1):175–187. doi: 10.1016/j.neuroscience.2006.07.036. [DOI] [PubMed] [Google Scholar]
- Klein R, Silos-Santiago I, Smeyne RJ, Lira SA, Brambilla R, Bryant S, Zhang L, Snider WD, Barbacid M. Disruption of the neurotrophin-3 receptor gene trkC eliminates la muscle afferents and results in abnormal movements. 1994 doi: 10.1038/368249a0. [DOI] [PubMed] [Google Scholar]
- Komiyama T, Carlson JR, Luo L. Olfactory receptor neuron axon targeting: intrinsic transcriptional control and hierarchical interactions. Nature neuroscience. 2004;7(8):819–825. doi: 10.1038/nn1284. [DOI] [PubMed] [Google Scholar]
- Komori T, Gyobu H, Ueno H, Kitamura T, Senba E, Morikawa Y. Expression of kin of irregular chiasm-like 3/mKirre in proprioceptive neurons of the dorsal root ganglia and its interaction with nephrin in muscle spindles. Journal of Comparative Neurology. 2008;511(1):92–108. doi: 10.1002/cne.21838. [DOI] [PubMed] [Google Scholar]
- Komori T, Gyobu H, Ueno H, Kitamura T, Senba E, Morikawa Y. Expression of kin of irregular chiasm-like 3/mKirre in proprioceptive neurons of the dorsal root ganglia and its interaction with nephrin in muscle spindles. Journal of Comparative Neurology. 2008;511(1):92–108. doi: 10.1002/cne.21838. [DOI] [PubMed] [Google Scholar]
- Leijon S, Magnusson AK. Physiological characterization of vestibular efferent brainstem neurons using a transgenic mouse model. PLoS One. 2014;9(5):e98277. doi: 10.1371/journal.pone.0098277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Ginty DD. The structure and organization of lanceolate mechanosensory complexes at mouse hair follicles. Elife. 2014;3:e01901–e01901. doi: 10.7554/eLife.01901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang X, Song M-R, Xu Z, Lanuza GM, Liu Y, Zhuang T, Chen Y, Pfaff SL, Evans SM, Sun Y. Isl1 is required for multiple aspects of motor neuron development. Molecular and Cellular Neuroscience. 2011;47(3):215–222. doi: 10.1016/j.mcn.2011.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lumsden A. The cellular basis of segmentation in the developing hindbrain. Trends in neurosciences. 1990;13(8):329–335. doi: 10.1016/0166-2236(90)90144-y. [DOI] [PubMed] [Google Scholar]
- Lumsden A, Keynes R. Segmental patterns of neuronal development in the chick hindbrain. Nature. 1989;337(6206):424–428. doi: 10.1038/337424a0. [DOI] [PubMed] [Google Scholar]
- Lumsden A, Krumlauf R. Patterning the vertebrate neuraxis. Science. 1996;274(5290):1109–1115. doi: 10.1126/science.274.5290.1109. [DOI] [PubMed] [Google Scholar]
- Luo W, Enomoto H, Rice FL, Milbrandt J, Ginty DD. Molecular identification of rapidly adapting mechanoreceptors and their developmental dependence on ret signaling. Neuron. 2009;64(6):841–856. doi: 10.1016/j.neuron.2009.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcus RC, Mason CA. The first retinal axon growth in the mouse optic chiasm: axon patterning and the cellular environment. The Journal of neuroscience. 1995;15(10):6389–6402. doi: 10.1523/JNEUROSCI.15-10-06389.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marina N, Abdala AP, Trapp S, Li A, Nattie EE, Hewinson J, Smith JC, Paton JFR, Gourine AV. Essential role of Phox2b-expressing ventrolateral brainstem neurons in the chemosensory control of inspiration and expiration. The Journal of Neuroscience. 2010;30(37):12466–12473. doi: 10.1523/JNEUROSCI.3141-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marquardt T, Ashery-Padan R, Andrejewski N, Scardigli R, Guillemot F, Gruss P. Pax6 is required for the multipotent state of retinal progenitor cells. Cell. 2001;105(1):43–55. doi: 10.1016/s0092-8674(01)00295-1. [DOI] [PubMed] [Google Scholar]
- Matsumoto S, Konishi H, Maeda R, Kiryu-Seo S, Kiyama H. Expression analysis of the regenerating gene (Reg) family members Reg-IIIβ and Reg-IIIλ in the mouse during development. Journal of Comparative Neurology. 2012;520(3):479–494. doi: 10.1002/cne.22705. [DOI] [PubMed] [Google Scholar]
- McEvilly RJ, Erkman L, Luo L, Sawchenko PE, Ryan AF, Rosenfeld MG. Requirement for Brn-3.0 in differentiation and survival of sensory and motor neurons. Nature. 1996;384(6609):574–577. doi: 10.1038/384574a0. [DOI] [PubMed] [Google Scholar]
- McKay IJ, Lewis J, Lumsden A. Organization and development of facial motor neurons in the kreisler mutant mouse. European Journal of Neuroscience. 1997;9(7):1499–1506. doi: 10.1111/j.1460-9568.1997.tb01504.x. [DOI] [PubMed] [Google Scholar]
- McNeill DS, Sheely CJ, Ecker JL, Badea TC, Morhardt D, Guido W, Hattar S. Development of melanopsin-based irradiance detecting circuitry. Neural Dev. 2011;6:10–1186. doi: 10.1186/1749-8104-6-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mesnage B, Gaillard S, Godin AG, Rodeau J-L, Hammer M, Von Engelhardt J, Wiseman PW, De Koninck Y, Schlichter R, Cordero-Erausquin M. Morphological and functional characterization of cholinergic interneurons in the dorsal horn of the mouse spinal cord. Journal of Comparative Neurology. 2011;519(16):3139–3158. doi: 10.1002/cne.22668. [DOI] [PubMed] [Google Scholar]
- Mikaels Å, Livet J, Westphal H, De Lapeyrière O, Ernfors P. A dynamic regulation of GDNF-family receptors correlates with a specific trophic dependency of cranial motor neuron subpopulations during development. European Journal of Neuroscience. 2000;12(2):446–456. doi: 10.1046/j.1460-9568.2000.00924.x. [DOI] [PubMed] [Google Scholar]
- Mu X, Fu X, Beremand PD, Thomas TL, Klein WH. Gene-regulation logic in retinal ganglion cell development: Isl1 defines a critical branch distinct from but overlapping with Pou4f2. Proceedings of the National Academy of Sciences. 2008;105(19):6942–6947. doi: 10.1073/pnas.0802627105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mu X, Silos-Santiago I, Carroll SL, Snider WD. Neurotrophin receptor genes are expressed in distinct patterns in developing dorsal root ganglia. The journal of neuroscience. 1993;13(9):4029–4041. doi: 10.1523/JNEUROSCI.13-09-04029.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narayanan CH, Narayanan Y. Neural crest and placodal contributions in the development of the glossopharyngeal-vagal complex in the chick. The Anatomical Record. 1980;196(1):71–82. doi: 10.1002/ar.1091960108. [DOI] [PubMed] [Google Scholar]
- Nomaksteinsky M, Kassabov S, Chettouh Z, Stoeklé H-C, Bonnaud L, Fortin G, Kandel ER, Brunet JF. Ancient origin of somatic and visceral neurons. BMC Biol. 2013;11:53–53. doi: 10.1186/1741-7007-11-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oury F, Murakami Y, Renaud J-S, Pasqualetti M, Charnay P, Ren S-Y, Rijli FM. Hoxa2-and rhombomere-dependent development of the mouse facial somatosensory map. Science. 2006;313(5792):1408–1413. doi: 10.1126/science.1130042. [DOI] [PubMed] [Google Scholar]
- Pak MW, Pinto LH, Mangini NJ, Vanable JW, Giolli RA, Gregory KM. Retinopretectal and accessory optic projections of normal mice and the OKN-defective mutant mice beige, beige-J, and pearl. Journal of Comparative Neurology. 1987;258(3):435–446. doi: 10.1002/cne.902580311. [DOI] [PubMed] [Google Scholar]
- Pan L, Deng M, Xie X, Gan L. ISL1 and BRN3B co-regulate the differentiation of murine retinal ganglion cells. Development. 2008;135(11):1981–1990. doi: 10.1242/dev.010751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan L, Yang Z, Feng L, Gan L. Functional equivalence of Brn3 POU-domain transcription factors in mouse retinal neurogenesis. Development. 2005;132(4):703–712. doi: 10.1242/dev.01646. [DOI] [PubMed] [Google Scholar]
- Pattyn A, Hirsch M, Goridis C, Brunet JF. Control of hindbrain motor neuron differentiation by the homeobox gene Phox2b. Development. 2000;127(7):1349–1358. doi: 10.1242/dev.127.7.1349. [DOI] [PubMed] [Google Scholar]
- Pattyn A, Morin X, Cremer H, Goridis C, Brunet JF. Expression and interactions of the two closely related homeobox genes Phox2a and Phox2b during neurogenesis. Development. 1997;124(20):4065–4075. doi: 10.1242/dev.124.20.4065. [DOI] [PubMed] [Google Scholar]
- Pattyn A, Morin X, Cremer H, Goridis C, Brunet JF. The homeobox gene Phox2b is essential for the development of autonomic neural crest derivatives. Nature. 1999;399(6734):366–370. doi: 10.1038/20700. [DOI] [PubMed] [Google Scholar]
- Paxinos G, Huang X-F, Sengul G, Watson C. Organization of brainstem nuclei 2012 [Google Scholar]
- Peleshok JC, Ribeiro-da-Silva A. Delayed reinnervation by nonpeptidergic nociceptive afferents of the glabrous skin of the rat hindpaw in a neuropathic pain model. Journal of Comparative Neurology. 2011;519(1):49–63. doi: 10.1002/cne.22500. [DOI] [PubMed] [Google Scholar]
- Pfaff SL, Mendelsohn M, Stewart CL, Edlund T, Jessell TM. Requirement for LIM homeobox gene Isl1 in motor neuron generation reveals a motor neuron–dependent step in interneuron differentiation. Cell. 1996;84(2):309–320. doi: 10.1016/s0092-8674(00)80985-x. [DOI] [PubMed] [Google Scholar]
- Quina LA, Tempest L, Hsu Y-WA, Cox TC, Turner EE. Hmx1 is required for the normal development of somatosensory neurons in the geniculate ganglion. Developmental biology. 2012;365(1):152–163. doi: 10.1016/j.ydbio.2012.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice FL, Fundin BT, Arvidsson J, Aldskogius H, Johansson O. Comprehensive immunofluorescence and lectin binding analysis of vibrissal follicle sinus complex innervation in the mystacial pad of the rat. Journal of Comparative Neurology. 1997;385(2):149–184. [PubMed] [Google Scholar]
- Rose MF, Ren J, Ahmad KA, Chao H-T, Klisch TJ, Flora A, Greer JJ, Zoghbi HY. Math1 is essential for the development of hindbrain neurons critical for perinatal breathing. Neuron. 2009;64(3):341–354. doi: 10.1016/j.neuron.2009.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross CA, Ruggiero DA, Reis DJ. Projections from the nucleus tractus solitarii to the rostral ventrolateral medulla. Journal of Comparative Neurology. 1985;242(4):511–534. doi: 10.1002/cne.902420405. [DOI] [PubMed] [Google Scholar]
- Satomi H, Takahashi K, Ise H, Yamamoto T. Identification of the superior salivatory nucleus in the cat as studied by the HRP method. Neuroscience Letters. 1979;14(2):135–139. doi: 10.1016/0304-3940(79)96137-8. [DOI] [PubMed] [Google Scholar]
- Schwaller B, Dick J, Dhoot G, Carroll S, Vrbova G, Nicotera P, Pette D, Wyss A, Bluethmann H, Hunziker W, et al. Prolonged contraction-relaxation cycle of fast-twitch muscles in parvalbumin knockout mice. American Journal of Physiology-Cell Physiology. 1999;276(2):C395–C403. doi: 10.1152/ajpcell.1999.276.2.C395. [DOI] [PubMed] [Google Scholar]
- Schwarz DWF, Satoh K, Schwarz IE, Hu K, Fibiger HC. Cholinergic innervation of the rat’s labyrinth. Experimental brain research. 1986;64(1):19–26. doi: 10.1007/BF00238197. [DOI] [PubMed] [Google Scholar]
- Scott MM, Williams KW, Rossi J, Lee CE, Elmquist JK. Leptin receptor expression in hindbrain Glp-1 neurons regulates food intake and energy balance in mice. The Journal of clinical investigation. 2011;121(6):2413–2413. doi: 10.1172/JCI43703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semba K, Egger MD. The facial “motor” nerve of the rat: control of vibrissal movement and examination of motor and sensory components. Journal of Comparative Neurology. 1986;247(2):144–158. doi: 10.1002/cne.902470203. [DOI] [PubMed] [Google Scholar]
- Shi M, Kumar SR, Motajo O, Kretschmer F, Mu X, Badea TC. Genetic interactions between Brn3 transcription factors in retinal ganglion cell type specification. PloS one. 2013;8(10):e76347. doi: 10.1371/journal.pone.0076347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon H, Lumsden A. Rhombomere-specific origin of the contralateral vestibulo-acoustic efferent neurons and their migration across the embryonic midline. Neuron. 1993;11(2):209–220. doi: 10.1016/0896-6273(93)90179-u. [DOI] [PubMed] [Google Scholar]
- Song M-R, Shirasaki R, Cai C-L, Ruiz EC, Evans SM, Lee S-K, Pfaff SL. T-Box transcription factor Tbx20 regulates a genetic program for cranial motor neuron cell body migration. Development. 2006;133(24):4945–4955. doi: 10.1242/dev.02694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spector AC, Glendinning JI. Linking peripheral taste processes to behavior. Current opinion in neurobiology. 2009;19(4):370–377. doi: 10.1016/j.conb.2009.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studer M, Lumsden A, Ariza-McNaughton L, Bradley A, Krumlauf R. Altered segmental identity and abnormal migration of motor neurons in mice lacking Hoxb-1. Nature. 1996;384:630–634. doi: 10.1038/384630a0. [DOI] [PubMed] [Google Scholar]
- Stuesse SL, Fish SE. Projections to the cardioinhibitory region of the nucleus ambiguus of rat. Journal of Comparative Neurology. 1984;229(2):271–278. doi: 10.1002/cne.902290211. [DOI] [PubMed] [Google Scholar]
- Thompson AM, Schofield BR. Afferent projections of the superior olivary complex. Microscopy research and technique. 2000;51(4):330–354. doi: 10.1002/1097-0029(20001115)51:4<330::AID-JEMT4>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- Thor S, Ericson J, Brännström T, Edlund T. The homeodomain LIM protein Isl-1 is expressed in subsets of neurons and endocrine cells in the adult rat. Neuron. 1991;7(6):881–889. doi: 10.1016/0896-6273(91)90334-v. [DOI] [PubMed] [Google Scholar]
- Tiveron MC, Pattyn A, Hirsch MR, Brunet JF. Role of Phox2b and Mash1 in the generation of the vestibular efferent nucleus. Developmental biology. 2003;260(1):46–57. doi: 10.1016/s0012-1606(03)00213-6. [DOI] [PubMed] [Google Scholar]
- Tiveron M-C, Hirsch M-R, Brunet JF. The expression pattern of the transcription factor Phox2 delineates synaptic pathways of the autonomic nervous system. The Journal of neuroscience. 1996;16(23):7649–7660. doi: 10.1523/JNEUROSCI.16-23-07649.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tritsch NX, Yi E, Gale JE, Glowatzki E, Bergles DE. The origin of spontaneous activity in the developing auditory system. Nature. 2007;450(7166):50–55. doi: 10.1038/nature06233. [DOI] [PubMed] [Google Scholar]
- Turner EE, Jenne KJ, Rosenfeld MG. Brn-3.2: a Brn-3-related transcription factor with distinctive central nervous system expression and regulation by retinoic acid. Neuron. 1994;12(1):205–218. doi: 10.1016/0896-6273(94)90164-3. [DOI] [PubMed] [Google Scholar]
- Uesaka T, Nagashimada M, Yonemura S, Enomoto H. Diminished Ret expression compromises neuronal survival in the colon and causes intestinal aganglionosis in mice. J Clin Invest. 2008;118(5):1890–1898. doi: 10.1172/JCI34425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varela-Echavarri A, Pfaff SL, Guthrie S. Differential expression of LIM homeobox genes among motor neuron subpopulations in the developing chick brain stem. Molecular and Cellular Neuroscience. 1996;8(4):242–257. doi: 10.1006/mcne.1996.0061. [DOI] [PubMed] [Google Scholar]
- Wang J, Chi F-l, Xin Y, Regner MF. The distribution of vestibular efferent neurons receiving innervation of secondary vestibular afferent nerves in rats. The Laryngoscope. 2013;123(5):1266–1271. doi: 10.1002/lary.23847. [DOI] [PubMed] [Google Scholar]
- Wang S, Shi Y, Shu S, Guyenet PG, Bayliss DA. Phox2b-expressing retrotrapezoid neurons are intrinsically responsive to H+ and CO2. The Journal of Neuroscience. 2013;33(18):7756–7761. doi: 10.1523/JNEUROSCI.5550-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warr WB, Beck JE. Multiple projections from the ventral nucleus of the trapezoid body in the rat. Hearing research. 1996;93(1):83–101. doi: 10.1016/0378-5955(95)00198-0. [DOI] [PubMed] [Google Scholar]
- Watson C, Kirkcaldie M, Paxinos G. The brain: an introduction to functional neuroanatomy: Access Online via Elsevier. 2010. [Google Scholar]
- White JS, Warr BW. The dual origins of the olivocochlear bundle in the albino rat. Journal of Comparative Neurology. 1983;219(2):203–214. doi: 10.1002/cne.902190206. [DOI] [PubMed] [Google Scholar]
- Xiang M, Gan L, Li D, Chen ZY, Zhou L, O’Malley JBW, Klein W, Nathans J. Essential role of POU-domain factor Brn-3c in auditory and vestibular hair cell development. Proc Natl Acad Sci U S A. 1997;94(17):9445–9450. doi: 10.1073/pnas.94.17.9445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang M, Gan L, Zhou L, Klein WH, Nathans J. Targeted deletion of the mouse POU domain gene Brn-3a causes selective loss of neurons in the brainstem and trigeminal ganglion, uncoordinated limb movement, and impaired suckling. Proceedings of the National Academy of Sciences. 1996;93(21):11950–11955. doi: 10.1073/pnas.93.21.11950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang M, Gao W-Q, Hasson T, Shin JJ. Requirement for Brn-3c in maturation and survival, but not in fate determination of inner ear hair cells. Development. 1998;125(20):3935–3946. doi: 10.1242/dev.125.20.3935. [DOI] [PubMed] [Google Scholar]
- Xiang M, Zhou L, Macke JP, Yoshioka T, Hendry SH, Eddy RL, Shows TB, Nathans J. The Brn-3 family of POU-domain factors: primary structure, binding specificity, and expression in subsets of retinal ganglion cells and somatosensory neurons. The Journal of neuroscience. 1995;15(7):4762–4785. doi: 10.1523/JNEUROSCI.15-07-04762.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang M, Zhou L, Peng YW, Eddy RL, Shows TB, Nathans J. Brn-3b: a POU domain gene expressed in a subset of retinal ganglion cells. Neuron. 1993;11(4):689–701. doi: 10.1016/0896-6273(93)90079-7. [DOI] [PubMed] [Google Scholar]
- Yamout A, Spec A, Cosmano J, Kashyap M, Rochlin MW. Neurotrophic factor receptor expression and in vitro nerve growth of geniculate ganglion neurons that supply divergent nerves. Developmental neuroscience. 2004;27(5):288–298. doi: 10.1159/000086708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou M, Li S, Klein WH, Xiang M. Brn3a/Pou4f1 regulates dorsal root ganglion sensory neuron specification and axonal projection into the spinal cord. Developmental biology. 2012;364(2):114–127. doi: 10.1016/j.ydbio.2012.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zylka MJ, Rice FL, Anderson DJ. Topographically distinct epidermal nociceptive circuits revealed by axonal tracers targeted to Mrgprd. Neuron. 2005;45(1):17–25. doi: 10.1016/j.neuron.2004.12.015. [DOI] [PubMed] [Google Scholar]


